Abstract
dUTPase superfamily enzymes generate dUMP, the obligate precursor for de novo dTTP biosynthesis, from either dUTP (monofunctional dUTPase, Dut) or dCTP (bifunctional dCTP deaminase/dUTPase, Dcd:dut). In addition, the elimination of dUTP by these enzymes prevents harmful uracil incorporation into DNA. These two beneficial outcomes have been thought to be related. Here we determined the relationship between dTTP biosynthesis (dTTP/dCTP balance) and the prevention of DNA uracilation in a mycobacterial model that encodes both the Dut and Dcd:dut enzymes, and has no other ways to produce dUMP. We show that, in dut mutant mycobacteria, the dTTP/dCTP balance remained unchanged, but the uracil content of DNA increased in parallel with the in vitro activity-loss of Dut accompanied with a considerable increase in the mutation rate. Conversely, dcd:dut inactivation resulted in perturbed dTTP/dCTP balance and two-fold increased mutation rate, but did not increase the uracil content of DNA. Thus, unexpectedly, the regulation of dNTP balance and the prevention of DNA uracilation are decoupled and separately brought about by the Dcd:dut and Dut enzymes, respectively. Available evidence suggests that the discovered functional separation is conserved in humans and other organisms.
Introduction
Proper control of the intracellular concentration of deoxyribonucleoside-5-triphosphates (dNTPs), the building blocks of DNA, is critically important for efficient and high-fidelity DNA replication and genomic stability1, 2. Three of the four canonical dNTPs are synthesized from their respective ribonucleoside diphosphate (NDP) counterparts3. The direct precursor for dTTP, however, is missing from the ribonucleoside pool and is synthesized via separate routes (Fig. 1).
Figure 1.
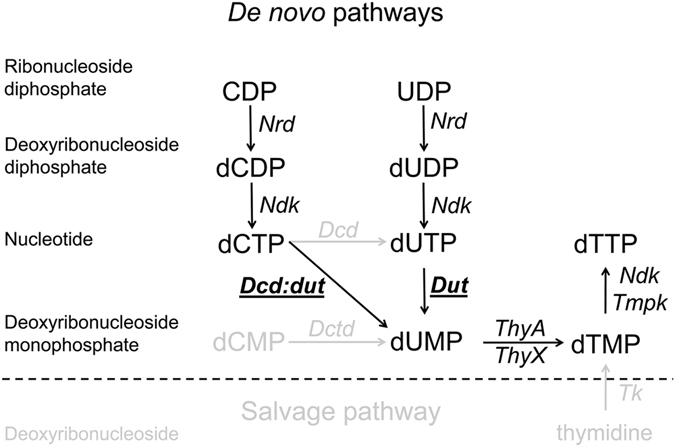
dTTP biosynthesis pathways and enzymes. Processes present in Mycobacteria are shown in black. Most organisms encode for additional de novo and salvage pathways that are shown in grey. Abbreviations: Dcd – dCTP deaminase, Dut – dUTPase, Dctd – dCMP deaminase, Dcd:dut – bifunctional dCTP deaminase: dUTPase, Nrd – Nucleoside diphosphate reductase, Ndk – Nucleoside diphosphate kinase, Tmpk – dTMP kinase, Tk – Thymidine kinase, ThyA,ThyX – thymidylate synthases.
The de novo synthesis of dTTP occurs through uracil base-containing precursors: dUMP is the direct input into the thymidylate synthase reaction (Fig. 1). In most organisms, the main dUMP supply is provided by the deamination of a cytosine deoxyribonucleotide (dCMP or dCTP) while other possible routes, e.g. the dephosphorylation of dUDP, are considered to be minor supplements4–6. When cytosine deamination occurs at the triphosphate level, the resulting dUTP is then converted into dUMP. The enzymes that catalyze these conversions belong to the dUTPase superfamily comprising dCTP deaminase (Dcd), dUTPase (Dut) and the bifunctional dCTP deaminase/dUTPase (Dcd:dut) (Fig. 1). These enzymes share the same quaternary structure as shown in Fig. 2A.
Figure 2.

(A) Superposition of the quaternary structures of the M. tuberculosis dUTPase (Dut) depicted in green (PDB ID:2PY4) and the M. tuberculosis bifunctional dCTP deaminase/dUTPase enzymes (Dcd:dut) depicted in yellow (PDB ID:2QLP). Note the identical organization of the enzyme core of the homotrimers. Both structures contain the non-hydrolysable substrate analog α-β-imido-dUTP (dUPNPP) in the active sites. (B) Enlarged view of the active site of M. tuberculosis Dut showing the C-terminal arm in green. The side chains of the amino acids in case of point mutations and C-terminal arm truncation are shown with atomic colored stick representation similarly to the dUPNPP molecule and with green cartoon representation, respectively. The catalytic water is shown as a red sphere while the yellow sphere denotes the Mg2+ ion that coordinates the nucleotide.
In addition to dUMP production, the dUTPase reaction also serves to eliminate excess dUTP to prevent uracil incorporation into DNA in place of thymine7, 8. Although not mutagenic when replacing thymine, the uracil in DNA is considered to be an error and induces uracil-excision repair mechanisms9. In high dUTP/dTTP ratios, however, DNA polymerases keep re-incorporating dUTP and the repair process becomes overwhelmed. Dut is ubiquitous and essential in most investigated cases10–16. Recently, novel functions of Dut emerged in gene expression regulation as well15, 17–20. Our genetic experiments also suggested that the mycobacterial Dut has a yet unknown but essential moonlighting function11.
In summary, Dut catalyzes the break-down of dUTP to dUMP and with this action it potentially takes part i) in dTTP biosynthesis, ii) in the maintenance of low dUTP/dTTP ratio to prevent uracil incorporation into DNA and iii) in interactions with regulatory proteins. The various roles now attributed to Dut and the large amount of knock-out and knock-down data on the dUTPase superfamily enzymes in various genetic backgrounds create a confusing picture of the contribution of Dut to the physiological processes in which it may be involved. As dTTP biosynthesis is an essential process and a major target in several current drug therapies, it is important to pinpoint those pathways in which Dut is a key contributing enzyme.
We therefore set-out to dissect the contributions of dUTP-hydrolyzing enzymes, Dut and Dcd:dut, to dTTP biosynthesis and to the prevention of DNA uracilation. For this reason, we searched for a simple model in which the obligatory dTTP precursor, dUMP, is produced exclusively by Dut and Dcd:dut in lack of salvage pathways and dCMP deamination (Fig. 1). This favorable set of conditions naturally occurs in the genus Mycobacteria 11, 21. Due to the exclusive biosynthetic role of Duts in these organisms, they present potential targets for drug development, as well. Earlier mutagenesis studies found the presence of the bifunctional Dcd:dut to be dispensable for growth in M. tuberculosis 10, 22 while the intact Dut protein is essential in Mycobacteria 10, 11, 22.
In the present study, we created M. tuberculosis Dut mutant proteins in which the enzyme activity is gradually tuned down. We then carried out genetic experiments in the fast-growing M. smegmatis in which we created the same Dut mutations and also included an inactive dcd:dut mutant strain in the experiments. We found that the dUTPase activity of either Dut or Dcd:dut can support cell growth. The double mutant M. smegmatis strain lacking the complete dUTPase activity, however, is inviable. We investigated the mutation-induced effects including dNTP pool changes, the mutation rate and the uracil content of DNA in M. smegmatis strains conferring various Dut and Dcd:dut mutants. Unexpectedly, the lack of Dut activity did not influence the biosynthesis of dTTP. We arrived to the conclusion that dTTP biosynthesis and the maintenance of genomic integrity by dUTP elimination are under differential control.
Results
Tuning down the activity of M. tuberculosis Dut
On the basis of our previous investigations on the human and Escherichia coli (E. coli) Duts23, 24, we planned and created three mutants of the M. tuberculosis Dut enzyme by site-directed mutagenesis. These mutants were chosen to represent enzymatic activity loss from one order of magnitude to the practical inactivity.
The D83N substitution aims at compromising the coordination of the catalytic water by mutating the catalytic aspartate residue (Asp90 in E. coli Dut) (Fig. 2B). In effect, this mutant presents an extremely low catalytic activity (Fig. 3A, Table 1) similarly to what was observed in the E. coli enzyme24. The determination of the Michaelis constant (KM), however, is uncertain due to the limitations of the activity measurements at such low activities. The KM could be better estimated in this case from the dissociation constant (Kd) of the protein complexed with the non-hydrolysable substrate analog α, β-imido-dUTP (dUPNPP). We measured the Kd of the D83N.dUPNPP complex to be similar to that of the WT.dUPNPP complex (Table 1, Fig. S1B). Its catalytic efficiency being 0.0002 compared to the wt means that the D83N Dut is a practically inactive mutant.
