Abstract
We examined tolerance mechanisms in patients receiving HLA-mismatched combined kidney and bone marrow transplantation (CKBMT) that led to transient chimerism under a previously-published non-myeloablative conditioning regimen (Immune Tolerance Network study ITN036). Polychromatic flow cytometry (FCM) and high throughput sequencing of TCRβ hypervariable regions of DNA from peripheral blood T regulatory cells (Tregs) and CD4 non-Tregs revealed marked early enrichment of regulatory T cells (CD3+CD4+CD25highCD127lowFoxp3+) in blood that resulted from peripheral proliferation (Ki67+), possibly new thymic emigration (CD31+) and, in one tolerant subject, conversion from non-Tregs. Among recovering conventional T cells, central memory CD4+ and CD8+ cells predominated. A large fraction of the T cell clones detected in post-transplant biopsy specimens by TCR sequencing were detected in the peripheral blood and were not donor-reactive. Our results suggest that enrichment of Tregs by new thymic emigration and lymphopenia-driven peripheral proliferation in the early post-transplant period may contribute to tolerance following CKBMT. Furthermore, most conventional T cell clones detected in immunologically quiescent post-transplant biopsies appear to be circulating cells in the microvasculature rather than infiltrating T cells.
Introduction
Induction of mixed chimerism by non-myeloablative conditioning followed by bone marrow transplantation (BMT) is a powerful means of inducing allograft tolerance in animal models (1). Our group has previously demonstrated long-term acceptance of HLA-mismatched renal allografts without maintenance immunosuppression (>5 years) in seven of ten patients following CKBMT. Four subjects remain free of immunosuppression for >6–>13 years, while three required reinstitution of immunosuppression after 6–7 years because of recurrent disease or chronic rejection (2). Transplantation tolerance induced with durable mixed chimerism is associated with central deletion of donor-reactive T cells during thymic maturation (1). However, in the patients undergoing CKBMT, multilineage donor chimerism was only short-lived (≤21 days) (3, 4), suggesting a role for other tolerance mechanisms. In these patients, we previously observed a striking enrichment of CD4+CD25+ cells and a significant increase in CD4+CD25highCD127low cells at 6 months (5). Here we demonstrate marked enrichment of CD3+CD4+CD25highCD127lowFoxp3+ Tregs very early after CKBMT and provide evidence that these cells originate both from the thymus and from peripheral expansion. T cell receptor sequencing of circulating Tregs reflected the early enrichment identified via flow cytometry and enabled tracking of Treg clones pre- and post-transplant. TCR sequencing demonstrated that many T cell clones detected within allograft biopsies were also detectable in the peripheral blood and very few were donor-reactive. CKBMT is thus associated with marked changes in both regulatory and effector cells which might be involved in the tolerance-inducing potential and complications of CKBMT, respectively.
Patients and Methods
Study protocol
All studies were performed with IRB approval. Five patients treated according to the ITN036 regimen were included in this study; the protocol and results have been reported (2). Patients 1, 2 and 4 successfully discontinued immunosuppression in the first year and allograft function has remained stable for more than 6 years. Patient 3 lost the allograft due to thrombotic microangiopathy. Patient 5 rejected the graft following discontinuation of immunosuppression (2, 6).
Flow cytometry (FCM)
Polychromatic FCM was performed using a LSR II flow cytometer (BD Biosciences, San Jose, CA, USA), as described (4) and detailed in Supplemental Methods.
Analysis of demethylation status of Treg-specific demethylated region (TSDR)
Genomic DNA was isolated using the DNeasy blood and tissue kit (Qiagen, Germantown, MD, USA). The protocol for cultured cells was followed. Bisulfite treatment (7) and analysis of demethylation status (8) was performed by Epiontis GmbH (Berlin, Germany) as described (see Supplemental Methods for details). In female subjects an adjustment by a factor of 2 was made to compensate for obligate Barr body X chromosome methylation.
T cell culturing from renal allograft biopsy specimens
Protocol kidney biopsies were obtained at 6, 12 and 24 months and one half of a core was digested to a single cell suspension and cultured with anti-CD3 antibody (OKT3, 90 ng/mL), IL-2 (100 U/ml), irradiated allogeneic EBV-transformed B cells from a healthy donor and PBMCs from multiple healthy donors, with or without rapamycin (1 μg/ml) to prevent the expansion of non-Tregs while allowing expansion of Tregs (9). Expanded cell lines were further cultured with IL-2 alone. Expanded lines were phenotyped (CD4, CD8, CD25, CD127, Foxp3) and cryopreserved.
T cell repertoire β-chain length distribution analysis
Methods for RNA isolation, reverse transcription, PCR, spectratyping and data analysis are detailed in the Supplemental Methods section.
High throughput T cell receptor β-chain sequence analysis
Genomic DNA was isolated from sorted Tregs, CD4 non-Tregs, CD8s, available biopsy specimens, and cell lines pooled from each subject by time point using the Qiagen DNeasy Blood and Tissue Kit. DNA was frozen at −20°C and shipped on dry ice to Adaptive Biotechnologies for high-throughput TCRB CDR3 sequencing. The TCR sequencing data were retrieved from Adaptive’s immunoSEQ software and analyzed as described (10).
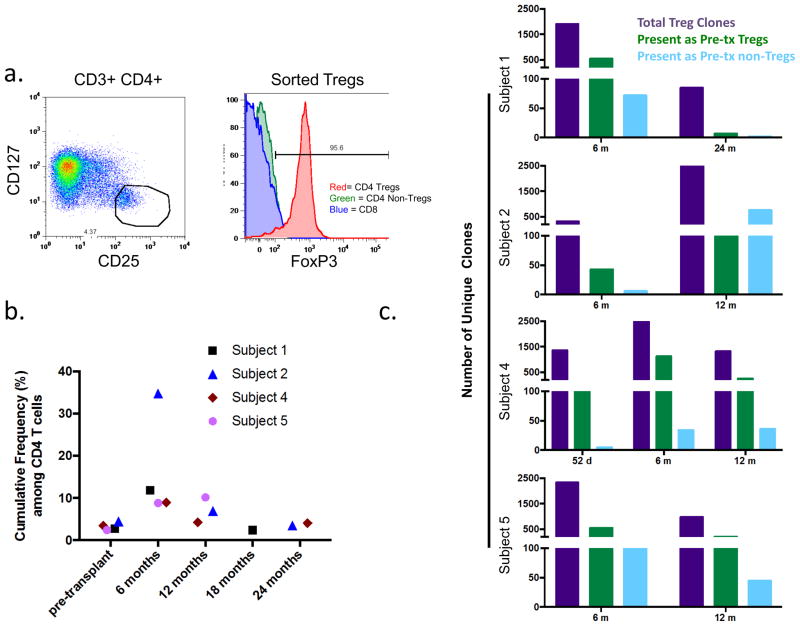
For analysis of circulating Tregs, CD3+CD4+CD127lowCD25high Tregs were sorted from frozen PBMCs after thawing and overnight resting in 200 units/ml IL-2 (Figure 5a). Simultaneously, CD4 non-Treg and CD3+CD4− (CD8) populations were sorted. We defined Treg clones as those detected in sorted Treg populations at more than twice frequency than that in both the CD8 and CD4 nonTreg sorted populations. Clones detected in sorted Treg populations that did not meet these criteria were considered to reflect sorting error. All sorted Treg clones from all time points were pooled to define the Treg clone set for which samples from a given patient were interrogated for the analysis shown in Figure 5b/Table S1.
