Abstract
The protein Crumbs is a determinant of apical-basal cell polarity and plays a role in apoptosis of epithelial cells and their protection against photodamage. Using the squid-vibrio system, a model for development of symbiotic partnerships, we examined the modulation of the crumbs gene in host epithelial tissues during initiation and maintenance of the association. The extracellular luminous symbiont Vibrio fischeri colonizes the apical surfaces of polarized epithelia in deep crypts of the Euprymna scolopes light organ. During initial colonization each generation, symbiont harvesting is potentiated by the biochemical and biophysical activity of superficial ciliated epithelia, which are several cell layers from the crypt epithelia where the symbionts reside. Within hours of crypt colonization, the symbionts induce the cell-death mediated regression of the remote superficial ciliated fields. However, the crypt cells directly interacting with the symbiont are protected from death. In the squid host, we characterized the gene and encoded protein during light-organ morphogenesis and in response to symbiosis. Features of the protein sequence and structure, phylogenetic relationships, and localization patterns in the eye supported assignment of the squid protein to the Crumbs family. In situ hybridization revealed that the crumbs transcript shows opposite expression at the onset of symbiosis in the two different regions of the light organ: elevated levels in the superficial epithelia were attenuated whereas low levels in the crypt epithelia were turned up. Although a rhythmic association in which the host controls the symbiont population over the day-night cycle begins in the juvenile upon colonization, cycling of crumbs was evident only in the adult organ with peak expression coincident with maximum symbiont population and luminescence. Our results provide evidence that crumbs responds to symbiont cues that induce developmental apoptosis and to symbiont population dynamics correlating with luminescence-based stress throughout the duration of the host-microbe association.
Keywords: apoptosis, cephalopod, eye, photophore, photoreceptor, squid vibrio, symbiosis
Introduction
As a regulator of apical-basal polarity (Wodarz et al. 1995) and localizer of adherens junctions (Izaddoost et al. 2002), the Crumbs protein maintains individual cell and cell-cell integrity in epithelial tissues. Providing these critical roles, Crumbs serves as a major determinant of cell life and death in eukaryotic organisms (Laprise 2011). Dysfunction of the protein can have devastating effects and is associated with some of the most intractable diseases (Bulgakova and Knust 2009). Further, a functional protein is essential for normal development and the maintenance of integrity in mature tissues; for example, Crumbs is crucial for the healthy structure and function of the retina and morphogenesis of the light-sensitive photoreceptor cells within (den Hollander et al. 1999; Johnson et al. 2002; Pellikka et al. 2002).
A transcriptional database derived from the light organ of the bobtail squid Euprymna scolopes (Fig. 1a, b) revealed a crumbs gene transcript (es-crumbs) (see Chun et al. 2006). The organ serves as a home for the luminous bacterial symbiont Vibrio fischeri. The host squid, which is a nocturnal predator in the shallow sand flats of the Hawaiian archipelago, uses the light produced by the bacterial partner to camouflage in a behavior called counterillumination (Jones and Nishiguchi 2004). Each generation the symbiont is acquired from the environment and induces morphogenesis of the light organ over the first hours to days of the symbiosis (for review see McFall-Ngai 2014). Briefly, a nascent light organ bearing superficial ciliated fields develops in the embryonic squid (Fig. 1a, b). These fields potentiate recruitment of the symbiont from the surrounding seawater. Once recruited as a small aggregate on the ciliated surface, the symbiont cells migrate into deep crypts lined by epithelia, where they take up residence along the apical surfaces of these host cells (Lamarcq and McFall-Ngai 1998; Heath-Heckman et al. 2016). Following the initiation of symbiosis, the light organ proceeds through a developmental process in which the cells of these fields undergo apoptosis, ultimately leading to the complete regression of this superficial tissue (Fig. 1c). Cell-envelope molecules presented by V. fischeri, specifically lipid A and tracheal cytotoxin (TCT), derivatives of lipopolysaccharide (LPS) and peptidoglycan, respectively, are the principal triggers of this process. These factors are members of a class of molecules called MAMPs, or microbe-associated molecular patterns, which signal to animal cells that they are interacting with microbes. A secondary morphogen, V. fischeri luminescence, accelerates the developmental process.
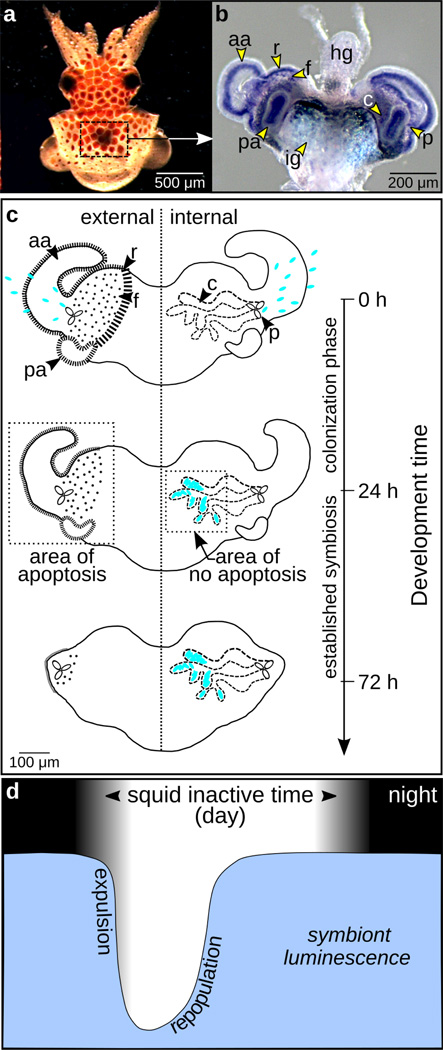
Figure 1.
Euprymna scolopes-Vibrio fischeri system. (a) Juvenile E. scolopes in which the light organ, bounded by the box, is visible through the dorsal surface. (b) Juvenile light organ surrounded by the ink gland (ig) and attached to the hindgut (hg). This light organ shows labeling of the developmental gene pax6 (Peyer et al. 2014), enhancing the visibility of the morphological structures described in the following. (c) Line drawing of the developing light organ in the newly hatched squid. Right side: Harvested from the seawater each generation, V. fischeri (shown as blue ovals) enters through pores (p) on either side of the light organ and ultimately resides along the apical surfaces of polarized epithelia in the crypt spaces (c) by 24 h post colonization. Left side: The superficial tissues of the light organ include the anterior appendages (aa), posterior appendages (pa), and the ciliated ridges and field (r and f). In contrast to the crypts, these superficial tissues undergo apoptosis immediately following symbiont colonization, a process that is largely complete within 96 h post colonization. (d) Level of symbiont luminescence corresponding with the nocturnal activity of the squid during evening hours (no scale). To control symbiont population, the squid expels the majority of the bacteria daily at dawn. The remaining bacterial cells repopulate the light organ during the day when the squid is inactive.
The symbiont factors, MAMPS and luminescence, signal from the crypts and trigger apoptosis in the superficial tissues (Fig. 1c: appendages, ciliated regions; Doino and McFall-Ngai 1995), which are several cell layers away from the crypts themselves. Interestingly, the crypt cells that interact directly with these symbiont factors are not triggered to undergo apoptosis. Thus, early development of the organ offers an interesting subject for the study of the crumbs gene, as the juvenile light-organ epithelia experience both apoptosis on the surface and exposure to MAMPs and light in the crypts, activities that typically control crumbs transcription in different ways in other systems (Pichaud 2014).
In addition to triggering morphogenesis, the symbiont induces the onset of a daily rhythm in the association in which the host expels the symbiont at dawn to control overpopulation and then promotes its regrowth over the subsequent 6-h period (Fig. 1d); the symbiont population is thus restored before the animal emerges to forage at night (Boettcher et al. 1996). The host tissues also have a concomitant rhythm in which the apical surfaces of the epithelial cells lose their microvilli as the symbionts are expelled at dawn and repolarize with the regrowth of the microbial partner (Wier et al. 2010). Interestingly, this effacement and regrowth is a feature of both the light-organ crypt epithelia and the squid’s photoreceptor cells (Heath-Heckman et al. 2016); squids and their relatives renew their microvillous-based photoreceptor cells daily (Young 1967). The effacement in both cases occurs as a response to the light cue of dawn. This convergence in the diel dynamics of the microvillous epithelium of the eye and light organ add to a growing list of similarities between these two tissues (Montgomery and McFall-Ngai 1992; Crookes et al. 2004; Tong et al. 2009; Peyer et al. 2014).
