Abstract
Tomato is the most economically important horticultural crop used as a model to study plant biology and particularly fruit development. Knowledge obtained from tomato research initiated improvements in tomato and, being transferrable to other such economically important crops, has led to a surge of tomato-related research and published literature. We developed DES-TOMATO knowledgebase (KB) for exploration of information related to tomato. Information exploration is enabled through terms from 26 dictionaries and combination of these terms. To illustrate the utility of DES-TOMATO, we provide several examples how one can efficiently use this KB to retrieve known or potentially novel information. DES-TOMATO is free for academic and nonprofit users and can be accessed at http://cbrc.kaust.edu.sa/des_tomato/, using any of the mainstream web browsers, including Firefox, Safari and Chrome.
Introduction
The Solanaceae family is a major plant family comprising several economically important crop species such as potato (Solanum tuberosum), eggplant (Solanum melongena), tomato (Solanum lycopersicum), peppers (Capsicum annuum) and chili peppers (Capsicum frutescens). Globally, cultivated tomato is the most important horticultural crop, with an annual production of approximately 164 million tons, and with a value of about $US 60 bn (FAOSTAT, 2013). Because of its value as a food source, tomato has been a target for crop breeding programs focused on traits that contribute to lower production costs, higher quality fruit with extended shelf-life, and sustainable production with higher yield1. Tomato, like many other domesticated crops, has suffered a drastic erosion of genetic variation. Thus, wild tomato species have been widely used in breeding programs to increase genetic variation especially for stress tolerance1, 2. All of the 13 known wild tomato species3, 4 are diploid, can be crossed with cultivated tomato and are important for the evolutionary history of the Solanum section Lycopersicon clade5–7. Due to tomato’s unique features, such as its sympodial shoot, compound leaves, and fleshy fruit, this species has become an established model to study plant biology and particularly fruit development8.
The availability of the reference genome of S. lycopersicum ‘Heinz 1706’9, the identification of millions of single-nucleotide polymorphisms (SNPs)10–12, and the launch of the ‘150 tomato genome re-sequencing project’ (http://www.tomatogenome.net/)5 together with the SNP data from other 360 tomato accessions13, have paved the way for a myriad of genomic studies in tomato and its wild relatives. The large volume of data generated from these studies further prompt the development of tomato-related resources such as the TOMATOMICS database (http://bioinf.mind.meiji.ac.jp/tomatomics/index.php)14, the Micro-Tom mutant database - TOMATOMA (http://tomatoma.nbrp.jp/)15, and the Plant Omics Data Center (PODC; http://bioinf.mind.meiji.ac.jp/podc/)16, which includes core gene expression information for tomato and other species. The tomato-related research addresses different topics, such as stress tolerance17, 18, plant-pathogen interactions19, transcriptional control of biological processes20, 21 and fruit biology22, 23. This plethora of information is becoming overwhelming. Thus, proper insights into metadata are critical to allow a straightforward way to analyze and establish associations within tomato-related literature. The Sol Genomics Network (SGN) (http://solgenomics.net)24 presents a clade-oriented database for the Solanaceae family. However, the SGN is only able to retrieve information previously curated by the Solanaceae consortium, such as genes, quantitative trait loci, and computationally predicted gene family members. As such, the most appealing ingredient in the SGN, which is manual curation, is also its most limiting aspect. The SGN depends on past curation and therefore can only capture a small part of information.
Several text-mining derived knowledgebases (KBs) that explore topic-specific literature and focus on “term associations”/“terms co-occurrence”, have been developed for life science topics25–39. Text-mining in these KBs is restricted to titles and abstracts from PubMed records, which is beneficial for extracting a significant portion of useful information. However, the increasing availability of full-text articles in electronic form is expanding sources of information. For example, when comparing the distribution of information contained in full-text articles versus abstracts, Shah, et al.40 recommended the use of full-text articles instead of just abstracts for extraction of keywords. Also, Schuemie, et al.41, who reported that, although abstracts have the highest information density, results sections have the highest information coverage. In plant sciences, however, text-mining has not been fully exploited35. These include, for example, textual data on Arabidopsis in combination with an integrated network approach42, the Ondex data integration platform (http://www.ondex.org/index.shtml), designed to identify key protein-stress associations43, and VESPA mining, a platform to access data information contained in documents (in this case printed bulletins) to explore pest and crops interactions44. In addition, HRGRN resource (http://plantgrn.noble.org/hrgrn/) enables the exploration of regulatory networks in Arabidopsis (i.e. signaling transduction, metabolism and gene regulation) through a graph search-empowered integrative database45. Nevertheless, and while effective in identifying topic-specific associations, the previous use of text-mining in plant sciences, to our knowledge, tends to have a relatively narrow scope.
To enable users to make a more thorough exploration of the information related to tomato and its close relatives, we developed a topic-specific KB, DES-TOMATO, with an upgraded text-mining methodology similar to46. Our KB uses a dictionary-based approach in which enriched terms and phrases (referred to as terms from here on) belonging to different thematic categories (e.g. pathways, genes, taxonomy, etc.) are pre-compiled to form the basis for indexing text. Terms can be atomic, when the data source provides only one name variation for the entity in question, or they can have a number of synonymous words/phrases that are normalized to the same internal identifier within our knowledgebase. These internal identifiers allow for the universal identification of the term (e.g. through its EntrezGene gene ID, NCBI Taxonomy ID, etc.), and for complementing text-mined information with data from external sources if needed. This dictionary approach allows the user to focus on entities of their interest as defined from commonly used authoritative sources such as ChEBI47 and EntrezGene48. Our KB aims to discover associations between enriched terms, where these terms are searched for in titles and abstracts (from PubMed Wheeler, et al.49) as well as full-length articles allowed for text-mining (from PubMed Central Wheeler, et al.49). Moreover, due to the importance of tomato as a model for the study of plant-pathogen interactions, relevant dictionaries have been included so that users can explore the tomato-associated viral, bacterial, archaeal, and fungal species, as well as their genes and pathways involved in the biotic stress response. The KB also enables users to explore abiotic stress responses.
DES-TOMATO is a resource designed to assist in the exploration, analysis and discovery of tomato-related information inferred through the integration of several data sources. We demonstrate the effectiveness of DES-TOMATO in finding useful associations by presenting four case studies. These examples demonstrate how users can, with ease and speed, identify putative candidate genes, build a network of gene regulation for a specific trait, generate topic-specific hypotheses and explore enriched pathways. To our knowledge, this is the only KB derived through literature text-mining that has a comprehensive information exploration capabilities dedicated to the Lycopersicon section of the Solanaceae.
Systems and Methods
DES-TOMATO is a topic-specific literature exploration system, designed to be visual, intuitive and interactive, and was generated using the Dragon Exploration System v2.0 (DES v2.0). DES was originally developed by VBB and AR and subsequently improved in various ways.
The knowledgebase is implemented and hosted on a CentOS-7 operating system. It uses Apache 2.4.6 as a web server. The literature repository is hosted on a MongoDB 2.6.11 database, and the KB index and related tables are hosted on a PostgreSQL 9.2.15 database. DES-TOMATO uses a Lucene text index for fast querying of the literature. Different components of the KB were developed using various programming languages/tools, namely: Java (openjdk 1.8.0_91), C/C++ (gcc 4.8.5), Perl v5.16.3, PHP 5.4.16, JavaScript, and JQuery 3.0.0.
DES-TOMATO is functional across major web-browsers on Linux, Windows, and Mac OS platforms. It was specifically tested for Firefox, Chrome and Safari. The only feature that we are aware of, which is functional only on Firefox, is the network export function. DES-TOMATO was not tested for hand-held devices, and is not currently intended for such use.
The workflow used within DES to create a KB such as DES-TOMATO comprises the following steps (Fig. 1): 1/data imports and normalization into DES unified schema for dictionaries; 2/indexing of literature repositories using the said dictionaries, and using the resulting index for preliminary data cleaning; 3/preparation of literature corpus via querying of PubMed and PubMed Central articles; 4/extracting term-document mapping information from the global index (created in step 2) that are specific to the corpus in context (defined in step 3); 5/creation of the KB by applying various analysis tasks, including statistical enrichment of terms, extraction and enrichment of pairs, and integrating these data with relevant external resources.
Figure 1.
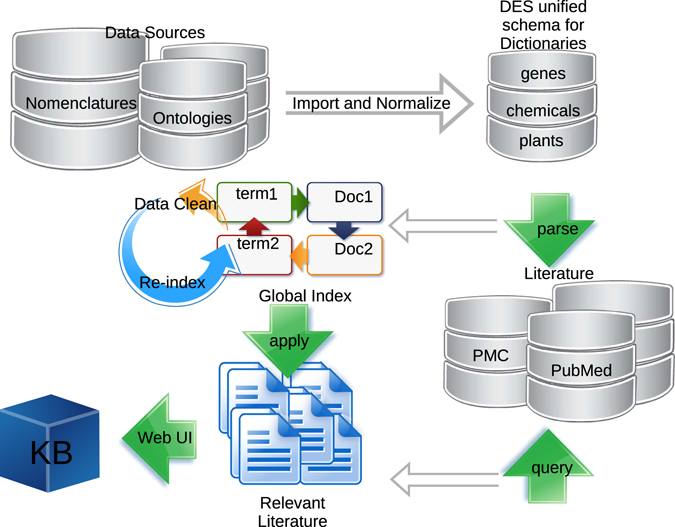
Workflow used within DES to create a KB such as DES-TOMATO.
Preparing the literature corpus
To create DES-TOMATO, we first queried our local literature repository, a MongoDB repository hosting PubMed and PubMed Central articles, backed up by a Lucene text index for fast query servicing. The following DES-TOMATO query was used to incorporate all tomato species: [tomato* OR lycopersicum OR lycopersicon OR ((Solanum OR S.) AND (esculentum OR pimpinellifolium OR pennellii OR sitiens OR habrochaites OR neorickii OR cheesmaniae OR galapagense OR peruvianum OR arcanum OR chilense OR huaylasense OR juglandifolium)]. This retrieved 22,647 articles. The query was made on data updated on August 30, 2016.