Figure 5.
a. Illustrative sorting gates for isolation of Tregs (CD3+CD4+CD25highCD127−). Intracellular FoxP3 staining confirms high percentage of FoxP3 expression among sorted Tregs not seen in CD4 non-Tregs and CD8 T cells from the same sample. b. Cumulative frequency of Treg clones pooled from all Treg samples for a given patient found in the unstimulated CD4 samples. Clones defined by amino acid sequence of CDR3 with associated Vβ and Jβ gene. Unique clone numbers shown in Table S1. c. Assessment of pre-transplant sorted non-Treg CD4 cells (blue bars) and Tregs (green bars) for TCRs detected in the indicated sorted post-transplant Treg populations (purple bars) (nucleotide level analysis). Number of non-Treg/Treg CD4 clones identified pre-transplant: Subject 1 123,266/6,592; Subject 2 114,011/5,891; Subject 4 130,542/107,540; Subject 5 179,754/24,925.
Genomic DNA was extracted from protocol biopsies frozen in Optimal Cutting Temperature compound, after thawing and extensive washing with PBS, using Qiagen DNeasy Blood & Tissue kit and subjected to the Adaptive Immunoseq TCR sequencing platform. Clone sets were compared with sorted CD4 and CD8 blood samples and Treg clone sets. TCR clones identified from the pooled cell lines and biopsy specimens were compared with the previously-defined TCR sequences of donor-reactive clones (10). Computational and statistical analysis were performed as described previously (10).
Statistical Analysis
Statistical analysis was performed using Microsoft Excel (Microsoft Corporation, Redmond, WA) and GraphPad Prism (GraphPad Software, La Jolla, CA). A two-tailed paired T-test was applied for comparisons of data at different time points, except for analysis of natural killer cells, where a two-tailed, two-sample equal variance T-test was applied, since pre-transplant data were not available for Patient 1.
Results
Enrichment for effector/memory T cells early post-transplant
Lymphocyte recovery in these subjects has been reported (4), including marked T cell lymphopenia immediately following transplantation (0–12 cells/μL by Day 10), a transient increase in counts between Week 2 and 3 and a subsequent decline that was followed by gradual T cell recovery in all patients (4). There was as an initial increase in CD4/CD8 ratio that peaked on Day 7, followed by inverted CD4/CD8 ratios by Day 14 (4).
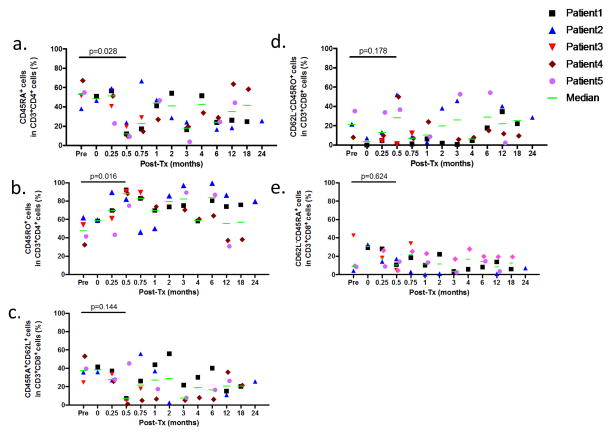
We further analyzed expression of CD45RA, CD45RO and CD62L on CD4+ and CD8+ cells (Figure 1a–b and Figure 1c–e, respectively). CD4+ cells showed a relative reduction of naïve cells on Day 14 compared to pre-transplant (defined as CD45RA+, p=0.028)(Figure 1a) with a corresponding increase in CD45RO+CD4+ cells (p=0.016)(Figure 1b). Changes in proportions of CD8+ naïve (CD45RA+CD62L+)(Figure 1c), effector memory (CD62L−CD45RO+)(Figure 1d) and effector memory RA cells (defined as CD62L−CD45RA+)(Figure 1e) were variable and did not achieve statistical significance.
Figure 1.
a. The percentages of CD45RA+ (naïve CD4+ T cells) and b. CD45RO+ (memory CD4+ T cells) in CD3+CD4+ cells. c. CD45RA+CD62L+ (naïve CD8+ T cells), d. CD45RO+CD62L− (effector memory CD8+ T cells), e. CD45RA+CD62L− (effector memory RA CD8+ T cells) in CD3+CD8+ cells are depicted for Patients 1 to 5.
Activation of CD4 and CD8 lymphocytes early post-transplantation
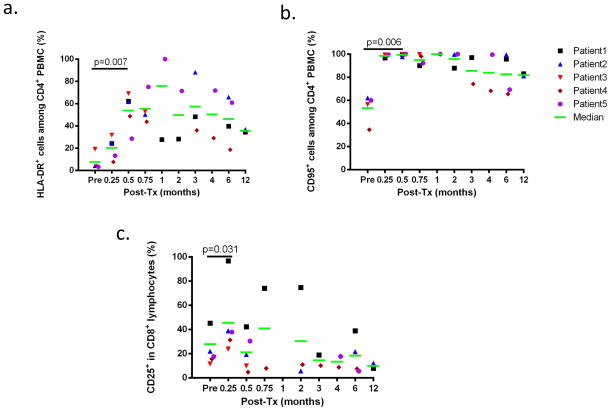
Both CD4+ and CD8+ populations showed evidence for activation early after CKBMT. CD4+ cells upregulated HLA-DR and CD95 at Day 14 (p=0.007 and p=0.006, respectively)(Figure 2a and 2b) and CD8+ cells upregulated CD25 at Day 7 (p=0.031)(Figure 2c). Duration and peak levels of HLA-DR (among CD4+ cells) and CD25 upregulation (in CD8+ cells) varied substantially between patients and persisted for up to 6 months (Figure 2a,c).
Figure 2.
a. The percentages of HLA-DR+ cells among total CD4+ cells are depicted for Patients 1 to 5. b. The percentages CD95+ in total CD4+ cells are depicted for patients 1 to 5. c. The percentages of CD25+ cells among total CD8+ cells are depicted for patients 1 to 5.
