Abstract
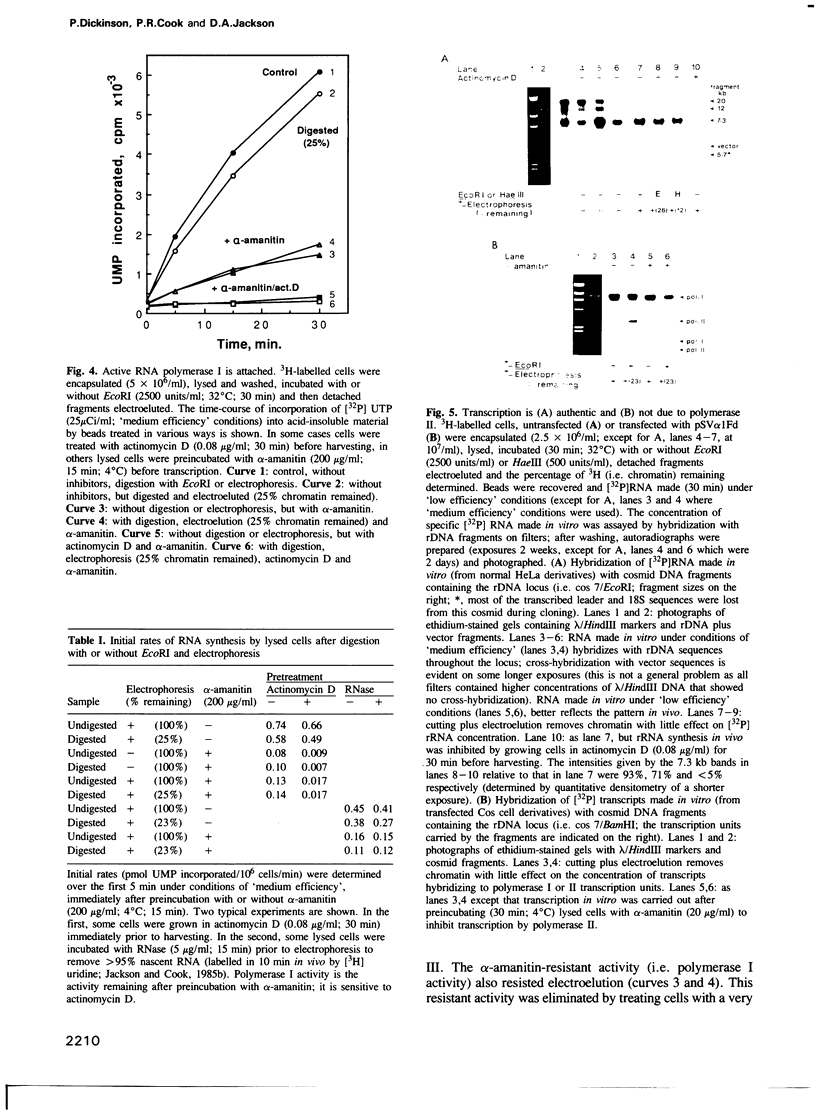
We have investigated whether active RNA polymerase I, the enzyme responsible for transcribing ribosomal RNA, is immobilized by attachment to a large subnuclear structure in HeLa cells. As unphysiological salt concentrations induce artifacts, we have used isotonic conditions throughout the preparative and analytic procedures. Cells are encapsulated in agarose microbeads and lysed in Triton and a 'physiological' buffer; then soluble proteins and RNA diffuse out through the agarose pores to leave encapsulated chromatin. This can be manipulated without aggregation but is accessible to molecular probes; it retains the replicational and transcriptional activities of the living cell. After treatment with a restriction endonuclease, most chromatin can be removed from beads by electrophoresis: then active ribosomal genes and polymerase I remain behind. Active ribosomal genes are very accessible to nuclease digestion whilst the rest are even more inaccessible than inactive globin genes. Our observations confirm the complex organization of rDNA within nucleoli and are compatible with transcription occurring at fixed sites. A model for transcription involving an attached polymerase is presented.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benavente R., Rose K. M., Reimer G., Hügle-Dörr B., Scheer U. Inhibition of nucleolar reformation after microinjection of antibodies to RNA polymerase I into mitotic cells. J Cell Biol. 1987 Oct;105(4):1483–1491. doi: 10.1083/jcb.105.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchsenius S., Bonven B., Leer J. C., Westergaard O. Nuclease-sensitive regions on the extrachromosomal r-chromatin from Tetrahymena pyriformis. Eur J Biochem. 1981 Jul;117(2):245–250. doi: 10.1111/j.1432-1033.1981.tb06329.x. [DOI] [PubMed] [Google Scholar]
- Briggs D., Jackson D., Whitelaw E., Proudfoot N. J. Direct demonstration of termination signals for RNA polymerase II from the sea urchin H2A histone gene. Nucleic Acids Res. 1989 Oct 25;17(20):8061–8071. doi: 10.1093/nar/17.20.8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Karrer K. M. Chromatin structure of the ribosomal RNA genes of Tetrahymena thermophila as analyzed by trimethylpsoralen crosslinking in vivo. J Mol Biol. 1980 Feb 5;136(4):395–416. doi: 10.1016/0022-2836(80)90397-6. [DOI] [PubMed] [Google Scholar]
- Conconi A., Widmer R. M., Koller T., Sogo J. M. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989 Jun 2;57(5):753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- Cook P. R. A general method for preparing intact nuclear DNA. EMBO J. 1984 Aug;3(8):1837–1842. doi: 10.1002/j.1460-2075.1984.tb02056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Mapping sequences in loops of nuclear DNA by their progressive detachment from the nuclear cage. Nucleic Acids Res. 1980 Jul 11;8(13):2895–2906. doi: 10.1093/nar/8.13.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P. R. The nucleoskeleton and the topology of transcription. Eur J Biochem. 1989 Nov 20;185(3):487–501. doi: 10.1111/j.1432-1033.1989.tb15141.x. [DOI] [PubMed] [Google Scholar]
