Abstract
Purpose
Determine the efficacy of a 45-amino acid Gp2 domain, engineered to bind to epidermal growth factor receptor (EGFR), as a positron emission tomography (PET) probe of EGFR in a xenograft mouse model.
Methods
The EGFR-targeted Gp2 (Gp2-EGFR) and a non-binding control were site-specifically labeled with 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) chelator. Binding affinity was tested towards human EGFR and mouse EGFR. Biological activity on downstream EGFR signaling was examined in cell culture. DOTA-Gp2 molecules were labeled with 64Cu and intravenously injected (0.6–2.3 MBq) into mice bearing EGFRhigh (n=7) and EGFRlow (n=4) xenografted tumors. PET/computed tomography (CT) images were acquired at 45 min, 2 h, and 24 h. Dynamic PET (25 min) was also acquired. Tomography results were verified with gamma counting of resected tissues. Two-tailed t tests with unequal variances provided statistical comparison.
Results
DOTA-Gp2-EGFR bound strongly to human (KD = 7 ± 5 nM) and murine (KD = 29 ± 6 nM) EGFR, and non-targeted Gp2 had no detectable binding. Gp2-EGFR did not agonize EGFR nor antagonize EGF-EGFR. 64Cu-Gp2-EGFR tracer effectively localized to EGFRhigh tumors at 45 minutes (3.2 ± 0.5 %ID/g). High specificity was observed with significantly lower uptake in EGFRlow tumors (0.9 ± 0.3 %ID/g, p < 0.001), high tumor-to-background ratios (11 ± 6 tumor:muscle, p < 0.001). Non-targeted Gp2 tracer had low uptake in EGFRhigh tumors (0.5 ± 0.3 %ID/g, p < 0.001). Similar data was observed at 2 h and tumor signal was retained at 24 h (2.9 ± 0.3 %ID/g).
Conclusion
An engineered Gp2 PET imaging probe exhibited low background and target-specific EGFRhigh tumor uptake at 45 min, with tumor signal retained at 24 h post-injection, and compared favorably with published EGFR PET probes for alternative protein scaffolds. These beneficial in vivo characteristics, combined with thermal stability, efficient evolution, and small size of the Gp2 domain validate its use as a future class of molecular imaging agents.
Keywords: Gp2 domain protein scaffold, epidermal growth factor receptor imaging, micro-positron emission tomography, murine model
Graphical Abstract
Introduction
Molecular cancer therapeutics have provided effective treatments for many cancers, yet are typically characterized by efficacy on only a subset of patients, even within a type of cancer as defined by tissue1, 2. Personalized or precision medicine via molecular characterization to differentiate responders from non-responders can aid patient outcomes3. Epidermal growth factor receptor (EGFR) overexpression is present in many cancer types4–10, correlates with differentiation, reduced disease-free and overall survival, and is an independent prognostic indicator of poor survival in colorectal cancer patients11, 12. EGFR amplification is predictive of response to cetuximab in wild-type KRAS metastatic colorectal cancer patients13–15. In HER2-positive primary breast cancer, EGFR overexpression – but not copy number – is a poor prognostic factor and predictive of response to trastuzumab16. The current biopsy/immunohistochemistry approach to EGFR characterization is invasive and does not account for spatiotemporal heterogeneity, most notably differential expression between primary tumors and metastases17–19. Positron emission tomography (PET) targeting EGFR could inform personalized treatment plans by enabling identification, localization, and characterization of primary tumors and metastases, while being non-invasive, quantitative, and sensitive to picomolar quantities. PET based imaging has been clinically useful for other receptors, such as imaging estrogen receptor for breast cancer20, 21.
Numerous scaffolds have been explored as molecular PET tracers of EGFR. Therapeutic monoclonal antibodies (~150 kDa) have been radiolabeled to visualize EGFR in vivo but slow clearance results in high background and liver signal and necessitates late imaging times that elevate patient dose22–27. 94-residue fibronectin domains28, 29, 58-residue affibodies30–34, 120-residue nanobodies35–37 and 400-residue Fab fragments38 have provided good tumor-to-background ratios at early time points (?4 h) via nuclear imaging due to their fast clearance, better extravasation, and increased tissue penetration compared to antibodies39–42. Additional scaffolds have been used for other targets43. Small molecule inhibitors44–49 and natural EGF ligand50 provide molecular characterization but are not biologically passive.
We recently developed the 45-residue Gp2 domain as a small, stable protein scaffold that has been successfully evolved towards multiple targets with high affinity (0.2–18 nM Kd) while retaining thermal stability (65–80°C)51. The Gp2 scaffold contains a framework of a single alpha-helix and three beta-strands, and two solvent-exposed loops that form the diversified paratope. Thermal stability, lack of cysteine, and presence of a single lysine residue distant from the proposed paratope provide ease of chemical conjugation of imaging moieties through amine or thiol chemistry. Additionally, the small size and straightforward structure enable direct chemical synthesis. The two Gp2 variants used here are Gp2-EGFR2.2.3, which was previously evolved to bind to EGFR with 18 ± 8 nM affinity, and EGFR non-binding control, Gp2-rIgG3.2.3, which previously evolved to bind to an irrelevant control (rabbit IgG; notably the molecule does not cross-react with murine IgG) (herein referred to as Gp2-EGFR and Gp2-nb). These variants share 70% sequence identity (Table S1).
We hypothesize that the small size of Gp2 will provide high tumor uptake with fast blood clearance enabling high contrast images at early time points. The ease of evolution and synthesis combined with high thermal stability and different paratope topology may provide a useful tool as an alternative imaging agent to available molecules. In particular, variant Gp2-EGFR2.2.3 has 18±8 nM affinity for cell-surface EGFR with a midpoint of thermal denaturation of 71°C. The current study evaluates the ability of this scaffold to function as molecular PET agent in a small animal model.
Experimental Section
Protein production and DOTA conjugation
Gp2 domains were produced recombinantly in E. coli as described previously51. Briefly, one liter of LB medium with 50 mg/L kanamycin was inoculated with 5 mL of overnight BL21(DE3) E. coli culture carrying the pET-Gp2-His6 plasmid, grown at 37 °C to an optical density (600 nm) of 0.6-1.5 units, and induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside for 20-24 hours at 30 °C. Cells were pelleted, resuspended in 10 mL of lysis buffer (50 mM sodium phosphate (pH 8.0), 0.5 M NaCl, 5% glycerol, 5 mM 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate, and 25 mM imidazole), and underwent four freeze-thaw cycles. The soluble fraction was isolated by centrifugation at 12,000 g for 10 min. Gp2 was purified by metal affinity chromatography on a HisPur resin (Pierce, Thermo Fisher Scientific). Purified Gp2, 30-60 µM, in PBS containing 150 mM imidazole was mixed with 25 to 50-fold molar excess 10 mg/mL DOTA-NHS-ester (Macrocyclics) in dimethyl sulfoxide and allowed to react at room temperature for 1 h. The reaction was quenched with excess 1 M Tris pH 8.0, purified on a PD-10 column (GE Healthcare), and evaluated by matrix-assisted laser desorption ionization mass spectrometry.
Size Exclusion Chromatography
Protein solutions in 100 mM sodium acetate at pH 5.0 were filtered with a 0.2 µM filter to remove any particulates. 200 µL of 40 µM DOTA-Gp2 was loaded onto an AKTA primeplus (GE Healthcare Bio-Sciences) and with a Superdex 75 10/300 GL column. The mobile phase was 100 mM sodium acetate at pH 5.0 flowing at 0.5 mL/min.
Cell growth
A431 epidermoid carcinoma were kindly provided by Dr. Daniel Vallera (University of Minnesota). MDA-MB-435 cells, which have similarities to a melanoma cell line but also show evidence of breast cancer lineage52, were kindly provided by Dr. Tim Starr (University of Minnesota). Cells were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum at 37 °C in humidified air with 5% CO2.
Affinity measurement
Cells to be used in flow cytometry were detached using trypsin for a shorter time (3–5 min) than recommended. Detached cells were washed and labeled with Gp2 at varying concentrations for 15–30 min at 4 °C. Cells were pelleted and washed with PBSA (PBS + 0.1% w/v BSA), then labeled with fluorescein-conjugated rabbit anti-His6 antibody (Abcam ab1206) for 15 min at 4 °C. Fluorescence was analyzed on a C6 Accuri flow cytometer (BD Biosciences). The equilibrium dissociation constant, KD, was identified by minimizing the sum of squared errors assuming a 1:1 binding interaction.
Affinity of Gp2 towards soluble murine EGFR ectodomain (Sino Biological) was determined using Gp2 displayed on the yeast surface as described previously51.
Western Blot Analysis
A431 cells were grown to approximately 60% confluency, washed with PBS and incubated in serum-free medium overnight at 37 °C in humidified air with 5% CO2. The next day, cells were washed with PBS and exposed to four different conditions at 37 °C: (1) PBS for 20 min; (2) 5 nM DOTA-Gp2-EGFR for 20 min; (3) 5 nM epidermal growth factor (Gemini Bio Products) for 20 min; or (4) 5 nM DOTA-Gp2-EGFR for 30 min, washed with PBS, followed by 5 nM epidermal growth factor for 20 min. Cells were detached from the plate by mechanical shearing in RIPA buffer (PBS with 1% v/v Triton X-100, 0.5% w/v sodium deoxycholate, 0.1% w/v sodium dodecyl sulfate). Cells were lysed through rotation at 4 °C for 30 min in RIPA buffer. After centrifuging at 15,000g for 15 min at 4 °C the supernatant was collected and protein concentration was determined with a Pierce BCA assay kit (Thermo Scientific).
