Abstract
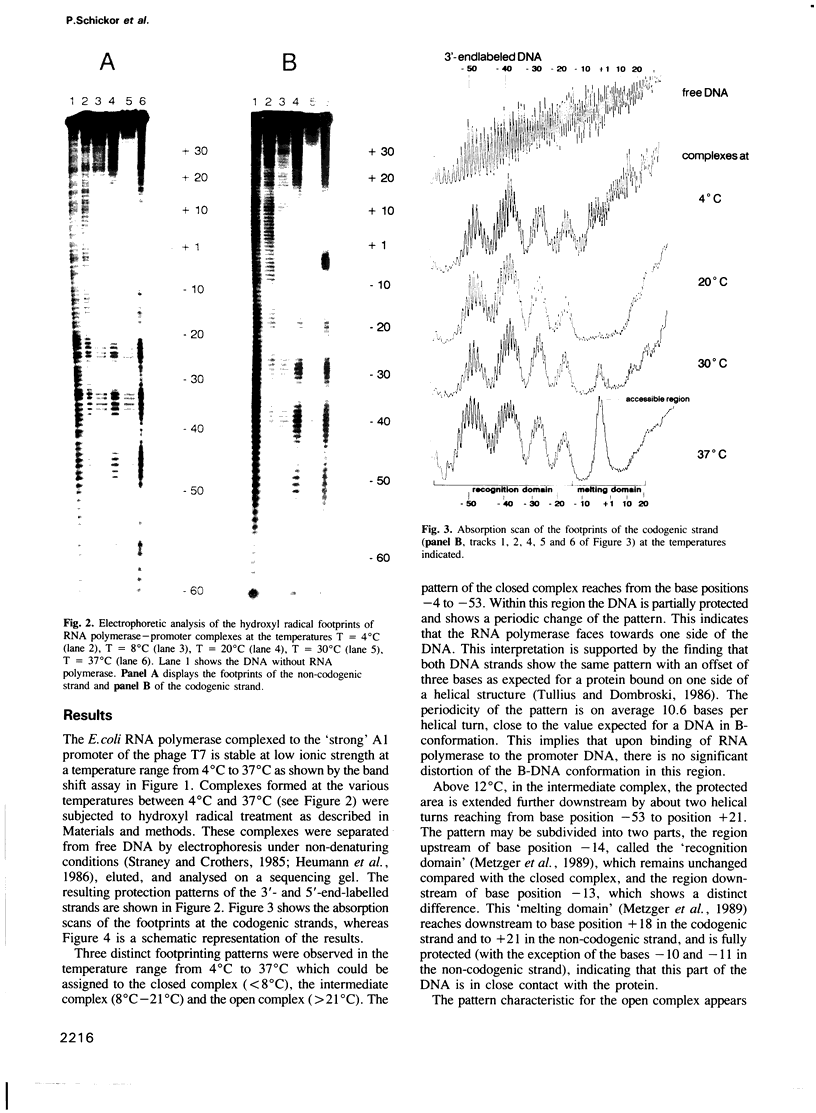
Three characteristic footprinting patterns resulted from probing the Escherichia coli RNA polymerase T7 A1 promoter complex by hydroxyl radicals in the temperature range between 4 degrees C and 37 degrees C. These were attributed to the closed complex, the intermediate complex and the open complex. In the closed complex, the RNA polymerase protects the DNA only at one side over five helical turns. In the intermediate complex, the range of the protected area is extended further downstream by two helical turns. This region of the DNA helix is fully protected, indicating that the RNA polymerase wraps around the DNA between base positions -13 and +20. In the open complex, a stretch between base positions -7 and +2, which was fully protected in the intermediate complex, becomes accessible towards hydroxyl radicals but only in the codogenic strand, indicating that the DNA strands are unwound. Our data suggest that only the DNA downstream of the promoter is involved in this unwinding process.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amouyal M., Buc H. Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and the UV5 sites of Escherichia coli. J Mol Biol. 1987 Jun 20;195(4):795–808. doi: 10.1016/0022-2836(87)90485-2. [DOI] [PubMed] [Google Scholar]
- Buc H., McClure W. R. Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter. Evidence for a sequential mechanism involving three steps. Biochemistry. 1985 May 21;24(11):2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- Buckle M., Buc H. Fine mapping of DNA single-stranded regions using base-specific chemical probes: study of an open complex formed between RNA polymerase and the lac UV5 promoter. Biochemistry. 1989 May 16;28(10):4388–4396. doi: 10.1021/bi00436a040. [DOI] [PubMed] [Google Scholar]
- Collis C. M., Molloy P. L., Both G. W., Drew H. R. Influence of the sequence-dependent flexure of DNA on transcription in E. coli. Nucleic Acids Res. 1989 Nov 25;17(22):9447–9468. doi: 10.1093/nar/17.22.9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darst S. A., Kubalek E. W., Kornberg R. D. Three-dimensional structure of Escherichia coli RNA polymerase holoenzyme determined by electron crystallography. Nature. 1989 Aug 31;340(6236):730–732. doi: 10.1038/340730a0. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper H. B., Hearst J. E. A topological model for transcription based on unwinding angle analysis of E. coli RNA polymerase binary, initiation and ternary complexes. Cell. 1982 May;29(1):81–90. doi: 10.1016/0092-8674(82)90092-7. [DOI] [PubMed] [Google Scholar]
