Abstract
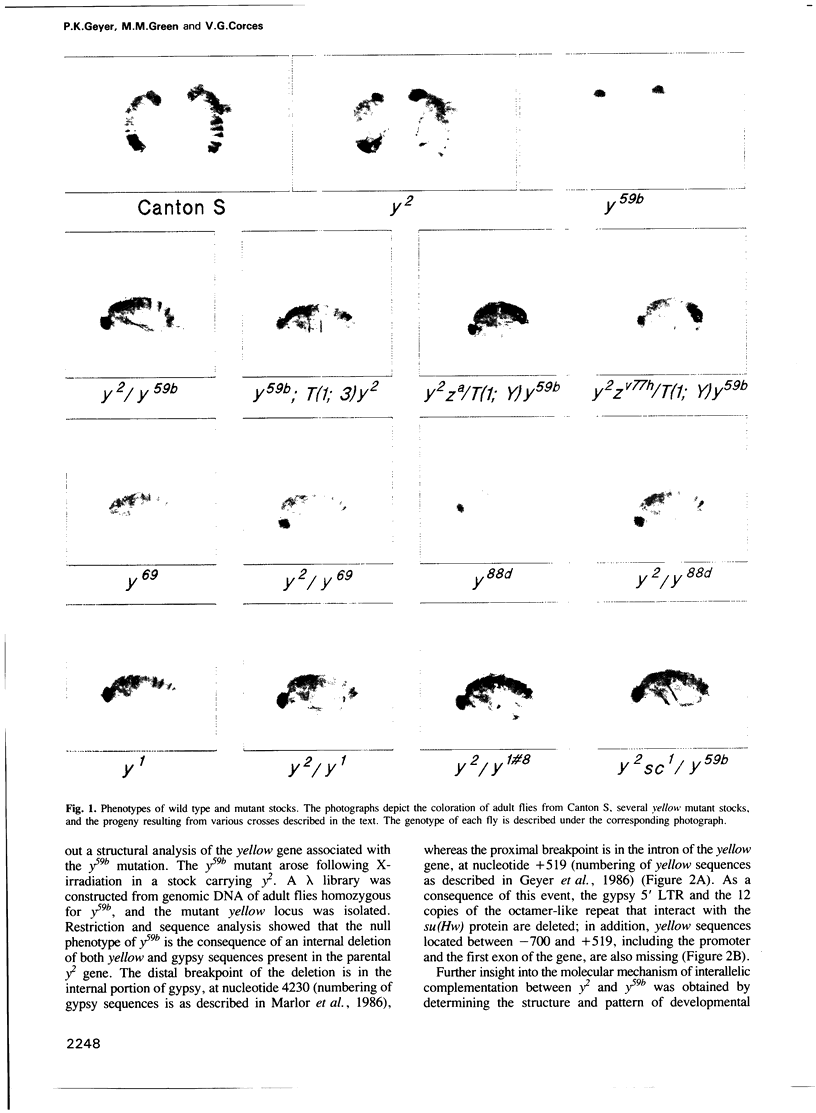
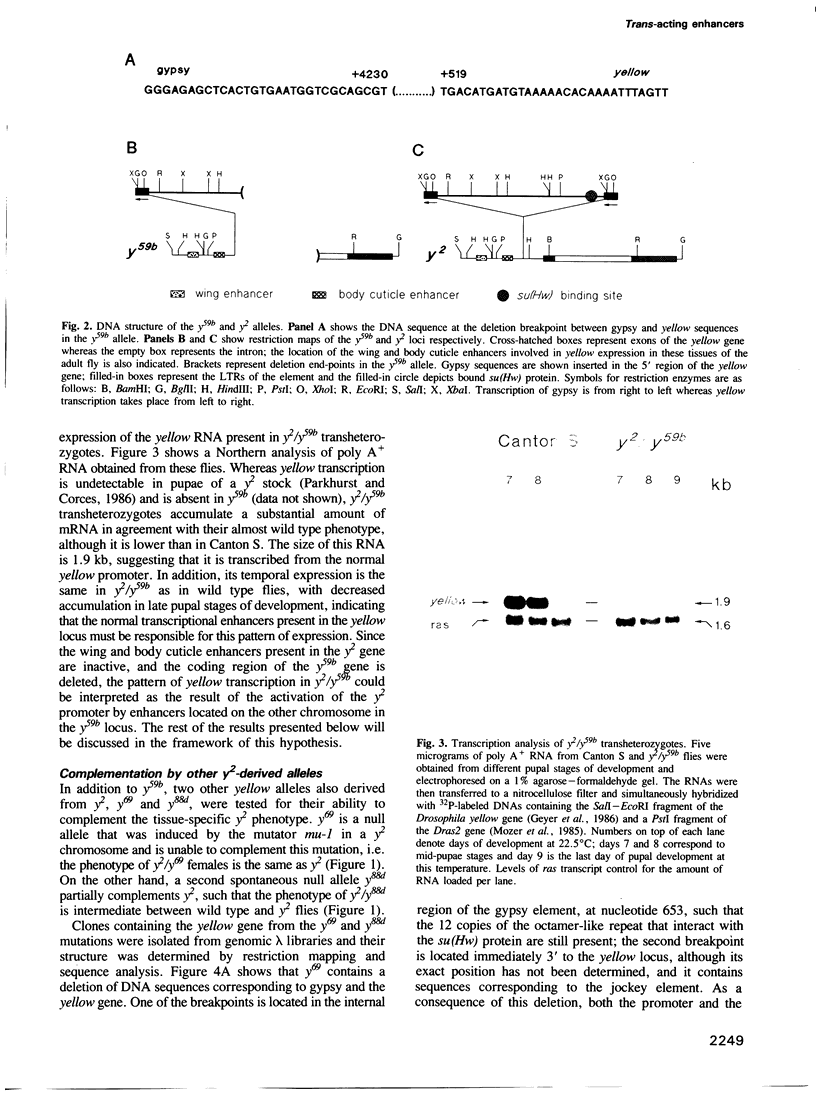
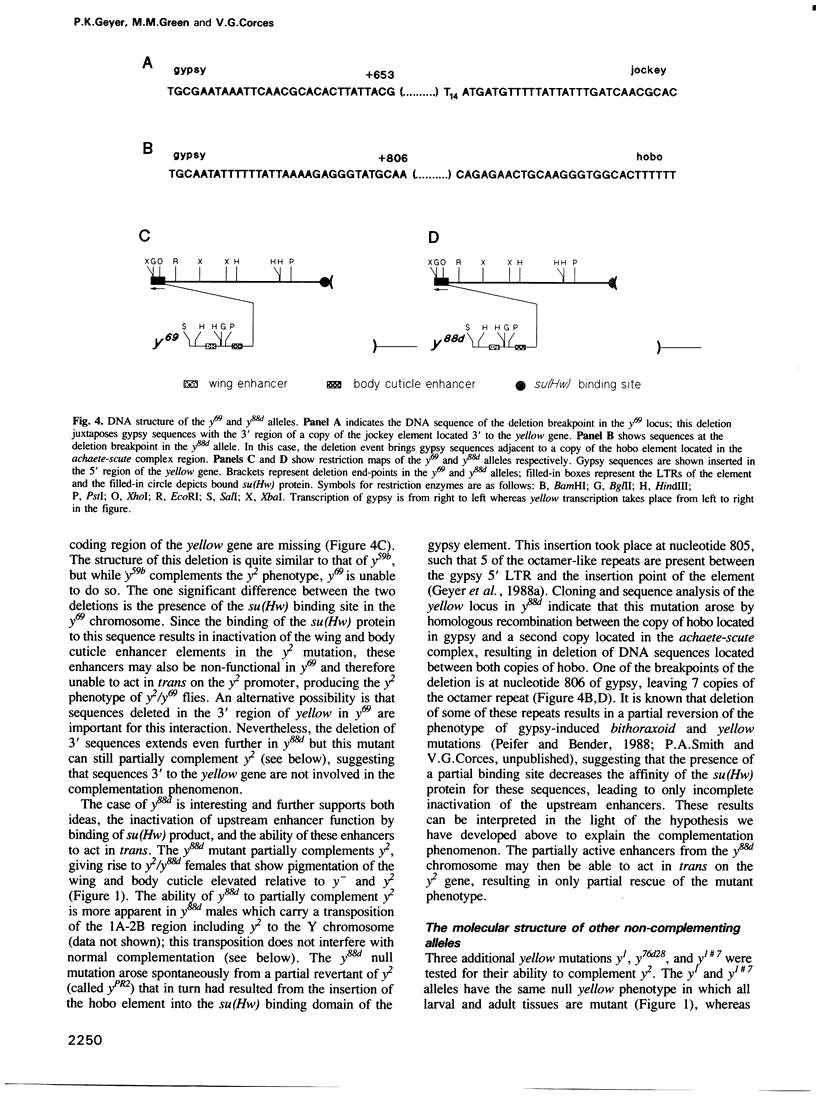
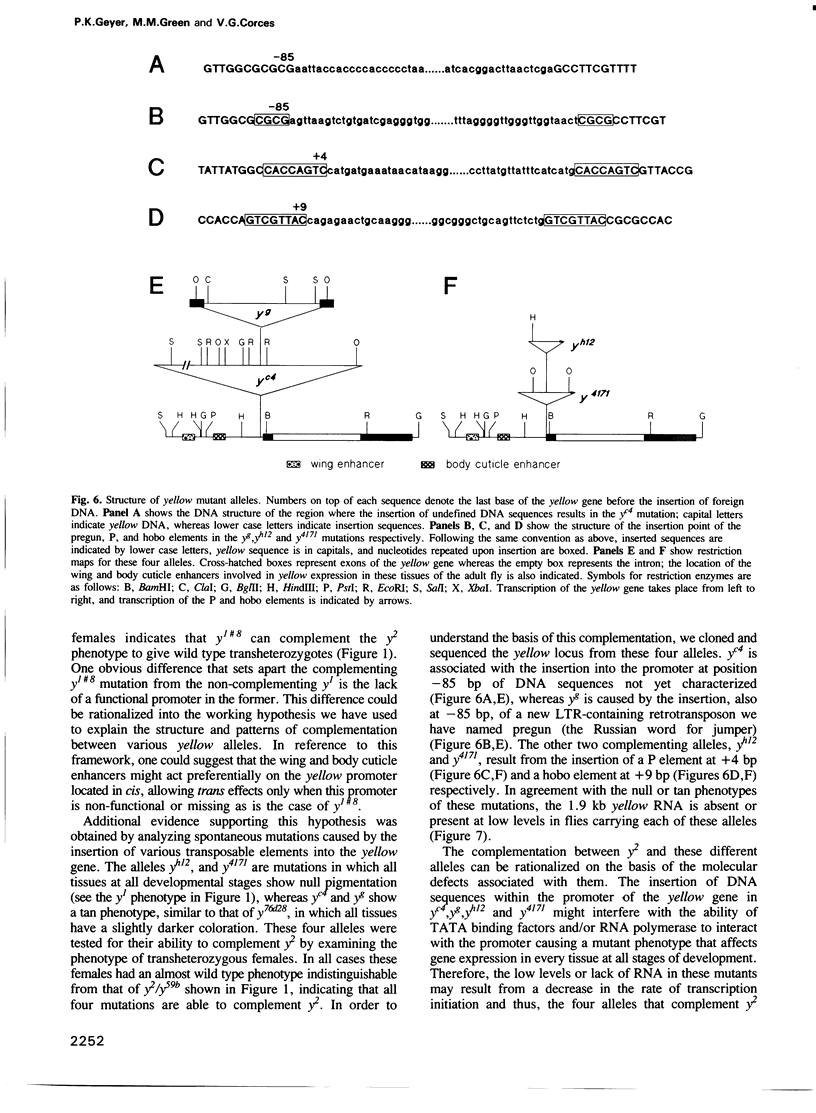
The y2 mutation resulted from the insertion of the gypsy element into the X-linked yellow locus of Drosophila melanogaster. As a consequence of this insertion, transcriptional enhancers that control the expression of the yellow gene in the wings and body cuticle of adult flies are unable to act on the yellow promoter, resulting in a tissue-specific phenotype characterized by mutant coloration in these structures. Some yellow null alleles (yn) are able to complement the y2 phenotype giving rise to near wild type y2/yn females. The molecular structure of the yellow locus in complementing and noncomplementing mutations was determined by cloning and sequencing the various alleles examined. From the information obtained in these studies, we propose a model suggesting that the complementing wild type phenotype of y2/yn flies might be due to the ability of functional wing and body cuticle transcriptional enhancers located in the yn locus to act in trans on the promoter of the yellow gene found in the y2-containing chromosome. Furthermore, this transactivation is abolished by the presence of an intact promoter in cis, suggesting that promoter competition between the yellow genes located on each homolog precludes the activation in trans by transcriptional enhancers in favour of cis effects on their own promoter.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atchison M. L. Enhancers: mechanisms of action and cell specificity. Annu Rev Cell Biol. 1988;4:127–153. doi: 10.1146/annurev.cb.04.110188.001015. [DOI] [PubMed] [Google Scholar]
- Babu P., Selvakumar K. S., Bhosekar S. Studies on transvection at the bithorax complex in Drosophila melanogaster. Mol Gen Genet. 1987 Dec;210(3):557–563. doi: 10.1007/BF00327212. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Bickel S., Benson M., Pirrotta V., Tjian R. Zeste encodes a sequence-specific transcription factor that activates the Ultrabithorax promoter in vitro. Cell. 1988 Jun 3;53(5):713–722. doi: 10.1016/0092-8674(88)90089-x. [DOI] [PubMed] [Google Scholar]