Whole-cell lysates (60 µg) were boiled in 5X Laemmli loading buffer at 95°C for 5 minutes, separated by 8% SDS-PAGE, transferred to PVDF membrane and subjected to indicated immunoblotting analyses according to manufacturer guidelines. The primary antibodies bind phosphorylated AKT serine 473 (#9271 Cell Signaling Technology), total AKT (#9272), phosphorylated EGFR tyrosine 1068 (#2234), total EGFR (#2232S) and actin (#A3853 Sigma-Aldrich) were incubated overnight at 4 °C. After washing with Tris-buffered saline with Tween-20 (50mM Tris, 150mM sodium chloride and 0.05% Tween-20), the membrane was further immunoblotted with either anti-rabbit horseradish peroxidase-conjugated antibody (#NA934V GE Healthcare Life Science) or anti-mouse horseradish peroxidase-conjugated (#170-6516 Biorad) secondary antibody for 1 h at 37 °C.
Internalization
Gp2-EGFR and Gp2-nb in PBS with 150 mM imidazole was allowed to react with fluorescein isothiocyanate in DMSO (3 mg/mL) at 100x molar excess at room temperature for 1 h. The reaction was quenched with excess 1 M Tris buffer pH 8, purified on a Zeba Spin Desalting Column 7K molecular weight cutoff (ThermoFisher). Fluorescein conjugation was verified by matrix-assisted laser desorption ionization mass spectrometry.
A431 and MDA-MB-435 cells were grown and detached as above. Cells were labeled with 100 nM fluorescein conjugated Gp2 at 37 °C for 0.5 and 1 h, followed by incubation with 0.2 M acetic acid, 0.5 M NaCl pH 2 for 5 min to strip extracellular binding. Fluorescence was detected by flow cytometry. Internalization was calculated by normalizing the change in fluorescence signal over time to fluorescence signal of A431 cells labeled with 100 nM fluorescein-Gp2-EGFR at 4 °C for 0.5 h.
Copper chelation and purification
64CuCl2 (UW-Madison) was diluted into 150 µL of 100 mM sodium acetate pH 5.0 and pH adjusted to pH 5.0. Approximately 50 MBq of the 64CuCl2 was added to 100 µL DOTA-Gp2 in 100 mM sodium acetate pH 5.0 at 30-60 µM. The mixture was allowed to incubate at 47 °C for 1 h and purified by PD-10 column equilibrated with 10 mM sodium acetate pH 5.0 in order to remove unchelated copper.
Radio TLC
1 µL of 64Cu-Gp2 was spotted on filter paper and a mobile phase of PBS was applied for 20 minutes. An AR-2000 radio-thin layer chromatography scanner (Eckert & Ziegler) scanned and analyzed the filter paper for migration of radioactive peaks. Comparison of scans before and after PD-10 purification showed removal of the peak near the solvent front (the unconjugated 64Cu) while retaining the less mobile peak (64Cu-Gp2), which cold PD-10 purifications along with SDS-PAGE and binding assays have shown to contain highly pure Gp2.
Tumor inoculation
Eight week old female (Foxn1nu/Foxn1nu) mice (Jackson Laboratory) were anesthetized with 1.5% isoflurane in 1 mL/min O2 and subcutaneously injected with 10 million MDA-MB-435 cells in 50% v/v Matrigel Matrix (Corning) in one shoulder. After 4 weeks, the mice were injected with two million A431 cells in 50% Matrigel Matrix into the opposite shoulder. Xenografted tumors were grown to 5-10 mm in diameter (approximately two weeks for A431 and six weeks for MDA-MB-435).
EGFR expression quantification
To quantify EGFR expression within in vivo xenografted tumor cells, GentleMACS dissociator C Tubes (Miltenyi Biotec) were used to generate single cell suspensions from excised tumors. Receptor expression was quantified by flow cytometry with Quantum Simply Cellular anti-mouse IgG calibration beads (Bang’s Laboratories), using Gp2-EGFR and/or mouse anti-EGFR antibody (Abcam ab30) at 1 µM, followed by secondary labeling with fluorescein conjugated rabbit anti-His6 (Abcam ab1206) or AlexaFluor 647 conjugated goat anti-mouse IgG (ThermoFisher), respectively. The cell population from the A431 tumor was approximated as two normally distributed subpopulations.
PET imaging – static and dynamic
All procedures performed in studies involving animals were in accordance with the ethical standards of the University of Minnesota and approved by the Institutional Animal Care and Use Committee. Mice were anesthetized with 1.5% isoflurane in 1 mL/min O2 and tail vein injected with approximately 0.6 to 2.3 MBq of 64Cu-Gp2 as measured by a Atomlab 100 dosimeter with a setting of 50.2. Five-minute static PET scans were performed at 45 min, 2 h and 24 h after injection using an Inveon micro-PET/CT (Siemens). The PET energy cutoffs were 350-650 keV with a timing window of 3.438 ns. The PET images were reconstructed with an OSEM2D method using 4 iterations of Fourier rebinning. PET images were smoothed with a 1×1×1 voxel Guassian filter. The CT used 340 projections of 80 kV at 500 µA with 200 ms exposure over 384 s of total scan time with an effective pixel size of 98.3 µm. The CT was reconstructed using the Feldkamp algorithm with a Shepp-Logan filter. The preceding methods are included in the Inveon Acquisition Workplace software (Siemens). A second batch of independently produced, DOTA-conjugated, 64Cu chelated, and purified 64Cu-Gp2-EGFR injected into another set of tumor inoculated mice validated the results of the other 45 min and 2 h PET/CT scans.
PET images were quantified using the Inveon Research Workplace software (Siemens). Using the CT as an anatomical guide, the volume of 10-20 mm3 that resulted in the maximum average PET signal for that tissue was selected. The anterior end of the liver was selected to avoid noise from kidney signal. The posterior leg furthest from any bladder signal was chosen to represent muscle background.
Tissue gamma counting
After imaging, mice were euthanized by cervical dislocation under isoflurane anesthesia. Blood, bone, brain, heart, large intestine, kidneys, liver, lungs, muscle, pancreas, skin, spleen, stomach, and tumors were resected, weighed, and had their activity measured by a CRC-25W (Capintec) gamma counter averaged over 45 seconds. The CRC-25W collected counts from all windows and was calibrated through serial dilutions based on the dose reported by the Atomlab 100 dosimeter used to measure injected dose. Renal radiation dose was calculated with the Medical Internal Radiation dose method.
Statistics
Comparisons between two samples were determined using a two-tailed student’s t-test for unequal variances. P-values are stated where relevant. Data were presented as average ± standard deviation.
Results
Gp2 Production and Conjugation
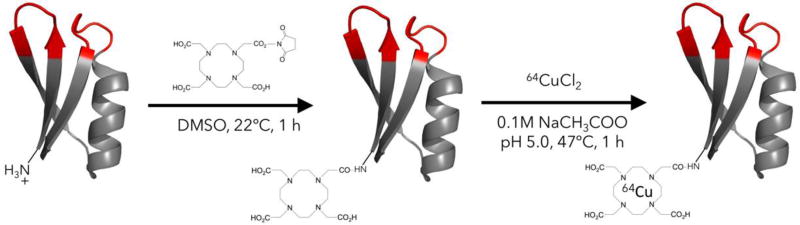
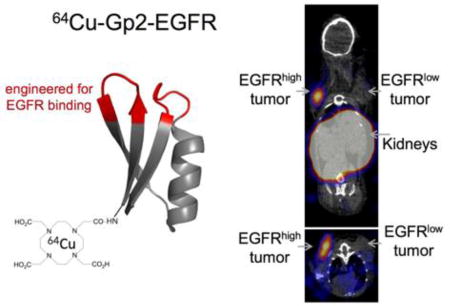
EGFR-binding Gp2-EGFR and non-binding control Gp2-nb, both containing a C-terminal His-6 tag, were produced in the soluble fraction of E. coli and purified by immobilized metal affinity chromatography. Purity was verified with SDS-PAGE and molecular weight was verified by matrix assisted laser desorption ionization mass spectrometry (Gp2-EGFR expected: 6873, actual: 6869; Gp2-nb expected: 6228, actual: 6226). The copper chelator 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) was conjugated to the N-terminal lysine residue distal from the proposed paratope in the Gp2 scaffold framework (Fig. 1). Mass spectrometry was used to verify an average labeling of 0.83 DOTA per molecule for Gp2-EGFR and 1.1 DOTA per molecule for Gp2-nb (Fig. S1). Size exclusion chromatography verified that DOTA-Gp2-EGFR (7.4 kDa) is dominantly monomeric, eluting at a comparable time to control proteins of a similar size (6.5 kDa aprotinin and 7.5 kDa affibody; Fig. S2).
Fig. 1. Gp2 conjugation.