Terms and dictionaries
Terms are compiled into thematic dictionaries. Terms can be atomic, when the data source provides only one name variation for the entity in question, or they can have a number of synonymous words/phrases that are normalized to the same internal identifier within our knowledgebase. These internal identifiers allow for the universal identification of the term (e.g. through its EntrezGene gene ID, NCBI Taxonomy ID, etc.), and for complementing text-mined information with data from external sources if needed.
Regarding the dictionaries of genes, we combine EntrezGene nomenclature (for genes) with UniProt nomenclature (for proteins) for a number of reasons. In literature, gene names or symbols are frequently used interchangeably with the names or symbol of their products. Thus, we also use UniProt nomenclature. These nomenclatures provide naming conventions that are the most used by the biomedical community in literature. When reporting results related to a particular gene/protein, it is customary to use the official name/symbol of the gene/protein or one of its aliases and EntrezGene and UniProt exhaustively provide these. EntrezGene also provides loci names for genes as unique identifiers within a species, which are also heavily used in text. There is an initiative by the Tomato Genome Consortium50 to introduce a standardized annotation for gene loci following Arabidopsis type identifiers, that is, these loci names have a general format of the type such as Solyc00g005440.1. Although we intended to use these identifiers, our search for ‘Solyc’ type identifiers in the whole of PubMed produced no hits, which may be partly due to their relatively recent adoption.
Dictionary selection and curation is one of the most important tasks in our KB building process. To ensure relevance and comprehensiveness, we imported 19 relevant dictionaries from the pre-existing DES v2.0 vocabularies. Furthermore, we compiled seven additional theme-specific dictionaries, namely: “Stress-related Vocabulary”, “Plant-related Vocabulary”, “Green Plants Genes (EntrezGene)”, “Solanaceae Genes (EntrezGene)”, “Green Plants (NCBI Taxonomy)”, “Solanaceae (NCBI Taxonomy)” and “Tomato Species (NCBI Taxonomy)” (see Table 1 for more details).
Table 1.
List of dictionaries used in DES-TOMATO.
| Dictionary | Enriched Unique Terms in the KB | Source |
|---|---|---|
| Chemicals/Compounds | ||
| Chemical Entities of Biological Interest (ChEBI) | 4561 | pre-existing in DES |
| Metabolites (MetaboLights) | 1556 | pre-existing in DES |
| Enzymes (IntEnz) | 1182 | pre-existing in DES |
| Toxins (T3DB) | 886 | pre-existing in DES |
| Antibiotics | 244 | pre-existing in DES |
| Industrially Important Enzymes (EC) | 215 | pre-existing in DES |
| Functional Annotation | ||
| Pathways (KEGG, Reactome, UniPathway, PANTHER) | 576 | pre-existing in DES |
| Biological Process (GO) | 1288 | pre-existing in DES |
| Molecular Function (GO) | 474 | pre-existing in DES |
| Cellular Component (GO) | 466 | pre-existing in DES |
| Genes/Proteins/Transcripts | ||
| Green Plants Genes (EntrezGene) | 16579 | newly compiled |
| Solanaceae Genes (EntrezGene) | 2994 | newly compiled |
| Bacteria Genes (EntrezGene) | 2879 | pre-existing in DES |
| Fungi Genes (EntrezGene) | 2758 | pre-existing in DES |
| Viruses Genes (EntrezGene) | 971 | pre-existing in DES |
| Archaea Genes (EntrezGene) | 536 | pre-existing in DES |
| Taxonomy | ||
| Green Plants (NCBI Taxonomy) | 5733 | newly compiled |
| Fungi (NCBI Taxonomy) | 2426 | pre-existing in DES |
| Bacteria (NCBI Taxonomy) | 1498 | pre-existing in DES |
| Viruses (NCBI Taxonomy) | 1109 | pre-existing in DES |
| Solanaceae (NCBI Taxonomy) | 297 | newly compiled |
| Source Microbes for Antibiotics | 113 | pre-existing in DES |
| Archaea (NCBI Taxonomy) | 40 | pre-existing in DES |
| Tomato Species (NCBI Taxonomy) | 15 | newly compiled |
| General | ||
| Plant-related Vocabulary | 2688 | newly compiled |
| Stress-related Vocabulary | 759 | newly compiled |
The following is a description of the process of importing/generating data for compiling dictionaries:
The general case
Irrespective of how the dictionary is generated, the importing and integration of a new dictionary into DES typically includes the following steps:
- Transforming the vocabulary data into a format that adheres to our local Term schema. This schema includes:
- a unique identifier for the term,
- a concept identifier shared by synonymous terms but unique across concepts,
- the English version of the term itself (so removing non-English nomenclatures if they exist), as well as
- metadata about the term, such as description, source (e.g. PO), the ID used by the source, (PO:0025002 for ‘basal root’), etc.
This is then used to update the dictionary set with the new data. New entries are checked for term redundancies within the same dictionary, in which case they are unified into one term with multiple source IDs.
An initial indexing is performed to see how the newly imported dictionaries match the literature, (e.g. which terms did actually have mentions, and how frequently across the whole PubMed and the whole (allowed for text-mining) PubMed Central documents). This information also provides the basis for dictionary cleaning, as it is often the case that promiscuous terms from thematic dictionaries appear with high false positive rates due to the high frequencies of their use usually as common English words. Such terms we generally have excluded. An example is term “content”: one of the synonyms for PATO:0000025.
Once the dictionary data is cleaned, another re-indexing occurs so that the index and the subsequent analyses are built around reasonably clean dictionary data.
We eliminated ambiguous terms from the dictionaries where possible. The problem of ambiguous words that might blur the outcome of a search, is a well-known challenge in the field of text-mining and natural language processing, because it is inherent to language51. Even in manual analysis of text, human interpretation is the key to disambiguate the meaning. In the case of DES-TOMATO, this disambiguation is left to the user and his/her knowledge and skills. Furthermore, this problem is more relevant to some dictionaries (types of biological entities) than others, e.g. gene names/abbreviations coinciding with disease names/abbreviations, or some ontologies containing some semantically broad terms. However, to reduce the proportion of these cases in DES-TOMATO, we carried out stringent term pre-processing steps: 1/initial data cleaning of the most frequent promiscuous terms, 2/eliminating terms shorter than three characters that have no synonyms in the same document, and 3/statistical enrichment, which filters out an additional good proportion of common and highly promiscuous terms.
The “Plant-related Vocabulary”
The “Plant-related Vocabulary” incorporates terms from a number of ontologies (see Table 2), which in some cases (e.g. FLOPO) are in turn, partially or completely, composed of information from other ontologies.
Table 2.
Plant-related ontologies used to compile the “Plant-related Vocabulary”.
| Terms or Phrases | Definition |
|---|---|
| Enriched Terms | Biological terms or keywords (e.g. lycopene, peroxidase activity, Solanum pimpinellifolium, etc.) used to mine the literature and organized into thematic dictionaries |
| Enriched Term Pairs | Connection/association (possibly biological) between two terms that is inferred based on the co-occurrence of these terms (e.g. signaling and salicylic acid; lycopene and carotenoids; Solanum lycopersicum and begomovirus, etc.) |
| Hypothesis | New connection of terms; a starting point for possible further investigation (e.g. AGO5 and ‘DNA methylation’; SNI1 and ‘jasmonic acid’) |
| KOBAS Pathways | Enriched pathways that were identified by the set of genes and/or proteins extracted from tomato-based literature |
| Dictionary | A set of terms, which are categorized into themes (e.g. Pathways, Metabolites, or Genes) |
| Interactive tools | Definition |
| Network Viewer | A tool for the visualization of term associations as a graph of interlinked nodes |
| Term Co-occurrences | A list of all the enriched terms from all dictionaries that is potentially associated with the term in question. |
| Term Link Sources | A graph/pie chart that visualize the distribution of data sources (dictionaries) from which associations to the term in question are drawn |
Note that sometimes ontologies reuse and integrate entities from other ontologies/sources when appropriate, such as is the case for FLOPO and PTO ontologies.
The “Stress-related Vocabulary”
This vocabulary was built from scratch to account for certain terminology that we believe is important for this KB, but was lacking in the plant ontologies that we considered. For compiling the “Stress-related Vocabulary”, we created 19 categories of keywords: ‘Salt’, ‘Heat’, ‘Cold’, ‘Flood’, ‘Drought’, ‘Light’, ‘pH’, ‘Osmotic’, ‘Oxidative’, ‘Anaerobia’, ‘Anoxia’, ‘Hypoxia’, ‘Hyperoxia’, ‘Nitrosative’, ‘Physiology’, ‘Nutrients’, ‘Pathology’, ‘Growth’, and ‘Biotic’. In each category, we manually searched the literature and added keywords that are related to the category, with the condition that the keyword must not exist in any plant ontology. For example, under the ‘Osmotic’ and ‘Flood’ categories, we included terms ‘Osmoprotectant’ and ‘Submergence’, respectively. These two keywords are related to stress and they are not found in any of the other DES-TOMATO dictionaries. In total, 92 keywords from the literature for the 19 categories were identified. Concurrently, we created 23 keywords that act as prefixes, such as ‘tolerance to’ (e.g. tolerance to salt stress) and 7 keywords that act as suffixes, such as ‘tolerance’ (e.g. salt stress tolerance). We then computationally compiled these affixes to the 92 keywords that resulted in 2,760 new terms that we used in the text-mining process. Some of these combinations were not detected in text, either because they were not used or because they do not representing viable term combinations.