Figure 3.
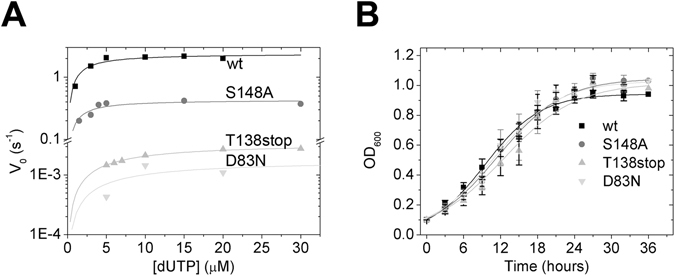
(A) Steady-state activity of wt and mutant Duts. Michaelis-Menten curves were measured using the phenol red pH indicator assay. Fitting the Michaelis-Menten equation to the curves yielded the following kcat and KM values: 1.22 ± 0.06 s−1 and 1.7 ± 0.5 μM for WT, 0.43 ± 0.04 s−1 and 1.5 ± 0.6 μM for S148A, 0.0035 ± 0.0001 s−1, 6.7 ± 0.4 μM for T138stop and 0.0013 ± 0.0005 s−1, 7.7 ± 6.7 μM for D83N mutant Dut. (B) In vitro growth analysis of wt and dut mutant M. smegmatis strains. The strains were grown in Lemco broth in shaking cultures for 2 days at 37 °C. Growth curves were prepared from (3*3) independent colonies from each mutation; means ± SD are plotted. Fitting the y = a/(1 + exp(−k*(x − xc))) equation to the curves yielded the following a, xc and k values: 0.94, 9.5 and 0.22 for WT, 1.04, 11.5 and 0.19 for S148A, 1.02, 12.4 and 0.17 for T138stop and 1.03, 11.0 and 0.19 for D83N Dut mutant strains.
Table 1.
Kinetic parameters of the M. tuberculosis Dut and Dcd:dut enzymes.
| Enzyme | kcat (s−1) | KM (μM) | Kd.dUPNPP (μM) | kcat/KM (M−1 s−1) | Efficiency |
|---|---|---|---|---|---|
| wt Dut | 1.22 ± 0.06 | 1.7 ± 0.5 | 0.9 ± 0.5 | 7.18E + 05 | 1 |
| S148A Dut | 0.43 ± 0.04 | 1.5 ± 0.6 | 1.8 ± 1.0 | 2.87E + 05 | 0.4 |
| T138stop Dut | 0.0035 ± 0.0001 | 6.7 ± 0.4 | 3.9 ± 1.3 | 5.22E + 02 | 0.0007 |
| D83N Dut | 0.0013 ± 0.0005 | 7.7 ± 6.7* | 1.5 ± 0.1 | 1.69E + 02* | 0.0002 |
| wt Dcd:dut dUTPase | 0.033 ± 0.008 | 12 ± 3 | — | 2.75 E + 03 | 0.004 |
| wt Dcd:dut dCTP deaminase | 0.022 ± 0.005 | 20 ± 12 | 1.10 E + 03 | NA | |
| A115F Dcd:dut | no activity | — | — | — | — |
*Data not reliable due to the limitations of the activity measurement.
NA: not applicable.
The efficient dUTPase catalysis requires conserved sequence motifs I-V from all three monomers of the Dut homotrimer7. Motifs I-IV constitute the active sites at the intermonomer clefts while motif V, located at the C-terminal arm, leaves the globular core of its monomer to associate with the neighboring active site and shield it from the solvent (C-terminal arm shown in green in Fig. 2B). This P-loop-like motif V changes conformation upon substrate binding and positions the phosphate chain of the nucleotide for efficient hydrolysis. The lack of this motif results in a nearly inactive enzyme in all investigated species23, 25–33. We created a mutant lacking conserved motif V by the truncation of the 154 amino acid long protein at position 138 (T138Stop). The T138Stop mutant exhibits 3-fold higher enzymatic activity than that of the D83N mutant the catalytic efficiency still being extremely low compared to the WT (Fig. 3A, Table 1). Both the Kd of the enzyme.dUPNPP complex (Fig. S1A) and the KM are 4-fold higher than that of the WT complex (Table 1).
We created the S148A mutation to distort one of the hydrogen bonding interactions between the P-loop-like motif and the γ–phosphate of the substrate dUTP (Fig. 2B). The S148A mutant showed one order of magnitude loss in the enzyme activity (Fig. 3A, Table 1) similarly to an analogous mutant in the human Dut23. The KM, in concert with the Kd, increased only 2-fold (Table 1, Fig. S1A).
The A115F mutation inactivates the Dcd:dut enzyme
We cloned and expressed the M. tuberculosis and M. smegmatis Dcd:dut enzymes and determined their steady-state kinetic parameters. In lack of significant difference between the behaviors of the M. tuberculosis and M. smegmatis Dcd:duts, we report the parameters for the M. tuberculosis enzyme for comparison with M. tuberculosis Dut constructs (Fig. 4A, Table 1). As expected21, the bifunctional enzyme is a relatively low-efficiency dUTPase compared to the monofunctional Dut (Table 1). In order to inactivate Dcd:dut without perturbing the overall structure of the enzyme, we introduced a bulky Phe into amino acid position 115 in place of an Ala (A115F) to prevent substrate binding to the active site. The Phe side chain in position 115 can only be accommodated within the active site cavity where it occupies the binding site of the uracil base (Fig. 4B). This mutant has the advantage that the cells keep synthetizing a structurally intact Dcd:dut protein. Fig. 4C shows that the Dcd:dut A115F does not exhibit enzyme activity. We have previously created this mutation in the structurally homologous human Dut and obtained an inactive but structurally intact mutant, as well34.
Figure 4.
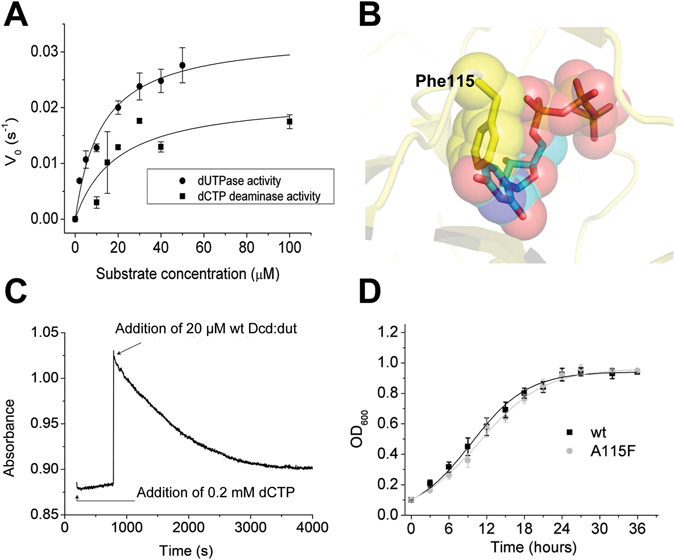
(A) Steady-state dUTPase and dCTP deaminase activity of wt Dcd:dut. The parameters yielded by fitting the Michaelis-Menten equation are shown in Table 1. The error represents the SD of 3 measurements. (B) The structural model of the active site of the M. tuberculosis Dcd:dut showing the steric conflict of the Phe115 side chain with the uracil ring of the substrate dUTP. (C) dCTP deaminase time course demonstrating the inactivity of the A115F Dcd:dut mutant. At t = 0, dCTP was added to a premix containing 0.02 mM A115F Dcd:dut. During ~500 s incubation time, no reaction (i.e. absorbance decrease) was detectable. The time course obtained upon the addition of the wt Dcd:dut enzyme confirmed that the assay was functional. (D) In vitro growth analysis of wt and Dcd:dut mutant M. smegmatis strains. The strains were grown in Lemco broth in shaking cultures for 2 days at 37 °C. Growth curves were prepared from (3*3) independent colonies from each mutation; means ± SD are plotted. Fitting the y = a/(1 + exp(−k*(x − xc))) equation to the curves yielded the following a, xc and k values: 0.94, 9.5 and 0.22 for the wt and 0.96, 10.8 and 0.19 for the A115F dcd:dut mutant strains.