Superimposed on this environmental-light influence in the crypts is the luminescence of the symbionts. Beginning immediately upon colonization of the juvenile, and continuing on through maturity, the light organ exhibits an additional daily rhythm in which the animal controls symbiont luminescence (Boettcher et al. 1995), with emission being highest in the evening when the animal is using bacterial light production in its camouflaging behavior (Fig. 1d).
The study of Crumbs within the light organ provides an opportunity to characterize evolutionarily-conserved cellular responses of animals in the context of initiation and persistence of a symbiotic association. Here we describe the es-crumbs transcript of E. scolopes, derive its phylogenetic relatedness to Crumbs protein family members in other animals, and explore the expression of the gene during the early development of the symbiosis and in the mature association. The results of our study suggest that es-crumbs responds to symbiont cues that induce apoptosis-mediated morphogenesis of the light organ in the juvenile squid and symbiont population dynamics and luminescence in the mature squid.
Methods
General methods
The breeding colony of E. scolopes was obtained from O‘ahu, Hawai‘i, USA, and maintained in the laboratory as described previously (Montgomery and McFall-Ngai 1993). We stored all tissues designated for PCR work in RNAlater (Thermo Fisher Scientific) for 24 h at 4°C and then at −80°C. We performed RNA extractions with the RNeasy Mini Kit (QIAGEN) and removed any contaminating DNA by DNase treatment with Ambion Turbo DNase Kit (Thermo Fisher Scientific). We measured RNA concentrations with a Qubit 2.0 fluorometer (Thermo Fisher Scientific) and tested the quality of the RNA by agarose gel electrophoresis. The primers (Integrated DNA Technologies) and probes (Molecular Instruments) used in the experiments that follow are listed in Table 1. All chemicals were from Sigma-Aldrich unless otherwise specified.
Table 1.
Primers for RACE-PCR, RT-PCR, qRT-PCR, and ISH, and probes for HCR-FISH.
| Gene | Primer Type | Direction | Sequence (5’ to 3’) |
|---|---|---|---|
| crumbs | 5’ RACE-PCR | Reverse | AGCACGGGTTCTCAATGTGGCACTAT |
| 5’ RACE-PCR nested | Reverse | CCATCATGCAGGCAAGGATTCAC | |
| 3’ RACE-PCR | Forward | GGAACCTACAGTCCCAGTCAGCAAGA | |
| 3’ RACE-PCR nested | Forward | CTCGTGTGGAACTCGGCAATGTTATG | |
| RT-PCR | Forward | ATTATCCGCCACCCTTCTGCAA | |
| RT-PCR | Reverse | AAGTCCTGCTCTTTGGTCTGCT | |
| qRT-PCR | Forward | TGTGGAACTCGGCAATGTTA | |
| qRT-PCR | Reverse | TCCTGCTCTTTGGTCTGCTT | |
| ISH (antisense) | Forward | ATACTGCCACATTGAGAACCCGTGCT | |
| ISH (antisense) | Reverse (T7) | *T7 + CATACCATTGCCGAGTTCCACACGAG* | |
| ISH (sense) | Forward (T7) | *T7 + ATACTGCCACATTGAGAACCCGTGCT* | |
| ISH (sense) | Reverse | CATACCATTGCCGAGTTCCACACGAG | |
| HCR-FISH probe 1 | NA | CGACCGATAGAGATCACAGGTCGAAACAA ACCAACTGCACCCAACGACAT |
|
| HCR-FISH probe 2 | NA | GAAAATAAAATCAACCCGAGTGAGTCGCTG TCGTTGTTGGCAACGGTGGC |
|
| HCR-FISH probe 3 | NA | GACAGTTGGGGTAGATCTTGCATTCGTCGAT GTTTTGTTCACACCTACAC |
|
| rhodopsin | ISH (antisense) | Forward | CACCAGCCAACATGTTCATC |
| ISH (antisense) | Reverse (T7) | T7 + CCGATAGCCCATAGGACAGA | |
| ISH (sense) | Forward (T7) | T7 + CACCAGCCAACATGTTCATC | |
| ISH (sense) | Reverse | CCGATAGCCCATAGGACAGA | |
| 40s ribosomal | qRT-PCR | Forward | AATCTCGGCGTCCTTGAGAA |
| qRT-PCR | Reverse | GCATCAATTGCACGACGAGT | |
| serine HMT | qRT-PCR | Forward | GTCCTGGTGACAAGAGTGCAATGA |
| qRT-PCR | Reverse | TTCCAGCAGAAAGGCACGATAGGT | |
| M13 | Sequencing | Forward | GTAAAACGACGGCCAG |
| Sequencing | Reverse | CAGGAAACAGCTATGAC |
*T7 = TAATACGACTCACTATAGGG.
Sequence generation of the crumbs transcript
As only a portion of the es-crumbs transcript was available from the EST database (Chun et al. 2006), we performed RACE-PCR (Rapid Amplification of cDNA Ends – PCR) to obtain the full sequence using GeneRacer cDNA amplification kit (Thermo Fisher Scientific). We used RNA from ~50 juvenile light organs to produce RACE-ready cDNA, and RNA from ~25 eyes to determine whether the same isoform occurs in both tissues. The remaining procedural details, including PCR, cloning, and purification, are described in a previous study (Peyer et al. 2014).
Sequence alignment and phylogenetic reconstruction
To determine the extent to which functional domains were conserved, we aligned the C-terminus portion of the translated EsCrumbs protein sequence with those of other representative species. This C-terminus portion in other organisms includes the transmembrane section and intracellular region featuring the FERM (4.1/Ezrin/Radixin/Moesin) and PDZ (PSD-95/Discs large/ZO-1) domains. In the alignment, we used model organisms from the Deuterostomia (zebrafish, Danio rerio, mouse, Mus musculus, and human, Homo sapiens) and Ecdysozoa (fruit fly, Drosophila melanogaster) superphyla. In addition, we included one Lophotrochozoa representative, the owl limpet Lottia gigantea, whose genome contains a sequence with similarity to EsCrumbs. We generated the alignments with CLC Sequence Viewer software (CLC Bio, QIAGEN). We also used the Simple Modular Architecture Research Tool (SMART; http://smart.embl-heidelberg.de/) to identify the presence and position of functional domains.
In addition, we tested for orthology between EsCrumbs and previously published sequences. To locate sequences similar to EsCrumbs, we searched the NCBI database using BLASTP (e-value < 1e-20) with default parameter settings. We also included the L. gigantea sequence, which we obtained using BlastX analysis against the genome. For the phylogenetic reconstruction, we used the 150 C-terminus amino acids of the sequences, which included evolutionarily-conserved residues while excluding ambiguous regions. We aligned the sequences in CLC Sequence Viewer software (CLC Bio, QIAGEN). After applying Gblocks to remove nonconserved regions, we performed maximum-likelihood phylogenetic analysis using the software program PhyML 3.1/3.0 assuming a WAG (Whelan and Goldman) model (Dereeper et al. 2008, 2010). We estimated support for bipartitions by examining the consensus of 500 bootstrapped maximum-likelihood trees.