Post-processing and indexing
Terms in the aforementioned dictionaries were then mined in the retrieved articles, highlighted and color-coded according to dictionary. This process is enabled by the back-end index that matches terms to their occurrences, up to the character level, within the mined articles. In total, 9,499,592 terms from 26 dictionaries were used to index the literature corpus in DES-TOMATO. A term is defined as enriched when it is overrepresented in DES-TOMATO documents as compared to all PubMed and all PubMed Central articles (for which text-mining is allowed) from our local repository. We used a false discovery rate (FDR) < 0.05, which was calculated based on the Benjamini–Hochberg procedure to correct for multiplicity testing. Terms in all dictionaries are normalized, i.e. names, symbols and synonyms referring to the same concept are represented by a single entity when analyzed. This process allowed us to identify 52,886 unique terms that are statistically enriched (FDR <= 0.05) in tomato-related documents and present in DES-TOMATO. We further identified 1,388,952 enriched unique term pairs (FDR <= 0.05) formed from the 52,886 statistically enriched terms.
Additionally, by matching genes and proteins enriched in DES-TOMATO to other resources beyond the KB literature corpus, in this case KOBAS52, we found hits to: 1/930 Bacterial pathways, of which 677 are statistically enriched (FDR <= 0.05), 2/427 Archaeal pathways, of which 90 are statistically enriched (FDR <= 0.05), 3/523 Fungi pathways, of which 86 are statistically enriched (FDR <= 0.05), and 4/1,747 Plant pathways, of which 488 are statistically enriched (FDR <= 0.05).
Results
Indirect assessment of the quality of extracted information
It is difficult to provide a global assessment of the quality of extracted information by DES-TOMATO KB. In an attempt to provide an independent assessment of the quality of associations identified by KB, we evaluated the quality of the gene pairs extracted by the KB by comparing them to their functional similarity, where functions of the genes are obtained from an independent data source. Specifically, we computed the semantic similarity of gene pairs based on their GO annotations using the Semantic Measures Library (SML)53. We hypothesize that a strong correlation between our extracted associations between gene pairs and their functional similarity is reflective of the quality of the data in DES-TOMATO and its analysis approach. Essentially, we propose that a correct association between two genes in DES-TOMATO will generally be reflected by the two genes’ sharing similar GO annotations, although some gene pairs may also be associated in a manner not reflected by GO term similarities. In other words, we performed an assessment of the quality of extracted tomato gene-gene associations under strict conditions.
EntrezGene IDs for normalized genes were mapped to identifiers in the agriGO annotation54. Starting with a total of 16,056 Solanaceae gene pairs, we removed all gene pairs between genes that are in another Solanaceae species, and retained 13,139 pairs in which at least one of the genes is present in tomato. Selecting pairs in which both genes are present in tomato produced a set of 3,975 pairs of which 2,227 had an agriGO annotation for both genes in each pair. We use only these 2,227 pairs in the assessment by semantic similarity. Here we used default parameters (lin_resnik_bma) with the aspect parameter set to GLOBAL. Of the 2,227 tomato gene pairs, 575 (26%) had maximum possible semantic similarity (value of 1.0), which means that genes in these pairs have identical GO annotations. Table 3 lists some examples from this set. In Table 4, we show the percentage of identified pairs of genes at different semantic similarity thresholds.
Table 3.
Examples of gene-gene associations identified in KB with semantic similarity equal to 1.0.
| Gene Symbol/Description | Gene Symbol/Description | Common Annotations |
|---|---|---|
| SERK3A/ID: 100736467 somatic embryogenesis receptor kinase 3 A [Solanum lycopersicum (tomato)] | LOC101259548/ID: 101259548 leucine-rich repeat receptor-like serine/threonine/tyrosine-protein kinase SOBIR1 [Solanum lycopersicum (tomato)] | “protein kinase activity”;“molecular_function”;“GO:0004672” “protein binding”;“molecular_function”;“GO:0005515” “ATP binding”;“molecular_function”;“GO:0005524” “protein phosphorylation”;“biological_process”;“GO:0006468” |
| PHYF ID: 101259349 phytochrome F [Solanum lycopersicum (tomato)] | PHYB1 ID: 101262847 phytochrome B1 [Solanum lycopersicum (tomato)] | genes are involved in photoreceptor activity (GO:0009881) |
| APX2 ID: 778224 cytosolic ascorbate peroxidase 2 [Solanum lycopersicum (tomato)] | LOC101264261 ID: 101264261 L-ascorbate peroxidase 3, peroxisomal [Solanum lycopersicum (tomato)] | “peroxidase activity”;“molecular_function”;“GO:0004601” “peroxidase activity”;“molecular_function”;“GO:0004601” “response to oxidative stress”;“biological_process”;“GO:0006979” “heme binding”;“molecular_function”;“GO:0020037” “oxidation-reduction process”;“biological_process”;“GO:0055114” |
Table 4.
The change of the number of gene pairs according to the change of required semantic similarity level.
| Semantic Similarity | Number of Gene Pairs | Percentage (out of 2,227) |
|---|---|---|
| >=0.4 | 1098 | 49% |
| >=0.45 | 991 | 45% |
| >=0.5 | 943 | 42% |
| >=0.55 | 913 | 41% |
| >=0.6 | 875 | 39% |
| >=0.65 | 832 | 37% |
| >=0.7 | 794 | 36% |
| >=0.75 | 760 | 34% |
| >=0.8 | 697 | 31% |
| >=0.85 | 674 | 30% |
| >=0.9 | 613 | 28% |
| >=0.95 | 579 | 26% |
| =1 | 575 | 26% |
Furthermore, results shown in Supplementary Material (distribution of high similarity pairs across FDR rank) demonstrate that the higher the FDR rank of a gene pair, the more likely it would have a high similarity rank. This shows the usefulness of the enrichment measure we use in DES-TOMATO. Therefore, our system not only extracts gene pairs through co-occurrence, it also has a robust means for ranking, or prioritizing, these associations.
It is important to note that for a number of pairs suggested by DES-TOMATO it was not possible to calculate the similarity score due to either one or both of the tomato genes in the pair lacking GO annotation in agriGO (as mentioned above). These gene pairs, which were false positives in our stringent assessment, should not be considered as unrelated. In fact, we manually evaluated a number of these ‘inconclusive’ pairs and found that some do have an association that was not reflected in the semantic similarity (see examples in Table 5). Unfortunately, manual curation of the entire dataset is beyond our means.
Table 5.
Some examples of gene-gene associations that have functional association but do not have semantic similarity.
| Gene Symbol/Description | Gene Symbol/Description | Common Annotations | Reference |
|---|---|---|---|
| IAA3/ID: 543540 IAA3 protein [Solanum lycopersicum (tomato)] | EXP2/ID: 543582 expansin [Solanum lycopersicum (tomato)] | Volatile Organic Compounds (albuterol and 1,3- propanediole) were shown to promote lateral root formation that correlates with an increase in levels of EXP2 and IAA3 in the roots of tomato plants | 124 |
| MAF1/ID: 543586 MFP1 attachment factor 1 [Solanum lycopersicum (tomato)] | FPP/ID: 543699 filament-like plant protein [Solanum lycopersicum (tomato)] | Filament-like plant proteins (FPP) belongs to a family of long coiled-coil proteins that interacts with the nuclear envelope-associated protein, MAF1 | 125 |
| LOC543607/ID: 543607 pirin [Solanum lycopersicum (tomato)] | DAD1/ID: 543753 dad-1 protein [Solanum lycopersicum (tomato)] | Both DAD1 and pirin are mediators of programmed cell death in plants. However, DAD1 was shown to interact with BCL2 family members, while pirin plays more of a downstream role as it forms a NF-kB, BCL3, Pirin complex that is capable of modulating NF-kB-driven gene expression through interaction with an NF-kB DNA-binding site. | 126 |
Using one of the most challenging text-mining entities (genes/proteins), we have demonstrated that the quality of the associations in our KB is reasonably reliable and by extension we extrapolate that entities and associations in the other dictionaries in the KB are also reasonably reliable.
Navigating the KB
The users of DES-TOMATO can explore and find relevant information in the literature, based on enriched terms. The content of this KB can be explored via links (described in detail by Salhi et al.34 under names in brackets), which include “Enriched Terms” [Concepts], “Enriched Term Pairs” [Associated Concepts], “Explore Hypotheses” [Hypothesis Explorer], and “KOBAS Pathways” [KOBAS pathways]. By navigating these links, users can view enriched terms via several types of ranking options and/or by restricting the FDR to zoom in on an enriched subset of interest. Moreover, users can access a menu with a right-click, which enables all terms to generate a “Network” view, “Term Co-occurrences” and “Term Link Sources” (refer to Table 6). It is important to note that users should always refer to organisms by their Latin name, namely for pathogens (except virus) and plant species. Case study examples are given below. We provided a detailed Manual that explains various functionalities of the DES-TOMATO and its use. Each page of the KB contains a link to “Help” for the fast instructions about how to use the page. In addition, we provided a quick start video on the “Home” page, which demonstrates basic functionalities of the KB.
Table 6.
Glossary.
| Ontology | Description |
|---|---|
| PO | Plant Ontology 127, 128: A structured vocabulary which incorporates: plant anatomy, morphology and growth and development. PO was developed as part of the Planteome project (License: http://planteome.org/License) |
| FLOPO | Flora Phenotype Ontology 129: an ontology of phenotypes reported in Floras. This ontology incorporates a number of entities from other ontologies, in addition to indigenous FLOPO entities. |
| PTO/TO | Plant Trait Ontology 128: A controlled vocabulary to describe phenotypic traits in plants. This ontology also incorporates classes from various other ontologies. |
| PECO/EO | Plant Environmental Conditions Ontology 128. This ontology describes the treatments, growing conditions, and/or study types used in plant biology experiments. |
| SPTO | Solanaceae Phenotype Ontology 130: Solanaceae crop phenotypes and traits, developed in collaboration with the research community, especially for breeder traits of agronomic importance. |
Case studies that substantiate the effectiveness of DES-TOMATO as a research supporting system
Example 1. “Enriched Terms” used for the exploration of genetic interactions underlying bacterial speck disease.
Here we explore the efficacy of DES-TOMATO in the exploration of plant-pathogen molecular interactions towards identifying the genetic components of resistance to bacterial speck (caused by Pseudomonas syringae) in the Solanaceae family. The genetic-basis for resistance to this disease was linked to the Pto gene55, 56.