Enrichment for Tregs
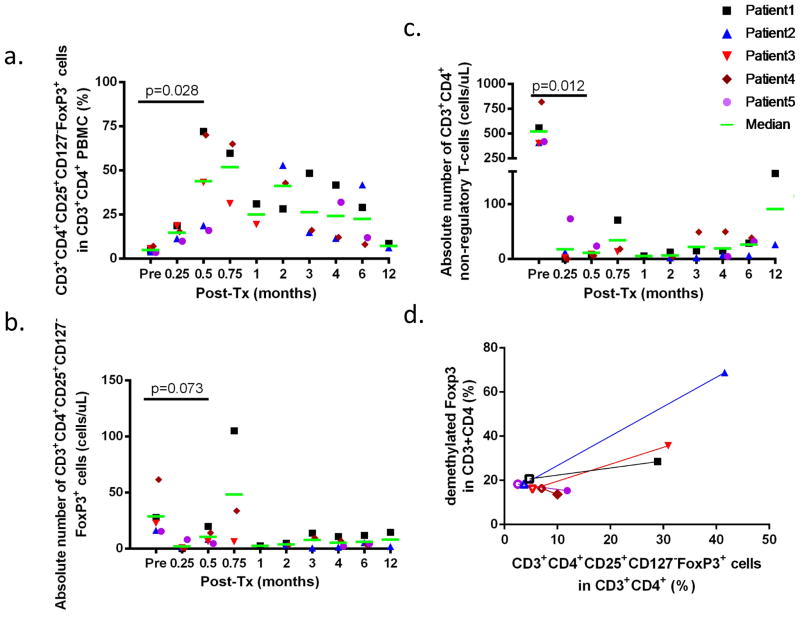
Percentages of CD25highCD127lowFoxp3+ T cells were dramatically increased as a proportion of CD3+CD4+ cells in the blood by 1 to 3 weeks after transplantation compared to pre-transplant levels (4.9±1.4%). Percentages of CD25highCD127lowFoxp3+ cells peaked at 71.8% (Day 14), 52.7% (Day 58), 42.9% (Day 14), 70.0% (Day 14), and 15.9% (Day 14) for Patients 1–5, respectively (p=0.028 at Day 14)(Figure 3a). The enrichment persisted for 12 months, 6 months, 1 month, 4 months and 6 months in Patients 1–5, respectively (Figure 3a). Although absolute numbers declined initially from the conditioning treatment, there was a partial recovery or even an increase in absolute numbers of CD3+CD4+CD25highCD127lowFoxp3+ T cells in the blood of all CKBMT recipients compared to pretransplant values by 2–3 weeks post-transplant (Figure 3b). In contrast to this early recovery of Tregs, conventional CD3+CD4+ cells recovered slowly (Figure 3c). These data suggest that there was either selective sparing, expansion or generation of CD3+CD4+CD25highCD127lowFoxp3+ regulatory T cells early following conditioning and CKBMT.
Figure 3.
a. Percentages of CD4+CD25+CD127−Foxp3+ cells in CD3+CD4+ cells are shown for Patients 1 to 5. b. Absolute number of CD4+CD25+CD127−Foxp3+ cells in the peripheral blood (cell per microliter) are shown for Patients 1 to 5. c. Absolute number of non-regulatory CD3+CD4+ cells in the peripheral blood (cell per microliter) are shown for Patients 1 to 5. d. Correlation plot for CD4+CD25+CD127−Foxp3+ cells in CD3+CD4+ cells and demethylated Foxp3 in CD3+CD4+ cells are depicted for Patients 1 to 5. Open symbols are values pretransplant (before conditioning) for patient 1 to 5 while closed symbols represent values at time d21 for patient 3 and d180 for patient 1, 2, 4 and 5.
As the stability of regulatory function is related to the demethylation status of the Treg-specific demethylated region of Foxp3 (TSDR-Foxp3), we determined methylation status of TSDR-Foxp3 before and after CKBMT (Figure 3d). In Subjects 1–3, the marked enrichment in CD3+CD4+CD25highCD127lowFoxp3+ T cells at 6 months (Subjects 1 and 2) and 3 weeks (Subject 3), respectively (Figure 3a) was accompanied by an increased percentage of demethylated TSDR-Foxp3 (Figure 3d). In contrast, Subjects 4 and 5 no longer showed enrichment in CD3+CD4+CD25highCD127lowFoxp3+ regulatory T cells at 6 months post-CKBMT (Figure 3a) and, consistently, had no increase in percentage of demethylated TSDR-Foxp3 (Figure 3d).
Phenotypic analysis of Tregs
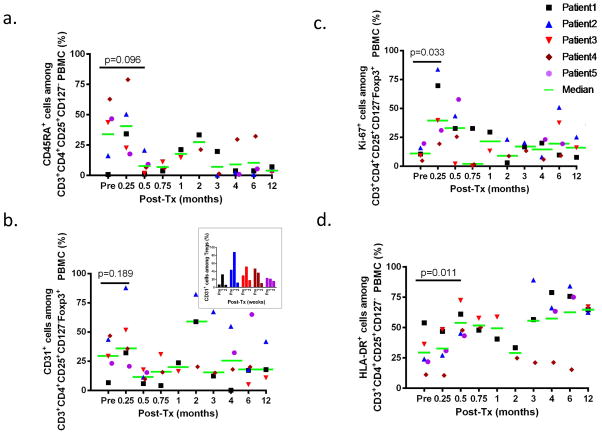
Further phenotypic analysis was performed using markers for activation and naïve/memory phenotype. In all 3 tolerant patients, Subjects 1, 2 and 4, there was an increase in the proportion of CD45RA+ (naïve or resting) Tregs at 1 week compared to pre-transplant (Figure 4a), whereas Subjects 3 and 5 showed reduced percentages of CD45RA+ Tregs in the same period. By 2 weeks post-transplant, most Tregs were CD45RA− in all subjects and this phenotype predominated thereafter.
Figure 4.
a. The percentages of CD45RA+ cells in CD4+CD25+CD127−Foxp3+ cells in CD3+CD4+ cells are shown for Patients 1 to 5. b. The percentages of CD31+ cells in CD4+CD25+CD127−Foxp3+ cells in CD3+CD4+ cells are shown for Patients 1 to 5. The inset shows the percentage of CD31+ cells in CD4+CD25+CD127−Foxp3+ cells from the pretransplant time point until week 2 post-transplant. c. The percentages of Ki-67+ cells in CD4+CD25+CD127−Foxp3+ cells in CD3+CD4+ cells are shown for Patients 1 to 5. d. The percentages of HLA-DR+ cells in CD4+CD25+CD127−Foxp3+ cells in CD3+CD4+ cells are shown for Patients 1 to 5.
We evaluated the expression of CD31, a marker of recent thymic emigrants (RTE) (11) and Ki-67, a marker of proliferating cells (12)(Figure 4b and 4c, respectively). At 7 days posttransplant, RTEs increased among Tregs compared to pretransplant in 3 out of 5 patients (Subjects 1,2, and 3) (p=0.189)(Figure 4b). At the same time, percentages of Ki-67+ Tregs also peaked and increased compared to pretransplant levels in 5 of 5 patients (p=0.032)(Figure 4c). Among non-Treg CD4+ T cells, CD31+ expression increased compared to pre-transplant in 2 out of 5 patients and did not differ strikingly from percentages among Tregs (Figure S1a, Figure 4b). A marked early increase in Ki-67 expression was also detected among CD8 cells (defined as CD3+CD4− PBMCs) and in non-Treg CD4+ T cells (defined as CD3+CD4+Foxp3− PBMCs (Figures S1b and S1c, respectively).