- Campuzano S., Carramolino L., Cabrera C. V., Ruíz-Gómez M., Villares R., Boronat A., Modolell J. Molecular genetics of the achaete-scute gene complex of D. melanogaster. Cell. 1985 Feb;40(2):327–338. doi: 10.1016/0092-8674(85)90147-3. [DOI] [PubMed] [Google Scholar]
- Chia W., Howes G., Martin M., Meng Y. B., Moses K., Tsubota S. Molecular analysis of the yellow locus of Drosophila. EMBO J. 1986 Dec 20;5(13):3597–3605. doi: 10.1002/j.1460-2075.1986.tb04688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Plon S. E., Wang J. C. The use of psoralen-modified DNA to probe the mechanism of enhancer action. Cell. 1986 May 23;45(4):567–574. doi: 10.1016/0092-8674(86)90288-6. [DOI] [PubMed] [Google Scholar]
- Davison D., Chapman C. H., Wedeen C., Bingham P. M. Genetic and physical studies of a portion of the white locus participating in transcriptional regulation and in synapsis-dependent interactions in Drosophila adult tissues. Genetics. 1985 Jul;110(3):479–494. doi: 10.1093/genetics/110.3.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn T. M., Hahn S., Ogden S., Schleif R. F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREEN M. M. Complementation at the yellow locus in Drosophila melanogaster. Genetics. 1961 Nov;46:1385–1388. doi: 10.1093/genetics/46.11.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart W. M. Synapsis-dependent allelic complementation at the decapentaplegic gene complex in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2636–2640. doi: 10.1073/pnas.79.8.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart W. M., Wu C. T. Interactions of zeste mutations with loci exhibiting transvection effects in Drosophila melanogaster. Genetics. 1982 Oct;102(2):179–189. doi: 10.1093/genetics/102.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer P. K., Corces V. G. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1987 Nov;1(9):996–1004. doi: 10.1101/gad.1.9.996. [DOI] [PubMed] [Google Scholar]