- Cook P. R. The nucleoskeleton: artefact, passive framework or active site? J Cell Sci. 1988 May;90(Pt 1):1–6. doi: 10.1242/jcs.90.1.1. [DOI] [PubMed] [Google Scholar]
- Cox R. F. Quantitation of elongating form A and B RNA polymerases in chick oviduct nuclei and effects of estradiol. Cell. 1976 Mar;7(3):455–465. doi: 10.1016/0092-8674(76)90176-8. [DOI] [PubMed] [Google Scholar]
- Davis A. H., Reudelhuber T. L., Garrard W. T. Varigated chromatin structures of mouse ribosomal RNA genes. J Mol Biol. 1983 Jun 15;167(1):133–155. doi: 10.1016/s0022-2836(83)80038-2. [DOI] [PubMed] [Google Scholar]
- Fakan S., Puvion E. The ultrastructural visualization of nucleolar and extranucleolar RNA synthesis and distribution. Int Rev Cytol. 1980;65:255–299. doi: 10.1016/s0074-7696(08)61962-2. [DOI] [PubMed] [Google Scholar]
- Fleischmann G., Pflugfelder G., Steiner E. K., Javaherian K., Howard G. C., Wang J. C., Elgin S. C. Drosophila DNA topoisomerase I is associated with transcriptionally active regions of the genome. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6958–6962. doi: 10.1073/pnas.81.22.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D. S., Elgin S. C. Localization of specific topoisomerase I interactions within the transcribed region of active heat shock genes by using the inhibitor camptothecin. Mol Cell Biol. 1987 Jan;7(1):141–148. doi: 10.1128/mcb.7.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocke E., Bonven B. J., Westergaard O. A site and strand specific nuclease activity with analogies to topoisomerase I frames the rRNA gene of Tetrahymena. Nucleic Acids Res. 1983 Nov 25;11(22):7661–7678. doi: 10.1093/nar/11.22.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Smith V. A., Grummt F. Amino acid starvation affects the initiation frequency of nucleolar RNA polymerase. Cell. 1976 Mar;7(3):439–445. doi: 10.1016/0092-8674(76)90174-4. [DOI] [PubMed] [Google Scholar]
- Guldner H. H., Szostecki C., Vosberg H. P., Lakomek H. J., Penner E., Bautz F. A. Scl 70 autoantibodies from scleroderma patients recognize a 95 kDa protein identified as DNA topoisomerase I. Chromosoma. 1986;94(2):132–138. doi: 10.1007/BF00286991. [DOI] [PubMed] [Google Scholar]
- Henderson A. S., Warburton D., Atwood K. C. Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3394–3398. doi: 10.1073/pnas.69.11.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S. L., Ryan K., Sollner-Webb B. The promoter-proximal rDNA terminator augments initiation by preventing disruption of the stable transcription complex caused by polymerase read-in. Genes Dev. 1989 Feb;3(2):212–223. doi: 10.1101/gad.3.2.212. [DOI] [PubMed] [Google Scholar]
- Henderson S., Sollner-Webb B. A transcriptional terminator is a novel element of the promoter of the mouse ribosomal RNA gene. Cell. 1986 Dec 26;47(6):891–900. doi: 10.1016/0092-8674(86)90804-4. [DOI] [PubMed] [Google Scholar]
- Jackson D. A., Cook P. R. A cell-cycle-dependent DNA polymerase activity that replicates intact DNA in chromatin. J Mol Biol. 1986 Nov 5;192(1):65–76. doi: 10.1016/0022-2836(86)90464-x. [DOI] [PubMed] [Google Scholar]