Furthermore, there was a significant increase in the expression of HLA-DR among CD3+CD4+CD25highCD127lowFoxp3+ T cells at 2 weeks post-transplant (p 0.011)(Figure 4d). Collectively, these data suggest that the early recovery of Tregs in patients receiving this regimen reflected a combination of Treg expansion with a possible contribution of migration from the thymus.
High throughput T cell receptor (TCR) sequencing of regulatory T cells
We performed high-throughput TCRβ CDR3 sequencing on genomic DNA extracted from sorted pre- and post-transplant (CD3+CD4+CD127lowCD25high) Tregs (Figure 5a). Because of the small size of the Treg population, sufficient cell numbers were limited to a few time points in Subjects 1, 2, 4, and 5; however, hundreds or thousands of unique clones were identified in most samples (Table 1). We then interrogated circulating CD4 T cells at various times for these Treg clones. Figure 5b shows the cumulative frequency of all Treg clones identified in the circulating total CD4 T cell populations: these mirror the flow cytometry findings, showing enrichment of Tregs in all patients at 6 months post-transplant compared to pre-transplant, most strikingly in Subject 2. Treg enrichment in tolerant Subjects 1, 2, and 4 did not persist after 12 months post-transplant, consistent with FCM results in Figure 3.
Table 1.
T cell clones from sorted peripheral blood T regulatory cells.
| Template Count | Unique Clones | |
|---|---|---|
| Subject 1 pre-transplant | 10184 | 6592 |
| Subject 1 6 months post-transplant | 3243 | 1909 |
| Subject 1 24 months post-transplant | 107 | 85 |
| Subject 2 pre-transplant | 7425 | 5891 |
| Subject 2 6 months post-transplant | 466 | 322 |
| Subject 2 12 months post-transplant | 10681 | 4292 |
| Subject 4 pre-transplant | 114915 | 107540 |
| Subject 4 52 days post-transplant | 1526 | 1365 |
| Subject 4 6 months post-transplant | 8928 | 5695 |
| Subject 4 12 months post-transplant | 1749 | 1323 |
| Subject 5 pre-transplant | 36170 | 24925 |
| Subject 5 6 months post-transplant | 4196 | 2329 |
| Subject 5 12 months post-transplant | 1647 | 983 |
Template count refers to the calculated number of productive TCRs sequenced.
TCR sequencing enabled interrogation of pre-transplant cell populations for the Treg clones detected post-transplant (Figure 5c). Some but not all of the Treg clones found at 6 months post-transplant were detected in the CD4 pre-transplant population for each of the patients, with the greatest number seen in Subject 4’s 6-month sample. While small numbers of Treg clones were identified as non-Tregs pre-transplant for Subjects 1,4 and 5, the overall low numbers of such clones do not point to prominent induction of Tregs from non-Tregs present prior to transplant (Figure 5C). For Subject 2, at 6 months post-transplant a similar pattern was seen; however a remarkably high number of Treg clones detected at 12 months in this patient appeared to be induced Tregs that were detectable in the non-Treg population pre-transplant (Figure 5c).
Analysis of intragraft T cells
Consistent with the detection of FoxP3 in long-term protocol biopsies (3), we hypothesized that intragraft Tregs might play a role in long-term tolerance. TCRs in serial post-transplant biopsy samples were therefore analyzed. The pathologic biopsy data are summarized in Table S2: in general, lymphocyte infiltrates were absent or scant, except in Patient 3 at the time of graft loss and in Patient 5 at 12 months, whose biopsy contained infiltrates consistent with treated acute cellular rejection (Table S2). We initially performed spectratyping on biopsies from Subject 2 at day 0 and at 6, 12, and 24 months post-transplant. While few Vβ families were identified on day 0, the post-transplant biopsies revealed polyclonal repertoires (Figure S2). The absence of most Vβ in the naïve donor graft biopsies may reflect the ex vivo perfusion performed before biopsies were obtained, which preceded reperfusion, resulting in purging of T cells present in the microcirculation at the time of biopsy.
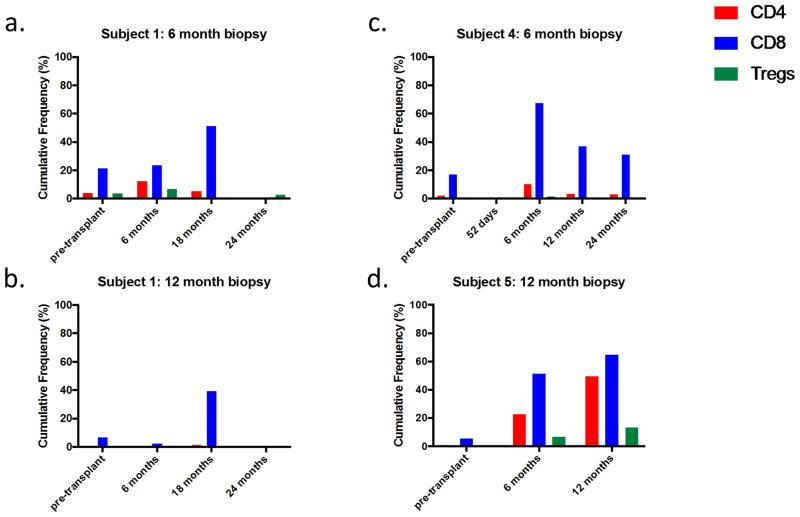
To investigate the T cell repertoire of intragraft T cells in greater depth, we performed high throughput TCRβ CDR3 sequencing on the remaining biopsy specimens for Subjects 1, 4, and 5 and compared the data with previously-generated sequence databases of pre- and post-transplant peripheral blood CD4 and CD8 T cells and donor-specific clones described in Morris et al. (10) and with the TCR sequences of circulating Tregs. Hundreds or thousands of unique T cell clones were identified in each biopsy specimen, including a mixture of CD4 and CD8 clones, as defined by their identification in a sorted circulating CD4 or CD8 T cell population (Table 2). In Subjects 1 and 4, who were tolerant, a high proportion (20–45% in CD4s, 11–35% in CD8s) of clones in the biopsies were also detected in the pre-transplant and/or post-transplant circulating CD4 and CD8 T cell populations (Table 2). Furthermore, the cumulative frequency within the peripheral circulation of clones also detected in the biopsies was strikingly high, particularly among CD8s, accounting for greater than 40% of the circulating CD8 population at least one post-transplant time point for each patient (Figure 6). The numbers of overlapping clones shared between biopsies and PBMCs at the same time point were comparable to the overlap between PBMC populations sampled at different time points (Table S3), consistent with the possibility that T cells in biopsies were largely within the microcirculation. More than 200 clones in each biopsy were identifiable as Tregs on the basis of sorted circulating Treg populations (Table 2), by frequency within 3 of 4 the biopsies accounting for 2.52%, 14.75%, and 4.51% of clones for subjects 1 (6 months post-transplant), 4 (6 months post-transplant) and 5 (12 months post-transplant), respectively.