The hydrolysis activity of either Dut or Dcd:dut supports the growth of M. smegmatis
We aimed to investigate the effect of dUTPase activity loss in the living cell. We therefore created the above mutations (S148A, T138Stop and D83N) within the genome of M. smegmatis. The M. tuberculosis and M. smegmatis Duts share 85% amino acid sequence identity (100% within the conserved motifs)11 and thus it is expected that the two enzymes behave similarly. In a previous paper, we established a method in which the disruption of the endogenous dut was rescued by a functional (complementing) copy of dut inserted into the genome on an integrating vector11. We used this scheme and introduced the mutations into the complementing copy of M. smegmatis dut. We obtained M. smegmatis strains that carried the mutant dut in a dut knock-out background (i.e. no wt copy present). Successful allele exchange was verified by Southern blot analysis (Fig. S2) and the mutations on the complementing dut copy were verified by sequencing of the appropriate genome region.
All three dut mutant strains were viable and unexpectedly, showed no growth defects when grown in liquid culture under stress-free conditions (Fig. 3B). This result suggests that the fully functional Dcd:dut in these dut mutant strains produces enough dUMP for the synthesis of more than limiting amounts of dTTP.
We also created a M. smegmatis strain carrying the non-functional A115F dcd:dut in a wt dut background. We constructed the A115F dcd:dut strain by allele exchange of the endogenous dcd:dut gene to a GFP-tagged copy carrying a point mutation. The A115F mutation was introduced either in wt or in inactive dut (D83N) background. Successful allele exchange was verified by Southern blot analysis (Fig. S2), and the mutation was verified by the sequencing of the appropriate genome region. The A115F dcd:dut strain encoding wt dut was viable and its growth rate was similar to that of the wt when grown in liquid culture (Fig. 4D). In contrast, we could not obtain any double mutant strains carrying both the dut D83N and the dcd:dut A115F mutations. In these cells, the dUTPase activity is completely abolished due to the fact that only inactive Duts are encoded. However, these inactive proteins are structurally intact and could still potentially mediate functions that are independent from their enzymatic activity or can operate in an inactive state (e.g. the essential surface loop in Dut11 and examples from other species15, 18–20, 35). This implies that the dUTPase activity is essential for viability and reinforces that the dUTPase activity has exclusive role in dTTP biosynthesis in Mycobacteria (cf. Fig. 1).
Mutator phenotype of the dut and dcd:dut mutant strains
As we could not reveal any obvious defects in the enzymatically compromised dut or dcd:dut mutant M. smegmatis strains, we investigated possible long term effects of these mutations. We measured the mutation rates in each of the mutant strains36, 37. We found that the mutation rates increased remarkably in the dut mutant strains (7-fold, 9-fold and 15-fold in the S148A, T138stop and D83N dut mutant strains, respectively). The mutation rate of the dcd:dut A115F strain appeared two-fold higher than the wt, however, the difference did not prove to be significant (Fig. 5A).
Figure 5.
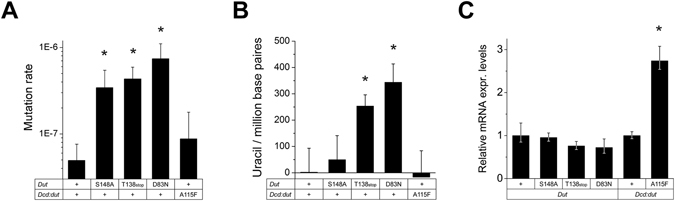
The effect of dut and dcd:dut mutations on the mutation rate, DNA uracilation and gene expression. (A) Mutation rates of mutant M. smegmatis strains. Means ± SE are calculated from (3*3) independent colonies from each mutation. Note that mutation rates of dut mutant strains directly correlate with the in vitro activity-loss of the corresponding mutant enzyme. Significance levels: P = 0.000115 for S148A, P = 0.000034 for T138stop and P =0.000004 for D83N. “+” denotes the wt enzyme. (B) Genomic uracil content of the mutant strains compared to the wt strain. Uracil contents were calculated from three independent strains from each mutant and normalized to the wt strain; means ± SE are plotted. Note that the genomic uracil content of dut mutant strains directly correlates with the in vitro activity-loss of the corresponding mutant enzyme. Significance levels: P = 0.074 for T138stop, P = 0.018 for D83N. (C) Quantitation of dut and dcd:dut expression levels in the wt and mutant M. smegmatis strains. mRNA levels were calculated from three independent strains from each mutant and normalized to their respective wt strain; means ± SE are plotted. Significance level: P = 0.029 for the A115F mutant.
Dut mutant strains accumulate uracil in their genome
We also measured the genomic uracil content of the mutant strains by a q-PCR based method developed in our laboratory38. This assay is based on the fact that the Pfu polymerase does not amplify uracil-containing template DNA while other polymerases do (e.g. Taq). It is therefore possible to calculate the relative uracil content of the sample on the basis of PCR efficiencies driven parallel by Pfu and by Taq polymerases. We found that dut mutants have elevated genomic uracil content compared to the wt strain and that the increase in uracil content correlates with the in vitro measured activity loss (50, 250 and 340 uracil/million base pairs in the S148A, T138stop and D83N dut mutant strains, respectively.) (Fig. 5B). For comparison, the 340 uracil/million base pairs genomic uracil content matches that measured in the ung- E. coli and the ung- MEF cells deficient in uracil misincorporation repair. The dut-/ung- E. coli strain contains even 20-times more uracils in its genome38. We could not detect any change in the genomic uracil content in the A115F dcd:dut mutant strain compared to the wt (Fig. 5B).
To investigate if these marked effects are not simply due to an underlying difference in gene expression, we measured the mRNA levels of the various constructs. The expression of the T138stop and D83N dut constructs in the M. smegmatis cells seems to decrease compared to the wt, however, the difference proved not to be significant (Fig. 5C). On the other hand, the expression level of the A115F dcd:dut construct increased significantly compared to the wt (Fig. 5C). This finding emphasizes the differential effects in DNA uracilation and mutagenicity between the dut and dcd:dut mutations even more.
Pyrimidine nucleotide pool changes in the dut and dcd:dut mutant strains
To reveal the mechanism of uracil accumulation and mutation rate changes in our mutant strains, we measured the pyrimidine nucleotide pools in each strain. We used a DNA polymerase-based method to determine the concentration of dUTP, dTTP and dCTP in cell extracts39, 40. The method is based on the incorporation of radiolabeled dATP into a nucleotide-specific template limited by the concentration of the quantifiable dNTP. The concentration of dUTP in the wt strain proved to be too low to be accurately quantified in our assay (<0.5 pmol/108 cells). We found that the concentration of dUTP became significantly elevated in the T138stop and D83N dut mutants (Fig. 6A). The dTTP:dUTP ratio changed from the >>10:1 ratio in the wt to 5:1 in the S148A dut mutant strain and to 4:1 in the T138stop and D83N dut mutant strains. The dTTP and dCTP concentrations remained quasi unchanged and consequently, the dTTP:dCTP ratio also remained in the wt range. However, the total pyrimidine content increased moderately but significantly in the D83N dut strain (Fig. 6, dCTP and dTTP concentrations increased 1.6- and 1.4-fold, respectively). Interestingly, however, the A115F dcd:dut strain had a normal low concentration of dUTP in its nucleotide pool while the dTTP:dCTP ratio became greatly imbalanced (Fig. 6 inset). The dCTP concentration of the A115F cell extract increased more than 6-fold while the dTTP concentration decreased to half of the wt (Fig. 6A).
Figure 6.
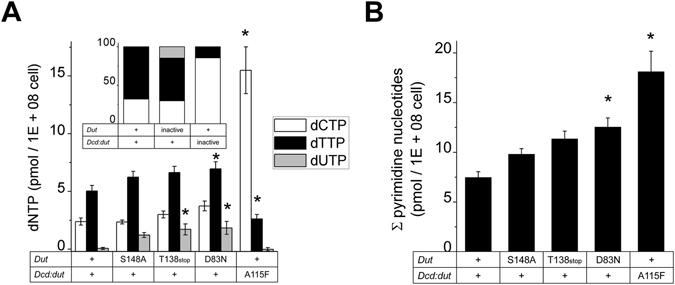
The effect of dut and dcd:dut mutations on the pyrimidine nucleotide pool. (A) dTTP, dCTP and dUTP concentrations were measured using a DNA polymerase-based method in all mutant strains. Mutant enzymes are indicated, “+” denote the wt enzyme. Means ± SE are calculated from 12 data points for each mutation. The dUTP concentration in the wt and in the A115F mutant falls out of the measurement range (<0.5 pmol/108 cells). Significance levels: P = 0.063 for the T138stop dUTP level, P = 0.095 for the D83N dTTP level, P = 0.047 for the D83N dUTP level, P = 0.000012 for the A115F dCTP level, P = 0.066 for the A115F dTTP level. The inset shows a comparison of the ratios of pyrimidine nucleotides within the wt and inactive mutant strains. (B) The change in total pyrimidine concentration in function of the mutation carried by each strain. Means ± SE are plotted. Significance levels: P = 0.05959 for D83N and P = 0.0028 for A115F.