We started exploring DES-TOMATO by clicking “Enriched Terms” (Fig. 2, Step 1), we, then, searched the list for ‘Pseudomonas syringae’, and generated a network with the right-click menu (Fig. 2, Step 2). On the network page, we selected “Solanaceae genes” and “Plant-related Vocabulary” from the dictionaries top-menu, then populated the network starting from the ‘Pseudomonas syringae’ node using the ‘Expand from the term’ right-click menu. Afterwards, we removed redundant terms, generic terms, and all “Plant-related Vocabulary” terms except ‘Disease resistance’ using the ‘Remove highlighted’ right-click menu (Fig. 2, Step 3). Using the “Solanaceae genes” dictionary only, a second round of network expansion was performed on all nodes obtained in Step 3, followed by a third round of expansion from the resulting ‘Pto’ node. The resulting network was simplified by removing nodes with a single link (Fig. 2, Step 4).
Figure 2.
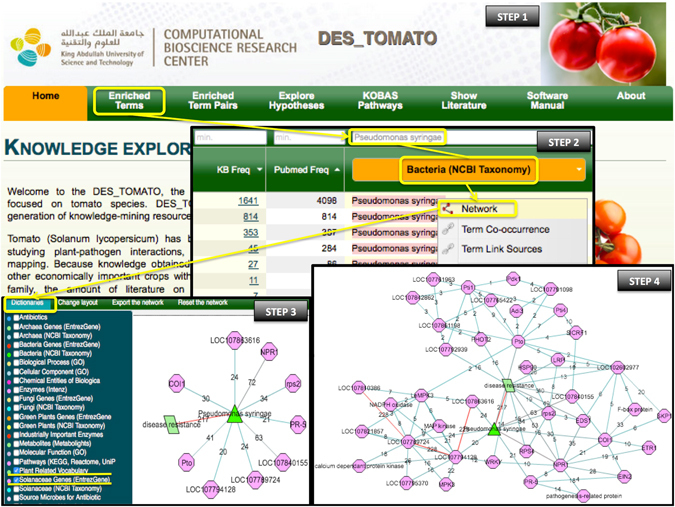
Step-by-step illustration of how DES-TOMATO can be used to identify components of genetic resistance for P. syringae (marked in yellow). The pink octagons represent the “Solanaceae Genes” dictionary; the dark green triangles represent the “Bacteria (NCBI Taxonomy)” dictionary; and the pale green trapezoids represent “Plant-related Vocabulary” dictionary. The edge color is distributed across a color spectrum from hot/red (high frequency co-occurrence/strong association) to cold/blue (small number of co-occurrences, weaker association). The numbers on the edges provide the number of publications that link the associated nodes.
The final network is clearly divided into two sub-networks; one is centered on ‘Pto’ while the other is centered on ‘NPR1’ (Fig. 2), which is consistent with previous knowledge. Upon infection, Pto detects the cognate AvrPto bacterial effector proteins, triggering a signal transduction cascade55, 57. Additionally, it is known that Pdk1 regulates Adi3 activity together with Pto58–60, and the loss of Adi3-mediated cell death suppression is believed to contribute, through MAPKKKα signaling, to the resistance response upon P. syringae infection61, 62. Similar relevant connections can be made by expanding from the other Pto-associated genes (not shown).
On the other hand, NPR1 is a master immune regulator that indirectly drives transcription of PR genes in response to the immune signal salicylic acid (SA), eliciting a defense response63. Additionally, Coronatine-insensitive 1 (COI1), inhibits jasmonate (JA) signaling-dependent process that is known to impair SA-mediated pathogen defense responses64. This pathway is hijacked by various P. syringae strains expressing the phytotoxin coronatine (COR), which mimics a bioactive JA conjugate to suppress immune responses through interactions with COI165, 66. Other noteworthy PRR genes associated with P. syringae include:
-
i)
R gene Resistance to P. syringae 2 (rps2), which encodes an NB-LRR protein involved in the recognition of the P. syringae effector AvrRpt267, 68;
-
ii)
R gene Resistance to P. syringae (rps4), which cooperates with Ralstonia solanacearum 1 (RRS1), to recognize the P. syringae effector AvrRps469; and
-
iii)
Two PR genes, PR5 and PR1 (LOC107840155).
Through this example we demonstrate the ability of DES-TOMATO to effectively identify key factors underpinning systems of interest. In this case, DES-TOMATO enables the construction of complex networks representing the genetic interactions underlying plant-pathogen responses with relative ease and speed, with little prior knowledge. This approach identified many well-characterized components as well as less evident connections, such as the one between COI1 and SGT1 (only hypothesized in Meldau, et al.70, yet not experimentally shown), which can used as suggestions for future investigations.
Example 2. “Enriched Term Pairs” used to explore “Na+/H+ antiporter” associated gene for the discovery of a putative candidate gene involved in salinity tolerance.
The accumulation of toxic levels of sodium in the cytosol is the main cause of salinity stress in plants, and cells cope through an efficient cytosolic Na+ homeostasis mechanism (e.g. Na+/H+ antiporters)71. To explore potential genes involved in this process, we start by clicking “Enriched Term Pairs” (Fig. 3, Step 1). This opens a page with two columns listing associated terms from all dictionaries. In the first dictionary (term A), we filtered the name for ‘Na+/H+ antiporter’ while in the second dictionary (term B), we selected the “Solanaceae genes” dictionary from the drop-down menu (Fig. 3, Step 2). The first two enriched term pairs are SOS1 and NHX1 genes, which are widely known in the literature to be involved in salinity response, meanwhile the third hit was ‘ATPase’. ATPases are proton pumps that are essential for establishing the proton gradient that powers the transport of Na+ by Na+/H+ antiporters across the plasma membrane and the tonoplast71, 72. Salinity stress induces the expression of H+-ATPases in both the tonoplast and the plasma membrane73, 74; thus, we chose to expand our search through ‘ATPase’. We right clicked on ‘ATPase’, and selected “Network” (Fig. 3, Step 2). In the new window, we selected ‘ATPase’ and expanded the association using the “Solanaceae Genes” dictionary (Fig. 3, Step 3). To focus the network, we removed redundant terms using the right click menu. Next, we searched PubMed for the other genes captured by the network and found the following:
-
i)
LOC107803903, which encodes the ‘zinc transporter 5-like’ in Nicotiana tabacum.
-
ii)
HSP90, which encodes the ‘Heat Shock Protein 90’ that has been reported to be involved in heat stress in tomato75;
-
iii)
HSP70, which encodes the ‘Heat Shock Protein 70’ from S. lycopersicum. HSP70 was proposed to act together with HSP90, at least, under heat stress75;
-
iv)
LOC107766295, which encodes for the ‘Heat Shock cognate 70 kDa protein 2-like’ from N. tabacum;
-
v)
PPA1, which encodes the soluble inorganic pyrophosphatase-like from S. tuberosum;
-
vi)
14-3-3 protein family, which is known to bind to several signaling proteins, namely activating the auto-inhibited plasma membrane H+-ATPases76;
-
vii)
SOS1, which is a gene known to be involved in salinity response, and abundantly described in the tomato literature77;
-
viii)
LHA2, which encodes for a plasma membrane H+-ATPase with higher expression in hypocotyls and leaves78; and
-
ix)
LHA4, which encodes for a plasma membrane H+-ATPase with higher expression in roots and hypocotyls78.
Figure 3.
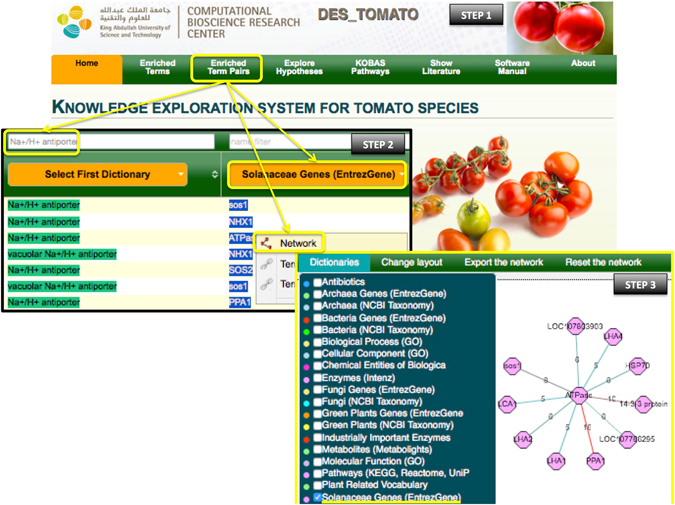
Step-by-step illustration of how DES-TOMATO can be used to find relevant candidate genes involved in salinity tolerance by focusing on Na+ homeostasis and plasma membrane H+-ATPases (in yellow). In the network, the pink octagons represent the “Solanaceae Genes” dictionary. The edge color is distributed across a color spectrum from hot/red (high frequency co-occurrence/strong association) to cold/blue (small number of co-occurrences, weaker association). The numbers on the edges provide the number of publications that link the associated nodes.
As an example, we then focused on LHA4 in tomato and by matching its sequence by BLAST79 against the NCBI nt database, we found that LHA4 is homologous to AHA2 in A. thaliana. AHA2’s overexpression has been suggested to improve salinity tolerance80. AHA2 was also shown to be phosphorylated upon salt stress81. However, and despite the growing amount of evidence, little is known about the role of AHA2 (Arabidopsis) in salinity stress. This example demonstrates how DES-TOMATO can facilitate an easy review of dictionary terms associated with a term of interest.
Example 3. Using “Explore Hypotheses” to demonstrate how topic-specific hypothesis can be generated and tested.