Table 2.
Overlapping T cell clones from biopsy samples compared to peripheral blood T cells pre- and post-transplant, donor-reactive TCR as described in Morris et al. 2015 (10), and circulating T regulatory cell repertoires.
| Biopsy Samples | Subject 1 | Subject 4 | Subject 5 | |
|---|---|---|---|---|
| 6 months | 12 months | 6 months | 12 months | |
| Total unique clones |

|
|
3020 |
|
| Pre-transplant CD4 PBMCs (in blood) | 1164 | 45 | 667 | 257 |
| Pre-transplant CD8 PBMCs (in blood) | 750 | 39 | 594 | 176 |
| All post-transplant CD4 PBMCs (in blood) | 1620 | 91 | 1237 | 1523 |
| All post-transplant CD8 PBMCs (in blood) | 959 | 65 | 1009 | 463 |
| Pre- or Post-transplant CD4 PBMCs (in blood) (% of total unique clones) | 2170 (20.18%) | 107 (29.97%) | 1359 (45%) | 1649 (13.3%) |
| Pre- or Post-transplant CD8 PBMCs (in blood) (% of total unique clones) | 1219 (11.34%) | 69 (19.33%) | 1067 (35.33%) | 554 (4.48%) |
| Donor-reactive CD4s (defined pre-transplant) (% = sum frequency) | 126 (1.23%) | 1 (0.24%) | 42 (0.87%) | 9 (0.02%) |
| Donor-reactive CD8s (defined pre-transplant) (% = sum frequency) | 30 (0.30%) | 0 | 21 (1.24%) | 17 (0.17%) |
| T regulatory clones (pooled peripheral blood Treg clones) (% = sum frequency) | 237 (2.52%) | 5 (2.57%) | 215 (14.75%) | 225 (4.51%) |
| Cell line overlap (same time point) | 9 | 61 | 7 | 6 |
Available post-transplant time point samples for each patient are pooled in the “all post-transplant” rows.
 samples analyzed on level of amino acid + V&J gene; all other analyzed on nucleotide level.
samples analyzed on level of amino acid + V&J gene; all other analyzed on nucleotide level.
Figure 6.
Cumulative frequency in circulation of clones found both in the indicated biopsy and in unstimulated CD4 (red), CD8 (blue), and Treg (green) TCR populations in circulating PBMCs at the indicated times (clones defined by amino acid sequence of CDR3 with associated Vβ and Jβ gene).
The availability of alloreactive TCR sequencing data on our subjects allowed us to interrogate the biopsy data for previously- defined donor-reactive TCRs (10). Although some donor-reactive CD4 and CD8 clones were identified in each of the biopsies (Table 2), they accounted for less than 1.5% by frequency of the biopsy TCR clones. Additionally, while the CD4 to CD8 ratios of pre-transplant-detected clones found in both the circulation and within the biopsy were similar (>1:1 for all subjects), this ratio was markedly altered for donor-reactive clones found only within the 12-month post-transplant biopsy of the rejector Subject 5 (9 CD4/17 CD8)(Table 2). This finding is consistent with the notion that clones in tolerated grafts largely represented circulating T cells, whereas infiltrating donor-reactive T cells, enriched for CD8 clones, may have persisted despite treatment and contributed to the clones detected in Patient 5’s allograft, which was ultimately rejected.
Since lymphocytes were sparse in protocol biopsies of tolerated allografts (3)(Table S2), we initially developed a technique for polyclonally expanding T cells from small pieces of core biopsies, generating numerous cell lines. Expression of host-specific class I HLA antigens revealed that these were of recipient origin (Figure S3). In addition, these cell lines were characterized by FCM for expression of CD3, CD4, and CD8 (Table S4), with some further analyzed for Treg markers. The phenotype of some lines was consistent with a Treg lineage (Table S4, Figure S4). TCR sequencing results from pooled cell lines demonstrated oligoclonality (Table S5) and there was little overlap with clones detected in biopsies or the donor-reactive repertoire (Table 2).
Discussion
The mechanisms of tolerance achieved via HLA-haploidentical CKBMT associated with transient chimerism are not fully understood (2–5). In some but not all CKBMT recipients, suppressive tolerance mechanisms could be implicated in donor-specific hypo/unresponsiveness at 6 months to 1 year, but no evidence for suppressive mechanisms in the persisting donor-specific unresponsiveness was seen at later time points (≥ 18 months)(5). Using high-throughput sequencing of TCR beta chain CDR3 regions and a pre-transplant MLR to identify donor-reactive clones, we recently obtained evidence for gradual deletion of these clones over time only in tolerant CKBMT recipients and not in those who failed tolerance or received conventional transplants (10).
We have previously reported enrichment of regulatory T cells in recipients of allogeneic BMT with similar non-myeloablative conditioning (13) and in the first series of CKBMT patients (5). However, these studies were performed on frozen samples with only a limited number of time points available for evaluation. Tregs were also implicated by high levels of Foxp3 expression relative to effector cell RNA in the kidneys of CKBMT patients (3). Here, we report that following CKBMT, Tregs are relatively spared from siplizumab (MEDI-507)-based, T cell-depleting conditioning therapy and provide evidence that new emigrants from the thymus as well as peripheral expansion may contribute to marked Treg enrichment among peripheral CD4 T cells early post-transplant. While both Treg and non-Treg CD4 populations included CD45RA+ and CD31+ cells in the first weeks post-transplant, consistent with either a wave of new thymic emigration, we cannot rule out the possibility that RTE CD4 T cells are spared from depletion by the conditioning regimen and have not yet undergone sufficient lymphopenia-driven expansion to convert to the memory phenotype by 1–2 weeks. Notably, the period of Treg enrichment corresponds to the early period during which suppression could be implicated in the donor-specific unresponsiveness observed in vitro (5).
Although Foxp3 is a robust marker for murine Tregs, Foxp3 can be induced by activation in naïve human conventional CD4+ T cells (14). Tregs express high levels of CD25 (15, 16) and lack cell surface CD127 (IL-7 receptor α-chain) expression (17, 18). We used the combination of Foxp3, CD127 and CD25 to identify Tregs, which may be contaminated to a minor degree by non-regulatory Foxp3-expressing CD4+CD45RO+Foxp3lowCD127low/intermediate effector cells (19). Stable and high Foxp3 expression is a feature of natural (thymus-derived) Tregs and is required for suppressive function (20, 21) and its genetic locus is demethylated in stably Foxp3+ natural Tregs (19), in contrast to TFGβ-induced Tregs (22). Our TSDR demethylation analysis shows a strong correlation with the percentage of CD25+Foxp3+CD127low cells among peripheral CD4+ cells, consistent with the interpretation that these represent expansions of natural Tregs.