Light-organ colonization with V. fischeri
In the juvenile light organ, we examined gene expression in response to symbiosis (see following sections). In preparation for experiments, we handled the treatments as follows. Immediately after hatching, we distributed the juveniles among different groups: aposymbiotic (uncolonized), symbiotic with wild-type luminous V. fischeri, and symbiotic with a V. fischeri mutant (Δlux) defective in light production (Bose et al. 2008). The terms ‘symbiotic’ and ‘aposymbiotic’ refer to squid exposed to environmental bacteria with and without V. fischeri, respectively. To determine host tissue response to bacterial luminescence, we compared light organs colonized by wild-type V. fischeri to those aposymbiotic and those colonized with Δlux V. fischeri. In a similar manner, to determine the effect of the bacteria alone on host tissue, we compared light organs colonized by either wild-type or Δlux V. fischeri to those aposymbiotic. For the bacterial colonizations, we followed an established protocol using 5000 colony-forming units of V. fischeri per mL of artificial seawater (Ruby and Asato 1993). We confirmed colonization of the light organ with wild-type V. fischeri with a TD 20/20 luminometer (Turner Designs). In our experiments, we sampled the light organs at a time point of 24 h post colonization, which corresponded with dusk when the symbiont population was high (Fig. 1d).
In situ hybridization
We used in situ hybridization (ISH) to examine es-crumbs expression in response to symbiosis. Our comparisons included light organs that were aposymbiotic, symbiotic with wild-type, or symbiotic with Δlux V. fischeri at the 24-h time point. By employing colorimetric ISH with Nitro Blue Tetrazolium and 5-bromo-4-chloro-3’-indolyl phosphate substrates as described in previous studies (Lee et al. 2009; Peyer et al. 2014), we visualized transcript expression primarily in surface tissues of the light organ (see Fig. 1b, c appendages, ciliated ridges, pores), though the crypts are also visible with the overlying tissue being somewhat transparent. To determine if transcript expression differed among the three colonization conditions, we simultaneously terminated all reactions following signal development; controls in which we exposed tissues to sense probes were also included. As a complementary experiment, we examined histological samples of the light organ and eye using paraffin-section ISH methods as previously described (Peyer et al. 2014). In the eye, we localized es-crumbs along with es-rhodopsin to confirm that both genes occur in regions of the photoreceptors similar to other animals (Kingston et al. 2015; Hsu and Jensen 2010). We included rhodopsin because it is a common reference gene in the eye. We also observed es-crumbs expression in embryos during early eye morphogenesis (stage 20, ~2/3 through embyogenesis) using whole-mount colorimetric ISH.
In addition to viewing surface tissues of the light organ with colorimetric ISH, we used hybridization chain reaction-fluorescent ISH (HCR-FISH) to co-localize es-crumbs expression and either wild-type or Δlux V. fischeri labeling with 16S ribosomal RNA in crypt epithelia. Our methods followed those described in a previous study (Nikolakakis et al. 2015) and we used a total of three crumbs probes (Molecular Instruments; Table 1) that had minimal sequence similarity with other host transcripts. We analyzed our samples on a Zeiss LSM 510 confocal microscope and normalized the gain of all symbiotic light organs to that of aposymbiotic light organs at which transcript signal was just barely detectable.
For each gene studied in the above experiments, we performed two replicate trials (N = 12 to 20 total animals per condition per gene). We collected all animals for each replicate from a single clutch so as to minimize variation in expression patterns due to genetic effects.
Quantitative real-time PCR
By qRT-PCR, we examined differential regulation of es-crumbs in the juvenile light organ in response to the onset of symbiosis and in the mature light organ over the day-night cycle. For all experiments, we followed previously established guidelines (Bustin et al. 2010). We collected the juveniles from several clutches immediately after hatching and divided them equally among different conditions, i.e., 24-h aposymbiotic and symbiotic with either wild-type or Δlux V. fischeri. As a control, we also included newly hatched juveniles (at 0 h). The qRT-PCR reactions of interest contained cDNA from the different colonization conditions, each with five biological replicates and three technical replicates. To obtain five independent biological replicates, we performed these collections on five independent days (N = 20 to 30 juveniles per condition per replicate). Our primers had an amplicon size of 125 bp with efficiency criteria between 98 and 102%. We normalized the expression of es-crumbs to the geometric mean of the expression of two housekeeping genes, ribosomal 40s and serine HMT. We used the comparative method (ΔΔCq method) (Pfaffl 2001) to analyze the data. In addition to the juveniles, we collected mature squid for examining es-crumbs expression in the light organ at four time points over the day-night cycle (N = 2 to 6 light organs per time point). By using the mature light organs we also removed any effects of the developmental process (Fig. 1c) on gene expression. All subsequent qRT-PCR steps are detailed in previous studies featuring juvenile (Peyer et al. 2014) and mature light organs (Heath-Heckman et al. 2013).
Using the statistical package R, we tested whether differential regulation by qRT-PCR differed among light-organ conditions for the juvenile and mature squid. For the juvenile light organ, we used an analysis of variance (ANOVA) to test whether differential regulation of es-crumbs depended on colonization condition and replicate. The model for normalized expression, E, as the dependent variable was:
where independent variables were colonization condition (xc) and replicate (xr). The variable xr represented the five independent biological replicates, each consisting of the three pooled technical replicates. For the mature light organ, we used an ANOVA to test whether the fold change in regulation of es-crumbs depended on time point over the day-night cycle and replicate. The model for fold change, F, as the dependent variable was:
where independent variables were time point (xt) and replicate (xr). The variable xr represented two to six independent biological replicates, each consisting of the two pooled technical replicates. In both analyses, maximum likelihood parameter estimates were β0, β1, and β2. To obtain normal distributions, we used Box-Cox transformations in R using maximum likelihood to determine the optimal power transformations for E and F. To obtain the models that best fit the data, we used a model-selection approach with backward selection and an F-test to evaluate the significance of removing replicate as a factor (Crawley 2008). If the response variables did not depend significantly on replicate, we removed this factor from each model. When applicable, we performed Tukey post-hoc pairwise comparisons for E and F between the different colonization conditions and time points, respectively. We treated all variables as fixed effects.
Results
Characterization of EsCrumbs
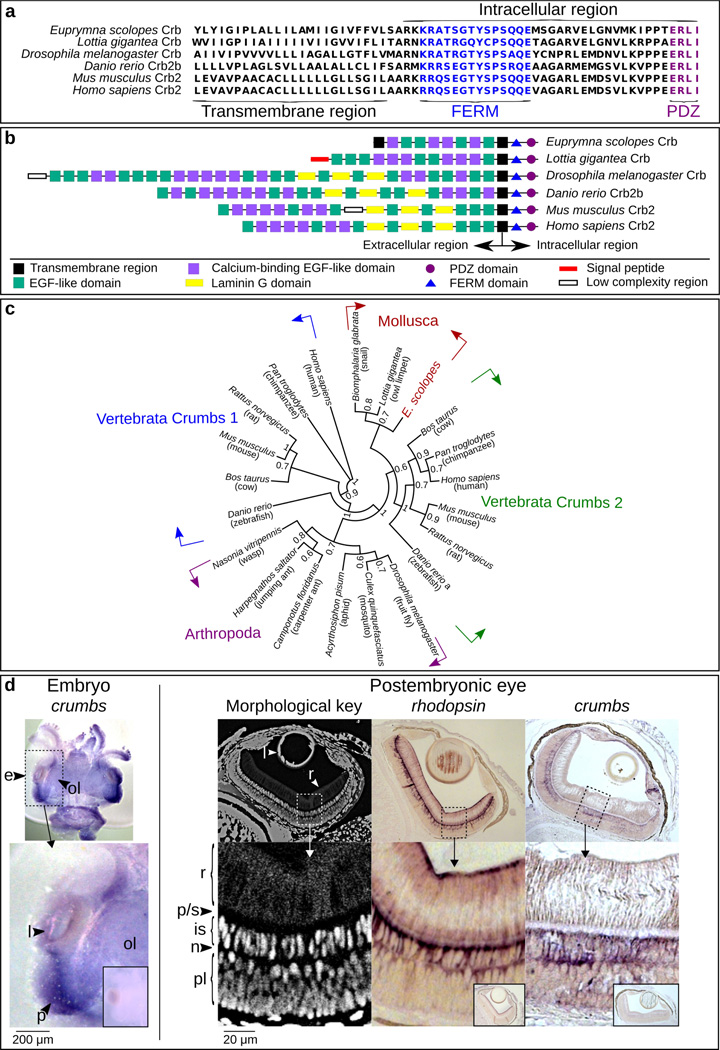
After obtaining the full-length es-crumbs in the light organ, we determined by standard PCR that the same isoform is also expressed in the eye. To verify that this transcript belongs to the Crumbs protein family, we: 1) performed sequence alignments with members of the family from other animals; 2) characterized domain structure; 3) defined the phylogenetic relatedness of EsCrumbs to other members of the family; and 4) determined whether es-crumbs localizes in the eye tissues as is typical of this transcript in other systems (Hsu and Jensen 2010). Crumbs is a transmembrane protein, consisting of a short intracellular region (37 amino acids) (reviewed in Tepass 2012) and a relatively long extracellular region. The intracellular portion contains two highly conserved FERM and PDZ domains with known functions (reviewed in Bilder et al. 2003; Tepass et al. 2001; Tepass 2012) in structuring adherens junctions (Izaddoost et al. 2002) and apical membranes (Wodarz et al. 1995; Tepass 2012), and both domains were present in EsCrumbs (Fig. 2a). The extracellular region, although less conserved as a whole among animals, is generally characterized by a series of epidermal growth factor-like (EGF) domains, laminin G (LamG) domains, and a signal-peptide domain. While EsCrumbs lacked the LamG and signal peptide domains, the EGF-like and calcium-binding EGF-like domains were predicted by SMART (Fig. 2b; http://smart.embl-heidelberg.de/). The location of the stop codon revealed that EsCrumbs is shorter than that of the Crb1 and Crb2 proteins in other organisms, due to a shortened extracellular region, but longer than that of the Crb3 protein (e.g., in H. sapiens). The prediction of a second, terminal transmembrane domain suggests that EsCrumbs forms an extracellular loop.