Plant growth is affected by various abiotic stress conditions in which abscisic acid (ABA) biosynthesis is a major hub. To generate a hypothesis on this topic, we used the “Explore Hypotheses” tool, which opens a page with two columns listing associated enriched terms from all dictionaries (Fig. 4, Step 1). The first dictionary (term A) was filtered with ‘ABA biosynthesis’ while for the second dictionary (term C), we selected “Green Plants Genes” dictionary from the drop-down menu, after which we clicked ‘test’ for hppd (Fig. 4, Step 2). This generated a hypothesis that hppd may be linked to ABA biosynthesis via the linking term LOC107839360 (term B), also known as carotenoid 9,10(9’,10’)-cleavage dioxygenase 1-like.
Figure 4.
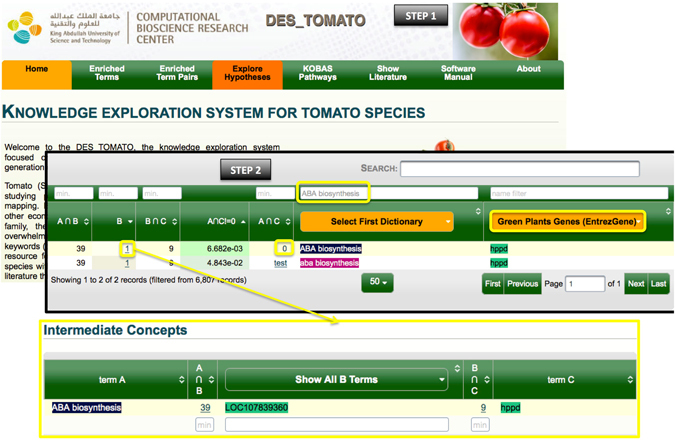
A simple demonstration of how a use “Explore hypotheses”. Boxed in yellow are the criteria used to direct or test the hypotheses generated.
The hppd gene encodes the enzyme p-hydroxyphenylpyruvate dioxygenase that acts as an oxireductase on pyruvate carriers. To our knowledge, current literature provides no direct link between p-hydroxyphenylpyruvate dioxygenase and ABA biosynthesis. But interestingly, pyruvate carriers have recently been implicated in ABA signaling82. In Arabidopsis, the putative mitochondrial pyruvate carrier, NRGA1, is a negative regulator of guard cell ABA signaling through the alleviation of ABA effect. This suggests that NRGA1 is responsible for the maintenance of optimal stomatal aperture during drought stress82. Here we show that by using “Explore Hypotheses”, we were able to conjecture that p-hydroxyphenylpyruvate dioxygenase (encoded by hppd) may act on the NRGA1 pyruvate carrier and consequently may indirectly interact with ABA. Further studies are required to validate this hypothesis.
Example 4. Exploring S. lycorpersicum enriched pathways using “KOBAS Pathways”.
Here we demonstrate how users can easily access the supplementary information from the KOBAS database52 using DES-TOMATO. First, we clicked on “KOBAS Pathways” (top menu) and selected ‘Solanum lycorpersicum’ from the “taxonomy for enrichment” drop-down menu. By selecting Benjamini-Hochberg correction and a significance level of 0.05 (View Enrichment Filters button), we obtain five enriched pathways (Fig. 5): (1) carotenoid biosynthesis; (2) brassinosteroid biosynthesis; (3) zeatin biosynthesis; (4) cysteine and methionine metabolism; and (5) butanoate metabolism. All of these pathways have been described in tomato as major contributors to plant and fruit development, fruit ripening and pathogen-resistance83–89. To further understand why these pathways are statistically enriched in tomato literature, we provide a brief and simple description for each.
Carotenoid biosynthesis. Carotenoids are colored pigments present in all plant tissues, and their formation is highly regulated. Lycopene is the major carotenoid in tomato. During fruit ripening, lycopenes’ concentration increases enormously90. The regulation of carotenoids biosynthesis in tomato and other major genes (e.g. phytoene synthase - Psy and and phytoene desaturase -Pds) that are involved in this process have been extensively studied84, 90–92;
Brassinosteroid biosynthesis. Brassinosteroids are steroidal hormones that are essential for plant growth and development, and are also involved in stress-response mechanisms93, 94. Castasterone is a precursor in the brassinosteroid biosynthesis pathway, which is the product of a cytochrome P450-catalyzed conversion reaction from 6-deoxocastasterone. The cytochrome P450 and its Dwarf encoding gene have been extensively studied in tomato fruit development85, 95–97;
Zeatin biosynthesis. Zeatins are plant-growth hormones that belong to the cytokinins family. They regulate cell division and expansion and delay senescence. In tomato, changes in root-synthesized zeatins have been implicated in stress-responses86, 88, and fruit development89;
Cysteine and methionine metabolism. Methionine is an essential amino acid, and is the precursor of ethylene. Ethylene is a plant hormone that is involved in several processes in plant life-cycle including seed germination, root hair development, flower senescence and fruit ripening87. In tomato, biosynthesis of ethylene has been extensively studied due to its importance in controlling fruit ripening83, 87, 98;
Butanoate metabolism. Gamma-aminobutyric acid (GABA) is a non-protein amino acid, and a major plant-growth regulator99. GABA levels undergo drastic fluctuations during fruit development, by increasing during the mature green stage, and rapidly decreasing during the ripening stage100, 101.
Figure 5.
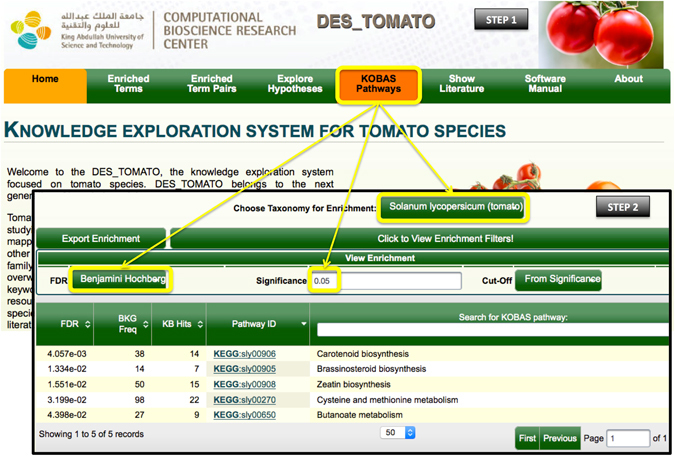
A simple demonstration of how S. lycorpersicum enriched pathways can be explored using “KOBAS Pathways”. Boxed in yellow are the criteria adapted for this exploration process.
Discussion
General Comments
Text-mining will not replace other types of computational data analysis in the biomedical field, the same way computational methods in general will not replace laboratory experiments and clinical research. However, text-mining should be considered as complementary to other (experimental and computational) approaches. The information obtained through text-mining, in many cases, cannot be obtained through other means in any simple manner102. Indeed, text-mining approaches have been deployed to complement other lines of investigation or as stand-alone tools for gaining quick insights. There are several reports where text-mined data alone were used to correctly infer links between concepts, e.g. Smalheiser and Swanson correctly inferred a link between Alzheimer’s disease and indomethacin103, 104 and Wren et al. correctly inferred a link between chlorpromazine and the progression of cardiac hypertrophy105. Text-mining was also used in conjunction with gene expression analysis to show that sphingosine 1-phosphate independently regulates glioblastoma cell invasiveness through urokinase-type plasminogen activators106, 107. Similarly, text-mining was also used with other types of data-mining to successfully identify disease genes in Wilms’ tumor108. Moreover, text-mining was successfully used to identify protein-protein interactions (see e.g. refs 36 and 37), transcription factor associations38, and methylated genes in various diseases and species39, 109. Thus, text-mining approaches are increasingly playing a role in a number of biomedical problems110 from pharmacogenomics111 (for the extraction of relations between drugs, genes and diseases), to precision medicine and drug repositioning26.
Limitations
DES-TOMATO generally has the same limitations as other existing text-mining-based resources. Here we list some of the most common constraints: 1/text-mining-based resources are confined to information presented in electronically available documents; 2/some documents are protected by copyright from text-mining; 3/all text-mining systems are far from being able to extract all useful information from available texts; 4/peer-reviewed literature contains errors that are often propagated in different articles and automated text-mining information extraction cannot correct for such errors. This field undoubtedly requires significant improvements. Additionally, an association in DES-TOMATO does not specify the type of relationship among the extracted pairs of entities, e.g. co-occurrence of terms does not necessarily imply direct or physical interaction between paired terms.
Coverage is also affected by the common practice of authors to report only on what are deemed as the most relevant data. For example, papers reporting on genomic studies related to gene expression data, describe only a handful of genes in the text, while the bulk of experimental results are deposited separately from the published articles. In DES-TOMATO, dictionaries cover 3,050 Solanaceae species and all of their 300,973 non-redundant genes. This was necessary in order to maximize coverage of the tomato genes and their potential homologs. However, only 297 species (10%) and 2,994 genes (1%) were enriched in the text, which is not surprising.
The question now becomes, given the constraints imposed on the information that can be extracted from text, is it even worth using it? We believe the answer is yes, for the very fact that the type of information in the published scientific literature in the vast majority of cases conveys what researchers considered the most important facts regarding the topic of interest. The vast majority of scientific studies start by reviewing literature on the topic of interest and not by delving directly into the analysis of experimental data. However, due to limitations in terms of coverage and sometimes uncertainty of the quality of automatically extracted information through text-mining, the resulting data presented to the user are mainly advisory, aimed to guide exploration and draw attention to linked concepts. Domain knowledge and expertise are required for the interpretation of linked concepts, equally as they are required for the interpretation of experimental results.
Concluding Remarks
Recent biotechnological advances have unleashed a tsunami of scientific literature that has become overwhelming for researchers. Even for the topic-specific literature insight, the volume of information is huge. To meet this challenge, we developed the DES-TOMATO KB that is focused on tomato species and its close relatives. DES-TOMATO performs the critical task of rapidly and comprehensively sifting through more than 20 thousands topic-specific publications and extracting relevant knowledge, both established and possibly novel. The current release comprises mined text elements from 22,647 tomato-related articles, in which 52,886 statistically enriched terms from 26 relevant dictionaries were identified, together with 1,388,952 statistically enriched pairs of these terms.