- Gamper H. B., Hearst J. E. Size of the unwound region of DNA in Escherichia coli RNA polymerase and calf thymus RNA polymerase II ternary complexes. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):455–461. doi: 10.1101/sqb.1983.047.01.054. [DOI] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R. Overexpression and purification of the sigma subunit of Escherichia coli RNA polymerase. Gene. 1983 Dec;26(2-3):109–118. doi: 10.1016/0378-1119(83)90180-4. [DOI] [PubMed] [Google Scholar]
- Heumann H., Lederer H., Baer G., May R. P., Kjems J. K., Crespi H. L. Spatial arrangement of DNA-dependent RNA polymerase of Escherichia coli and DNA in the specific complex. A neutron small angle scattering study. J Mol Biol. 1988 May 5;201(1):115–125. doi: 10.1016/0022-2836(88)90443-3. [DOI] [PubMed] [Google Scholar]
- Heumann H., Lederer H., Kammerer W., Palm P., Metzger W., Baer G. Large-scale preparation of a DNA fragment containing the strong promoter A1 of the phage T7. Biochim Biophys Acta. 1987 Jul 14;909(2):126–132. doi: 10.1016/0167-4781(87)90034-0. [DOI] [PubMed] [Google Scholar]
- Heumann H., Metzger W., Niehörster M. Visualization of intermediary transcription states in the complex between Escherichia coli DNA-dependent RNA polymerases and a promoter-carrying DNA fragment using the gel retardation method. Eur J Biochem. 1986 Aug 1;158(3):575–579. doi: 10.1111/j.1432-1033.1986.tb09793.x. [DOI] [PubMed] [Google Scholar]
- Heumann H., Ricchetti M., Werel W. DNA-dependent RNA polymerase of Escherichia coli induces bending or an increased flexibility of DNA by specific complex formation. EMBO J. 1988 Dec 20;7(13):4379–4381. doi: 10.1002/j.1460-2075.1988.tb03336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer B., Müller D., Köster H. The pathway of E. coli RNA polymerase-promoter complex formation as visualized by footprinting. Nucleic Acids Res. 1985 Aug 26;13(16):5995–6013. doi: 10.1093/nar/13.16.5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Buc H., Spassky A., Wang J. C. Mapping of single-stranded regions in duplex DNA at the sequence level: single-strand-specific cytosine methylation in RNA polymerase-promoter complexes. Proc Natl Acad Sci U S A. 1983 May;80(9):2544–2548. doi: 10.1073/pnas.80.9.