Tregs include several subsets distinguishable by their expression of CD45RA and CD45RO (23). Tolerant patients showed a significant increase in the proportion of CD45RA+ Tregs early post-transplant, which may be naïve or resting Tregs (19), which express low levels of Foxp3 and also express CD31, suggesting they include recent thymic emigrants (19). The CKBMT recipients showed an increase in CD31+ cells among Tregs in 3 out of 5 patients in the early post-transplant period, consistent with but not definitively indicative of a thymic origin. In mice, de novo intrathymic generation of antigen-specific Tregs occurred after peripheral injection of antigen under proinflammatory conditions (24). A similar phenomenon might occur in patients undergoing CKBMT in association with the inflammatory “engraftment syndrome” seen early post-transplant in association with expanded recipient CD8+ T cells in the glomeruli without a rejection pattern (25). The marked increase in Ki-67 expression suggests an important role for lymphopenia-driven peripheral expansion in the enrichment of Tregs in the immediate post-CKBMT period. It is possible that tissue resident Tregs may also contribute to this enrichment, as suggested by studies in lymphopenic mice (26). The peripheral CD8+ T cells in these patients showed very early upregulation of CD25, even before BMT was administered, suggesting a marked effect of the conditioning itself. The studies presented here show that high levels of activated CD8+ T cells persisted for at least one week post-transplant and in one case persisted for up to 6 months.
TCR sequencing enabled us to directly investigate whether the clones expanding post-transplant were present pre-transplant. We validated that sorted CD4+CD127−CD25high Tregs were FoxP3+ and, to ensure the clones we defined as Tregs were not sort contaminants, we removed from consideration clones that were detected at similar frequencies in sorted non-Treg populations. Strikingly, many of the Tregs found at 6 months post-transplant were present pre-transplant. Our data did not suggest a strong component of Treg induction from non-Tregs detected pre-transplant, with the exception of Patient 2 at 12 months. In all cases, Treg enrichment following CKBMT with this protocol appears to reflect relative sparing of pre-existing Tregs compared to effector CD4 cells, combined with post-transplant lymphopenia-driven expansion and de novo generation in the thymus. Human Tregs have been shown to relatively resistant to cyclophosphamide (27), which, along with siplizumab (13) may contribute to Treg enrichment. Rabbit ATG, which was used to treat engraftment syndrome in some patients, has been reported to enrich Tregs in humans (28).
Thymic irradiation in our conditioning regimen depletes pre-existing thymocytes and makes space for de novo thympoiesis. Consistent with the notion that an early wave of thymopoiesis may have followed this treatment, peripheral CD4+ T cells included substantial proportions of naïve (CD45RA+) cells at one week and it was not until 2 weeks post-transplant that these shifted largely to a CD45RO+ memory-type population, presumably due to lymphopenia-driven expansion. Studies in humanized mice have shown that naïve human T cells expand and convert to effector/memory phenotype when placed in a lymphopenic environment (29). Additionally, memory T cells may be relatively resistant to mAb-induced depletion, resulting in selective sparing and expansion from a small residual pool of peripheral T cells (30).
Following antigenic stimulation, CD45RA+Foxp3low Tregs become CD45RO+Foxp3high “effector” Tregs, which are highly proliferative and strongly suppressive (31). HLA-DR identifies a terminally differentiated, highly suppressive subpopulation of effector Tregs (31, 32). The marked enrichment in CD4+CD45RO+Foxp3+CD127lowCD45RA−HLA-DR+ Tregs that we observed following CKBMT may reflect antigen- and/or lymphopenia-driven enrichment of these effector-type Tregs (33–39), consistent with the increased Ki67 expression among Tregs early post-transplant. While we do not yet have an established method of identifying the donor-specific Treg repertoire to assess enrichment for these cells, our TCR tracking study suggested that antigenic pressure from the graft combined with lymphopenia promotes expansion of donor-specific T cells early following kidney transplantation (10).
High-throughput sequencing allowed assessment of the hypothesis that Tregs accumulated in the transplanted kidney, as pre-clinical and clinical studies leading to the CKBMT protocol suggested a role for the allograft itself in promoting tolerance (5, 13, 40). Long-term protocol biopsies revealed absent or minimal lymphoid infiltrates (3). This absence of lymphoid infiltrates contrasts with observations in a murine model in which MHC-mismatched renal allografts are spontaneously accepted in a Treg-dependent fashion, where accumulations of Tregs is seen around the cortical arteries (41) (42). While some Treg clones were found within the biopsies, the most abundant graft T cell clones were prominent in the circulation and were not donor-reactive; the extensive clonal overlap between biopsies and circulating T cells is consistent with the degree of overlap expected from samples of the size tested from a single population. These findings and the contrasting absence of many Vβ families in pre-transplant biopsies from avascular, perfused kidneys, support the notion that TCR sequences from the transplanted kidney largely reflect circulating cells within the kidney’s microvasculature. However, our definition of donor-reactive clones only applies to those detected pre-transplant and our data do not rule out the possibility that new donor-specific clones that emerge from the thymus post-transplant may be included in the biopsy specimens (10).
Our sequencing analysis suggested a lack of utility of generating T cell lines via polyclonal expansion to characterize graft-infiltrating T cells. Because of the paucity of lymphocytes in biopsy samples, we polyclonally expanded T cells from small biopsy specimens. However, TCRβ sequencing of these cell lines revealed little overlap with clones identified directly from whole biopsy specimens and included many fewer clones, suggestive of selective clonal expansion rather than reflecting the diversity of the initial population.
In summary, our studies demonstrate enrichment for Tregs in the circulation early post-transplant in CKBMT recipients and suggest that peripheral expansion, possibly in combination with early de novo generation in the thymus may be largely responsible for this enrichment, which may play a significant role in the initial tolerance achieved with CKBMT and may contribute to the quiescent state that results in eventual deletion of donor-reactive T cells.
Supplementary Material
Figure S1: Expression of CD31 and Ki-67 in non-regulatory T cells
Figure S2: Number of Vβ detected in biopsies at different time points
Figure S3: Representative cell line sample for each patient
Figure S4: Representative cell line with Treg phenotype
Table S1: T cell clones defined as Tregs within pre- and post-transplant circulating sorted Treg T cell populations
Table S2: Pathologic evaluation of allograft biopsies in Subjects 1–5
Table S3: Comparison of the number of unique clones shared between biopsy and PBMCs (same time point) versus PBMCs at disparate time points for Subjects 1, 4, and 5.