DES-TOMATO has various tools that enable users to perform complex tasks including querying for enriched terms or pairs of terms, building and testing hypotheses based on transitive associations, identifying enriched KOBAS pathways based on list of genes and proteins identified in the KB corpus. Using the network viewer, results can be visualized and further developed by successively expanding upon terms of interest using selected dictionaries; thus, offering a highly flexible exploration experience. In addition, publications that substantiate enrichment of a term or an association are readily accessible to the user. DES-TOMATO exceeds other discovery platforms in plant sciences (such as SGN and HRGRN), through the use of a literature text-mining methodology that enables: 1) computational assignment of terms-to-publication associations (i.e. independent of gene identifiers); 2) very comprehensive coverage of information not easily or not at all available in other tomato-related databases; 3) straightforward and regular updates with new publications to ensure the KB remains current and relevant.
DES-TOMATO is a unique information/knowledge exploration system in plant sciences. It was built to explore and generate useful information using a broad set of topic-related dictionaries that provide the user the flexibility to examine various questions. DES-TOMATO also provides a user-friendly interface, and an extensive instructional material to facilitate the navigation through the KB. Altogether, we hope that DES-TOMATO will be a useful tool for supporting tomato-related research questions112.
Electronic supplementary material
Acknowledgements
The computational analysis for this study was performed on Dragon and Snapdragon compute clusters of the Computational Bioscience Research Center (CBRC) at King Abdullah University of Science and Technology (KAUST). This work has been supported by the King Abdullah University of Science and Technology (KAUST) Office of Sponsored Research (OSR) under Awards No URF/1/2302 and No URF/1/1976-02, and KAUST Base Research Fund (BAS/1/1606-01-01) to VBB.
Author Contributions
A.S., S.N. and M.E. contributed equally to this work. V.B.B. and M.T. conceived the study; V.B.B., M.E. and S.N. designed the study; A.S. conducted the main technical development; A.R., M.K. and B.M. worked on some aspects of technical implementation; M.E., S.N., S.B., R.R. and M.J.L.M. updated dictionaries; M.E., S.N., S.B. and M.J.L.M. developed the examples; V.B.B., A.S., M.E., S.N., S.B., R.R., M.J.L.M, M.T. and R.H. contributed to writing the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Adil Salhi, Sónia Negrão and Magbubah Essack contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-05448-0
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bai YL, Lindhout P. Domestication and breeding of tomatoes: What have we gained and what can we gain in the future? Annals of Botany. 2007;100:1085–1094. doi: 10.1093/aob/mcm150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rick, C. M. & Chetelat, R. T. Utilization of related wild species for tomato improvement. Acta Horticulturae, 21–38 (1995).
- 3.Peralta, I. E., Spooner, D. M. & Knapp, S. Taxonomy of tomatoes: a revision of wild tomatoes (Solanum section Lycopersicon) and their outgroup relatives in sections Juglandifolia and Lycopersicoides. Systematic Botany Monographs84 (2008).
- 4.Spooner DM, Peralta IE, Knapp S. Comparison of AFLPs with other markers for phylogenetic inference in wild tomatoes [Solanum L. section Lycopersicon (Mill.) Wettst.] Taxon. 2005;54:43–61. doi: 10.2307/25065301. [DOI] [Google Scholar]
- 5.Tomato Genome Sequencing C, et al. Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant Journal. 2014;80:136–148. doi: 10.1111/tpj.12616. [DOI] [PubMed] [Google Scholar]
- 6.Foolad MR. Genome mapping and molecular breeding of tomato. International Journal of Plant Genomics. 2007;2007:ID64358. doi: 10.1155/2007/64358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimura S, Sinha N. Tomato (Solanum lycopersicum): a model fruit-bearing crop. CSH Protocols. 2008;3:1–9. doi: 10.1101/pdb.emo105. [DOI] [PubMed] [Google Scholar]
- 8.Meissner R, et al. A new model system for tomato genetics. The Plant Journal. 1997;12:1465–1472. doi: 10.1046/j.1365-313x.1997.12061465.x. [DOI] [Google Scholar]
- 9.Consortium TTG. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamilton JP, et al. Single nucleotide polymorphism discovery in cultivated tomato via sequencing by synthesis. The Plant Genome. 2012;5:17–29. doi: 10.3835/plantgenome2011.12.0033. [DOI] [Google Scholar]
- 11.Sim S-C, et al. Development of a large SNP genotyping array and generation of high-density genetic maps in tomato. PLoS One. 2012;7:e40563. doi: 10.1371/journal.pone.0040563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sim S-C, et al. High-density SNP genotyping of tomato (Solanum lycopersicum L.) reveals patterns of genetic variation due to breeding. PloS One. 2012;7:e45520. doi: 10.1371/journal.pone.0045520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin T, et al. Genomic analyses provide insights into the history of tomato breeding. Nature Genetics. 2014;46:1220–1226. doi: 10.1038/ng.3117. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi M, et al. Genome-wide analysis of intraspecific DNA polymorphism in ‘Micro-Tom’, a model cultivar of tomato (Solanum lycopersicum) Plant and Cell Physiology. 2013;55:445–454. doi: 10.1093/pcp/pct181. [DOI] [PubMed] [Google Scholar]
- 15.Shikata M, et al. TOMATOMA update: phenotypic and metabolite information in the Micro-Tom mutant resource. Plant and Cell Physiology. 2016;57:e11–e11. doi: 10.1093/pcp/pcv194. [DOI] [PubMed] [Google Scholar]
- 16.Ohyanagi H, et al. Plant Omics Data Center: an integrated web repository for interspecies gene expression networks with NLP-based curation. Plant and Cell Physiology. 2014;56:e9. doi: 10.1093/pcp/pcu188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuartero J, Fernández-Muñoz R. Tomato and salinity. Scientia Horticulturae. 1999;78:83–125. doi: 10.1016/S0304-4238(98)00191-5. [DOI] [Google Scholar]
- 18.Sabehat A, Weiss D, Lurie S. The correlation between heat-shock protein accumulation and persistence and chilling tolerance in tomato fruit. Plant Physiol. 1996;110:531–537. doi: 10.1104/pp.110.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arie T, Takahashi H, Kodama M, Teraoka T. Tomato as a model plant for plant-pathogen interactions. Plant Biotechnology. 2007;24:135–147. doi: 10.5511/plantbiotechnology.24.135. [DOI] [Google Scholar]
- 20.Li Z, et al. Genome-wide Identification and analysis of the MYB transcription factor superfamily in Solanum lycopersicum. Plant and Cell Physiology. 2016;57:1657–1677. doi: 10.1093/pcp/pcw091. [DOI] [PubMed] [Google Scholar]
- 21.Thagun C, et al. Jasmonate-responsive ERF transcription factors regulate steroidal glycoalkaloid biosynthesis in tomato. Plant and Cell Physiology. 2016;57:961–975. doi: 10.1093/pcp/pcw067. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda H, et al. Dynamic metabolic regulation by a chromosome segment from a wild relative during fruit development in a tomato introgression line, IL8-3. Plant and Cell Physiology. 2016;57:1257–1270. doi: 10.1093/pcp/pcw075. [DOI] [PubMed] [Google Scholar]
- 23.Takayama M, et al. Tomato glutamate decarboxylase genes SlGAD2 and SlGAD3 play key roles in regulating gamma-aminobutyric acid Levels in tomato (Solanum lycopersicum) Plant and Cell Physiology. 2015;56:1533–1545. doi: 10.1093/pcp/pcv075. [DOI] [PubMed] [Google Scholar]
- 24.Pujar A, et al. From manual curation to visualization of gene families and networks across Solanaceae plant species. Database. 2013;2013:bat028. doi: 10.1093/database/bat028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawe AS, et al. DESTAF: a database of text-mined associations for reproductive toxins potentially affecting human fertility. Reproductive Toxicology. 2012;33:99–105. doi: 10.1016/j.reprotox.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 26.Essack M, Radovanovic A, Bajic VB. Information exploration system for sickle cell disease and repurposing of hydroxyfasudil. PLoS One. 2013;8:e65190. doi: 10.1371/journal.pone.0065190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Essack M, et al. DDEC: Dragon database of genes implicated in esophageal cancer. BMC Cancer. 2009;9:219. doi: 10.1186/1471-2407-9-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaur M, et al. Database for exploration of functional context of genes implicated in ovarian cancer. Nucleic Acids Research. 2009;37:D820–823. doi: 10.1093/nar/gkn593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwofie SK, et al. Dragon exploratory system on hepatitis C virus (DESHCV) Infection, Genetics and Evolution. 2011;11:734–739. doi: 10.1016/j.meegid.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Kwofie SK, Schaefer U, Sundararajan VS, Bajic VB, Christoffels A. HCVpro: hepatitis C virus protein interaction database. Infection, Genetics and Evolution. 2011;11:1971–1977. doi: 10.1016/j.meegid.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Maqungo M, et al. DDPC: Dragon Database of Genes associated with Prostate Cancer. Nucleic Acids Research. 2011;39:D980–985. doi: 10.1093/nar/gkq849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sagar S, et al. DDESC: Dragon database for exploration of sodium channels in human. BMC genomics. 2008;9:622. doi: 10.1186/1471-2164-9-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sagar S, Kaur M, Radovanovic A, Bajic VB. Dragon exploration system on marine sponge compounds interactions. Journal of cheminformatics. 2013;5:11. doi: 10.1186/1758-2946-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salhi A, et al. DESM: portal for microbial knowledge exploration systems. Nucleic Acids Research. 2016;44:D624–633. doi: 10.1093/nar/gkv1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bajic VB, et al. Dragon Plant Biology Explorer. A text-mining tool for integrating associations between genetic and biochemical entities with genome annotation and biochemical terms lists. Plant Physiol. 2005;138:1914–1925. doi: 10.1104/pp.105.060863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chowdhary R, et al. PIMiner: a web tool for extraction of Protein Interactions from Biomedical Literature. International journal of data mining and bioinformatics. 2013;7:450–462. doi: 10.1504/IJDMB.2013.054232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chowdhary R, et al. Context-specific protein network miner–an online system for exploring context-specific protein interaction networks from the literature. PLoS One. 2012;7:e34480. doi: 10.1371/journal.pone.0034480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan H, et al. Dragon TF Association Miner: a system for exploring transcription factor associations through text-mining. Nucleic acids research. 2004;32:W230–W234. doi: 10.1093/nar/gkh484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raies AB, Mansour H, Incitti R, Bajic VB. Combining position weight matrices and document-term matrix for efficient extraction of associations of methylated genes and diseases from free text. PloS one. 2013;8:e77848. doi: 10.1371/journal.pone.0077848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah PK, Perez-Iratxeta C, Bork P, Andrade MA. Information extraction from full text scientific articles: where are the keywords? BMC bioinformatics. 2003;4:20. doi: 10.1186/1471-2105-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuemie MJ, et al. Distribution of information in biomedical abstracts and full-text publications. Bioinformatics (Oxford, England) 2004;20:2597–2604. doi: 10.1093/bioinformatics/bth291. [DOI] [PubMed] [Google Scholar]
- 42.Van Landeghem S, De Bodt S, Drebert ZJ, Inzé D, Van de Peer Y. The potential of text mining in data integration and network biology for plant research: a case study on Arabidopsis. The Plant Cell. 2013;25:794–807. doi: 10.1105/tpc.112.108753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hassani-Pak K, et al. Enhancing data integration with text analysis to find proteins implicated in plant stress response. Journal of Integrative Bioinformatics. 2010;7:121. doi: 10.2390/biecoll-jib-2010-121. [DOI] [PubMed] [Google Scholar]
- 44.Turenne, N., Andro, M., Corbière, R. & Phan, T. T. Open data platform for knowledge access in plant health domain: VESPA Mining. arXiv preprint arXiv:1504.06077 (2015).