Purified Gp2 was conjugated with the N-hydroxysuccinimidyl ester of the chelator DOTA then radiolabeled with 64Cu
EGFR Binding
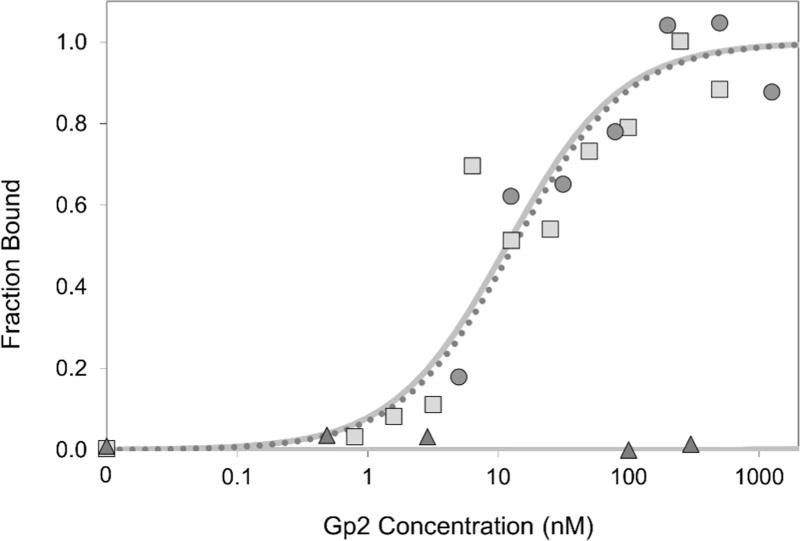
Gp2-EGFR binding affinity towards cellular EGFR was previously found to be 18 ± 8 nM51. The effect of DOTA conjugation on binding affinity was examined by labeling EGFR-expressing A431 epidermoid carcinoma cells with varying levels of DOTA-Gp2-EGFR. DOTA conjugation did not significantly change ligand affinity (7 ± 5 nM; Fig. 2). The non-binding control DOTA-Gp2-nb showed no detectable binding up to 300 nM on A431 cells (Fig. 2). Preclinical imaging experiments with EGFR targeted Gp2 were carried out in mice, so the affinity of Gp2-EGFR towards murine EGFR was examined. Yeast displaying Gp2-EGFR were labeled with varying levels of recombinantly-produced murine EGFR extracellular domain, which revealed an affinity of 29 ± 6 nM (Fig. S3).
Fig. 2. Affinity titration.
A431 cells were labeled with DOTA-Gp2 domains (squares, DOTA-Gp2-EGFR; triangles, DOTA-Gp2-nb) at the indicated concentrations. Binding was detected by fluorescein-conjugated anti-His6 antibody via flow cytometry. Fluorescence signal is normalized between minimal and maximal fluorescence. One representative titration of triplicate experiments is presented. A representative Gp2-EGFR titration curve is also included for comparison (circles, dotted line). The equilibrium dissociation constant for DOTA-Gp2-EGFR, assuming a 1:1 binding model, is 7±5 nM
Biological Activity
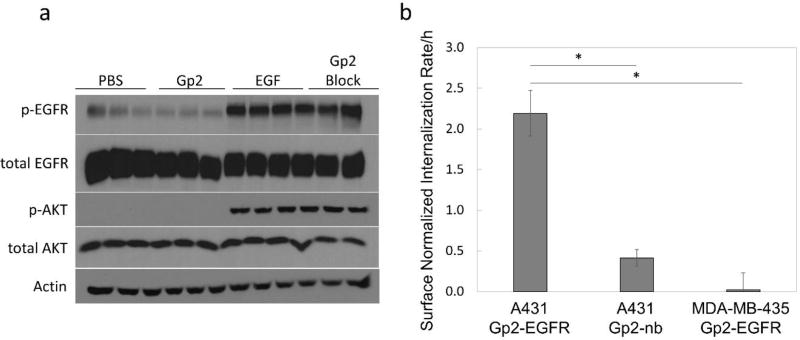
The effect of DOTA-Gp2-EGFR binding on the EGFR signaling pathway was determined by Western blot to detect phosphorylated AKT (p-AKT at S473), a downstream protein kinase, and phosphorylated EGFR (p-EGFR at Y1068) (Fig. 3a). A431 cells labeled with 5 nM DOTA-Gp2-EGFR show no change in p-AKT or p-EGFR compared to PBS only control suggesting that DOTA-Gp2-EGFR is not agonistic to EGFR, DOTA-Gp2-EGFR is also not antagonistic, as blocking the A431 cells with 5 nM Gp2 before addition of 5 nM epidermal growth factor showed no change in level of p-AKT or p-EGFR compared to EGF only control.
Fig. 3. Biological Activity.
(a) A431 cells were labeled with four different conditions in triplicate: PBS only, 5 nM DOTA-Gp2-EGFR in PBS, 5 nM epidermal growth factor (EGF) in PBS, or 5 nM DOTA-Gp2-EGFR followed by 5 nM EGF (Gp2 block). Cells were lysed and separated by SDS-PAGE. Blotting was done to detect phosphorylated AKT (S473), a protein kinase in the EGFR signaling pathway, and phosphorylated EGFR (Y1068), as well as total amounts of the two proteins and actin to verify similar total protein concentration. DOTA-Gp2-EGFR is neither agonistic, since it does not activate the EGFR pathway, nor antagonistic, since it does not block activation when EGF is present. (b) A431 and MDA-MB-435 cells were labeled with 100 nM fluorescein conjugated Gp2 at 37 °C for 0.5 and 1 h, followed by incubation with acid for 5 min to strip extracellular binding. Internalization was calculated by normalizing the change in fluorescence signal over time to fluorescence signal of A431 cells labeled with 100 nM fluorescein-Gp2-EGFR at 4 °C for 0.5 h. Error bars represent standard deviation for n = 3 biological replicates. P < 0.001 is indicated by *.
The ability of A431 cells to internalize Gp2 was examined through flow cytometry of cells grown in tissue plate culture. Fluorescein was conjugated to Gp2-EGFR (0.45 fluorescein/protein) and Gp2-nb (0.61 fluorescein/protein) through amine chemistry and used to label A431 and MDA-MB-435 cells at 37 °C for up to 1 h. At 0.5 and 1 h, cells were acid stripped and the increase in signal over time was used to calculate internalization rate (Fig 3b). Fluorescein-Gp2-EGFR rapidly internalized into A431 cells (2.2±0.3-fold of saturated surface EGFR per hour) compared to control cells (MDA-MB-435, p<0.001) and control non-binder (fluorescein-Gp2-nb, p<0.001).
Copper chelation and purification
Radioactive 64Cu was incubated with DOTA-Gp2 for 1 h at 47°C. Free 64Cu was separated by size exclusion chromatography resulting in, on average, 93% purity. Labeling efficiency was 29%, perhaps due to the low number of DOTA per Gp2 (to assure site specific conjugation) or modest protein concentration. Based on historical yields from non-radioactive DOTA-Gp2 purifications, the specific activity of chelated protein was 0.6-1.1 MBq/nmol. Radiolabeled DOTA-Gp2 variants are referred to as 64Cu-Gp2-EGFR or 64Cu-Gp2-nb.