- Jackson D. A., Cook P. R. A general method for preparing chromatin containing intact DNA. EMBO J. 1985 Apr;4(4):913–918. doi: 10.1002/j.1460-2075.1985.tb03718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. A., Cook P. R., Patel S. B. Attachment of repeated sequences to the nuclear cage. Nucleic Acids Res. 1984 Sep 11;12(17):6709–6726. doi: 10.1093/nar/12.17.6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. A., Cook P. R. Transcription occurs at a nucleoskeleton. EMBO J. 1985 Apr;4(4):919–925. doi: 10.1002/j.1460-2075.1985.tb03719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. A., Cook P. R. Visualization of a filamentous nucleoskeleton with a 23 nm axial repeat. EMBO J. 1988 Dec 1;7(12):3667–3677. doi: 10.1002/j.1460-2075.1988.tb03248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. A., Dickinson P., Cook P. R. The size of chromatin loops in HeLa cells. EMBO J. 1990 Feb;9(2):567–571. doi: 10.1002/j.1460-2075.1990.tb08144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. A., McCready S. J., Cook P. R. Replication and transcription depend on attachment of DNA to the nuclear cage. J Cell Sci Suppl. 1984;1:59–79. doi: 10.1242/jcs.1984.supplement_1.5. [DOI] [PubMed] [Google Scholar]
- Jackson D. A., Yuan J., Cook P. R. A gentle method for preparing cyto- and nucleo-skeletons and associated chromatin. J Cell Sci. 1988 Jul;90(Pt 3):365–378. doi: 10.1242/jcs.90.3.365. [DOI] [PubMed] [Google Scholar]
- Jordan E. G., McGovern J. H. The quantitative relationship of the fibrillar centres and other nucleolar components to changes in growth conditions, serum deprivation and low doses of actinomycin D in cultured diploid human fibroblasts (strain MRC-5). J Cell Sci. 1981 Dec;52:373–389. doi: 10.1242/jcs.52.1.373. [DOI] [PubMed] [Google Scholar]
- Jordan G. At the heart of the nucleolus. Nature. 1987 Oct 8;329(6139):489–490. doi: 10.1038/329489a0. [DOI] [PubMed] [Google Scholar]
- Keppel F. Transcribed human ribosomal RNA genes are attached to the nuclear matrix. J Mol Biol. 1986 Jan 5;187(1):15–21. doi: 10.1016/0022-2836(86)90402-x. [DOI] [PubMed] [Google Scholar]
- Lucchini R., Reeder R. H. A test of 'polymerase handover' as a mechanism for stimulating initiation by RNA polymerase I. Nucleic Acids Res. 1989 Jan 11;17(1):373–387. doi: 10.1093/nar/17.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum F. S., Maden B. E. Human 18 S ribosomal RNA sequence inferred from DNA sequence. Variations in 18 S sequences and secondary modification patterns between vertebrates. Biochem J. 1985 Dec 15;232(3):725–733. doi: 10.1042/bj2320725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Miller O. L., Jr Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell. 1976 Jun;8(2):305–319. doi: 10.1016/0092-8674(76)90014-3. [DOI] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. A termination site for Xenopus RNA polymerase I also acts as an element of an adjacent promoter. Cell. 1986 Dec 26;47(6):913–920. doi: 10.1016/0092-8674(86)90806-8. [DOI] [PubMed] [Google Scholar]