- 45.Dai X, Li J, Liu T, Zhao PX. HRGRN: a graph search-empowered integrative database of Arabidopsis signaling transduction, metabolism and gene regulation networks. Plant and Cell Physiology. 2016;57:e12–e12. doi: 10.1093/pcp/pcv200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salhi, A. et al. DES-ncRNA: A knowledgebase for exploring information about human micro and long noncoding RNAs based on literature-mining. RNA biology, 00–00 (2017). [DOI] [PMC free article] [PubMed]
- 47.Hastings J, et al. The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic acids research. 2013;41:D456–D463. doi: 10.1093/nar/gks1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Research. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wheeler DL, et al. Database resources of the National Center for Biotechnology. Nucleic Acids Research. 2003;31:28–33. doi: 10.1093/nar/gkg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bombarely A, et al. The Sol Genomics Network (solgenomics. net): growing tomatoes using Perl. Nucleic acids research. 2011;39:D1149–D1155. doi: 10.1093/nar/gkq866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajaraman, K. et al. In Information Processing and Living Systems 687–694 (World Scientific, 2005).
- 52.Xie C, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Research. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harispe S, Ranwez S, Janaqi S, Montmain J. The semantic measures library and toolkit: fast computation of semantic similarity and relatedness using biomedical ontologies. Bioinformatics (Oxford, England) 2014;30:740–742. doi: 10.1093/bioinformatics/btt581. [DOI] [PubMed] [Google Scholar]
- 54.Du, Z., Zhou, X., Ling, Y., Zhang, Z. & Su, Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic acids research, gkq310 (2010). [DOI] [PMC free article] [PubMed]
- 55.Pedley KF, Martin GB. Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annual Review of Phytopathology. 2003;41:215–243. doi: 10.1146/annurev.phyto.41.121602.143032. [DOI] [PubMed] [Google Scholar]
- 56.Thapa SP, Miyao EM, Davis RM, Coaker G. Identification of QTLs controlling resistance to Pseudomonas syringae pv. tomato race 1 strains from the wild tomato, Solanum habrochaites LA1777. Theoretical and Applied Genetics. 2015;128:681–692. doi: 10.1007/s00122-015-2463-7. [DOI] [PubMed] [Google Scholar]
- 57.Zhou J, Tang X, Martin GB. The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis‐element of pathogenesis‐related genes. The EMBO Journal. 1997;16:3207–3218. doi: 10.1093/emboj/16.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Devarenne TP, Ekengren SK, Pedley KF, Martin GB. Adi3 is a Pdk1‐interacting AGC kinase that negatively regulates plant cell death. The EMBO journal. 2006;25:255–265. doi: 10.1038/sj.emboj.7600910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Avila J, et al. The β-subunit of the SnRK1 complex is phosphorylated by the plant cell death suppressor Adi3. Plant Physiol. 2012;159:1277–1290. doi: 10.1104/pp.112.198432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Z-Y, et al. A novel role for Arabidopsis CBL1 in affecting plant responses to glucose and gibberellin during germination and seedling development. PloS One. 2013;8:e56412. doi: 10.1371/journal.pone.0056412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Devarenne TP, Martin GB. Manipulation of plant programmed cell death pathways during plant-pathogen interactions. Plant Signaling and Behavior. 2007;2:188–190. doi: 10.4161/psb.2.3.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ek-Ramos MJ, et al. The tomato cell death suppressor Adi3 is restricted to the endosomal system in response to the Pseudomonas syringae effector protein AvrPto. PLoS One. 2014;9:e110807. doi: 10.1371/journal.pone.0110807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Withers J, Dong X. Posttranslational modifications of NPR1: a single protein playing multiple roles in plant immunity and physiology. PLoS Pathogens. 2016;12:e1005707. doi: 10.1371/journal.ppat.1005707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Torres Zabala M, et al. Novel JAZ co‐operativity and unexpected JA dynamics underpin Arabidopsis defence responses to Pseudomonas syringae infection. New Phytologist. 2016;209:1120–1134. doi: 10.1111/nph.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Geng X, Cheng J, Gangadharan A, Mackey D. The coronatine toxin of Pseudomonas syringae is a multifunctional suppressor of Arabidopsis defense. The Plant Cell. 2012;24:4763–4774. doi: 10.1105/tpc.112.105312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Geng X, Jin L, Shimada M, Kim MG, Mackey D. The phytotoxin coronatine is a multifunctional component of the virulence armament of Pseudomonas syringae. Planta. 2014;240:1149–1165. doi: 10.1007/s00425-014-2151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/S0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 68.Ntoukakis V, Saur IM, Conlan B, Rathjen JP. The changing of the guard: the Pto/Prf receptor complex of tomato and pathogen recognition. Current Opinion in Plant Biology. 2014;20:69–74. doi: 10.1016/j.pbi.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 69.Narusaka M, et al. Leucine zipper motif in RRS1 is crucial for the regulation of Arabidopsis dual resistance protein complex RPS4/RRS1. Scientific Reports. 2016;6:18702. doi: 10.1038/srep18702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meldau S, Baldwin IT, Wu J. For security and stability: SGT1 in plant defense and development. Plant Signaling and Behavior. 2011;6:1479–1482. doi: 10.4161/psb.6.10.17708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tester M, Davenport R. Na+ tolerance and Na+ transport in higher plants. Annals of Botany. 2003;91:503–527. doi: 10.1093/aob/mcg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Serrano R. Structure and function of plasma membrane ATPase. Annual Review of Plant Biology. 1989;40:61–94. doi: 10.1146/annurev.pp.40.060189.000425. [DOI] [Google Scholar]
- 73.Golldack D, Dietz K-J. Salt-induced expression of the vacuolar H+-ATPase in the common ice plant is developmentally controlled and tissue specific. Plant Physiol. 2001;125:1643–1654. doi: 10.1104/pp.125.4.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Niu X, Narasimhan ML, Salzman RA, Bressan RA, Hasegawa PM. NaCl regulation of plasma membrane H+-ATPase gene expression in a glycophyte and a halophyte. Plant Physiol. 1993;103:713–718. doi: 10.1104/pp.103.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hahn A, Bublak D, Schleiff E, Scharf KD. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. The Plant Cell. 2011;23:741–755. doi: 10.1105/tpc.110.076018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Palmgren MG. Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annual Review of Plant Physiology and Plant Molecular Biology. 2001;52:817–845. doi: 10.1146/annurev.arplant.52.1.817. [DOI] [PubMed] [Google Scholar]
- 77.Olias R, et al. The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell and Environment. 2009;32:904–916. doi: 10.1111/j.1365-3040.2009.01971.x. [DOI] [PubMed] [Google Scholar]
- 78.Ewing NN, Bennett AB. Assessment of the number and expression of P-type H+-ATPase genes in tomato. Plant Physiol. 1994;106:547–557. doi: 10.1104/pp.106.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boratyn GM, et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Research. 2013;41:W29–33. doi: 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Munns R. Genes and salt tolerance: bringing them together. New Phytologist. 2005;167:645–663. doi: 10.1111/j.1469-8137.2005.01487.x. [DOI] [PubMed] [Google Scholar]
- 81.Vialaret J, et al. Phosphorylation dynamics of membrane proteins from Arabidopsis roots submitted to salt stress. PROTEOMICS. 2014;14:1058–1070. doi: 10.1002/pmic.201300443. [DOI] [PubMed] [Google Scholar]
- 82.Li C-L, Wang M, Ma X-Y, Zhang W. NRGA1, a putative mitochondrial pyruvate carrier, mediates ABA regulation of guard cell ion channels and drought stress responses in Arabidopsis. Molecular Plant. 2014;7:1508–1521. doi: 10.1093/mp/ssu061. [DOI] [PubMed] [Google Scholar]
- 83.Katz YS, Galili G, Amir R. Regulatory role of cystathionine-γ-synthase and de novo synthesis of methionine in ethylene production during tomato fruit ripening. Plant Molecular Biology. 2006;61:255–268. doi: 10.1007/s11103-006-0009-8. [DOI] [PubMed] [Google Scholar]
- 84.Giuliano G, Bartley GE, Scolnik PA. Regulation of carotenoid biosynthesis during tomato development. The Plant Cell. 1993;5:379–387. doi: 10.1105/tpc.5.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Montoya T, et al. Patterns of Dwarf expression and brassinosteroid accumulation in tomato reveal the importance of brassinosteroid synthesis during fruit development. The Plant Journal. 2005;42:262–269. doi: 10.1111/j.1365-313X.2005.02376.x. [DOI] [PubMed] [Google Scholar]