Murine model micro-PET/CT and tissue biodistribution
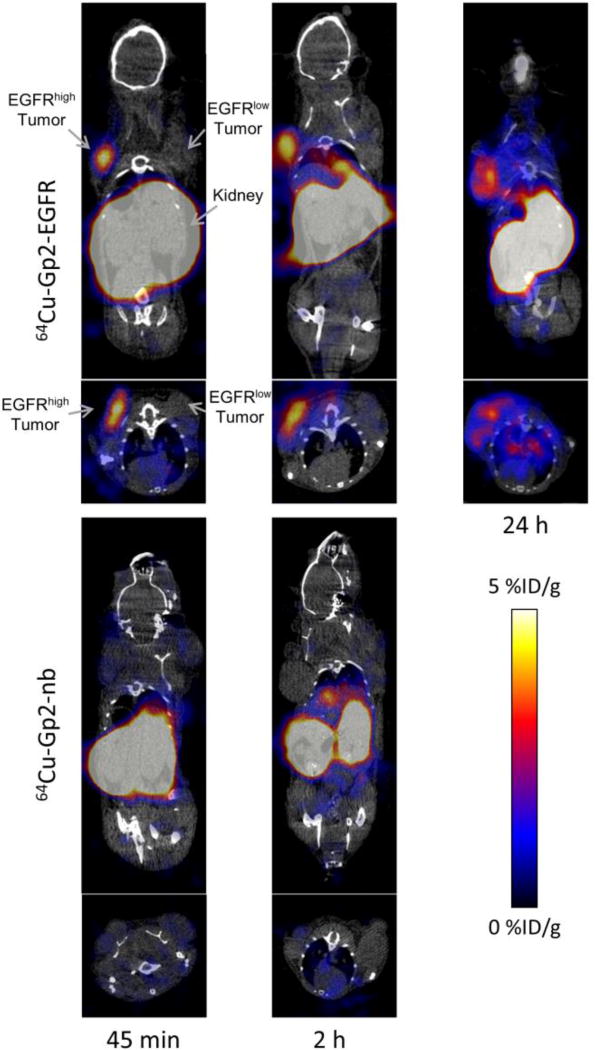
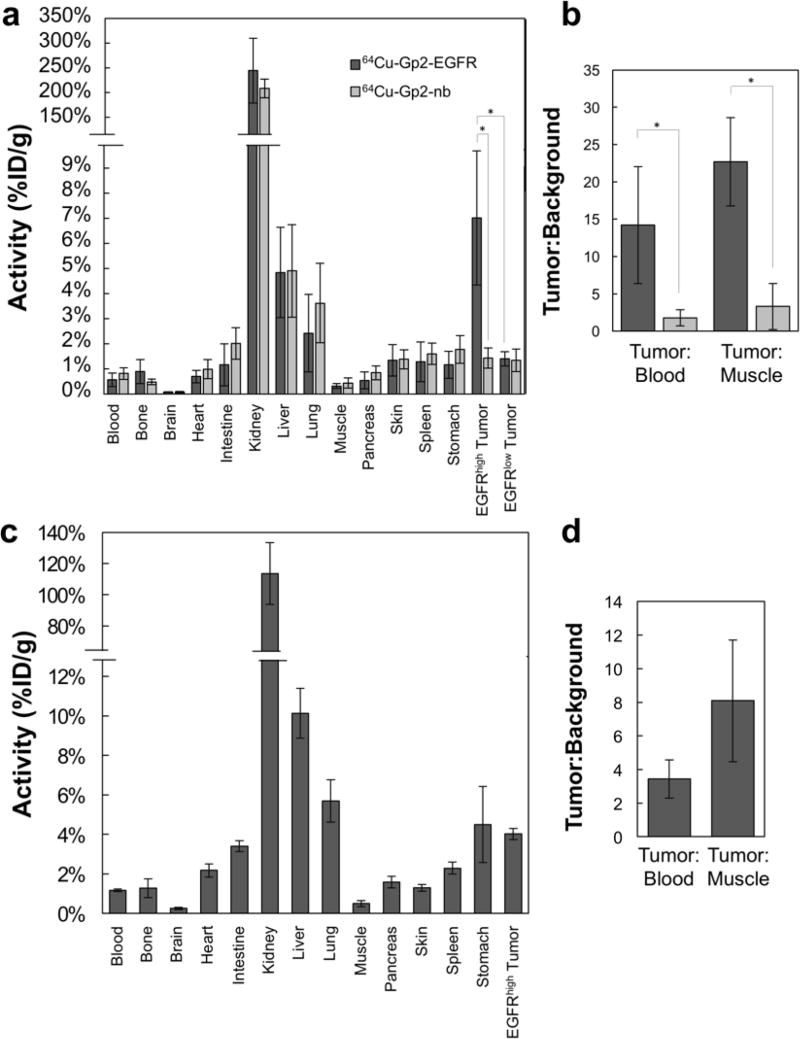
The efficacy of the Gp2 domain was evaluated in a murine model with xenografted human tumor lines. To assess specificity EGFRhigh A431 tumors (mean: 5.2×105 EGFR/cell; 75th percentile: 1.9×106) and EGFRlow MDA-MB-435 tumors (mean and 75th percentile: < 4×103 EGFR/cell) (Fig. S4) were simultaneously evaluated. A non-binding Gp2 domain was tested in parallel. 64Cu-Gp2 was injected via the tail vein into mice harboring dual tumors. PET/CT was performed at 45 min and 2 h. 64Cu-Gp2-EGFR effectively localized to A431 tumors highly expressing EGFR (3.2 ± 0.5 %ID/g) and cleared from background (11 ± 6 tumor:background ratio, p < 0.001) as early as 45 minutes after injection (Fig. 4). Targeting was molecularly specific as EGFRlow MDA-MB-435 tumors had demonstrably lower signal (0.9 ± 0.3 %ID/g, p < 0.001). Moreover, the non-targeted control 64Cu-Gp2-nb exhibited lower signal in EGFRhigh tumors (0.5 ± 0.3 %ID/g, p < 0.001). As for most small protein imaging agents, high kidney signal is observed (78 ±16 %ID/g) resulting from renal processing. Similar imaging is observed at 2 h where 64Cu-Gp2-EGFR uptake to EGFRhigh tumors was 3.2 ± 0.6 %ID/g and 12 ± 4 tumor:background (p = 0.006). Specificity is retained at 2 h as EGFRlow tumors had low uptake (0.7 ± 0.2 %ID/g, p = 0.009) and the non-targeted control had lower signal in EGFRhigh tumors (0.7 ± 0.3 %ID/g, p = 0.007). While early time point imaging is the preferred translational route, we acknowledge that for alternative applications, such as targeted therapy, and biological safety concerns the behavior of engineered proteins at later times is relevant. Even with the fast clearance, preferential EGFRhigh tumor signal from 64Cu-Gp2-EGFR is still evident at 24 h (2.9 ± 0.3 %ID/g) with high tumor:background (8 ± 6, p = 0.009).
Fig. 4. PET/CT imaging.
Coronal and axial micro-PET/CT images of anesthetized athymic nude mice bearing subcutaneously xenografted A431 tumors (EGFRhigh) in the left shoulder and MDA-MB-435 tumors (EGFRlow) in the right shoulder. The mouse in the 24 h image lacks a MDA-MB-435 tumor. Mice were injected by tail vein with 0.6-2.6 MBq of either 64Cu-Gp2-EGFR (top row) or, the non-targeted control, 64Cu-Gp2-nb (bottom row). Five minute static PET scans followed by CT scans were acquired at 45 min (left), 2 h (middle), and 24 h (right, for targeted Gp2 only) post-injection. Image planes were selected such that both tumors appear in the image.
PET images were corroborated by ex vivo tissue gamma counting at 2 h and 24 h (Fig. 5). At 2 hours post injection, 64Cu-Gp2-EGFR localized significantly more to xenografted EGFRhigh tumors (7.0 ± 1.9 % ID/g) as compared to EGFRlow tumors (1.4 ± 0.3 % ID/g; p < 0.001). The targeted Gp2 had 14 ± 8 tumor-to-blood ratio and 23 ± 6 tumor-to-muscle at 2 h, compared to 1.8 ± 1 tumor-to-blood (p = 0.005) and 3.3 ± 3.1 tumor-to-muscle (p < 0.001) for the non-targeted Gp2. In addition, the non-targeted Gp2 showed significantly lower EGFRhigh tumor uptake with 1.4 ± 0.4 % ID/g (p = 0.001). Renal retention was high for the targeted (244 ±66 %ID/g) and non-targeted (208 ± 19 %ID/g) probes. Liver signal was modest for both (4.8 ± 1.8 and 4.9 ± 1.9 %ID/g). At 24 h the fast clearance leads to lower signal in most tissues, including EGFRhigh tumor (4.0 ± 0.3 %ID/g) and kidney (114 ± 20 %ID/g), with the exception of a notable increase in liver signal (10.1 ± 1.3 %ID/g). Tumor-to-blood and tumor-to-muscle ratios (3.4 ± 1.1, p = 0.002 and 8.1 ± 3.6, p < 0.001, respectively) indicate there is still preferential uptake to EGFRhigh tumor.
Fig. 5. Resected tissue gamma counting.
After PET/CT imaging, mice were euthanized and tissues were collected, weighed, and measured for activity. (A) The targeted 64Cu-Gp2-EGFR (dark gray) and non-targeted 64Cu-Gp2-nb (light gray) distribution is shown for the selected tissues at 2 h post-injection. (B) Ratios of tumor signal to relevant background signals in blood and muscle. The data is combined over two separate experiments, n = 4 for mice containing EGFRhigh and EGFRlow tumors and another n = 3 for mice containing only EGFRhigh tumors. Significance for important comparisons (p < 0.005) is denoted by *. (C and D) Biodistribution and tumor-to-background ratios of 64Cu-Gp2-EGFR in n = 3 mice at 24 h post-injection. Error bars represent standard deviation.
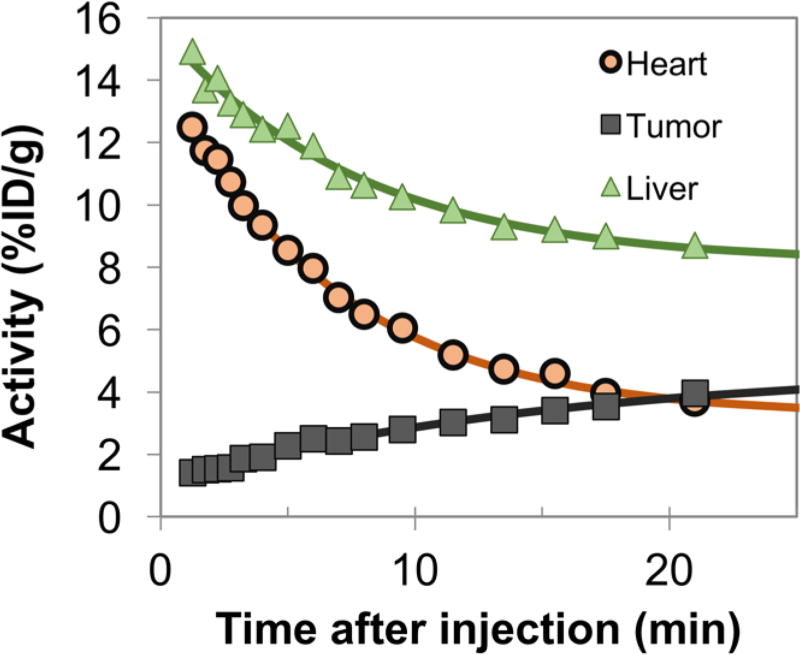
The rapid distribution and clearance of 64Cu-Gp2 evident at the 45 minute scan was more thoroughly investigated by 25-minute dynamic PET scans (Fig. 6). Using heart signal as a surrogate for probe blood levels, clearance half-time was revealed to be 3.2 ± 1.0 min, supporting the low accumulation in muscle background seen at 45 minutes post-injection.