- Geyer P. K., Green M. M., Corces V. G. Mutant gene phenotypes mediated by a Drosophila melanogaster retrotransposon require sequences homologous to mammalian enhancers. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8593–8597. doi: 10.1073/pnas.85.22.8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer P. K., Green M. M., Corces V. G. Reversion of a gypsy-induced mutation at the yellow (y) locus of Drosophila melanogaster is associated with the insertion of a newly defined transposable element. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3938–3942. doi: 10.1073/pnas.85.11.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer P. K., Richardson K. L., Corces V. G., Green M. M. Genetic instability in Drosophila melanogaster: P-element mutagenesis by gene conversion. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6455–6459. doi: 10.1073/pnas.85.17.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer P. K., Spana C., Corces V. G. On the molecular mechanism of gypsy-induced mutations at the yellow locus of Drosophila melanogaster. EMBO J. 1986 Oct;5(10):2657–2662. doi: 10.1002/j.1460-2075.1986.tb04548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. M. Genetic instability in Drosophila melanogaster: De novo induction of putative insertion mutations. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3490–3493. doi: 10.1073/pnas.74.8.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. M. Genetic instability in Drosophila melanogaster: the genetics of an MR element that makes complete P insertion mutations. Proc Natl Acad Sci U S A. 1986 Feb;83(4):1036–1040. doi: 10.1073/pnas.83.4.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Udvardy A., Schedl P. Novobiocin blocks the Drosophila heat shock response. J Mol Biol. 1985 May 5;183(1):13–29. doi: 10.1016/0022-2836(85)90277-3. [DOI] [PubMed] [Google Scholar]
- Harrison D. A., Geyer P. K., Spana C., Corces V. G. The gypsy retrotransposon of Drosophila melanogaster: mechanisms of mutagenesis and interaction with the suppressor of Hairy-wing locus. Dev Genet. 1989;10(3):239–248. doi: 10.1002/dvg.1020100313. [DOI] [PubMed] [Google Scholar]