These results clearly suggest two different roles for dcd:dut and dut in the regulation of pyrimidine nucleotide concentration and in the maintenance of genome integrity, respectively.
Discussion
Previous knock-out studies found dut essential and dcd:dut dispensable for Mycobacteria 10, 11, 22. We reported that a complete knockout of dut is lethal11. We also reported that lethality is not due to the loss of the catalytic function but is mediated by a surface loop of unknown function independently of the dUTPase activity11. A well-folded and enzymatically active Dut enzyme lacking only this 5 amino acid long mycobacterium-specific surface loop does not support growth11. These previous studies did not provide information about the in vivo function of the enzymes bearing dUTPase activity, dut and dcd:dut. It was not clear, for example, how dUTPase activity affects the dNTP pool and downstream genomic processes. In the present study, we combined enzymology with genetics to address the role of dUTPase activity within the mycobacterial cell.
The mutant enzymes we created to tune down enzyme activity behaved in a well predictable manner in our in vitro experiments. The S148A and the T138stop dut mutations that compromised one or all interactions of the P-loop-like motif with the phosphate chain of the substrate resulted in small or severe activity loss, respectively, accompanied by weaker binding of the substrate (proportionally increased Kd and KM, Table 1). The D83N mutation impaired the coordination of the catalytic water which left the binding of the substrate unaffected (Table 1). However, the subsequent catalytic reaction was compromised resulting in practical inactivity (Table 1).
The activity-compromised dut point mutant M. smegmatis strains exhibit an increased mutation rate that accompanies the increase in the genomic uracil content (20–150 folds, Fig. 5). Mycobacteria encode three uracil DNA glycosylase enzymes (ung, udgB and udgX)41–43 enabling effective uracil excision repair in these organisms42. This functional redundancy reinforces that the accumulation of uracil in DNA is an unwanted process. The potential reason why uracil could still accumulate in the DNA of our dut mutants is that DNA polymerases re-incorporate uracil despite the excision repair mechanism constantly excising it from the mycobacterial DNA. Polymerases usually do not distinguish between dTTP and dUTP and thus, they will likely incorporate dUTP from a nucleotide pool containing highly elevated dUTP concentrations (Fig. 6). Uracils (U·A pairs) are generally not considered as mutagenic compounds44 while there is a controversy in the literature about the mutagenicity of abasic sites44–46. However, constantly excised and re-incorporated uracils probably cause stress and genome instability in the bacteria. In an ndk, dut double mutant E. coli strain, increased dUTP levels and replication intermediates from the uracil excision process caused several thousand-fold elevations in the mutation rate. The Ung mutation, which enables stable incorporation of uracil into DNA, could only partially alleviate the mutagenic effect47. This suggests that among other possible mechanisms, an increased frequency of uracil repair processed by either short patch or long patch base excision repair (BER) mechanism48 may be responsible for the observed elevation in the mutation rates. In addition, the dut mutation had a modest effect on dCTP and dTTP levels47. In our experiments, the most severe D83N dut mutation also resulted in a modest increase of cellular pyrimidine levels. The overproduction of dNTPs in response to endogenous DNA damage is a general stress response3, 49, 50 and most likely serves to promote tolerance against genotoxic stress. The elevated dNTP levels, in turn, increase the mutation rate50–53 as the activity balance of DNA polymerases is transiently altered allowing the proofreading activity to decrease for the benefit of the nucleotide incorporating function (i.e. next nucleotide effect)52. These considerations are in agreement with our observations as well. The equally large increase in the mutation rates of the various dut mutants also suggests that this phenomenon is part of a general stress response and not specific to dUTPase activity loss. Interestingly, although the dTTP:dCTP ratio was highly imbalanced in the dcd:dut mutant strain, it displayed only two-fold increase in the mutation rate possibly indicating that dCTP is a relatively poor mutagenic precursor (Figs 5, 6). These results are in good accordance with previous literature data also reporting 2-fold increase in the mutation rate using the rifampicin resistance method in dcd mutant E. coli 46. However, Schaaper and Matthews also found that the experimental system greatly affects the observation of mutagenicity. In their study, the 2-fold increase in the mutation rate observed using the rifampicin resistance assay appeared much higher (<42-fold) using a different assay (only available for E. coli for the moment)46. Moreover, Kumar and his colleagues found that even mild dNTP pool imbalances were mutagenic in Saccharomyces cerevisiae. However, the mutagenic potential of different imbalances did not directly correlate with their extent53. Nordman and Wright proposed that dNTP imbalances were not responsible for increased mutation rate in the ndk, dut mutant E. coli 47. Our findings suggest that the dCTP:dTTP imbalance results in lower mutagenic potential than that of DNA uracilation.
Unexpectedly, the reduced dTTP concentration in the dcd:dut mutant strain did not limit the proliferation rate of the bacteria (Fig. 4D). This indicates that the activity of either of the two dUTPases is sufficient to support dTTP synthesis for the efficient growth of M. smegmatis. Our further results shown in Fig. 6 suggest, however, that dUMP production and dTTP biosynthesis are mainly under the control of the bifunctional Dcd:dut enzyme. The fact that Dcd:dut is a hundred-fold less efficient dUTPase than Dut (Table 1 and ref 21, 54) suggests that the mechanistic differences between the two dUTPase enzymes are more important than simply is their catalytic efficiency. Dcd:dut is able to bind dTTP which inhibits its activity55. This negative feedback inhibition allows for the regulation of the cellular dCTP:dTTP concentration ratio. Dut, however, can only accommodate dUTP and does not show any allosteric features56. Based on our results shown in Figs 5, 6, we propose that Dut is responsible for the efficient elimination of dUTP, while the role of the bifunctional Dcd:dut is to maintain the proper dCTP:dTTP ratio. There are clear advantages to such functional diversion. dUTP is constantly generated from dUDP by the nucleoside diphosphate kinase (Ndk) and also by spontaneous dCTP deamination in mycobacteria (and by other enzymatic pathways in eukaryotes and in some prokaryotes (Table 2). The accumulation of dUTP is efficiently prevented by the monofunctional Dut. The purpose here is to sanitize the dNTP pool from dUTP to prevent genome uracilation. The regulatory capabilities are built in the other, closely related, Dcd:dut enzyme that proved to be an inefficient dUTP sanitizer but keeps the pyrimidine DNA building blocks in a correct concentration ratio (Fig. 6). The preventive aspect of the bifunctional enzyme may be that dangerous dUTP is not released following the dCTP deamination reaction but is converted to dUMP on the same enzyme21, 57.
Table 2.
The effects of dUTPase modification in various organisms.
| Organism | Pathways | Phenotype | Rescue | dNTP pool/Mutagenicity | Ref. |
|---|---|---|---|---|---|
| E. coli | Dcd, Dut, Tk | KO lethal; Mutants: thymidine auxotroph, filamentous, hyperrec (nicks in DNA), prolonged generation time, increased sensitivity to 5′- FUs | Dcd-, ung- rescues synthetic lethality with pyrE, xth, recA, recBC but not with Tk | dUTP 10x up, dTTP 3x up, mutagenicity 5–15x up | 12, 59, 68–70 |
| M. smegmatis | Dcd:dut, Dut | Dut KO lethal; Mutants: normal generation time, high genomic U content | No rescue with active but loop(−) Dut | dUTP 20x up, mutagenicity ~15 × up | 11, this study |
| T. brucei | Dut, Tk | KO lethal; Mutants: cell cycle alterations, chromosome fragmentation, sensitive to MTX | Thymidine supply (ung- increases cytotoxicity) | dUTP 9x up, mutagenicity 9x up | 13, 71, 72 |
| C. elegans | Dctd, Dut, Tk | embrionic lethality; ATL1, RAD51 foci –> S-phase checkpoint activation | ung1, clk2 rescues, thymidine supplemented medium only partially | n.d. | 14 |
| D. melanogaster | Dctd, Dut, Tk? | lethality in early pupal stage, DNA strand breaks, U in DNA | n.d. | n.d. | 15 |
| S. cerevisiae | Dctd, Dut | dTMP auxotrophs (no Tk!), growth delay, cell cycle abnormalities | exogenous dTMP, ung inactivation rescues, APE inactivation does not | mostly AT –> CG mutations | 58, 73 |
| Human cell lines | Dctd, Dut, Tk | sensitization for FdUrd, even more with Tmk double silencing; decline in clonogenic survival, increase in DNA double strand breaks and in Tmk, Tk expression levels; genome instability, tumorigenesis; apoptosis in pancreatic beta cells | n.d. | variable, no significant change or n.d. | 67, 74–77 |
| Arabidopsis | Dctd, Dut, Tk | lethality or sterility, sensitive to 5FUs, 7-fold increase in homologous recombination events | critical partners unknown | n.d. | 16, 78 |
We compiled the available literature data on the in vivo effects of dUTPase activity loss in various organisms in Table 2. This comprehensive data set supports a similar partition between the dUTP eliminating and the dNTP balancing functions in other organisms that bear additional dTTP producing pathways, as well. In Saccharomyces cerevisiae and Caenorhabditis elegans (bearing dCMP deaminase and a salvage pathway), the inactivation of dut is lethal. The inhibition of ung, however, can rescue the observed phenotype in these organisms14, 58 indicating the deleterious effect of genome uracilation in the dut mutant. The numerous reports on E. coli dut mutants also indicate that the major effect of Dut inactivation is genome instability and not a short supply of dTTP (Table 2). While the dut-1/ung-1 mutant E. coli strain can be maintained, the dut-1 phenotype is lethal59. However, in Trypanosoma brucei, in which there is no dCTP/dCMP deaminase and dUMP production strongly depends on dUDP/dUTP hydrolysis, the inactivation of ung could not rescue dut silencing but instead increased the cytotoxic effects conveyed by the low dUMP levels13. Efficient dut silencing in human cell lines resulted in a decline in clonogenic survival due to genome instability (Table 2). All these literature data support the major role of dut in dNTP sanitizing. The regulation of dTTP concentration, however, seems to be exclusively committed to dCTP and/or dCMP transforming enzymes (Dcd, Dcd:dut, Dctd) which may be structurally unrelated from each other but are all allosterically regulated55, 57, 60–62. A further in-depth investigation of the de-coupling of these functions may shine light on mechanisms supporting the appearance of T in and the exclusion of U from DNA.