Figure 2.
Evidence for the presence of the Crumbs protein in E. scolopes. (a) Alignment of the C-terminus of EsCrumbs indicating the transmembrane region and the conserved FERM and PDZ domains within the intracellular region (see also Richard et al 2006). (b) Domain structures throughout the full EsCrumbs and the Crumbs proteins of other organisms as predicted by SMART (http://smart.embl-heidelberg.de/). (c) Phylogenetic position of EsCrumbs relative to those of other animal species. Supports are from 500 bootstrapped replicates. (d) Localization of es-crumbs during early development by in situ hybridization with either whole-mount or histological samples. Left: A stage 22 embryo (~1/2 through embryogenesis at 14 days) showing es-crumbs localized to the region of the eye (e). Features of the eye include the lens (l), optic lobe (ol), and region of development of the photoreceptors (p), indicated for reference. Right: Morphological key for the cephalopod eye, showing the rhabdoms (r), proximal region/supporting cell layer (p/s), inner segments (is), photoreceptor nuclei (n), and plexiform layer (pl). For comparative labeling in the eye, es-rhodopsin expression occurred at the extreme ends of the rhabdoms similar to the protein localization in another squid species (Doryteuthis pealeii; Kingston et al. 2015), but also extended into the inner segments. Labeling of es-crumbs occurred within the inner segments. The sense controls for the ISH experiments involving the embryo and postembryonic eye are indicated in the small boxed regions. Lens objective: 10×.
To our knowledge, EsCrumbs is the first to be fully described in the superphylum Lophotrochozoa, although we located candidate sequences in the L. gigantea and Biomphalaria glabrata (Mollusca:Gastropoda) genomes. The E. scolopes and L. gigantea sequences are more similar in length and domain structure to each other than to other organisms (Fig. 2b). Our phylogenetic analysis grouped EsCrumbs with the putative L. gigantea protein and that of the freshwater snail B. glabrata (Fig. 2c). Within the vertebrates, the Crumbs 1 and Crumbs 2 proteins separated into two distinct groups. The sequences in the Mollusca (Lophotrochozoa) clade fell sister to those in the Vertebrata (Deuterostomia) Crumbs 2 clade.
In situ hybridization (ISH) revealed labeling of es-crumbs in the eye (Fig. 2d). Specifically, with whole-mount colorimetric ISH, es-crumbs showed strong labeling in the embryos, especially around the region of the eye where the rhabdomeric photoreceptors develop. With paraffin-section ISH, es-crumbs localized within the inner segments of the photoreceptors in the juvenile eye, a location consistent with the Crumbs protein other organisms (D. rerio, Hsu and Jensen 2010).
Transcript expression in light-organ superficial tissues and its modulation with symbiosis
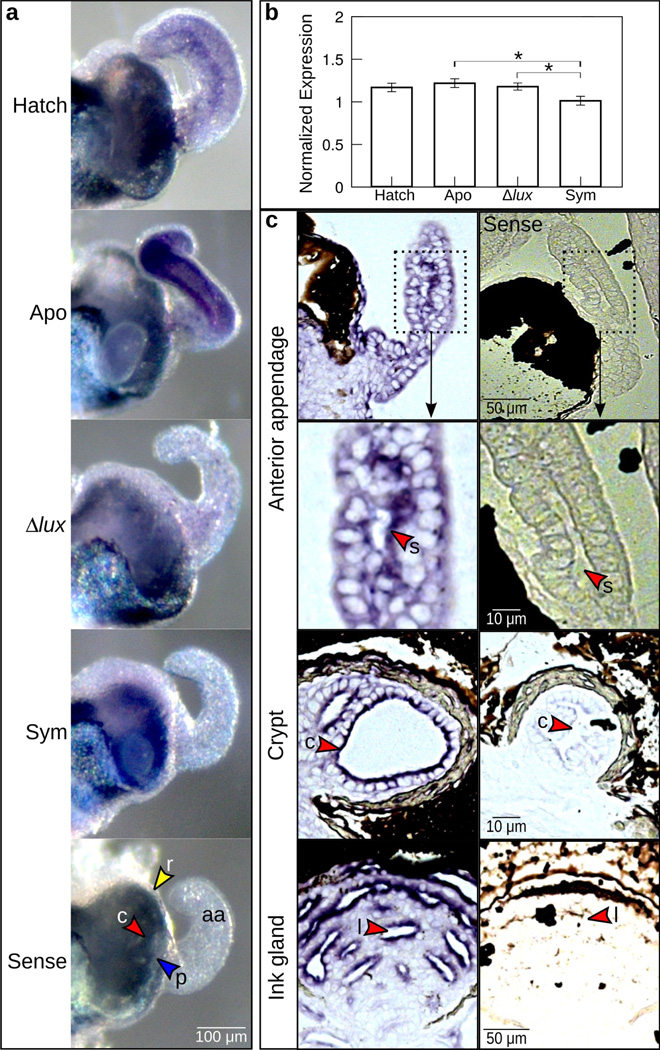
Within the superficial tissues of the light organ that undergo apoptosis (Fig. 1c), we observed es-crumbs expression by ISH in the light organs of newly hatched (Fig. 3a, ‘Hatch’) and 24-h aposymbiotic (exposed to other environmental bacteria, but not V. fischeri; Fig. 3a ‘Apo’) juvenile squid. Labeling was most vivid in the anterior appendages and around the pores. Throughout these same regions, we observed a loss of es-crumbs expression in response to symbiont colonization of the light-organ crypts (Fig. 3a ‘Sym’). In organs colonized by Δlux V. fischeri, es-crumbs expression was still detectable in the anterior appendages and pores, although the signal was much less pronounced than in aposymbiotic light organs (Fig. 3a ‘Δlux’). These patterns suggest that in the surface tissues, es-crumbs expression is turned down in response to colonization of the crypts by V. fischeri, although luminescence might be required for full attenuation of the signal.
Figure 3.