- Hiraizumi Y. Spontaneous recombination in Drosophila melanogaster males. Proc Natl Acad Sci U S A. 1971 Feb;68(2):268–270. doi: 10.1073/pnas.68.2.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. A., Mahadevan S., Struhl K. Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature. 1988 Jun 16;333(6174):635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- Jack J. W., Judd B. H. Allelic pairing and gene regulation: A model for the zeste-white interaction in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1368–1372. doi: 10.1073/pnas.76.3.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman T. C., Tasaka S. E., Suzuki D. T. The interaction of two complex loci, zeste and bithorax in Drosophila melanogaster. Genetics. 1973 Oct;75(2):299–321. doi: 10.1093/genetics/75.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschytz E., Hareven D. Heterochromatin markers: arrangement of obligatory heterochromatin, histone genes and multisite gene families in the interphase nucleus of D. melanogaster. Chromosoma. 1982;86(4):443–455. doi: 10.1007/BF00330120. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Marlor R. L., Parkhurst S. M., Corces V. G. The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol Cell Biol. 1986 Apr;6(4):1129–1134. doi: 10.1128/mcb.6.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Meng Y. B., Chia W. Regulatory elements involved in the tissue-specific expression of the yellow gene of Drosophila. Mol Gen Genet. 1989 Jul;218(1):118–126. doi: 10.1007/BF00330574. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mazo A. M., Mizrokhi L. J., Karavanov A. A., Sedkov Y. A., Krichevskaja A. A., Ilyin Y. V. Suppression in Drosophila: su(Hw) and su(f) gene products interact with a region of gypsy (mdg4) regulating its transcriptional activity. EMBO J. 1989 Mar;8(3):903–911. doi: 10.1002/j.1460-2075.1989.tb03451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozer B., Marlor R., Parkhurst S., Corces V. Characterization and developmental expression of a Drosophila ras oncogene. Mol Cell Biol. 1985 Apr;5(4):885–889. doi: 10.1128/mcb.5.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müeller-Storm H. P., Sogo J. M., Schaffner W. An enhancer stimulates transcription in trans when attached to the promoter via a protein bridge. Cell. 1989 Aug 25;58(4):767–777. doi: 10.1016/0092-8674(89)90110-4. [DOI] [PubMed] [Google Scholar]