Table S4: CD4, CD8, and FoxP3 phenotyping of expanded intragraft T cell line
Table S5: T cell clones from cultured cell lines compared peripheral blood T cells pre- and post-transplant, donor-reactive TCR as described in Morris et al. 2015 (10), and circulating T regulatory cell repertoires
Acknowledgments
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number # UM1 AI109565. The study was also supported by NIAID grant #RO1 AI084074 and NIDDK grant #RO1 DK106436. B.S. was supported by a fellowship from the American Society of Transplantation. S.D.W. was supported by a Kidney Research Student Scholar Grant from the American Society of Nephrology and by an HONORS award from the American Society of Hematology. FACS was performed in the CCTI Flow Cytometry Core, supported in part by the Office of the Director, National Institutes of Health under awards S10RR027050 and S10OD020056. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Ms. TeShima Brennen for expert assistance with manuscript preparation. We also thank Dr. Y. Shen for helpful review of the manuscript.
Abbreviations
- ATG
anti-thymocyte globulin
- BMT
bone marrow transplantation
- CKBMT
combined kidney/bone marrow transplantation
- IMGT
International ImMunoGeneTic information system
- FCM
flow cytometry
- PBMC
peripheral blood mononuclear cell
- RTE
recent thymic emigrants
- TCR
T cell receptor
- Treg
regulatory T cell
- TSDR
Treg-specific demethylated region
- WBC
white blood cell
Footnotes
Specific Contributions: Ben Sprangers, Susan DeWolf, Thomas Savage, and Tatsuaki Morokata performed experiments, analyzed data and wrote the manuscript. Aleksandar Obradovic analyzed TCR sequencing data. Samuel LoCascio coordinated sample acquisition, processed samples and analyzed data. Brittany Shonts, Julien Zuber, and Ravi Shah performed analyses of the cell lines. Heather Morris analyzed data. Frederic I. Preffer and David M. Dombkowski provided expert advice on and performed flow cytometry. Emmanuel Zorn participated in the design of the protocol for T cell expansion from protocol biopsies. Sven Olek performed TSDR analysis. Valeria Steshenko performed and Robert Winchester supervised and analyzed TCR spectratype analysis and sequencing. Robert Colvin performed pathology analysis and provided specimens. Tatsuo Kawai designed and carried out the clinical protocol and patient care. Laurence Turka participated in experimental design. Megan Sykes participated in the design of the clinical protocol, participated in and oversaw design of all laboratory studies, data analysis and interpretation, and wrote the manuscript.
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Sykes M. Mechanisms of tolerance induced via mixed chimerism. Front Biosci. 2007;12:2922–2934. doi: 10.2741/2282. [DOI] [PubMed] [Google Scholar]
- 2.Kawai T, Sachs DH, Sprangers B, Spitzer TR, Saidman SL, Zorn E, et al. Long-Term Results in Recipients of Combined HLA-Mismatched Kidney and Bone Marrow Transplantation Without Maintenance Immunosuppression. Am J Transplant. 2014;14:1599–1611. doi: 10.1111/ajt.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawai T, Cosimi AB, Spitzer TR, Tolkoff-Rubin N, Suthanthiran M, Saidman SL, et al. HLA-mismatched renal transplantation without maintenance immunosuppression. N Engl J Med. 2008;358:353–361. doi: 10.1056/NEJMoa071074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Locascio SA, Morokata T, Chittenden M, Preffer FI, Dombkowski DM, Andreola G, et al. Mixed chimerism, lymphocyte recovery, and evidence for early donor-specific unresponsiveness in patients receiving combined kidney and bone marrow transplantation to induce tolerance. Transplantation. 2010;90:1607–1615. doi: 10.1097/TP.0b013e3181ffbaff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreola G, Chittenden M, Shaffer J, Cosimi AB, Kawai T, Cotter P, et al. Mechanisms of donor-specific tolerance in recipients of haploidentical combined bone marrow/kidney transplantation. Am J Transplant. 2011;11:1236–1247. doi: 10.1111/j.1600-6143.2011.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawai T, Sachs DH, Sykes M, Cosimi AB. HLA-mismatched renal transplantation without maintenance immunosuppression. N Engl J Med. 2013;368:1850–1852. doi: 10.1056/NEJMc1213779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olek A, Oswald J, Walter J. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 1996;24:5064–5066. doi: 10.1093/nar/24.24.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wieczorek G, Asemissen A, Model F, Turbachova I, Floess S, Liebenberg V, et al. Quantitative DNA methylation analysis of FOXP3 as a new method for counting regulatory T cells in peripheral blood and solid tissue. Cancer Res. 2009;69:599–608. doi: 10.1158/0008-5472.CAN-08-2361. [DOI] [PubMed] [Google Scholar]
- 9.Battaglia M, Stabilini A, Migliavacca B, Horejs-Hoeck J, Kaupper T, Roncarolo MG. Rapamycin promotes expansion of functional CD4+CD25+FOXP3+ regulatory T cells of both healthy subjects and type 1 diabetic patients. J Immunol. 2006;177:8338–8347. doi: 10.4049/jimmunol.177.12.8338. [DOI] [PubMed] [Google Scholar]
- 10.Morris H, DeWolf S, Robins H, Sprangers B, Locascio SA, Shonts BA, et al. Tracking donor-reactive T cells: Evidence for clonal deletion in tolerant kidney transplant patients. Sci Transl Med. 2015;7:272ra10. doi: 10.1126/scitranslmed.3010760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimmig S, Przybylski GK, Schmidt CA, Laurisch K, Mowes B, Radbruch A, et al. Two subsets of naive T helper cells with distinct T cell receptor excision circle content in human adult peripheral blood. J Exp Med. 2002;195:789–794. doi: 10.1084/jem.20011756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–1715. [PubMed] [Google Scholar]
- 13.Shaffer J, Villard J, Means TK, Alexander S, Dombkowski D, Dey BR, et al. Regulatory T-cell recovery in recipients of haploidentical nonmyeloablative hematopoietic cell transplantation with a humanized anti-CD2 mAb, MEDI-507, with or without fludarabine. Exp Hematol. 2007;35:1140–1152. doi: 10.1016/j.exphem.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavin MA, Torgerson TR, Houston E, DeRoos P, Ho WY, Stray-Pedersen A, et al. Single-cell analysis of normal and FOXP3-mutant human T cells: FOXP3 expression without regulatory T cell development. Proc Natl Acad Sci U S A. 2006;103:6659–6664. doi: 10.1073/pnas.0509484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allan SE, Crome SQ, Crellin NK, Passerini L, Steiner TS, Bacchetta R, et al. Activation-induced FOXP3 in human T effector cells does not suppress proliferation or cytokine production. Int Immunol. 2007;19:345–354. doi: 10.1093/intimm/dxm014. [DOI] [PubMed] [Google Scholar]
- 16.Baecher-Allan C, Viglietta V, Hafler DA. Inhibition of human CD4(+)CD25(+high) regulatory T cell function. J Immunol. 2002;169:6210–6217. doi: 10.4049/jimmunol.169.11.6210. [DOI] [PubMed] [Google Scholar]