Methods
Bacterial strains, media and growth conditions
M. smegmatis mc 2 155 was grown in Lemco medium (broth) or in Lemco with the addition of 15 g L−1 Bacto agar (solid). Kanamycin was added at 20 μg/ml, hygromycin B at 100 μg/ml, gentamicin at 10 μg/ml, and streptomycin at 20 μg/ml final concentration. For sucrose selection, 5% (wt/v) sucrose was included in the medium. X-Gal (5-bromo-4-chloro-3-indolyl-b- D- galactopyranoside) was used at 40 μg/ml.
Mutagenesis, cloning and gene expression
All recombinant proteins were expressed in E. coli BL21(DE3)pLysS cells using the M. tuberculosis dut gene (Rv2697c) and the M. smegmatis and M. tuberculosis dcd:dut genes (MSMEG_0678 and Rv0321 respectively). The M. tuberculosis pTBdcd7 Dcd:dut expression plasmid was kindly provided by Martin Willemoes. The site-directed mutagenesis of Dut was carried out according to the Stratagene QuikChange site-directed mutagenesis instructions and verified by sequencing of both strands. The recombinant Dut carrying an N-terminal hexa-His tag was cloned into pET19-b vector, and the recombinant M. smegmatis Dcd:dut amplified from p2NIL_dcdWT and p2NIL_dcdA115F (created in this study) was cloned into pET45-b vector with restriction sites BamHI and HindIII (Table S1). Both proteins were expressed in E. coli BL21(DE3)pLysS cells. For protein overexpression, the cells were grown to an OD600 of 0.4, treated with 0.5 mM isopropyl-b-D-thiogalactopyranoside (IPTG) at 37 °C for 3 hours for Dut and at 30 °C for 6 hours for Dcd:dut expression.
Protein purification
Pellets of cells expressing Dut were lysed in a buffer containing 50 mM TRIS pH 7.5, 100 mM NaCl, 0.5 mM EDTA, 1 mM DTT, 0.1 mM PMSF and EDTA-free protease inhibitor (Roche). Dcd:dut expressing cells were lysed in a buffer containing 20 mM HEPES pH 7.5, 100 mM NaCl, 5 mM MgCl2, 10 mM ß-ME, 0.1% v/v TRITON-X-100, ca. 10 μg/ml RNase, ca. 100 μg/ml DNase, 5 mM benzamidine, 0.1 mg/ml lysozyme and EDTA-free protease inhibitor (Roche). Cell suspensions were sonicated (3 × 60 s) and centrifuged (15550g, 30 min). The final Dut and M. smegmatis Dcd:dut supernatant after cell extraction was loaded onto a Ni-NTA column (Novagen) and purified according to the Novagen protocol. The M. tuberculosis Dcd:dut was purified on Q-Sepharose (GE Healthcare) anion-exchange column, followed by gel filtration on a Superdex 75 column (GE Healthcare) using an AKTA Explorer purifier. The purity of the protein preparation was analyzed by SDS-PAGE. Protein concentration was measured using the Bradford method (Bio-Rad Protein Assay) and by UV absorbance (λ280 = 8480 M−1 cm−1 for H145W M. tuberculosis Dut and its mutant enzymes, and λ280 = 9970 M−1 cm−1 for the M. smegmatis Dcd:dut and the A115F mutant enzyme, λ280 = 11460 M−1 cm−1 for the M. tuberculosis Dcd:dut and the A115F mutant enzyme) and is given in monomers.
Steady-state colorimetric dUTPase assay
Protons released in the dUTPase reaction were detected by phenol red pH indicator in 1 mM HEPES pH 7.5 buffer also containing 100 mM KCl, 40 μM phenol red (Merck) and 5 mM MgCl2. A Specord 200 (Analytic Jena, Germany) spectrophotometer and 10 mm path length thermostatted cuvettes were used at 20 oC for measuring dUTPase activity of the wt and mutant enzymes. The absorbance was recorded at 559 nm. V0 was extracted from the raw absorbance vs. time curves followed by fitting the Michaelis-Menten equation to the V0 vs. substrate concentration steady-state curves using Origin 7.5 (OriginLab Corp., Northampton, MA).
dCTP deaminase activity measurements
The dCTP deaminase activity was measured in a continuous spectrophotometric assay using the difference in the molar extinction coefficients between deoxycytidine and deoxyuridine (Δε286 = 3240 M−1 cm−1). The absorbance was recorded at 286 nm. The assay was buffered with 20 mM HEPES pH 7.5 also containing 100 mM NaCl and 5 mM MgCl2. The reaction was initiated by the addition of deoxicitidine triphosphate into the enzyme containing premix. A Specord 200 (Analytic Jena, Germany) spectrophotometer and 10 mm path length thermostatted quartz cuvettes were used at 20 °C. V0 was extracted from the row absorbance vs. time curves followed by fitting the Michaelis-Menten equation to the V0 vs. substrate concentration steady-state curves using Origin 7.5 (OriginLab Corp., Northampton, MA).
Construction of the dut mutant strains
All genetic experiments were carried out in M. smegmatis mc 2 155 using M. smegmatis genes (MSMEG_2765 dut and MSMEG_0678 dcd:dut) for complementation. Dut KO SCO cells were used11 to construct our dut mutant strains. The mutant dut containing complementing vectors were created by the QuikChange method (Stratagene) using the vector pGem-dut11 as template. Mutant strains were constructed by electroporating the Dut KO SCO strains with the appropriate complementing plasmids. Double crossovers (DCOs) carrying the mutant duts were selected by colony PCR (Table S1) and verified by Southern blot (Fig. S2) and by sequencing the appropriate genome region. Three parallel strains from each mutant were chosen and used for forward experiments. Primers used for cloning, mutagenesis and screening are compiled in Table S1.
Construction of the dcd:dut mutant strains
Dut KO strains carrying the wt11 or the D83N mutant complementing dut copy were used to construct the A115F dcd:dut mutant and the D83N dut/A115F dcd:dut double mutant strains, respectively. A 3.5 kb fragment containing the dcd:dut gene and its flanking regions was cloned into p2NIL using HindIII restriction sites (Table S1) generating p2NIL_dcdWT. The A115F mutant dcd:dut containing vector was created by a modified QuikChange method63. A green fluorescent protein from pLL19264 was C-terminally fused to the dcd:dut. The 6.1 kb PacI cassette carrying the lacZ and sacB selection markers from pGOAL17 was cloned into the sole PacI site of p2NIL to yield p2NIL_dcdA115F. p2NIL_dcdA115F was electroporated into electrocompetent cells. DCOs carrying the mutant dcd:dut were selected by colony PCR and verified by Southern blot (Fig. S2), then finally by sequencing the appropriate genome region. Three parallel strains were chosen and used for forward experiments. While the dcd:dut allele exchange worked in the wt dut background, it did not work in the D83N dut background even after several trials. Primers used for cloning, mutagenesis and screening are compiled in Table S1.