Fig. 6. Dynamic PET scans.
25 minute dynamic PET scans were acquired on anesthetized mice containing xenografted EGFRhigh tumors. The average signal within ~15 mm3 regions, guided by an anatomical CT scan, is presented. Data were fit assuming exponential kinetics. The clearance half-time within the heart (predominantly blood pool) was t1/2 = 3.2 ± 1.0 min (n=2 mice).
Discussion
Other small scaffolds have been successfully used for in vivo imaging previously but drawbacks, such as the relatively larger size of fibronectins (11 kDa)53 and DARPins (20 kDa)54, or the difficultly of broad evolution and presence of cysteines in knottins55 and cyclic peptides56, has driven the search for additional scaffolds. Cysteine-free Affibodies57 have gone to smaller size (58 amino acids) and their helical paratope has yielded high affinity binders, however they are typically severely destabilized after mutation58. Gp2 domains push the size even smaller (45-49 amino acids), have thus far remained highly thermally stable after mutation, and provide a vastly different paratope structure compared to Affibodies. Beyond its previous characterization for high-affinity, EGFR-specific binding51, further biophysical evaluation of Gp2-EGFR in the current study revealed that it is well-suited for use in molecular imaging. Though selected solely for EGFR ectodomain binding, the current Gp2 variant is neither agonistic nor antagonistic (Fig. 3a). This enables passive imaging – unlike radiolabeled EGF or bivalent, crosslinking-compatible antibodies – which is preferred to avoid impacting EGF signaling cascades. Additionally, Gp2-EGFR is internalized into A431 cells (Fib. 3b). Internalization potentially allows for an accumulation of signal in target tissues over time, but may not be highly relevant for Gp2 due to the rapid clearance of the small agent, which has the benefit of reducing background. Primary amine / N-hydroxysuccinimidyl chemistry was selected for conjugation at the N-terminal lysine distal to the evolved loops (Fig. 1). As hoped, DOTA conjugation did not hinder binding affinity (18 ± 8 nM as Gp2-EGFR to 7 ± 5 nM as DOTA-Gp2-EGFR). Importantly, Gp2-EGFR exhibits cross-reactive binding to murine EGFR, which aids the validity of the murine model to assess the probe’s tumor selectivity relative to lower levels of EGFR expression in healthy tissue including liver. Modest liver accumulation was observed (4.8 ± 1.8 %ID/g at 2 h), which was due to physiological processing, not EGFR targeting, as the non-binding control exhibited equivalent hepatic retention (4.9 ± 1.8 %ID/g). This liver signal remains below the EGFRhigh tumor signal (1.5 ± 0.4 tumor:liver). Nevertheless, efforts are underway to mutate surface hydrophobic amino acids to increase Gp2 hydrophilicity, which effectively reduced liver signal for engineered fibronectin domains29.
The relevance of non-invasive EGFR detection in the clinic has led to development of many imaging probes, including a variety of small protein scaffolds. The increased extravasation and tissue penetration of protein scaffolds compared to larger proteins allows for high contrast early imaging resulting in lower patient dose. Multiple successes have been realized for EGFR previously. The beneficial properties of Gp2 domains as evolvable protein scaffolds, such as small size, lack of cysteines, and high thermal stability, do not guarantee successful translation to an imaging agent. However, these properties provide benefits during evolution, conjugation, administration, and biodistribution that are useful for imaging agents or therapeutics towards many targets. Due to the variations between labs, strict quantitative comparisons between scaffolds does not prove superiority. Moreover, comparisons across scaffolds must take care to acknowledge the context-dependent properties – affinity, charge, hydrophilicity – of individual protein variants. Nevertheless, the current data demonstrate that the Gp2 domain is a promising PET imaging agent for EGFR with potential benefits versus other probes, and further optimization of the affinity and biophysical properties of Gp2-EGFR could lead to a clinically effective PET imaging agent. 64Cu-Gp2-EGFR exhibits tumor accumulation (3.2 ± 0.5 %ID/g at 0.75 h via PET; 7.0 ± 1.9 %ID/g at 2 h via excised tissue) comparable to other small protein PET probes including fibronectin domains (3.4 ± 1.0 and 2.4 ± 1.0 %ID/g at 1 h)28 and affibodies (5.7 ± 0.6 and 9.7 ± 4.9 %ID/g at 1 h)30 as well as nanobodies for single-photon emission computed tomography (4.6 ± 0.4 %ID/g at 1 h)37. The dramatically lower uptake of 64Cu-Gp2-EGFR in EGFRlow tumors and non-binding control in EGFRhigh tumors was similarly observed for the fibronectin domain. For affibody, neither EGFRlow tumors nor non-targeted affibody were evaluated as controls. Blocking did yield a reduction, albeit incomplete (47%), in EGFRhigh tumor uptake. 64Cu-Gp2-EGFR exhibits high tumor:blood ratio (14 ± 8 at 2 h) because of rapid clearance (3.2 ± 1.0 min half-time). Conversely, affibody provides limited tumor:blood differentiation (1.2 ± 1.1 and 1.0 ± 0.1 at 1 h and 4 h) because of slower clearance (20 – 120 min half-time30, 59–62) while fibronectin is intermediate (8.9 ± 4.7 and 6.4 ± 4.3 at 1 h and 4 h28) with rapid clearance (2.1 ± 0.3 min half-time28). Tumor:muscle specificity is also strong for 64Cu-Gp2-EGFR (11 ± 6 at 0.75 h via PET; 23 ± 6 at 2 h via excised tissue), comparable to affibody (16 ± 7 at 1 h, 18 ± 4 at 4 h, both via excised tissue) and higher than fibronectin (8.6 ± 3.0 at 1 h via PET; 10 ± 4 and 4.2 ± 1.3 at 1 and 4 h via excised tissue).
The main disadvantage with Gp2 as an imaging agent is the high kidney signal due to partial renal retention during clearance, which is observed for most small protein scaffolds43. Dosimetry calculations indicate 3.0 mGy/MBq renal dose, which is 3% of the maximum tolerated dose for a 185 MBq injection thereby rendering this a minor concern clinically for non-renal tumors. Yet strategies exist to lower kidney signal. Modulation of charge has been shown to reduce renal uptake in fibronectin domains29, affibodies63, and knottins64. Preliminary data indicate an ability to modify charge on Gp2-EGFR while retaining activity. Additionally, alternative radiochemical conjugation has drastically reduced renal uptake of other small protein scaffolds62, 65–70. Specifically, transchelation from the DOTA chelator71 may account for some signal in the liver and kidney, and other chelators such as NOTA or PCTA have shown higher stability in vivo72. Notably, 64Cu (t1/2 = 12.7 h) was used in the current study to enable examination of distribution kinetics over short and long time periods, which is important for initial physiological characterization of this new protein scaffold. Yet, clinical use may benefit from a radioisotope with decay kinetics that align with the rapid distribution of the small Gp2 domain to reduce patient dose. Future studies with 18F (t1/2 = 110 min), 68Ga (t1/2 = 68 min), or 61Cu (t1/2 = 3.3 h) will be valuable for clinical translation. Evaluation on cells with intermediate EGFR expression will also be informative. It should be noted that, as with any synthetically engineered protein with non-human sequence components, immunogenicity of evolved molecules will need to be evaluated.
Overall, the performance of these initial Gp2 domains in vivo gives promise to the potential of Gp2-EGFR, and other targeted Gp2 domains, as molecular imaging agents.
Supplementary Material
Acknowledgments
We are grateful to Joanne Johnson of the Center for Clinical Imaging Research for assistance with PET/CT imaging and Dr. Blake Jacobson from the Department of Medicine at the University of Minnesota for assistance with ex vivo tumor analysis. This work was funded by the National Institutes of Health (EB019518 to BJH), Komen for the Cure (SAC110039 to DY), National Cancer Institute Cancer Center Support Grant P30 077598, and the University of Minnesota.