- Parkhurst S. M., Corces V. G. Interactions among the gypsy transposable element and the yellow and the suppressor of hairy-wing loci in Drosophila melanogaster. Mol Cell Biol. 1986 Jan;6(1):47–53. doi: 10.1128/mcb.6.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst S. M., Harrison D. A., Remington M. P., Spana C., Kelley R. L., Coyne R. S., Corces V. G. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 1988 Oct;2(10):1205–1215. doi: 10.1101/gad.2.10.1205. [DOI] [PubMed] [Google Scholar]
- Peifer M., Bender W. Sequences of the gypsy transposon of Drosophila necessary for its effects on adjacent genes. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9650–9654. doi: 10.1073/pnas.85.24.9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plon S. E., Wang J. C. Transcription of the human beta-globin gene is stimulated by an SV40 enhancer to which it is physically linked but topologically uncoupled. Cell. 1986 May 23;45(4):575–580. doi: 10.1016/0092-8674(86)90289-8. [DOI] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982 Oct 22;218(4570):348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C. Vectors for P element-mediated gene transfer in Drosophila. Nucleic Acids Res. 1983 Sep 24;11(18):6341–6351. doi: 10.1093/nar/11.18.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoji M., Worcel A. Chromatin assembly in Xenopus oocytes: in vivo studies. Cell. 1984 May;37(1):21–32. doi: 10.1016/0092-8674(84)90297-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana C., Harrison D. A., Corces V. G. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 1988 Nov;2(11):1414–1423. doi: 10.1101/gad.2.11.1414. [DOI] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Augereau P., Chambon P. The SV40 72 bp repeat preferentially potentiates transcription starting from proximal natural or substitute promoter elements. Cell. 1983 Feb;32(2):503–514. doi: 10.1016/0092-8674(83)90470-1. [DOI] [PubMed] [Google Scholar]
- Zachar Z., Chapman C. H., Bingham P. M. On the molecular basis of transvection effects and the regulation of transcription. Cold Spring Harb Symp Quant Biol. 1985;50:337–346. doi: 10.1101/sqb.1985.050.01.043. [DOI] [PubMed] [Google Scholar]