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudelka G. B., Harbury P., Harrison S. C., Ptashne M. DNA twisting and the affinity of bacteriophage 434 operator for bacteriophage 434 repressor. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4633–4637. doi: 10.1073/pnas.85.13.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnke G., Fritz H. J., Ehring R. Unusual properties of promoter-up mutations in the Escherichia coli galactose operon and evidence suggesting RNA polymerase-induced DNA bending. EMBO J. 1987 Feb;6(2):507–513. doi: 10.1002/j.1460-2075.1987.tb04782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnke G., Theres C., Fritz H. J., Ehring R. RNA polymerase and gal repressor bind simultaneously and with DNA bending to the control region of the Escherichia coli galactose operon. EMBO J. 1989 Apr;8(4):1247–1255. doi: 10.1002/j.1460-2075.1989.tb03498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
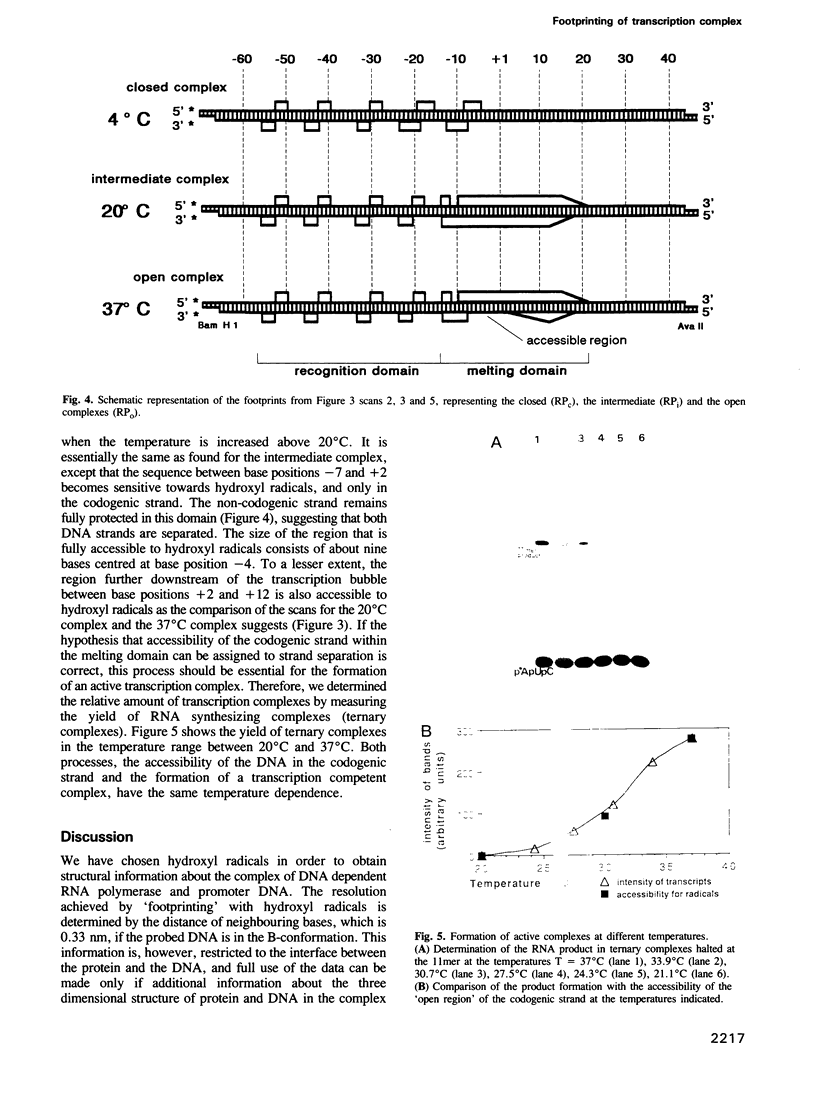
- Lederer H., May R. P., Kjems J. K., Baer G., Heumann H. Solution structure of a short DNA fragment studied by neutron scattering. Eur J Biochem. 1986 Nov 17;161(1):191–196. doi: 10.1111/j.1432-1033.1986.tb10141.x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisenberger O., Heumann H., Pilz I. Small-angle X-ray study of DNA-dependent RNA polymerase holoenzyme from Escherichia coli. FEBS Lett. 1981 Jan 12;123(1):22–24. doi: 10.1016/0014-5793(81)80010-5. [DOI] [PubMed] [Google Scholar]
- Metzger W., Schickor P., Heumann H. A cinematographic view of Escherichia coli RNA polymerase translocation. EMBO J. 1989 Sep;8(9):2745–2754. doi: 10.1002/j.1460-2075.1989.tb08416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe J. H., Burgess R. R., Record M. T., Jr Temperature dependence of the rate constants of the Escherichia coli RNA polymerase-lambda PR promoter interaction. Assignment of the kinetic steps corresponding to protein conformational change and DNA opening. J Mol Biol. 1985 Aug 5;184(3):441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]