Abbreviations
- EGFR
epidermal growth factor
- PET
positron emission tomography
- DOTA
1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid
- CT
computed tomography
- PBS
phosphate buffered saline
- SDS-PAGE
sodium dodecyl sulfate – polyacrylamide gel electrophoresis
Footnotes
Supporting Information
Mass spectrometry verification of DOTA conjugation, size exclusion chromatography results, murine EGFR affinity titration curve, EGFR receptor quantification
References
- 1.Hamburg MA, Collins FS. The Path to Personalized Medicine. N. Engl. J. Med. 2010;363(4):301–304. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 2.Scott AM, Wolchok JD, Old LJ. Antibody Therapy of Cancer. Nat. Rev. Cancer. 2012;12(4):278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 3.Kircher MF, Hricak H, Larson SM. Molecular Imaging for Personalized Cancer Care. Mol. Oncol. 2012;6(2):182–195. doi: 10.1016/j.molonc.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hynes NE, MacDonald G. ErbB Receptors and Signaling Pathways in Cancer. Curr. Opin. Cell Biol. 2009;21(2):177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 5.Shinojima N, Tada K, Shiraishi S, Kamiryo T, Kochi M, Nakamura H, Makino K, Saya H, Hirano H, Kuratsu J-I, Oka K, Ishimaru Y, Ushio Y. Prognostic Value of Epidermal Growth Factor Receptor in Patients with Glioblastoma Multiforme. Cancer Res. 2003;63(20):6962–6970. [PubMed] [Google Scholar]
- 6.Nieto Y, Nawaz F, Jones RB, Shpall EJ, Nawaz S. Prognostic Significance of Overexpression and Phosphorylation of Epidermal Growth Factor Receptor (EGFR) and the Presence of Truncated EGFRvIII in Locoregionally Advanced Breast Cancer. J. Clin. Oncol. 2007;25(28):4405–4413. doi: 10.1200/JCO.2006.09.8822. [DOI] [PubMed] [Google Scholar]
- 7.Lugli A, Iezzi G, Hostettler I, Muraro MG, Mele V, Tornillo L, Carafa V, Spagnoli G, Terracciano L, Zlobec I. Prognostic Impact of the Expression of Putative Cancer Stem Cell Markers CD133, CD166, CD44s, EpCAM, and ALDH1 in Colorectal Cancer. Br. J. Cancer. 2010;103(3):382–390. doi: 10.1038/sj.bjc.6605762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galizia G, Lieto E, Orditura M, Castellano P, Mura A La, Imperatore V, Pinto M, Zamboli A, De Vita F, Ferraraccio F. Epidermal Growth Factor Receptor (EGFR) Expression Is Associated with a Worse Prognosis in Gastric Cancer Patients Undergoing Curative Surgery. World J. Surg. 2007;31(7):1458–1468. doi: 10.1007/s00268-007-9016-4. [DOI] [PubMed] [Google Scholar]
- 9.Parra HS, Cavina R, Latteri F, Zucali PA, Campagnoli E, Morenghi E, Grimaldi GC, Roncalli M, Santoro A. Analysis of Epidermal Growth Factor Receptor Expression as a Predictive Factor for Response to Gefitinib (“Iressa”, ZD1839) in Non-Small-Cell Lung Cancer. Br. J. Cancer. 2004;91:208–212. doi: 10.1038/sj.bjc.6601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlomm T, Kirstein P, Iwers L, Daniel B, Steuber T, Walz J, Chun FHK, Haese A, Kollermann J, Graefen M, Huland H, Sauter G, Simon R, Erbersdobler A. Clinical Significance of Epidermal Growth Factor Receptor Protein Overexpression and Gene Copy Number Gains in Prostate Cancer. Clin. Cancer Res. 2007;13(22 Pt 1):6579–6584. doi: 10.1158/1078-0432.CCR-07-1257. [DOI] [PubMed] [Google Scholar]
- 11.Huang C-W, Tsai H-L, Chen Y-T, Huang C-M, Ma C-J, Lu C-Y, Kuo C-H, Wu D-C, Chai C-Y, Wang J-Y. The Prognostic Values of EGFR Expression and KRAS Mutation in Patients with Synchronous or Metachronous Metastatic Colorectal Cancer. BMC Cancer. 2013;13(1):599. doi: 10.1186/1471-2407-13-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rokita M, Stec R, Bodnar L, Charkiewicz R, Korniluk J, Smoter M, Cichowicz M, Chyczewski L, Niklinski J, Kozlowski W, Szczylik C. Overexpression of Epidermal Growth Factor Receptor as a Prognostic Factor in Colorectal Cancer on the Basis of the Allred Scoring System. Onco. Targets. Ther. 2013;6:967–976. doi: 10.2147/OTT.S42446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet J-B, Lecomte T, Rougier P, Lievre A, Landi B, Boige V, Ducreux M, Ychou M, Bibeau F, Bouché O, Reid J, Stone S, Penault-Llorca F. Analysis of PTEN, BRAF, and EGFR Status in Determining Benefit from Cetuximab Therapy in Wild-Type KRAS Metastatic Colon Cancer. J. Clin. Oncol. 2009;27(35):5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 14.Scartozzi M, Bearzi I, Mandolesi A, Pierantoni C, Loupakis F, Zaniboni A, Negri F, Quadri A, Zorzi F, Galizia E, Berardi R, Biscotti T, Labianca R, Masi G, Falcone A, Cascinu S. Epidermal Growth Factor Receptor (EGFR) Gene Copy Number (GCN) Correlates with Clinical Activity of Irinotecan-Cetuximab in K-RAS Wild-Type Colorectal Cancer: A Fluorescence in Situ (FISH) and Chromogenic in Situ Hybridization (CISH) Analysis. BMC Cancer. 2009;9:303. doi: 10.1186/1471-2407-9-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, Gambacorta M, Siena S, Bardelli A. Gene Copy Number for Epidermal Growth Factor Receptor (EGFR) and Clinical Response to antiEGFR Treatment in Colorectal Cancer: A Cohort Study. Lancet. Oncol. 2005;6(5):279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 16.Lee HJ, Seo AN, Kim EJ, Jang MH, Kim YJ, Kim JH, Kim S-W, Ryu HS, Park IA, Im S-A, Gong G, Jung KH, Kim HJ, Park SY. Prognostic and Predictive Values of EGFR Overexpression and EGFR Copy Number Alteration in HER2-Positive Breast Cancer. Br. J. Cancer. 2015;112(1):103–111. doi: 10.1038/bjc.2014.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scartozzi M, Bearzi I, Berardi R, Mandolesi A, Fabris G, Cascinu S. Epidermal Growth Factor Receptor (EGFR) Status in Primary Colorectal Tumors Does Not Correlate with EGFR Expression in Related Metastatic Sites: Implications for Treatment with EGFR-Targeted Monoclonal Antibodies. J. Clin. Oncol. 2004;22(23):4772–4778. doi: 10.1200/JCO.2004.00.117. [DOI] [PubMed] [Google Scholar]
- 18.Yarom N, Marginean C, Moyana T, Gorn-Hondermann I, Birnboim HC, Marginean H, Auer RC, Vickers M, Asmis TR, Maroun J, Jonker D. EGFR Expression Variance in Paired Colorectal Cancer Primary and Metastatic Tumors. Cancer Biol. Ther. 2014;10(5):416–421. doi: 10.4161/cbt.10.5.12610. [DOI] [PubMed] [Google Scholar]
- 19.Bozzetti C, Tiseo M, Lagrasta C, Nizzoli R, Guazzi A, Leonardi F, Gasparro D, Spiritelli E, Rusca M, Carbognani P, Majori M, Franciosi V, Rindi G, Ardizzoni A. Comparison between Epidermal Growth Factor Receptor (EGFR) Gene Expression in Primary Non-Small Cell Lung Cancer (NSCLC) and in Fine-Needle Aspirates from Distant Metastatic Sites. J. Thorac. Oncol. 2008;3(1):18–22. doi: 10.1097/JTO.0b013e31815e8ba2. [DOI] [PubMed] [Google Scholar]
- 20.Linden HM, Stekhova SA, Link JM, Gralow JR, Livingston RB, Ellis GK, Petra PH, Peterson LM, Schubert EK, Dunnwald LK, Krohn KA, Mankoff DA. Quantitative Fluoroestradiol Positron Emission Tomography Imaging Predicts Response to Endocrine Treatment in Breast Cancer. J. Clin. Oncol. 2006;24(18):2793–2799. doi: 10.1200/JCO.2005.04.3810. [DOI] [PubMed] [Google Scholar]
- 21.Peterson LM, Kurland BF, Schubert EK, Link JM, Gadi VK, Specht JM, Eary JF, Porter P, Shankar LK, Mankoff DA, Linden HM. A Phase 2 Study of 16α-[18F]-Fluoro-17β-Estradiol Positron Emission Tomography (FES-PET) as a Marker of Hormone Sensitivity in Metastatic Breast Cancer (MBC) Mol. Imaging Biol. 2014;16(3):431–440. doi: 10.1007/s11307-013-0699-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai W, Chen K, He L, Cao Q, Koong A, Chen X. Quantitative PET of EGFR Expression in Xenograft-Bearing Mice Using 64Cu-Labeled Cetuximab, a Chimeric Anti-EGFR Monoclonal Antibody. Eur. J. Nucl. Med. Mol. Imaging. 2007;34(6):850–858. doi: 10.1007/s00259-006-0361-6. [DOI] [PubMed] [Google Scholar]