Expression of es-crumbs throughout the light organ. (a) Whole light organs showing es-crumbs expression by whole-mount in situ hybridization. Conditions consisted of newly hatched juveniles (‘Hatch’) and 24-h juveniles aposymbiotic with no V. fischeri (‘Apo’), symbiotic with Δlux V. fischeri (‘Δlux’), and symbiotic with wild-type V. fischeri (‘Sym’). Tissues of the light organ showing es-crumbs signal included the anterior appendages (aa), ciliated ridges (r, yellow arrowheads), pores (p, blue arrowheads), or crypts (c, red arrowheads), and these features are shown in the ‘sense’ (control) image. (b) Regulation of es-crumbs by qRT-PCR in response to the different symbiotic conditions. Expression of es-crumbs was normalized to the geometric mean of the expression of two housekeeping genes, ribosomal 40s and serine HMT. Data consist of five biological replicates each with three technical replicates pooled per condition. Error bars are standard error of the mean (*, 0.01< P<0.05). (c) Histological sections of the light organ anterior appendage sinus (s) and crypt (c), and ink gland lumen (l) showing escrumbs localization by ISH (see red arrowheads). The images shown are from an animal colonized with Δlux V. fischeri after 24 h. The sense controls are shown along the right side.
The crypt tissues appeared to show the opposite pattern in comparison with the surface epithelia. Hatchling (Fig. 3a, ‘Hatch’) and aposymbiotic (Fig. 3a, ‘Apo’) animals had undetectable labeling in the crypts, whereas labeling was visible in the crypts of symbiotic animals (Fig. 3a, ‘Sym’, ‘Δlux’). Closer examination of transcript expression in the crypts by HCR-FISH was required, but confirmed these patterns (see ‘Transcript localization in light-organ crypts’ below).
At 24 h post colonization es-crumbs regulation differed significantly among the different symbiotic conditions (Fig. 3b) (ANOVA: F=6.2, df=3, P=0.01) and reflected the patterns of expression that we observed in superficial tissues with whole-mount colorimetric ISH. Specifically, es-crumbs was significantly down regulated in light organs with wild-type V. fischeri relative to those aposymbiotic (Tukey: P=0.01). Also, es-crumbs was significantly down regulated in light organs with wild-type V. fischeri as compared to those with Δlux V. fischeri, although to a lesser degree (Tukey: P<0.04). Aposymbiotic light organs and those with Δlux V. fischeri did not differ significantly from one another in es-crumbs regulation (Tukey: P=0.80). These data suggest that the turn down of transcript in the superficial epithelium is greater overall than the increase in transcript in the crypts.
Histological ISH analyses (Fig. 3c), although not suitable for quantification, showed labeling strongest along the apical surfaces of the light organ epithelia and adjoining tissues. Specifically, labeling of es-crumbs was apparent along the cells lining the anterior appendage sinuses and crypts (Fig. 3c). In addition, labeling occurred along the apical surfaces of the epithelia in the ink gland lumen (Fig. 3c), which would produce high background in the qRT-PCR results.
Transcript localization in light-organ crypts
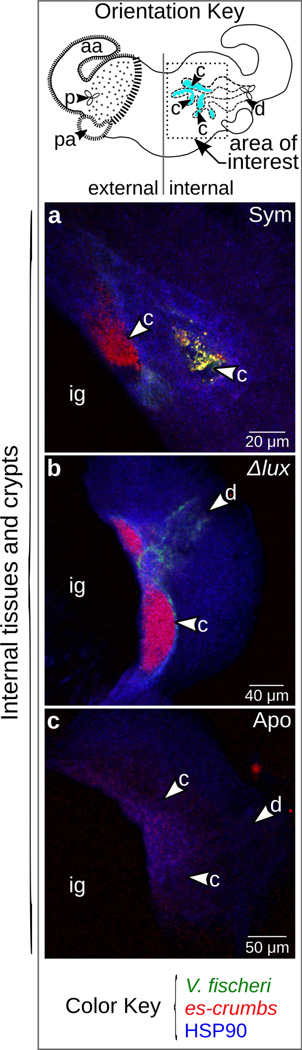
Because the crypts are deep tissues in the light organ, we used confocal microscopy to examine transcript localization more precisely. To determine if the transcript localized to crypt epithelia, we probed for labeling of es-crumbs along with V. fischeri cells (i.e., 16S ribosomal RNA) by HCR-FISH (Nikolakakis et al. 2015). These experiments indicated that the transcript co-localized with V. fischeri cells (Fig. 4a) and is expressed at higher levels in the crypts of animals colonized by V. fischeri (Fig. 4a, b) than those left uncolonized (Fig. 4c), confirming the patterns we observed with colorimetric ISH (Fig. 3a).
Figure 4.
Crumbs transcript expression in light-organ crypts. (a–c) Results from HCR-FISH experiments showing es-crumbs expression in the crypts of light organs symbiotic with wild-type V. fischeri (a; ‘Sym’) and Δlux V. fischeri (b; ‘Δlux’) as compared to an aposymbiotic light organ with no transcript expression (c; ‘Apo’). As landmark features, the ink gland (ig, see Fig. 1b), crypts (c), and duct (d) that connects the pores to the crypts are labeled. Heat Shock Protein 90 (HSP90) was used only as a counterstain. Lens objective: 40×.
Daily variation in es-crumbs transcript
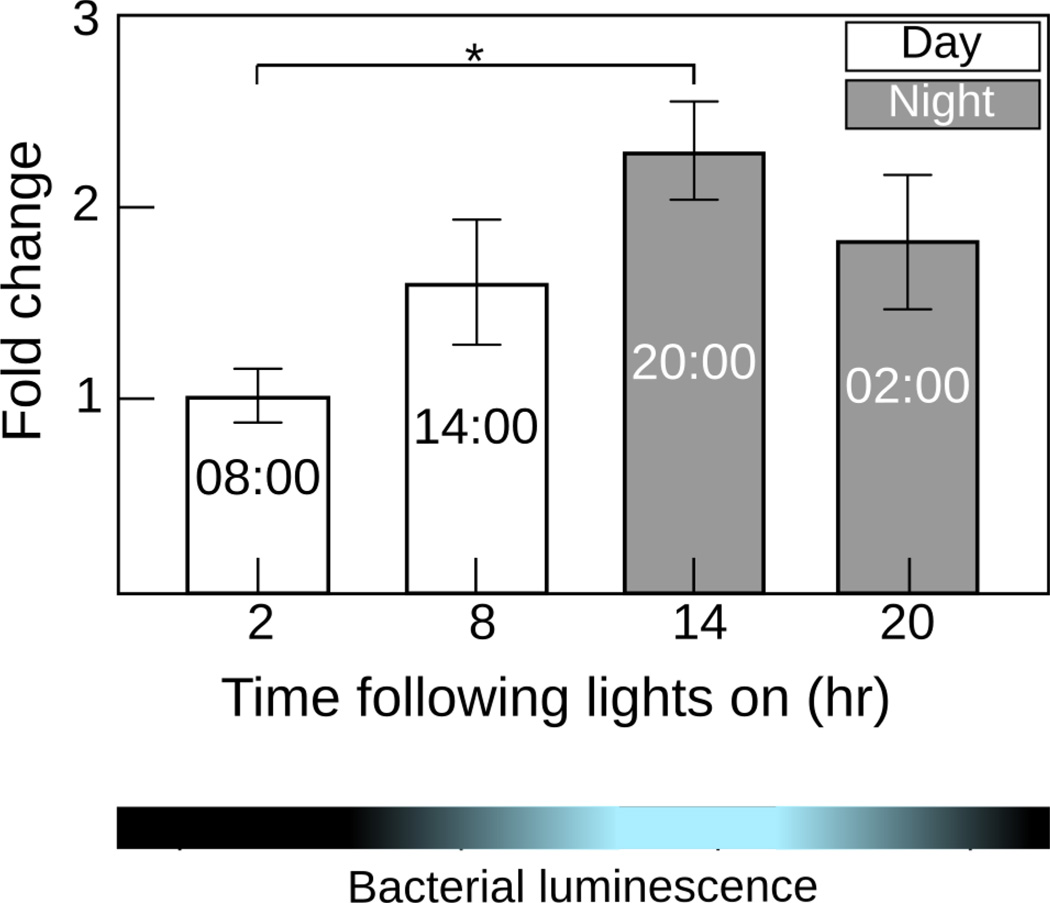
We examined es-crumbs regulation in the light organs of mature animals at four time points over the day-night cycle. In using mature light organs, we were able to observe changes in gene expression without any confounding effect of development that occurs in the juvenile (Fig. 1c). We found es-crumbs to differ significantly across this 24-h period (Fig. 5) (ANOVA: F=3.7, df=3, P=0.04). In general, this regulation corresponded with symbiont density and was significantly up regulated during the period of highest luminescence (night time point at ‘20:00’) relative to the period of lowest luminescence (day time point at ‘08:00’) (Tukey: P=0.03).