Growth assays
M. smegmatis mutant strains were grown in liquid media. OD600 was measured every 3 hours. Three parallel strains were used from each investigated strain (9 parallel from each mutation) in these experiments. For quantitative comparison, growth curves were fitted with the y = a/(1 + exp(−k*(x − xc))) equation that yielded the best fit keeping the function as simple as possible.
Determination of the spontaneous mutation rate
To determine the spontaneous mutation rates, three rifampicin sensitive independent colonies of each of the three strains/applied mutations were used to inoculate cultures that were grown at 37 °C, 150 rpm. Saturated cultures were serially diluted in sterile broth and plated onto agar plates to determine total CFUs or onto agar plates supplemented with 100 µg/ml rifampicin. The mutation rate for each of the 9 (3 × 3) independent cultures/applied mutation was determined as follows,
where m0 is the observed number of mutants at time point 0, mt is the observed number of mutants at the next time point, and N0 and Nt are the numbers of cells at time points 0 and t, respectively. The mean mutation rate was calculated for each mutant36.
Genomic DNA isolation
10 ml liquid culture was grown until OD600 = 0.5 and harvested. The cells were resuspended in 1 ml 10 mM Tris, pH 7.5 and 0.1 mm glass beads were added to 2 ml volume. The cells were disrupted by vortex and incubation on ice by turn. After centrifugation, the supernatant was manipulated routinely to purify DNA by phenol:chloroform:IAA (25:24:1) extraction followed by isopropanol precipitation.
Determination of the genomic uracil content
In order to quantify the uracil content of DNA, a real-time quantitative PCR-based assay was used38. Genomic DNA was isolated and digested with BamHI. DNA fragments of 5 kb were purified from gel. Real-time PCR was performed on a Mx3000P qPCR System (Agilent Technologies) using EvaGreen dye (Biotium) and PfuTurbo Hotstart DNA polymerase (Stratagene) and Mytaq Hotstart DNA polymerase (Bioline). A segment with 1017 base length defined by the primers (Table S41) was amplified during the PCR reaction. Two-fold dilution series were prepared from the DNA samples. Three parallel strains were used for each mutation in the experiments.
dNTP extraction
Exponential phase cells were grown with appropriate antibiotics until OD600 = 0.6. The total CFUs were determined for each culture, and cells were centrifuged for extraction. Washed pellets were extracted in 0.5 ml ice-cold 60% methanol overnight at −20 °C. Cells were removed by centrifugation (15–20 min, 13.000 rpm) the methanolic supernatant was boiled for 5 min and centrifuged. The supernatant containing the soluble dNTP fraction was vacuum-dried (Eppendorf) at 45 °C, 1h. Extracted dNTPs were dissolved in 50 μl dUTPase buffer (30 mM Tris-HCL, pH 7.5, 10 mM MgCl2, 50 mM NaCl, 1 mM EDTA) and stored at −80 °C.
The quantitation of dcd:dut and dut expression levels
Cells were grown in 50 ml liquid culture until saturation, washed in ice cold PBS and harvested by centrifugation (3100 g, 20 min). Bacterial pellets were resuspended in 1 ml Trizol (Life Technologies), and the cell wall was disrupted by repetitive vortexing with glass beads (6 × 1min). Nucleic acid recovered in the aqueous phase after addition of 0.2 ml chloroform was precipitated with the addition of 0.5 ml isopropanol. The RNA preparations were DNase-treated (10 min, 37 oC) and purified with the Nucleospin RNA Clean-up kit according to the instructions of the manufacturer. Mycobacterial RNA yield were assayed using the Nano-Drop ND-2000 Spectrophotometer (NanoDrop Technologies). RNA samples were amplified from 1 µg total RNA by random hexamer primers using the Transcriptor First Strand cDNA Synthesis Kit (Roche). The resulting cDNA was quantified by Quantitative PCR using EvaGreen (Bioline) and MyTaq PCR master mix (Bioline) in a Stratagene Mx3000P instrument. sigA (MSMEG_2758), an endogenous reference gene was used to normalize input cDNA concentration65. The relative expression ratios of the examined genes were calculated using the comparative Ct method (ΔΔCt). Primers used to measure cDNA of sigA, dcd and dut are compiled in Table S1.
Determination of the pyrimidine nucleotide pool size
The determination of the pyrimidine nucleotide pool size in each extract was based on DNA polymerase-catalyzed incorporation of radioactive dNTP into the synthetic oligonucleotide template method described in ref. 66. The reaction mixture (50 μl) contained Klenow buffer, 0.5 unit exonuclease negative Klenow-fragment (Fermentas), 0.25 μM dTTP/dCTP specific template, 0.25 μM primer (Table S1), 2.5 μM [3H] dATP (1,5 Ci/mmol) (American Radiolabeled Chemicals, Inc.) and 8 μl dNTP-extract or premixed dNTP for calibration. Calibration curve was prepared using 0, 0.1, 0.5, 1, 2, 4 and 8 pmol of each dNTPs/reaction mixture. Incubation was carried out for 60 min at 37 °C and the reaction mix was spotted onto DE81 paper. The papers were dried, washed (3 × 10 min) with 5% Na2HPO4, and rinsed once with distilled water and once with 95% ethanol. After drying, radioactivity on the papers was measured in a liquid scintillation counter (Beckman). In case of the dCTP measurement, we used Taq polymerase (RedTaq, Sigma), the incubation was carried out at 48 °C for 1 hour, as Klenow polymerase is capable of incorporating CTP and GTP from nucleotide extracts39.
dUTP concentration was measured according to Koehler et al.67. Half of the samples for dTTP measurement were treated with 40 ng recombinant Dut at 37 °C, 45 min. The Dut enzyme was precipitated with 60% methanol. The dUTP in half of the extract was enzymatically hydrolyzed while in the parallel sample it was not. Then dUTP concentration was measured using the same protocol as for dTTP determination.
Two-fold dilution series were prepared from cell extracts in the polymerase reactions. Four samples for each of the three parallel strains were used for each mutation in the experiments (i.e. 12 data points).
Statistical analysis
Statistical analysis was carried out using the STATISTICA.13 software. The non-parametric Kruskal–Wallis test or the one-way ANOVA test with Student-Newman-Keuls multiple comparison post-hoc test was used when samples passed the equal variance (Bartlett’s) criterion.