- 17.Liu W, Putnam AL, Xu-Yu Z, Szot GL, Lee MR, Zhu S, et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med. 2006;203:1701–1711. doi: 10.1084/jem.20060772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seddiki N, Santner-Nanan B, Martinson J, Zaunders J, Sasson S, Landay A, et al. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med. 2006;203:1693–1700. doi: 10.1084/jem.20060468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyara M, Yoshioka Y, Kitoh A, Shima T, Wing K, Niwa A, et al. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity. 2009;30:899–911. doi: 10.1016/j.immuni.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann P, Boeld TJ, Eder R, Huehn J, Floess S, Wieczorek G, et al. Loss of FOXP3 expression in natural human CD4+CD25+ regulatory T cells upon repetitive in vitro stimulation. Eur J Immunol. 2009;39:1088–1097. doi: 10.1002/eji.200838904. [DOI] [PubMed] [Google Scholar]
- 21.Allan SE, Song-Zhao GX, Abraham T, McMurchy AN, Levings MK. Inducible reprogramming of human T cells into Treg cells by a conditionally active form of FOXP3. Eur J Immunol. 2008;38:3282–3289. doi: 10.1002/eji.200838373. [DOI] [PubMed] [Google Scholar]
- 22.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell DJ, Koch MA. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat Rev Immunol. 2011;11:119–130. doi: 10.1038/nri2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zelenay S, Bergman ML, Paiva RS, Lino AC, Martins AC, Duarte JH, et al. Cutting edge: Intrathymic differentiation of adaptive Foxp3+ regulatory T cells upon peripheral proinflammatory immunization. J Immunol. 2010;185:3829–3833. doi: 10.4049/jimmunol.1001281. [DOI] [PubMed] [Google Scholar]
- 25.Farris AB, Taheri D, Kawai T, Fazlollahi L, Wong W, Tolkoff-Rubin N, et al. Acute renal endothelial injury during marrow recovery in a cohort of combined kidney and bone marrow allografts. Am J Transplant. 2011;11:1464–1477. doi: 10.1111/j.1600-6143.2011.03572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komatsu N, Hori S. Full restoration of peripheral Foxp3+ regulatory T cell pool by radioresistant host cells in scurfy bone marrow chimeras. Proc Natl Acad Sci U S A. 2007;104:8959–8964. doi: 10.1073/pnas.0702004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanakry CG, Ganguly S, Zahurak M, Bolanos-Meade J, Thoburn C, Perkins B, et al. Aldehyde dehydrogenase expression drives human regulatory T cell resistance to posttransplantation cyclophosphamide. Sci Transl Med. 2013;5:211ra157. doi: 10.1126/scitranslmed.3006960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gurkan S, Luan Y, Dhillon N, Allam SR, Montague T, Bromberg JS, et al. Immune reconstitution following rabbit antithymocyte globulin. Am J Transplant. 2010;10:2132–2141. doi: 10.1111/j.1600-6143.2010.03210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onoe T, Kalscheuer H, Chittenden M, Zhao G, Yang YG, Sykes M. Homeostatic expansion and phenotypic conversion of human T cells depend on peripheral interactions with APCs. J Immunol. 2010;184:6756–6765. doi: 10.4049/jimmunol.0901711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chace JH, Cowdery JS, Field EH. Effect of anti-CD4 on CD4 subsets I. Anti-CD4 preferentially deletes resting, naive CD4 cells and spares activated CD4 cells. J Immunol. 1994;152:405–412. [PubMed] [Google Scholar]
- 31.Baecher-Allan C, Brown JA, Freeman GJ, Hafler DA. CD4+CD25high regulatory cells in human peripheral blood. J Immunol. 2001;167:1245–1253. doi: 10.4049/jimmunol.167.3.1245. [DOI] [PubMed] [Google Scholar]
- 32.Baecher-Allan C, Wolf E, Hafler DA. MHC class II expression identifies functionally distinct human regulatory T cells. J Immunol. 2006;176:4622–4631. doi: 10.4049/jimmunol.176.8.4622. [DOI] [PubMed] [Google Scholar]
- 33.Tsuji M, Komatsu N, Kawamoto S, Suzuki K, Kanagawa O, Honjo T, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s patches. Science. 2009;323:1488–1492. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- 34.Komatsu N, Mariotti-Ferrandiz ME, Wang Y, Malissen B, Waldmann H, Hori S. Heterogeneity of natural Foxp3+ T cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc Natl Acad Sci U S A. 2009;106:1903–1908. doi: 10.1073/pnas.0811556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 36.Zheng SG, Wang J, Horwitz DA. Cutting edge: Foxp3+CD4+CD25+ regulatory T cells induced by IL-2 and TGF-beta are resistant to Th17 conversion by IL-6. J Immunol. 2008;180:7112–7116. doi: 10.4049/jimmunol.180.11.7112. [DOI] [PubMed] [Google Scholar]
- 37.Xu L, Kitani A, Fuss I, Strober W. Cutting edge: regulatory T cells induce CD4+CD25-Foxp3− T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-beta. J Immunol. 2007;178:6725–6729. doi: 10.4049/jimmunol.178.11.6725. [DOI] [PubMed] [Google Scholar]
- 38.Deknuydt F, Bioley G, Valmori D, Ayyoub M. IL-1beta and IL-2 convert human Treg into T(H)17 cells. Clin Immunol. 2009;131:298–307. doi: 10.1016/j.clim.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 39.Barthlott T, Kassiotis G, Stockinger B. T cell regulation as a side effect of homeostasis and competition. J Exp Med. 2003;197:451–460. doi: 10.1084/jem.20021387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gibbons C, Sykes M. Manipulating the immune system for anti-tumor responses and transplant tolerance via mixed hematopoietic chimerism. Immunol Rev. 2008;223:334–360. doi: 10.1111/j.1600-065X.2008.00636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyajima M, Chase CM, Alessandrini A, Farkash EA, Della PP, Benichou G, et al. Early acceptance of renal allografts in mice is dependent on foxp3(+) cells. Am J Pathol. 2011;178:1635–1645. doi: 10.1016/j.ajpath.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cook CH, Bickerstaff AA, Wang JJ, Nadasdy T, Della PP, Colvin RB, et al. Spontaneous renal allograft acceptance associated with “regulatory” dendritic cells and IDO. J Immunol. 2008;180:3103–3112. doi: 10.4049/jimmunol.180.5.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Expression of CD31 and Ki-67 in non-regulatory T cells
Figure S2: Number of Vβ detected in biopsies at different time points
Figure S3: Representative cell line sample for each patient
Figure S4: Representative cell line with Treg phenotype
Table S1: T cell clones defined as Tregs within pre- and post-transplant circulating sorted Treg T cell populations
Table S2: Pathologic evaluation of allograft biopsies in Subjects 1–5
Table S3: Comparison of the number of unique clones shared between biopsy and PBMCs (same time point) versus PBMCs at disparate time points for Subjects 1, 4, and 5.
Table S4: CD4, CD8, and FoxP3 phenotyping of expanded intragraft T cell line
Table S5: T cell clones from cultured cell lines compared peripheral blood T cells pre- and post-transplant, donor-reactive TCR as described in Morris et al. 2015 (10), and circulating T regulatory cell repertoires