Figure 5.
Regulation of es-crumbs in mature light organs sampled over the day-night cycle. The fold change on the y-axis represents data normalized to the time point of lowest expression. The blue and black bar denotes the cycle of V. fischeri luminescence within the light organ. Data consist of two to six biological replicates each with two technical replicates pooled per condition. Error bars for both graphs are standard error of the mean (*, 0.01< P<0.05).
Discussion
We isolated a crumbs gene in the E. scolopes light organ and examined its expression in response to symbiosis with the mutualistic partner V. fischeri. Several lines of evidence provided confidence that this putative es-crumbs sequence was a homolog of the crumbs genes found previously in the eyes of other organisms, including its domain structure, intracellular localization (in both eye and light organ), and phylogenetic relationships. In both the juvenile and mature squid, es-crumbs responded to symbiosis, with the two developmental stages offering different insights into the role of the protein in the light organ. Examination of the juvenile light organ revealed that the gene was oppositely expressed in different tissues depending on whether they were lost or preserved following symbiosis. Over the day-night cycle, expression of the gene in the mature light organ peaked with maximum symbiont population and luminescence, indicating a responsiveness to bacterial cues. The presence of the gene in the juvenile crypts colonized by both wild-type V. fischeri and a mutant strain defective in light production further supported that es-crumbs responds to microbial products. Our work is the first to describe crumbs host responses to bacterial cues in a mutualistic association and in an organism from the Lophotrochozoa superphylum.
The Crumbs protein is involved in development (Johnson et al. 2002) and apoptosis in other organisms (Johnson et al. 2002; Chartier et al. 2012; Aartsen et al. 2010), and might function similarly during postembryonic development of the E. scolopes light organ. As mentioned above (see Introduction), following the initiation of symbiosis, the light organ undergoes a developmental process in which the superficial tissues regress as their cells experience apoptosis (Fig. 1c) (McFall-Ngai 2014). During this process different host responses are triggered (Koropatnick et al. 2004, 2007) and a suite of genes are influenced by V. fischeri cues. For example, in superficial tissues of the light organ the expression of three eye specification genes is lost, including six (i.e., sine oculis) in response to V. fischeri luminescence, and pax6 and eya in response to bacterial products other than luminescence (Peyer et al. 2014). In the present study, we also observed loss of es-crumbs expression by ISH in superficial tissues following symbiont colonization, most notably within the anterior appendages (Fig. 3a). Although apoptosis results most dramatically in response to wild-type, luminescent V. fischeri, some cells also die in response to MAMPs from Δlux V. fischeri (McFall-Ngai et al. 2012). Thus, the influence of symbiosis on es-crumbs provides yet another example of the effect of V. fischeri cues on genes that have developmental roles. In a larger context, Crumbs is an upstream regulator of the Salvador/Warts/Hippo (SWH) pathway in D. melanogaster (Robinson et al. 2010; Chen et al 2010), which is a known regulator of apoptosis (Hamaratoglu et al. 2006). At the same time that es-crumbs is lost in superficial tissues, the transcript is expressed in the crypts (Fig. 4a, b). The protein might thus function in two different capacities in the developing light organ: its loss correlating with the degradation of superficial cells that are no longer needed for colonization, contrasting with its production in the protected crypt microvilli cells that are the direct recipients of symbiont stressors. The Crb1 protein is also important in the maintenance of the microvilli of mouse Muller glial cells (van de Pavert et al. 2007). In general, opposing dynamics in crumbs expression occur in other developing organisms when dramatic tissue remodeling is common (Pichaud 2014). Overall, our qRT-PCR results in the juveniles reflected the expression patterns we observed for es-crumbs in superficial tissues, i.e., down regulation especially in response to the wild-type V. fischeri (Fig. 3a, b), although the presence of es-crumbs in the crypts of symbiotic light organs (Fig. 4a, b) and in the ink gland likely opposed and diluted this trend to some degree (Fig. 3c).
In animal eyes the Crumbs protein protects against light-induced apoptosis (Johnson et al. 2002; Chartier et al. 2012; Aartsen et al. 2010). For example, in D. melanogaster eyes, crumbs mutants have resulted in shortened rhabdomeres, which upon exposure to light undergo degeneration, exhibiting the cytoplasm and nucleoplasm condensation that typifies apoptosis (Richard et al. 2006). Along similar lines, the Crumbs homolog 2 (Crb2) when over expressed in the zebrafish D. rerio enlarges the inner and outer segments of photoreceptor cells, suggesting that Crb2 serves a role in the size and circadian-based renewal of outer segment discs (Hsu and Jensen 2010). In the current study, peak expression of es-crumbs coincided with peak symbiont population, which correlates with luminescence (Fig. 5). Thus, squid host tissues respond to light and the rhythms of light presentation and Crumbs might be a candidate player in these photoreceptive responses. In other words, es-crumbs might encode more or less protein in response to symbiont population dynamics (Fig. 1d) depending on the requirements for protection from stressors presented by the bacteria, including luminescence. Similar expression patterns occur with other genes in E. scolopes in response to V. fischeri luminescence, such as cryptochrome (escry1) (Heath-Heckman et al. 2013), which is typically involved in circadian rhythms in other organisms. Although we have not investigated a relationship between escry1 and es-crumbs, and we are unaware of any interaction between the two genes in other organisms, their parallel patterns of expression offer an intriguing subject for future study.
Up regulation of es-crumbs at peak luminescence might serve to limit tissue damage from oxidative stress caused by the bacteria (for review see Schwartzman and Ruby 2016). In the vertebrate eye, the rods are heavy users of oxygen and their death can lead to the accumulation of oxidative stress in the retina (e.g., from NADPH oxidase), causing additional photoreceptor degeneration (e.g., in mice, Usui et al. 2009). The Crumbs protein is known to reduce oxidative stress from NADPH oxidase and limit light-induced photoreceptor damage in D. melanogaster (Chartier et al. 2012). Within the light organ, components of the inflammatory response, including peroxidase, nitric oxide, and NFκB, are activated during V. fischeri colonization (Weis et al. 1996; Davidson et al. 2004; Goodson et al. 2005), suggesting the presence of oxidative stress at least in certain tissues.
As the gene is expressed in response to Δlux V. fischeri that emit no light (Fig. 4b), EsCrumbs may also serve an additional protective role in cells presented with MAMPs. In general, cell polarity proteins appear to be common targets of micro-organisms (e.g., viruses in humans: Javier 2008; Helicobacter pylori: Reid et al. 2012). For example, H. pylori strains with the CagA protein perturb host cell-cell junctions and cell polarity, but over expression of the Crumbs protein in D. melanogaster limits this disruption (Reid et al. 2012). Other proteins associate with the Crumbs protein through the FERM (Moesin, βH-Spectin, Expanded,Yurt, Notch) and PDZ domains (Par6, Stardust, Bazooka, Scribble, Lin-7, Patj) (Bilder et al. 2003; Tepass et al. 2001; Tepass 2012), some of which interact with or are manipulated by microbes (e.g., Moesin, Spectrin, Notch, Par6). On human monocytes, the Moesin protein serves as a receptor of bacterial LPS (Tohme et al. 1999). Also, the Spectrin protein is depleted from HeLa cells by several bacterial species, including Enteropathogenic Escherichia coli, Salmonella enterica serovar Typhimurium, Listeria monocytogenes (Ruetz et al. 2011), and Shigella flexneri (Ruetz et al. 2012). In addition, Notch protein signaling in cultured cells is suppressed when exposed to LPS (Kim et al. 2008). Finally, from human brain endothelial cells, Neisseria meningitidis can sequester the Par6 protein (Coureuil et al. 2009; Join-Lambert et al. 2010). Interestingly, mutations in some cell polarity proteins, such as the Moesin and Lin7, are also linked to light-induced photoreceptor damage (Chorna-Ornan et al. 2005; Bachmann et al. 2008). Thus, EsCrumbs might be indirectly influenced by a collection of other V. fischeri cues through association with other cell polarity proteins.