- Miller O. L., Jr, Beatty B. R. Visualization of nucleolar genes. Science. 1969 May 23;164(3882):955–957. doi: 10.1126/science.164.3882.955. [DOI] [PubMed] [Google Scholar]
- Miller O. L., Jr Some ultrastructural aspects of genetic activity in eukaryotes. J Cell Sci Suppl. 1984;1:81–93. doi: 10.1242/jcs.1984.supplement_1.6. [DOI] [PubMed] [Google Scholar]
- Ness P. J., Koller T., Thoma F. Topoisomerase I cleavage sites identified and mapped in the chromatin of Dictyostelium ribosomal RNA genes. J Mol Biol. 1988 Mar 5;200(1):127–139. doi: 10.1016/0022-2836(88)90338-5. [DOI] [PubMed] [Google Scholar]
- Ochs R. L., Smetana K. Fibrillar center distribution in nucleoli of PHA-stimulated human lymphocytes. Exp Cell Res. 1989 Oct;184(2):552–557. doi: 10.1016/0014-4827(89)90354-6. [DOI] [PubMed] [Google Scholar]
- Ochs R., Lischwe M., O'Leary P., Busch H. Localization of nucleolar phosphoproteins B23 and C23 during mitosis. Exp Cell Res. 1983 Jun;146(1):139–149. doi: 10.1016/0014-4827(83)90332-4. [DOI] [PubMed] [Google Scholar]
- Pardoll D. M., Vogelstein B. Sequence analysis of nuclear matrix associated DNA from rat liver. Exp Cell Res. 1980 Aug;128(2):466–470. doi: 10.1016/0014-4827(80)90083-x. [DOI] [PubMed] [Google Scholar]
- Reeves R. Ribosomal genes of Xenopus laevis: evidence of nucleosomes in transcriptionally active chromatin. Science. 1976 Oct 29;194(4264):529–532. doi: 10.1126/science.973136. [DOI] [PubMed] [Google Scholar]
- Reimer G., Rose K. M., Scheer U., Tan E. M. Autoantibody to RNA polymerase I in scleroderma sera. J Clin Invest. 1987 Jan;79(1):65–72. doi: 10.1172/JCI112809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safrany G., Hidvegi E. J. New tandem repeat region in the non-transcribed spacer of human ribosomal RNA gene. Nucleic Acids Res. 1989 Apr 25;17(8):3013–3022. doi: 10.1093/nar/17.8.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U., Rose K. M. Localization of RNA polymerase I in interphase cells and mitotic chromosomes by light and electron microscopic immunocytochemistry. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1431–1435. doi: 10.1073/pnas.81.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Tower J. Transcription of cloned eukaryotic ribosomal RNA genes. Annu Rev Biochem. 1986;55:801–830. doi: 10.1146/annurev.bi.55.070186.004101. [DOI] [PubMed] [Google Scholar]
- Wachtler F., Hartung M., Devictor M., Wiegant J., Stahl A., Schwarzacher H. G. Ribosomal DNA is located and transcribed in the dense fibrillar component of human Sertoli cell nucleoli. Exp Cell Res. 1989 Sep;184(1):61–71. doi: 10.1016/0014-4827(89)90364-9. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Secondary structure maps of RNA: processing of HeLa ribosomal RNA. Proc Natl Acad Sci U S A. 1973 Oct;70(10):2827–2831. doi: 10.1073/pnas.70.10.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zackroff R. V., Goldman R. D. In vitro assembly of intermediate filaments from baby hamster kidney (BHK-21) cells. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6226–6230. doi: 10.1073/pnas.76.12.6226. [DOI] [PMC free article] [PubMed] [Google Scholar]