- Rosenberg S., Kadesch T. R., Chamberlin M. J. Binding of Escherichia coli RNA polymerase holoenzyme to bacteriophage T7 DNA. Measurements of the rate of open complex formation at T7 promoter A. J Mol Biol. 1982 Feb 15;155(1):31–51. doi: 10.1016/0022-2836(82)90490-9. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U. RNA polymerase unwinds an 11-base pair segment of a phage T7 promoter. Nature. 1979 Jun 14;279(5714):651–652. doi: 10.1038/279651a0. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Simpson R. B. The molecular topography of RNA polymerase-promoter interaction. Cell. 1979 Oct;18(2):277–285. doi: 10.1016/0092-8674(79)90047-3. [DOI] [PubMed] [Google Scholar]
- Spassky A., Kirkegaard K., Buc H. Changes in the DNA structure of the lac UV5 promoter during formation of an open complex with Escherichia coli RNA polymerase. Biochemistry. 1985 May 21;24(11):2723–2731. doi: 10.1021/bi00332a019. [DOI] [PubMed] [Google Scholar]
- Straney D. C., Crothers D. M. Intermediates in transcription initiation from the E. coli lac UV5 promoter. Cell. 1985 Dec;43(2 Pt 1):449–459. doi: 10.1016/0092-8674(85)90175-8. [DOI] [PubMed] [Google Scholar]
- Stöckel P., May R., Strell I., Cejka Z., Hoppe W., Heumann H., Zillig W., Crespi H. L. The subunit positions within RNA polymerase holoenzyme determined by triangulation of centre-to-centre distances. Eur J Biochem. 1980 Nov;112(2):419–423. doi: 10.1111/j.1432-1033.1980.tb07221.x. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter G., Zillig W., Palm P., Fuchs E. Initiation of DNA-dependent RNA synthesis and the effect of heparin on RNA polymerase. Eur J Biochem. 1967 Dec;3(2):194–201. doi: 10.1111/j.1432-1033.1967.tb19515.x. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Jacobsen J. H., Saucier J. M. Physiochemical studies on interactions between DNA and RNA polymerase. Unwinding of the DNA helix by Escherichia coli RNA polymerase. Nucleic Acids Res. 1977;4(5):1225–1241. doi: 10.1093/nar/4.5.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillig W., Zechel K., Halbwachs H. J. A new method of large scale preparation of highly purified DNA-dependent RNA-polymerase from E. coli. Hoppe Seylers Z Physiol Chem. 1970 Feb;351(2):221–224. doi: 10.1515/bchm2.1970.351.1.221. [DOI] [PubMed] [Google Scholar]