Conclusions
The Crumbs protein is a regulator cell polarity and is part of a pathway that influences development and apoptosis. In the developing E. scolopes light organ, the crumbs gene was lost in cells experiencing symbiont-induced apoptosis and expressed in tissues that are protected from cell death and are the direct recipients of symbiont stressors, including luminescence. In the mature squid, regulation of the gene also correlated with symbiont population over the day-night cycle, indicating host tissue response to microbial cues.
Acknowledgments
This work was supported by grants from the National Institutes of Health RO1-A150661 (to MM-N and EG Ruby), RO1-OD11024 (to EG Ruby and MM-N), and the National Science Foundation IOS 0817232 (to MM-N and EG Ruby). EACH-H was supported by NRSA T-32 GM07215. We are grateful for assistance in various ways from the following individuals: Nell Bekiares, Mahdi Belcaid, Fangmin Chen, Eric Koch, Ben Krasity, Natacha Kremer, Silvia Moriano-Gutierrez, Kiel Nikolakakis, and Julia Schwartzman. We also appreciate the thoughtful comments from two anonymous reviewers.
References
- Aartsen WM, van Cleef KW, Pellissier LP, Hoek RM, Vos RM, Blits B, Ehlert EM, Balaggan KS, Ali RR, Verhaagen J, Wijnholds J. GFAP-driven GFP expression in activated mouse Muller glial cells aligning retinal blood vessels following intravitreal injection of AAV2/6 vectors. PLoS One. 2010;5:12387. doi: 10.1371/journal.pone.0012387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann A, Grawe F, Johnson K, Knust E. Drosophila Lin-7 is a component of the Crumbs complex in epithelia and photoreceptor cells and prevents light-induced retinal degeneration. Eur J Cell Biol. 2008;87:123–136. doi: 10.1016/j.ejcb.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Bilder D, Schober M, Perrimon N. Integrated activity of PDZ protein complexes regulates epithelial polarity. Nat Cell Biol. 2003;5:53–58. doi: 10.1038/ncb897. [DOI] [PubMed] [Google Scholar]
- Boettcher KJ, Ruby EG. Detection and quantification of Vibrio fischeri autoinducer from symbiotic squid light organs. J Bacteriol. 1995;177:1053–1058. doi: 10.1128/jb.177.4.1053-1058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher KJ, Ruby EG, McFall-Ngai MJ. Bioluminescence in the symbiotic squid Euprymna scolopes is controlled by a daily biological rhythm. J Comp Physiol A. 1996;179:65–73. [Google Scholar]
- Bose JL, Rosenberg CS, Stabb EV. Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch Microbiol. 2008;190:169–183. doi: 10.1007/s00203-008-0387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgakova NA, Knust E. The Crumbs complex: from epithelial-cell polarity to retinal degeneration. J Cell Sci. 2009;122:2587–2596. doi: 10.1242/jcs.023648. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Beaulieu JF, Huggett J, Jaggi R, Kibenge FS, Olsvik PA, Penning LC, Toegel S. MIQE précis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol Biol. 2010;11:74. doi: 10.1186/1471-2199-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier FJ, Hardy ÉJ, Laprise P. Crumbs limits oxidase-dependent signaling to maintain epithelial integrity and prevent photoreceptor cell death. J Cell Biol. 2012;198:991–998. doi: 10.1083/jcb.201203083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, Halder G. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci U S A. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorna-Ornan I, Tzarfaty V, Ankri-Eliahoo G, Joel-Almagor T, Meyer NE, Huber A, Payre F, Minke B. Light-regulated interaction of Dmoesin with TRP and TRPL channels is required for maintenance of photoreceptors. J Cell Biol. 2005;171:143–152. doi: 10.1083/jcb.200503014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun CK, Scheetz TE, de Fatima Bonaldo M, Brown B, Clemens A, Crookes-Goodson WJ, Crouch K, DeMartini T, Eyestone M, Goodson MS, Janssens B, Kimbell JL, Koropatnick TA, Kucaba T, Smith C, Stewart JJ, Tong D, Troll JV, Webster S, Winhall-Rice J, Yap C, Casavant TL, McFall-Ngai MJ, Soares MB. An annotated cDNA library of juvenile Euprymna scolopes with and without colonization by the symbiont Vibrio fischeri. BMC Genomics. 2006;7:154. doi: 10.1186/1471-2164-7-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coureuil M, Mikaty G, Miller F, Lécuyer H, Bernard C, Bourdoulous S, Duménil G, Mège RM, Weksler BB, Romero IA, Couraud PO, Nassif X. Meningococcal type IV pili recruit the polarity complex to cross the brain endothelium. Science. 2009;325:83–87. doi: 10.1126/science.1173196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley MJ. The R Book. West Sussex: John Wiley; 2008. [Google Scholar]
- Crookes WJ, Ding LL, Huang QL, Kimbell JR, Horwitz J, McFall-Ngai MJ. Reflectins: the unusual proteins of squid reflective tissues. Science. 2004;303:235–238. doi: 10.1126/science.1091288. [DOI] [PubMed] [Google Scholar]
- Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. NO means ‘yes’ in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol. 2004;6:1139–11351. doi: 10.1111/j.1462-5822.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- den Hollander AI, ten Brink JB, de Kok YJ, van Soest S, van den Born LI, van Driel MA, van de Pol DJ, Payne AM, Bhattacharya SS, Kellner U, Hoyng CB, Westerveld A, Brunner HG, Bleeker-Wagemakers EM, Deutman AF, Heckenlively JR, Cremers FP, Bergen AA. Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12) Nat Genet. 1999;23:217–221. doi: 10.1038/13848. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Audic S, Claverie JM, Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doino JA, McFall-Ngai MJ. A Transient Exposure to Symbiosis-Competent Bacteria Induces Light Organ Morphogenesis in the Host Squid. Biol Bull. 1995;189:347–355. doi: 10.2307/1542152. [DOI] [PubMed] [Google Scholar]
- Goodson MS, Kojadinovic M, Troll JV, Scheetz TE, Casavant TL, Soares MB, McFall-Ngai MJ. Identifying components of the NF-κB pathway in the beneficial Euprymna scolopes-Vibrio fischeri light organ symbiosis. Appl Environ Microbiol. 2005;71:6934–6946. doi: 10.1128/AEM.71.11.6934-6946.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Heath-Heckman EA, Peyer SM, Whistler CA, Apicella MA, Goldman WE, McFall-Ngai MJ. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-vibrio symbiosis. MBio. 2013;4:e00167–13. doi: 10.1128/mBio.00167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath-Heckman EA, Foster J, Apicella MA, Goldman WE, McFall-Ngai M. Environmental cues and symbiont MAMPs function in concert to drive the daily remodeling of the crypt-cell brush border of the Euprymna scolopes light organ. Cell Microbiol. 2016 doi: 10.1111/cmi.12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Jensen AM. Multiple domains in the Crumbs Homolog 2a (Crb2a) protein are required for regulating rod photoreceptor size. BMC Cell Biol. 2010;11:60. doi: 10.1186/1471-2121-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaddoost S, Nam SC, Bhat MA, Bellen HJ, Choi KW. Drosophila Crumbs is a positional cue in photoreceptor adherens junctions and rhabdomeres. Nature. 2002;416:178–183. doi: 10.1038/nature720. [DOI] [PubMed] [Google Scholar]
- Javier RT. Cell polarity proteins: common targets for tumorigenic human viruses. Oncogene. 2008;27:7031–7046. doi: 10.1038/onc.2008.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Grawe F, Grzeschik N, Knust E. Drosophila crumbs is required to inhibit light-induced photoreceptor degeneration. Curr Biol. 2002;12:1675–1680. doi: 10.1016/s0960-9822(02)01180-6. [DOI] [PubMed] [Google Scholar]