- 23.Niu G, Li Z, Xie J, Le Q-T, Chen X. PET of EGFR Antibody Distribution in Head and Neck Squamous Cell Carcinoma Models. J. Nucl. Med. 2009;50(7):1116–1123. doi: 10.2967/jnumed.109.061820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niu G, Sun X, Cao Q, Courter D, Koong A, Le Q-T, Gambhir SS, Chen X. Cetuximab-Based Immunotherapy and Radioimmunotherapy of Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. 2010;16(7):2095–2105. doi: 10.1158/1078-0432.CCR-09-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ping Li W, Meyer LA, Capretto DA, Sherman CD, Anderson CJ. Receptor-Binding, Biodistribution, and Metabolism Studies of 64Cu-DOTA-Cetuximab, a PET-Imaging Agent for Epidermal Growth-Factor Receptor-Positive Tumors. Cancer Biother. Radiopharm. 2008;23(2):158–171. doi: 10.1089/cbr.2007.0444. [DOI] [PubMed] [Google Scholar]
- 26.Menke-van der Houven van Oordt CW, Gootjes EC, Huisman MC, Vugts DJ, Roth C, Luik AM, Mulder ER, Schuit RC, Boellaard R, Hoekstra OS, Dongen GAMS Van, Verheul HMW. Zr-Cetuximab PET Imaging in Patients with Advanced Colorectal Cancer. Oncotarget. 2015;6(30) doi: 10.18632/oncotarget.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharyya S, Kurdziel K, Wei L, Riffle L, Kaur G, Hill GC, Jacobs PM, Tatum JL, Doroshow JH, Kalen JD. Zirconium-89 Labeled Panitumumab: A Potential Immuno-PET Probe for HER1-Expressing Carcinomas. Nucl. Med. Biol. 2013;40(4):451–457. doi: 10.1016/j.nucmedbio.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hackel BJ, Kimura RH, Gambhir SS. Use of 64Cu-Labeled Fibronectin Domain with EGFR-Overexpressing Tumor Xenograft: Molecular Imaging. Radiology. 2012;263(1):179–188. doi: 10.1148/radiol.12111504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hackel BJ, Sathirachinda A, Gambhir SS. Designed Hydrophilic and Charge Mutations of the Fibronectin Domain: Towards Tailored Protein Biodistribution. Protein Eng. Des. Sel. 2012;25(10):639–647. doi: 10.1093/protein/gzs036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miao Z, Ren G, Hongguang L, Jiang L, Cheng Z. Small-Animal PET Imaging of Human Epidermal Growth Factor Receptor Positive Tumor with a 64Cu Labeled Affibody Protein. Bioconjug. Chem. 2010;21(5):947–954. doi: 10.1021/bc900515p. [DOI] [PubMed] [Google Scholar]
- 31.Miao Z, Ren G, Liu H, Qi S, Wu S, Cheng Z. PET of EGFR Expression with an 18F-Labeled Affibody Molecule. J. Nucl. Med. 2012;53(7):1110–1118. doi: 10.2967/jnumed.111.100842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nordberg E, Orlova A, Friedman M, Tolmachev V, Ståhl S, Nilsson FY, Glimelius B, Carlsson J. In Vivo and in Vitro Uptake of 111In, Delivered with the Affibody Molecule (ZEGFR:955)2, in EGFR Expressing Tumour Cells. Oncol. Rep. 2008;19(4):853–857. doi: 10.3892/or.19.4.853. [DOI] [PubMed] [Google Scholar]
- 33.Tolmachev V, Rosik D, Wållberg H, Sjöberg A, Sandström M, Hansson M, Wennborg A, Orlova A. Imaging of EGFR Expression in Murine Xenografts Using Site-Specifically Labelled Anti-EGFR 111In-DOTA-Z EGFR":2377 Affibody Molecule: Aspect of the Injected Tracer Amount. Eur. J. Nucl. Med. Mol. Imaging. 2010;37(3):613–622. doi: 10.1007/s00259-009-1283-x. [DOI] [PubMed] [Google Scholar]
- 34.Su X, Cheng K, Jeon J, Shen B, Venturin GT, Hu X, Rao J, Chin FT, Wu H, Cheng Z. Comparison of Two Site-Specifically 18F-Labeled Affibodies for PET Imaging of EGFR Positive Tumors. Mol. Pharm. 2014;11(11):3947–3956. doi: 10.1021/mp5003043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang L, Gainkam LOT, Caveliers V, Vanhove C, Keyaerts M, De Baetselier P, Bossuyt A, Revets H, Lahoutte T. SPECT Imaging with 99mTc-Labeled EGFR-Specific Nanobody for in Vivo Monitoring of EGFR Expression. Mol. Imaging Biol. 2008;10(3):167–175. doi: 10.1007/s11307-008-0133-8. [DOI] [PubMed] [Google Scholar]
- 36.Gainkam LOT, Keyaerts M, Caveliers V, Devoogdt N, Vanhove C, Van Grunsven L, Muyldermans S, Lahoutte T. Correlation between Epidermal Growth Factor Receptor-Specific Nanobody Uptake and Tumor Burden: A Tool for Noninvasive Monitoring of Tumor Response to Therapy. Mol. Imaging Biol. 2011;13(5):940–948. doi: 10.1007/s11307-010-0428-4. [DOI] [PubMed] [Google Scholar]
- 37.Gainkam LOT, Huang L, Caveliers V, Keyaerts M, Hernot S, Vaneycken I, Vanhove C, Revets H, De Baetselier P, Lahoutte T. Comparison of the Biodistribution and Tumor Targeting of Two 99mTc-Labeled Anti-EGFR Nanobodies in Mice, Using Pinhole SPECT/micro-CT. J. Nucl. Med. 2008;49(5):788–795. doi: 10.2967/jnumed.107.048538. [DOI] [PubMed] [Google Scholar]
- 38.Chakravarty R, Goel S, Valdovinos HF, Hernandez R, Hong H, Nickles RJ, Cai W. Matching the Decay Half-Life with the Biological Half-Life: ImmunoPET Imaging with44Sc-Labeled Cetuximab Fab Fragment. Bioconjug. Chem. 2014;25(12):2197–2204. doi: 10.1021/bc500415x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt MM, Wittrup KD. A Modeling Analysis of the Effects of Molecular Size and Binding Affinity on Tumor Targeting. Mol. Cancer Ther. 2009;8(10):2861–2871. doi: 10.1158/1535-7163.MCT-09-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zahnd C, Kawe M, Stumpp MT, de Pasquale C, Tamaskovic R, Nagy-Davidescu G, Dreier B, Schibli R, Binz HK, Waibel R, Plückthun A. Efficient Tumor Targeting with High-Affinity Designed Ankyrin Repeat Proteins: Effects of Affinity and Molecular Size. Cancer Res. 2010;70(4):1595–1605. doi: 10.1158/0008-5472.CAN-09-2724. [DOI] [PubMed] [Google Scholar]
- 41.Yuan F, Dellian M, Fukumura D, Leunig M, Berk DA, Torchilin VP, Jain RK. Vascular Permeability in a Human Tumor Xenograft: Molecular Size Dependence and Cutoff Size. Cancer Res. 1995;55(17):3752–3756. [PubMed] [Google Scholar]
- 42.Thurber GM, Wittrup KD. Quantitative Spatiotemporal Analysis of Antibody Fragment Diffusion and Endocytic Consumption in Tumor Spheroids. Cancer Res. 2008;68(9):3334–3341. doi: 10.1158/0008-5472.CAN-07-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stern LA, Case BA, Hackel BJ. Alternative Non-Antibody Protein Scaffolds for Molecular Imaging of Cancer. Curr. Opin. Chem. Eng. 2013;2(4):425–432. doi: 10.1016/j.coche.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Memon AA, Jakobsen S, Dagnaes-Hansen F, Sorensen BS, Keiding S, Nexo E. Positron Emission Tomography (PET) Imaging with [11C]-Labeled Erlotinib: A Micro-PET Study on Mice with Lung Tumor Xenografts. Cancer Res. 2009;69(3):873–878. doi: 10.1158/0008-5472.CAN-08-3118. [DOI] [PubMed] [Google Scholar]
- 45.Zhang MR, Kumata K, Hatori A, Takai N, Toyohara J, Yamasaki T, Yanamoto K, Yui J, Kawamura K, Koike S, Ando K, Suzuki K. [11C]Gefitinib ([11C]Iressa): Radiosynthesis, In Vitro Uptake, and In Vivo Imaging of Intact Murine Fibrosarcoma. Mol. Imaging Biol. 2010;12(2):181–191. doi: 10.1007/s11307-009-0265-5. [DOI] [PubMed] [Google Scholar]
- 46.Wang H, Yu J, Yang G, Song X, Sun X, Zhao S, Mu D. Assessment of 11C-Labeled-4-N-(3-Bromoanilino)-6,7-Dimethoxyquinazoline as a Positron Emission Tomography Agent to Monitor Epidermal Growth Factor Receptor Expression. Cancer Sci. 2007;98(9):1413–1416. doi: 10.1111/j.1349-7006.2007.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dai D, Li X-F, Wang J, Liu J-J, Zhu Y-J, Zhang Y, Wang Q, Xu W-G. Predictive Efficacy of 11C-PD153035 PET Imaging for EGFR-Tyrosine Kinase Inhibitor Sensitivity in Non-Small Cell Lung Cancer Patients. Int. J. Cancer. 2016;138(4):1003–1012. doi: 10.1002/ijc.29832. [DOI] [PubMed] [Google Scholar]