- 86.Ghanem ME, et al. Root-synthesized cytokinins improve shoot growth and fruit yield in salinized tomato (Solanum lycopersicum L.) plants. Journal of Experimental Botany. 2011;62:125–140. doi: 10.1093/jxb/erq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang KL-C, Li H, Ecker JR. Ethylene biosynthesis and signaling networks. The Plant Cell. 2002;14:S131–S151. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kudoyarova GR, Vysotskaya LB, Cherkozyanova A, Dodd IC. Effect of partial rootzone drying on the concentration of zeatin-type cytokinins in tomato (Solanum lycopersicum L.) xylem sap and leaves. Journal of Experimental Botany. 2007;58:161–168. doi: 10.1093/jxb/erl116. [DOI] [PubMed] [Google Scholar]
- 89.Matsuo, S., Kikuchi, K., Fukuda, M., Honda, I. & Imanishi, S. Roles and regulation of cytokinins in tomato fruit development. Journal of Experimental Botany (2012). [DOI] [PMC free article] [PubMed]
- 90.Ronen G, Cohen M, Zamir D, Hirschberg J. Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon‐cyclase is down‐regulated during ripening and is elevated in the mutantDelta. The Plant Journal. 1999;17:341–351. doi: 10.1046/j.1365-313X.1999.00381.x. [DOI] [PubMed] [Google Scholar]
- 91.Fraser PD, Truesdale MR, Bird CR, Schuch W, Bramley PM. Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression) Plant Physiol. 1994;105:405–413. doi: 10.1104/pp.105.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bramley PM. Regulation of carotenoid formation during tomato fruit ripening and development. Journal of Experimental Botany. 2002;53:2107–2113. doi: 10.1093/jxb/erf059. [DOI] [PubMed] [Google Scholar]
- 93.Shimada Y, et al. Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 2001;126:770–779. doi: 10.1104/pp.126.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou J, et al. H2O2 mediates the crosstalk of brassinosteroid and abscisic acid in tomato responses to heat and oxidative stresses. Journal of Experimental Botany. 2014;65:4371–4383. doi: 10.1093/jxb/eru217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bishop GJ, et al. The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proceedings of the National Academy of Sciences. 1999;96:1761–1766. doi: 10.1073/pnas.96.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lisso J, Altmann T, Müssig C. Metabolic changes in fruits of the tomato dx mutant. Phytochemistry. 2006;67:2232–2238. doi: 10.1016/j.phytochem.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 97.Srivastava A, Handa AK. Hormonal regulation of tomato fruit development: a molecular perspective. Journal of Plant Growth Regulation. 2005;24:67–82. doi: 10.1007/s00344-005-0015-0. [DOI] [Google Scholar]
- 98.Alexander L, Grierson D. Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. Journal of Experimental Botany. 2002;53:2039–2055. doi: 10.1093/jxb/erf072. [DOI] [PubMed] [Google Scholar]
- 99.Ramesh, S. A. et al. GABA signalling modulates plant growth by directly regulating the activity of plant-specific anion transporters. Nature Communications6 (2015). [DOI] [PMC free article] [PubMed]
- 100.Akihiro T, et al. Biochemical mechanism on GABA accumulation during fruit development in tomato. Plant and Cell Physiology. 2008;49:1378–1389. doi: 10.1093/pcp/pcn113. [DOI] [PubMed] [Google Scholar]
- 101.Takayama, M. & Ezura, H. How and why does tomato accumulate a large amount of GABA in the fruit? Frontiers in Plant Science6 (2015). [DOI] [PMC free article] [PubMed]
- 102.Pan, H. et al. In Discovering Biomolecular Mechanisms with Computational Biology 57–73 (Springer, 2006).
- 103.Smalheiser NR, Swanson DR. Indomethacin and Alzheimer’s disease. Neurology. 1996;46:583–583. doi: 10.1212/WNL.46.2.583. [DOI] [PubMed] [Google Scholar]
- 104.Dvir E, et al. DP‐155, a Lecithin Derivative of Indomethacin, is a Novel Nonsteroidal Antiinflammatory Drug for Analgesia and Alzheimer’s Disease Therapy. CNS drug reviews. 2007;13:260–277. doi: 10.1111/j.1527-3458.2007.00014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wren JD, Bekeredjian R, Stewart JA, Shohet RV, Garner HR. Knowledge discovery by automated identification and ranking of implicit relationships. Bioinformatics (Oxford, England) 2004;20:389–398. doi: 10.1093/bioinformatics/btg421. [DOI] [PubMed] [Google Scholar]
- 106.Natarajan J, et al. Text mining of full-text journal articles combined with gene expression analysis reveals a relationship between sphingosine-1-phosphate and invasiveness of a glioblastoma cell line. BMC bioinformatics. 2006;7:373. doi: 10.1186/1471-2105-7-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bryan L, et al. Sphingosine-1-phosphate and interleukin-1 independently regulate plasminogen activator inhibitor-1 and urokinase-type plasminogen activator receptor expression in glioblastoma cells: implications for invasiveness. Molecular Cancer Research. 2008;6:1469–1477. doi: 10.1158/1541-7786.MCR-08-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tiffin N, et al. Integration of text-and data-mining using ontologies successfully selects disease gene candidates. Nucleic acids research. 2005;33:1544–1552. doi: 10.1093/nar/gki296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Raies, A. B., Mansour, H., Incitti, R. & Bajic, V. B. DDMGD: the database of text-mined associations between genes methylated in diseases from different species. Nucleic acids research, gku1168 (2014). [DOI] [PMC free article] [PubMed]
- 110.Gonzalez GH, Tahsin T, Goodale BC, Greene AC, Greene CS. Recent advances and emerging applications in text and data mining for biomedical discovery. Briefings in bioinformatics. 2016;17:33–42. doi: 10.1093/bib/bbv087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sangkuhl K, Berlin DS, Altman RB, Klein TE. PharmGKB: understanding the effects of individual genetic variants. Drug metabolism reviews. 2008;40:539–551. doi: 10.1080/03602530802413338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Leser U, Hakenberg J. What makes a gene name? Named entity recognition in the biomedical literature. Briefings in bioinformatics. 2005;6:357–369. doi: 10.1093/bib/6.4.357. [DOI] [PubMed] [Google Scholar]
- 113.Kale NS, et al. MetaboLights: an open-access database repository for metabolomics data. Current protocols in bioinformatics/editoral board, Andreas D. Baxevanis… [et al.] 2016;53:14.13.11–14.13.18. doi: 10.1002/0471250953.bi1413s53. [DOI] [PubMed] [Google Scholar]
- 114.Fleischmann A, et al. IntEnz, the integrated relational enzyme database. Nucleic Acids Research. 2004;32:D434–437. doi: 10.1093/nar/gkh119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wishart D, et al. T3DB: the toxic exposome database. Nucleic Acids Research. 2015;43:D928–934. doi: 10.1093/nar/gku1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Alam I, et al. INDIGO - INtegrated data warehouse of microbial genomes with examples from the red sea extremophiles. PLoS One. 2013;8:e82210. doi: 10.1371/journal.pone.0082210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bairoch A. The ENZYME database in 2000. Nucleic Acids Research. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Consortium GO. Gene ontology consortium: going forward. Nucleic Acids Research. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kanehisa, M. In Data Mining for Systems Biology: Methods and Protocols (eds Hiroshi Mamitsuka, Charles DeLisi, & Minoru Kanehisa) 263–275 (Humana Press, 2013).
- 120.Croft D, et al. The Reactome pathway knowledgebase. Nucleic Acids Research. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mi H, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Research. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Morgat A, et al. UniPathway: a resource for the exploration and annotation of metabolic pathways. Nucleic Acids Research. 2011;40:D761–D769. doi: 10.1093/nar/gkr1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Federhen S. The NCBI Taxonomy database. Nucleic Acids Research. 2012;40:D136–143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tahir, H. A. et al. Plant growth promotion by volatile organic compounds produced by Bacillus subtilis SYST2. Frontiers in Microbiology8 (2017). [DOI] [PMC free article] [PubMed]
- 125.Chen L, et al. TCS1, a Microtubule-Binding Protein, Interacts with KCBP/ZWICHEL to Regulate Trichome Cell Shape in Arabidopsis thaliana. PLoS Genet. 2016;12:e1006266. doi: 10.1371/journal.pgen.1006266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hoeberichts FA, Woltering EJ. Multiple mediators of plant programmed cell death: interplay of conserved cell death mechanisms and plant‐specific regulators. Bioessays. 2003;25:47–57. doi: 10.1002/bies.10175. [DOI] [PubMed] [Google Scholar]
- 127.Cooper L, et al. The plant ontology as a tool for comparative plant anatomy and genomic analyses. Plant and Cell Physiology. 2013;54:e1. doi: 10.1093/pcp/pcs163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Walls RL, et al. Ontologies as integrative tools for plant science. American Journal of Botany. 2012;99:1263–1275. doi: 10.3732/ajb.1200222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hoehndorf, R. et al. The Flora Phenotype Ontology (FLOPO): tool for integrating morphological traits and phenotypes of vascular plants. Journal of Biomedical Semantics, Accepted for publication (2016). [DOI] [PMC free article] [PubMed]
- 130.Menda N, Buels RM, Tecle I, Mueller LA. A community-based annotation framework for linking solanaceae genomes with phenomes. Plant Physiol. 2008;147:1788–1799. doi: 10.1104/pp.108.119560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