- Join-Lambert O, Morand PC, Carbonnelle E, Coureuil M, Bille E, Bourdoulous S, Nassif X. Mechanisms of meningeal invasion by a bacterial extracellular pathogen, the example of Neisseria meningitidis. Prog Neurobiol. 2010;91:130–139. doi: 10.1016/j.pneurobio.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Jones BW, Nishiguchi MK. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda) Mar Biol. 2004;144:1151–1155. [Google Scholar]
- Kim MY, Park JH, Mo JS, Ann EJ, Han SO, Baek SH, Kim KJ, Im SY, Park JW, Choi EJ, Park HS. Downregulation by lipopolysaccharide of Notch signaling, via nitric oxide. J Cell Sci. 2008;121:1466–1476. doi: 10.1242/jcs.019018. [DOI] [PubMed] [Google Scholar]
- Kingston ACN, Wardill TJ, Hanlon RT, Cronin TW. An Unexpected Diversity of Photoreceptor Classes in the Longfin Squid, Doryteuthis pealeii. PlosOne. 2015;10(9):e0135381. doi: 10.1371/journal.pone.0135381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- Koropatnick TA, Kimbell JR, McFall-Ngai MJ. Responses of host hemocytes during the initiation of the squid-vibrio symbiosis. Biol Bull. 2007;212:29–39. doi: 10.2307/25066578. [DOI] [PubMed] [Google Scholar]
- Lamarcq LH, McFall-Ngai MJ. Induction of a gradual, reversible morphogenesis of its host's epithelial brush border by Vibrio fischeri. Infect Immun. 1998;66:777–785. doi: 10.1128/iai.66.2.777-785.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprise P. Emerging role for epithelial polarity proteins of the Crumbs family as potential tumor suppressors. J Biomed Biotechnol. 2011 doi: 10.1155/2011/868217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PN, McFall-Ngai MJ, Callaerts P, de Couet HG. Whole-mount in situ hybridization of Hawaiian bobtail squid (Euprymna scolopes) embryos with DIG-labeled riboprobes: II. Embryo preparation, hybridization, washes, and immunohistochemistry. Cold Spring Harb Protoc. 2009;11 doi: 10.1101/pdb.prot5322. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M, Heath-Heckman EAC, Gillette AA, Peyer SM, Harvie EA. The secret languages of coevolved symbioses: Insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Sem Immunol. 2012;24:3–8. doi: 10.1016/j.smim.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ. The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu Rev Microbiol. 2014;68:177–194. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery MK, McFall-Ngai MJ. The muscle-derived lens of a squid bioluminescent organ is biochemically convergent with the ocular lens. Evidence for recruitment of aldehyde dehydrogenase as a predominant structural protein. J Biol Chem. 1992;267:20999–21003. [PubMed] [Google Scholar]
- Montgomery MK, McFall-Ngai MJ. Embryonic development of the light organ of the sepiolid squid Euprymna scolopes Berry. Biol Bull. 1993;184:296–308. doi: 10.2307/1542448. [DOI] [PubMed] [Google Scholar]
- Nikolakakis K, Lehnert E, McFall-Ngai MJ, Ruby EG. Use of hybridization chain reaction-fluorescent in situ hybridization to track gene expression by both partners during initiation of symbiosis. Appl Environ Microbiol. 2015;81:4728–4735. doi: 10.1128/AEM.00890-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellikka M, Tanentzapf G, Pinto M, Smith C, McGlade CJ, Ready DF, Tepass U. Crumbs, the Drosophila homologue of human CRB1/RP12, is essential for photoreceptor morphogenesis. Nature. 2002;416:143–149. doi: 10.1038/nature721. [DOI] [PubMed] [Google Scholar]
- Peyer SM, Pankey MS, Oakley TH, McFall-Ngai MJ. Eye-specification genes in the bacterial light organ of the bobtail squid Euprymna scolopes, and their expression in response to symbiont cues. Mech Dev. 2014;131:111–126. doi: 10.1016/j.mod.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichaud F. Transcriptional regulation of tissue organization and cell morphogenesis: the fly retina as a case study. Dev Biol. 2014;385:168–178. doi: 10.1016/j.ydbio.2013.09.031. [DOI] [PubMed] [Google Scholar]
- Reid DW, Muyskens JB, Neal JT, Gaddini GW, Cho LY, Wandler AM, Botham CM, Guillemin K. Identification of genetic modifiers of CagA-induced epithelial disruption in Drosophila. Front Cell Infect Microbiol. 2012;2:24. doi: 10.3389/fcimb.2012.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M, Roepman R, Aartsen WM, van Rossum AGSH, den Hollander AI, Knust E, Wijnholds J, Cremers FPM. Towards understanding CRUMBS function in retinal dystrophies. Human Molecular Genetics. 2006;15:R235–R243. doi: 10.1093/hmg/ddl195. [DOI] [PubMed] [Google Scholar]
- Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, Asato LM. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;59:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- Ruetz T, Cornick S, Guttman JA. The spectrin cytoskeleton is crucial for adherent and invasive bacterial pathogenesis. PLoS One. 2011;6:e19940. doi: 10.1371/journal.pone.0019940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruetz TJ, Lin AE, Guttman JA. Shigella flexneri utilize the spectrin cytoskeleton during invasion and comet tail generation. BMC Microbiol. 2012;12:36. doi: 10.1186/1471-2180-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzman JA, Ruby EG. A conserved chemical dialog of mutualism: lessons from squid and vibrio. Microbes Infect. 2016;18:1–10. doi: 10.1016/j.micinf.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U, Tanentzapf G, Ward R, Fehon R. Epithelial cell polarity and cell junctions in Drosophila. Annu Rev Genet. 2001;35:747–784. doi: 10.1146/annurev.genet.35.102401.091415. [DOI] [PubMed] [Google Scholar]
- Tepass U. The apical polarity protein network in Drosophila epithelial cells: regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu Rev Cell Dev Biol. 2012;28:655–685. doi: 10.1146/annurev-cellbio-092910-154033. [DOI] [PubMed] [Google Scholar]
- Tohme ZN, Amar S, Van Dyke TE. Moesin functions as a lipopolysaccharide receptor on human monocytes. Infect Immun. 1999;67:3215–3220. doi: 10.1128/iai.67.7.3215-3220.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong D, Rozas NS, Oakley TH, Mitchell J, Colley NJ, McFall-Ngai MJ. Evidence for light perception in a bioluminescent organ. Proc Natl Acad Sci USA. 2009;106:9836–9841. doi: 10.1073/pnas.0904571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui S, Oveson BC, Lee SY, Jo YJ, Yoshida T, Miki A, Miki K, Iwase T, Lu L, Campochiaro PA. NADPH oxidase plays a central role in cone cell death in retinitis pigmentosa. J Neurochem. 2009;110:1028–1037. doi: 10.1111/j.1471-4159.2009.06195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Pavert SA, Sanz AS, Aartsen WM, Vos RM, Versteeg I, Beck SC, Klooster J, Seeliger MW, Wijnholds J. Crb1 is a determinant of retinal apical Muller glia cell features. Glia. 2007;55:1486–1497. doi: 10.1002/glia.20561. [DOI] [PubMed] [Google Scholar]
- Weis VM, Small AL, McFall-Ngai MJ. A peroxidase related to the mammalian antimicrobial protein myeloperoxidase in the Euprymna-Vibrio mutualism. Proc Natl Acad Sci USA. 1996;93:13683–13688. doi: 10.1073/pnas.93.24.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiasse RP, Schaefer AL, Koroleva I, Splinter-BonDurant S, Brown B, Manzella L, Snir E, Almabrazi H, Scheetz TE, Bonaldo MD, Casavant TL, Soares MB, Cronan JE, Reed JL, Ruby EG, McFall-Ngai MJ. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci USA. 2010;107:2259–2264. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Hinz U, Engelbert M, Knust E. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell. 1995;82:67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- Young RW. The renewal of photoreceptor cell outer segments. J Cell Biol. 1967;33:61–72. doi: 10.1083/jcb.33.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]