- 48.Bahce I, Smit EF, Lubberink M, Van Der Veldt AAM, Yaqub M, Windhorst AD, Schuit RC, Thunnissen E, Heideman DAM, Postmus PE, Lammertsma AA, Hendrikse NH. Development of [11C]erlotinib Positron Emission Tomography for in Vivo Evaluation of EGF Receptor Mutational Status. Clin. Cancer Res. 2013;19(1):183–193. doi: 10.1158/1078-0432.CCR-12-0289. [DOI] [PubMed] [Google Scholar]
- 49.Slobbe P, Windhorst AD, Walsum MS, van Schuit RC, Smit EF, Niessen HG, Solca F, Stehle G, van Dongen GAMS, Poot AJ. Development of [18F]afatinib as New TKI-PET Tracer for EGFR Positive Tumors. Nucl. Med. Biol. 2014;41(9):749–757. doi: 10.1016/j.nucmedbio.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 50.Kareem H, Sandström K, Elia R, Gedda L, Anniko M, Lundqvist H, Nestor M. Blocking EGFR in the Liver Improves the Tumor-to-Liver Uptake Ratio of Radiolabeled EGF. Tumor Biol. 2010;31(2):79–87. doi: 10.1007/s13277-009-0011-2. [DOI] [PubMed] [Google Scholar]
- 51.Kruziki MA, Bhatnagar S, Woldring DR, Duong VT, Hackel BJ. A 45-Amino-Acid Scaffold Mined from the PDB for High-Affinity Ligand Engineering. Chem. Biol. 2015;22(7):946–956. doi: 10.1016/j.chembiol.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chambers AF. MDA-MB-435 and M14 Cell Lines: Identical but Not M14 Melanoma? Cancer Res. 2009;69(13):5292–5293. doi: 10.1158/0008-5472.CAN-09-1528. [DOI] [PubMed] [Google Scholar]
- 53.Koide A, Bailey CW, Huang X, Koide S. The Fibronectin Type III Domain as a Scaffold for Novel Binding Proteins. J. Mol. Biol. 1998;284(4):1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 54.Tamaskovic R, Simon M, Stefan N, Schwill M, Plückthun A. Designed Ankyrin Repeat Proteins (DARPins) from Research to Therapy. Methods Enzymol. 2012;503:101–134. doi: 10.1016/B978-0-12-396962-0.00005-7. [DOI] [PubMed] [Google Scholar]
- 55.Moore SJ, Leung CL, Cochran JR. Knottins: Disulfide-Bonded Therapeutic and Diagnostic Peptides. Drug Discov. Today Technol. 2012;9(1):e3–e11. doi: 10.1016/j.ddtec.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 56.Heinis C, Rutherford T, Freund S, Winter G. Phage-Encoded Combinatorial Chemical Libraries Based on Bicyclic Peptides. Nat. Chem. Biol. 2009;5(7):502–507. doi: 10.1038/nchembio.184. [DOI] [PubMed] [Google Scholar]
- 57.Löfblom J, Feldwisch J, Tolmachev V, Carlsson J, Ståhl S, Frejd FY. Affibody Molecules: Engineered Proteins for Therapeutic, Diagnostic and Biotechnological Applications. FEBS Lett. 2010;584(12):2670–2680. doi: 10.1016/j.febslet.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 58.Hackel BJ. Alternative Protein Scaffolds for Molecular Imaging and Therapy. In: Cai W, editor. Engineering in Translational Medicine. Springer London; London: 2013. pp. 343–364. [Google Scholar]
- 59.Ahlgren S, Orlova A, Wallberg H, Hansson M, Sandstrom M, Lewsley R, Wennborg A, Abrahmsen L, Tolmachev V, Feldwisch J. Targeting of HER2-Expressing Tumors Using 111In-ABY-025, a Second-Generation Affibody Molecule with a Fundamentally Reengineered Scaffold. J. Nucl. Med. 2010;51(7):1131–1138. doi: 10.2967/jnumed.109.073346. [DOI] [PubMed] [Google Scholar]
- 60.Ahlgren S, Wallberg H, Tran TA, Widstrom C, Hjertman M, Abrahmsen L, Berndorff D, Dinkelborg LM, Cyr JE, Feldwisch J, Orlova A, Tolmachev V. Targeting of HER2-Expressing Tumors with a Site-Specifically 99mTc-Labeled Recombinant Affibody Molecule, ZHER2:2395, with C-Terminally Engineered Cysteine. J. Nucl. Med. 2009;50(5):781–789. doi: 10.2967/jnumed.108.056929. [DOI] [PubMed] [Google Scholar]
- 61.Kramer-Marek G, Kiesewetter DO, Martiniova L, Jagoda E, Lee SB, Capala J. [18F]FBEM-ZHER2:342–Affibody Molecule—a New Molecular Tracer for in Vivo Monitoring of HER2 Expression by Positron Emission Tomography - Springer. Eur. J. Nucl. Med. Mol. Imaging. 2007;35(5):1008–1018. doi: 10.1007/s00259-007-0658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng Z, De Jesus OP, Namavari M, De A, Levi J, Webster JM, Zhang R, Lee B, Syud FA, Gambhir SS. Small-Animal PET Imaging of Human Epidermal Growth Factor Receptor Type 2 Expression with Site-Specific 18F-Labeled Protein Scaffold Molecules. J. Nucl. Med. 2008;49(5):804–813. doi: 10.2967/jnumed.107.047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tran T, Engfeldt T, Orlova A, Sandström M, Feldwisch J, Abrahmsén L, Wennborg A, Tolmachev V, Karlström AE. Detection of HER2 Expression in Malignant Tumors. 2007;2:1956–1964. doi: 10.1021/bc7002617. [DOI] [PubMed] [Google Scholar]
- 64.Kimura RH, Teed R, Hackel BJ, Pysz MA, Chuang CZ, Sathirachinda A, Willmann JK, Gambhir SS. Pharmacokinetically Stabilized Cystine Knot Peptides That Bind Alpha-v-Beta-6 Integrin with Single-Digit Nanomolar Affinities for Detection of Pancreatic Cancer. Clin. Cancer Res. 2012;18(3):839–849. doi: 10.1158/1078-0432.CCR-11-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng Z, De Jesus OP, Kramer DJ, De A, Webster JM, Gheysens O, Levi J, Namavari M, Wang S, Park JM, Zhang R, Liu H, Lee B, Syud FA, Gambhir SS. 64Cu-Labeled Affibody Molecules for Imaging of HER2 Expressing Tumors. Mol. Imaging Biol. 2010;12(3):316–324. doi: 10.1007/s11307-009-0256-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ahlgren S, Orlova A, Rosik D, Sandström M, Sjöberg A, Baastrup B, Widmark O, Fant G, Feldwisch J, Tolmachev V. Evaluation of Maleimide Derivative of DOTA for Site-Specific Labeling of Recombinant Affibody Molecules. Bioconjug. Chem. 2008;19(1):235–243. doi: 10.1021/bc700307y. [DOI] [PubMed] [Google Scholar]
- 67.Tolmachev V, Friedman M, Sandström M, Eriksson TLJ, Rosik D, Hodik M, Ståhl S, Frejd FY, Orlova A. Affibody Molecules for Epidermal Growth Factor Receptor Targeting in Vivo: Aspects of Dimerization and Labeling Chemistry. J. Nucl. Med. 2009;50(2):274–283. doi: 10.2967/jnumed.108.055525. [DOI] [PubMed] [Google Scholar]
- 68.Tolmachev V, Nilsson FY, Widström C, Andersson K, Rosik D, Gedda L, Wennborg A, Orlova A. 111In-Benzyl-DTPA-ZHER2:342, an Affibody-Based Conjugate for in Vivo Imaging of HER2 Expression in Malignant Tumors. J. Nucl. Med. 2006;47(5):846–853. [PubMed] [Google Scholar]
- 69.Orlova A, Wållberg H, Stone-Elander S, Tolmachev V. On the Selection of a Tracer for PET Imaging of HER2-Expressing Tumors: Direct Comparison of a 124I-Labeled Affibody Molecule and Trastuzumab in a Murine Xenograft Model. J. Nucl. Med. 2009;50(3):417–425. doi: 10.2967/jnumed.108.057919. [DOI] [PubMed] [Google Scholar]
- 70.Kramer-Marek G, Kiesewetter DO, Capala J. Changes in HER2 Expression in Breast Cancer Xenografts after Therapy Can Be Quantified Using PET and (18)F-Labeled Affibody Molecules. J. Nucl. Med. 2009;50(7):1131–1139. doi: 10.2967/jnumed.108.057695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boswell CA, Sun X, Niu W, Weisman GR, Wong EH, Rheingold AL, Anderson CJ. Comparative in Vivo Stability of Copper-64-Labeled Cross-Bridged and Conventional Tetraazamacrocyclic Complexes. J. Med. Chem. 2004;47(6):1465–1474. doi: 10.1021/jm030383m. [DOI] [PubMed] [Google Scholar]
- 72.Ait-Mohand S, Fournier P, Dumulon-Perreault V, Kiefer GE, Jurek P, Ferreira CL, Benard F, Guerin B. Evaluation of 64Cu-Labeled Bifunctional Chelate-Bombesin Conjugates. Bioconjug. Chem. 2011;22(8):1729–1735. doi: 10.1021/bc2002665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.