Electronic supplementary material
Acknowledgements
We thank Drs Ildikó Pécsi, András Horváth and Gábor Merényi for constructive advice on mycobacterial strain construction and genomic uracil and dNTP pool measurements. We thank Prof. Camille Locht for mycobacterial vector pLL192. We also thank Judit E. Szabó and Prof. Mihály Kovács for their constructive comments and suggestions on the manuscript. This work was supported by the National Research, Development and Innovation Office, Hungary [OTKA K115993; K109486; K119493; NVKP_16-1-2016-0020], ICGEB CRP/HUN14-01, the European Commission FP7 Biostruct-X project [contract No. 283570]. RH is the recipient of a Postgraduate Research Fellowship of Gedeon Richter Plc. Hungary. JT is the recipient of the János Bolyai Research Scholarship of the Hungarian Academy of Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
Conceived and designed the experiments: R.H., A.L., E.V.S., B.G.V., J.T. Performed the experiments: R.H., A.L., E.V.S. Analyzed the data: R.H., A.L., E.V.S., J.T. Wrote the paper: R.H., A.L., E.V.S., B.G.V., J.T. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Rita Hirmondo, Anna Lopata and Eva Viola Suranyi contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-06206-y
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mathews, C. K. Deoxyribonucleotides as genetic and metabolic regulators. FASEB J. 1–9, doi:10.1096/fj.14-251249 (2014). [DOI] [PMC free article] [PubMed]
- 2.Kunz BA. Mutagenesis and deoxyribonucleotide pool imbalance. Mutat. Res. 1988;200:133–47. doi: 10.1016/0027-5107(88)90076-0. [DOI] [PubMed] [Google Scholar]
- 3.Nordlund P, Reichard P. Ribonucleotide reductases. Annu. Rev. Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi V, Pontis E, Reichard P. Regulation of pyrimidine deoxyribonucleotide metabolism by substrate cycles in dCMP deaminase-deficient V79 hamster cells. Mol. Cell. Biol. 1987;7:4218–24. doi: 10.1128/MCB.7.12.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neuhard J, Thomassen E. Deoxycytidine triphosphate deaminase: identification and function in Salmonella typhimurium. J. Bacteriol. 1971;105:657–65. doi: 10.1128/jb.105.2.657-665.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mollgard, H. & Neuhard, J. Deoxycytidylate Deaminase from Bacillus subtilis. J. Biol. Chem. (1978). [PubMed]
- 7.Vértessy BG, Tóth J. Keeping uracil out of DNA: physiological role, structure and catalytic mechanism of dUTPases. Acc. Chem. Res. 2009;42:97–106. doi: 10.1021/ar800114w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lari S-U, Chen C-Y, Vertéssy BG, Morré J, Bennett SE. Quantitative determination of uracil residues in Escherichia coli DNA: Contribution of ung, dug, and dut genes to uracil avoidance. DNA Repair (Amst). 2006;5:1407–20. doi: 10.1016/j.dnarep.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visnes T, et al. Uracil in DNA and its processing by different DNA glycosylases. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2009;364:563–8. doi: 10.1098/rstb.2008.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 11.Pecsi I, et al. The dUTPase enzyme is essential in Mycobacterium smegmatis. PLoS One. 2012;7:e37461. doi: 10.1371/journal.pone.0037461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.el-Hajj HH, Zhang H, Weiss B. Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli. J. Bacteriol. 1988;170:1069–75. doi: 10.1128/jb.170.3.1069-1075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castillo-Acosta VM, et al. Pyrimidine requirements in deoxyuridine triphosphate nucleotidohydrolase deficient Trypanosoma brucei mutants. Mol. Biochem. Parasitol. 2013;187:9–13. doi: 10.1016/j.molbiopara.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Dengg M, et al. Abrogation of the CLK-2 checkpoint leads to tolerance to base-excision repair intermediates. EMBO Rep. 2006;7:1046–51. doi: 10.1038/sj.embor.7400782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muha V, et al. Uracil-containing DNA in Drosophila: stability, stage-specific accumulation, and developmental involvement. PLoS Genet. 2012;8:e1002738. doi: 10.1371/journal.pgen.1002738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siaud N, et al. The SOS screen in Arabidopsis: a search for functions involved in DNA metabolism. DNA Repair (Amst). 2010;9:567–78. doi: 10.1016/j.dnarep.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 17.Tormo-Más MA, et al. Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature. 2010;465:779–82. doi: 10.1038/nature09065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ariza M-E, Williams MV. A human endogenous retrovirus K dUTPase triggers a TH1, TH17 cytokine response: does it have a role in psoriasis? J. Invest. Dermatol. 2011;131:2419–27. doi: 10.1038/jid.2011.217. [DOI] [PubMed] [Google Scholar]
- 19.Szabó JE, et al. Highly potent dUTPase inhibition by a bacterial repressor protein reveals a novel mechanism for gene expression control. Nucleic Acids Res. 2014;42:11912–20. doi: 10.1093/nar/gku882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leveles I, et al. Structure and enzymatic mechanism of a moonlighting dUTPase. Acta Crystallogr. D. Biol. Crystallogr. 2013;69:2298–308. doi: 10.1107/S0907444913021136. [DOI] [PubMed] [Google Scholar]
- 21.Helt SS, et al. Mechanism of dTTP inhibition of the bifunctional dCTP deaminase:dUTPase encoded by Mycobacterium tuberculosis. J. Mol. Biol. 2008;376:554–69. doi: 10.1016/j.jmb.2007.11.099. [DOI] [PubMed] [Google Scholar]
- 22.Griffin JE, et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pécsi I, et al. Nucleotide pyrophosphatase employs a P-loop-like motif to enhance catalytic power and NDP/NTP discrimination. Proc. Natl. Acad. Sci. USA. 2011;108:14437–42. doi: 10.1073/pnas.1013872108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barabás O, Pongrácz V, Kovári J, Wilmanns M, Vértessy BG. Structural insights into the catalytic mechanism of phosphate ester hydrolysis by dUTPase. J. Biol. Chem. 2004;279:42907–15. doi: 10.1074/jbc.M406135200. [DOI] [PubMed] [Google Scholar]
- 25.Vertessy BG. Flexible glycine rich motif of Escherichia coli deoxyuridine triphosphate nucleotidohydrolase is important for functional but not for structural integrity of the enzyme. Proteins. 1997;28:568–79. doi: 10.1002/(SICI)1097-0134(199708)28:4<568::AID-PROT10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 26.Nord J, Kiefer M, Adolph HW, Zeppezauer MM, Nyman PO. Transient kinetics of ligand binding and role of the C-terminus in the dUTPase from equine infectious anemia virus. FEBS Lett. 2000;472:312–6. doi: 10.1016/S0014-5793(00)01453-8. [DOI] [PubMed] [Google Scholar]
- 27.Shao H, et al. Characterization and mutational studies of equine infectious anemia virus dUTPase. Biochim. Biophys. Acta. 1997;1339:181–91. doi: 10.1016/S0167-4838(96)00229-4. [DOI] [PubMed] [Google Scholar]
- 28.Freeman L, et al. The flexible motif V of Epstein-Barr virus deoxyuridine 5′-triphosphate pyrophosphatase is essential for catalysis. J. Biol. Chem. 2009;284:25280–9. doi: 10.1074/jbc.M109.019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mol CD, Harris JM, McIntosh EM, Tainer JA. Human dUTP pyrophosphatase: uracil recognition by a beta hairpin and active sites formed by three separate subunits. Structure. 1996;4:1077–92. doi: 10.1016/S0969-2126(96)00114-1. [DOI] [PubMed] [Google Scholar]
- 30.Pecsi I, Leveles I, Harmat V, Vertessy BG, Toth J. Aromatic stacking between nucleobase and enzyme promotes phosphate ester hydrolysis in dUTPase. Nucleic Acids Res. 2010;38:7179–86. doi: 10.1093/nar/gkq584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takács E, Grolmusz VK, Vértessy BG. A tradeoff between protein stability and conformational mobility in homotrimeric dUTPases. FEBS Lett. 2004;566:48–54. doi: 10.1016/j.febslet.2004.04.039. [DOI] [PubMed] [Google Scholar]
- 32.Németh-Pongrácz V, et al. Flexible segments modulate co-folding of dUTPase and nucleocapsid proteins. Nucleic Acids Res. 2007;35:495–505. doi: 10.1093/nar/gkl1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mustafi D, Bekesi A, Vertessy BG, Makinen MW. Catalytic and structural role of the metal ion in dUTP pyrophosphatase. Proc. Natl. Acad. Sci. USA. 2003;100:5670–5. doi: 10.1073/pnas.1031504100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szabó JE, Takács E, Merényi G, Vértessy BG, Tóth J. Trading in cooperativity for specificity to maintain uracil-free DNA. Sci. Rep. 2016;6:24219. doi: 10.1038/srep24219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maiques E, et al. Another look at the mechanism involving trimeric dUTPases in Staphylococcus aureus pathogenicity island induction involves novel players in the party. Nucleic Acids Res. 2016;44:5457–5469. doi: 10.1093/nar/gkw317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.David HL. Probability distribution of drug-resistant mutants in unselected populations of Mycobacterium tuberculosis. Appl. Microbiol. 1970;20:810–814. doi: 10.1128/am.20.5.810-814.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pope CF, O’Sullivan DM, McHugh TD, Gillespie SH. A practical guide to measuring mutation rates in antibiotic resistance. Antimicrob. Agents Chemother. 2008;52:1209–14. doi: 10.1128/AAC.01152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horváth A, Vértessy BG. A one-step method for quantitative determination of uracil in DNA by real-time PCR. Nucleic Acids Res. 2010;38:e196. doi: 10.1093/nar/gkq815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferraro P, Franzolin E, Pontarin G, Reichard P, Bianchi V. Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res. 2010;38:e85. doi: 10.1093/nar/gkp1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martí R, Dorado B, Hirano M. Measurement of Mitochondrial dNTP Pools. Methods Mol. Biol. 2012;837:135–148. doi: 10.1007/978-1-61779-504-6_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar P, Bharti SK, Varshney U. Uracil excision repair in Mycobacterium tuberculosis cell-free extracts. Tuberculosis (Edinb). 2011;91:212–8. doi: 10.1016/j.tube.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Srinath T, Bharti SK, Varshney U. Substrate specificities and functional characterization of a thermo-tolerant uracil DNA glycosylase (UdgB) from Mycobacterium tuberculosis. DNA Repair (Amst). 2007;6:1517–28. doi: 10.1016/j.dnarep.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 43.Sang PB, Srinath T, Patil AG, Woo E-J, Varshney U. A unique uracil-DNA binding protein of the uracil DNA glycosylase superfamily. Nucleic Acids Res. 2015;43:8452–8463. doi: 10.1093/nar/gkv854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duncan BK, Weiss B. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J. Bacteriol. 1982;151:750–5. doi: 10.1128/jb.151.2.750-755.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glassner BJ, Rasmussen LJ, Najarian MT, Posnick LM, Samson LD. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc. Natl. Acad. Sci. USA. 1998;95:9997–10002. doi: 10.1073/pnas.95.17.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaaper, R. M. & Mathews, C. K. Mutational consequences of dNTP pool imbalances in E. coli Roel. DNA Repair (Amst). DNA Repair 12 (2013) 73–79, doi:10.1016/j.dnarep.2012.10.011 (2013). [DOI] [PMC free article] [PubMed]
- 47.Nordman J, Wright A. The relationship between dNTP pool levels and mutagenesis in an Escherichia coli NDP kinase mutant. Proc. Natl. Acad. Sci. USA. 2008;105:10197–202. doi: 10.1073/pnas.0802816105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sung JS, Mosbaugh DW. Escherichia coli uracil- and ethenocytosine-initiated base excision DNA repair: Rate-limiting step and patch size distribution. Biochemistry. 2003;42:4613–4625. doi: 10.1021/bi027115v. [DOI] [PubMed] [Google Scholar]
- 49.Gon, S. & Beckwith, J. Ribonucleotide Reductases: Influence of Environment on Synthesis and Activity. Antioxidants Redox Signal. 8 (2006). [DOI] [PubMed]
- 50.Davidson MB, et al. Endogenous DNA replication stress results in expansion of dNTP pools and a mutator phenotype. EMBO J. 2012;31:895–907. doi: 10.1038/emboj.2011.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahluwalia D, Bienstock RJ, Schaaper RM. Novel mutator mutants of E. coli nrdAB ribonucleotide reductase: Insight into allosteric regulation and control of mutation rates. DNA Repair (Amst). 2012;11:480–487. doi: 10.1016/j.dnarep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gon S, Napolitano R, Rocha W, Coulon S, Fuchs RP. Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. 2011;108:19311–19316. doi: 10.1073/pnas.1113664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar D, Viberg J, Nilsson AK, Chabes A. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 2010;38:3975–83. doi: 10.1093/nar/gkq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varga B, et al. Active site of mycobacterial dUTPase: structural characteristics and a built-in sensor. Biochem. Biophys. Res. Commun. 2008;373:8–13. doi: 10.1016/j.bbrc.2008.05.130. [DOI] [PubMed] [Google Scholar]
- 55.Johansson E, et al. Regulation of dCTP deaminase from Escherichia coli by nonallosteric dTTP binding to an inactive form of the enzyme. FEBS J. 2007;274:4188–98. doi: 10.1111/j.1742-4658.2007.05945.x. [DOI] [PubMed] [Google Scholar]
- 56.Tóth J, Varga B, Kovács M, Málnási-Csizmadia A, Vértessy BG. Kinetic mechanism of human dUTPase, an essential nucleotide pyrophosphatase enzyme. J. Biol. Chem. 2007;282:33572–82. doi: 10.1074/jbc.M706230200. [DOI] [PubMed] [Google Scholar]
- 57.Siggaard JHB, et al. Concerted bifunctionality of the dCTP deaminase-dUTPase from Methanocaldococcus jannaschii: a structural and pre-steady state kinetic analysis. Arch. Biochem. Biophys. 2009;490:42–9. doi: 10.1016/j.abb.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 58.Guillet M, Van Der Kemp PA, Boiteux S. dUTPase activity is critical to maintain genetic stability in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34:2056–66. doi: 10.1093/nar/gkl139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warner HR, Duncan BK, Garrett C, Neuhard J. Synthesis and metabolism of uracil-containing deoxyribonucleic acid in Escherichia coli. J. Bacteriol. 1981;145:687–95. doi: 10.1128/jb.145.2.687-695.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hou HF, Liang YH, Li LF, Su XD, Dong YH. Crystal Structures of Streptococcus mutans 2‴-Deoxycytidylate Deaminase and Its Complex with Substrate Analog and Allosteric Regulator dCTP × Mg2+ J. Mol. Biol. 2008;377:220–231. doi: 10.1016/j.jmb.2007.12.064. [DOI] [PubMed] [Google Scholar]
- 61.Marx A, Alian A. The First Crystal Structure of a dTTP-bound Deoxycytidylate Deaminase Validates and Details the Allosteric-Inhibitor Binding Site. J. Biol. Chem. 2015;290:682–690. doi: 10.1074/jbc.M114.617720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kadirvelraj R, Sennett NC, Polizzi SJ, Weitzel S, Wood ZA. Role of packing defects in the evolution of allostery and induced fit in human UDP-glucose dehydrogenase. Biochemistry. 2011;50:5780–5789. doi: 10.1021/bi2005637. [DOI] [PubMed] [Google Scholar]
- 63.Liu H, Naismith JH. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Srivastava V, et al. Macrophage-specific Mycobacterium tuberculosis genes: identification by green fluorescent protein and kanamycin resistance selection. Microbiology. 2007;153:659–66. doi: 10.1099/mic.0.2006/000547-0. [DOI] [PubMed] [Google Scholar]
- 65.Milano A, et al. The Mycobacterium tuberculosis Rv2358–furB operon is induced by zinc. Res. Microbiol. 2004;155:192–200. doi: 10.1016/j.resmic.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 66.Sherman PA, James A. Fyfe. Enzymatic Assay for Deoxyribonucleoside Triphosphates Using Synthetic Oligonucleotides as Template. Primers. Anal. Biochem. 1989;226:222–226. doi: 10.1016/0003-2697(89)90420-X. [DOI] [PubMed] [Google Scholar]
- 67.Koehler SE, Ladner RD. Small interfering RNA-mediated suppression of dUTPase sensitizes cancer cell lines to thymidylate synthase inhibition. Mol. Pharmacol. 2004;66:620–6. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 68.Hochhauser SJ, Weiss B. Escherichia coli mutants deficient in deoxyuridine triphosphatase. J. Bacteriol. 1978;134:157–66. doi: 10.1128/jb.134.1.157-166.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taylor AF, Weiss B. Role of exonuclease III in the base excision repair of uracil-containing DNA. J. Bacteriol. 1982;151:351–7. doi: 10.1128/jb.151.1.351-357.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kouzminova EA, Kuzminov A. Chromosomal fragmentation in dUTPase-deficient mutants of Escherichia coli and its recombinational repair. Mol. Microbiol. 2004;51:1279–95. doi: 10.1111/j.1365-2958.2003.03924.x. [DOI] [PubMed] [Google Scholar]
- 71.Castillo-Acosta VM, et al. Increased uracil insertion in DNA is cytotoxic and increases the frequency of mutation, double strand break formation and VSG switching in Trypanosoma brucei. DNA Repair (Amst). 2012;11:986–95. doi: 10.1016/j.dnarep.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 72.Castillo-Acosta VM, Estévez AM, Vidal AE, Ruiz-Perez LM, González-Pacanowska D. Depletion of dimeric all-alpha dUTPase induces DNA strand breaks and impairs cell cycle progression in Trypanosoma brucei. Int. J. Biochem. Cell Biol. 2008;40:2901–13. doi: 10.1016/j.biocel.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 73.Gadsden MH, McIntosh EM, Game JC, Wilson PJ, Haynes RH. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J. 1993;12:4425–31. doi: 10.1002/j.1460-2075.1993.tb06127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Studebaker AW, Lafuse WP, Kloesel R, Williams MV. Modulation of human dUTPase using small interfering RNA. Biochem. Biophys. Res. Commun. 2005;327:306–10. doi: 10.1016/j.bbrc.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 75.Merényi G, et al. Cellular response to efficient dUTPase RNAi silencing in stable HeLa cell lines perturbs expression levels of genes involved in thymidylate metabolism. Nucleosides. Nucleotides Nucleic Acids. 2011;30:369–90. doi: 10.1080/15257770.2011.582849. [DOI] [PubMed] [Google Scholar]
- 76.Dos Santos RS, et al. dUTPase (DUT) Is Mutated in a Novel Monogenic Syndrome With Diabetes and Bone Marrow Failure. Diabetes. 2017;66:1086–1096. doi: 10.2337/db16-0839. [DOI] [PubMed] [Google Scholar]
- 77.Chen C-W, et al. The Impact of dUTPase on Ribonucleotide Reductase-Induced Genome Instability in Cancer Cells. Cell Rep. 2016;16:1287–1299. doi: 10.1016/j.celrep.2016.06.094. [DOI] [PubMed] [Google Scholar]
- 78.Dubois E, et al. Homologous recombination is stimulated by a decrease in dUTPase in Arabidopsis. PLoS One. 2011;6:e18658. doi: 10.1371/journal.pone.0018658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


