Abstract
After a forest wildfire, the microbial communities have a transient alteration in their composition. The role of the soil microbial community in the recovery of an ecosystem following such an event remains poorly understood. Thus, it is necessary to understand the plant-microbe interactions that occur in burned soils. By high-throughput sequencing, we identified the main bacterial taxa of burnt holm-oak rhizosphere, then we obtained an isolate collection of the most abundant genus and its growth promoting activities were characterised. 16S rRNA amplicon sequencing showed that the genus Arthrobacter comprised more than 21% of the total community. 55 Arthrobacter strains were isolated and characterized using RAPDs and sequencing of the almost complete 16S rRNA gene. Our results indicate that isolated Arthrobacter strains present a very high genetic diversity, and they could play an important ecological role in interaction with the host plant by enhancing aerial growth. Most of the selected strains exhibited a great ability to degrade organic polymers in vitro as well as possibly presenting a direct mechanism for plant growth promotion. All the above data suggests that Arthrobacter can be considered as an excellent PGP rhizobacterium that may play an important role in the recovery of burned holm-oak forests.
Introduction
Wildfires are a recurring threat to the vegetation of the Mediterranean Basin. Moreover, the rural desertion since the 1960s1 along with climate change2 are causing an increase in forest fires. The effect of wildfires3, 4, the recovery of vegetation5, 6, the dynamic of microbial communities7–9 and nutrient cycling10, 11 after forest fires have been given considerable attention. Although understory vegetation could be recovered in a period of time of 20–100 years following a forest fire, microbial communities could recover much faster9, 12. However, microorganisms at the recovery stage have never been isolated and characterized on their plant interaction activities.
Microbial communities have a transient alteration in its composition with predominance of spore-forming and Gram-positive microorganisms after a wildfire7, 8, 10–12. In this sense, phylum Actinobacteria is a unique group of soil microorganisms that now we know are closely associated with plants and they have been isolated from different parts of plants of diverse genera13. Several Actinobacteria isolates have been reported to promote plant growth14–16. Another important trait, the ability to fix atmospheric nitrogen, has been reported in Frankia and other Actinobacteria genera: Arthrobacter 17, Streptomyces 18 and Propionibacterium 19. Arthrobacter has also been found to degrade a wide variety of compounds, including aromatic molecules, organochloride, pesticides, etc.20. In addition, several species of this genus are desiccation tolerant21, 22. Recently, it has been described how Actinobacteria genera such as Blastoccocus and Arthrobacter have a main role in the soil nitrogen cycle after a wildfire11.
Understanding the main role of the transient dominant microbial population, the new plant-microbe interactions in post-fire conditions as well as the ecological role of these microorganisms is likely to allow a new use as helpers in the recovery of burnt soils and ecosystem. Thus, it is necessary to carry out an adequate phylogenetic analysis based on deep-sequencing of 16S rRNA amplicons from the total prokaryotic communities to reveal the dominant microbial population to isolate them; furthermore “in vitro” and “in planta” evaluation will allow the correct selection of the microbial strains to be used as plant growth promoting (PGP) rhizobacteria, in order to obtain the desired effect of revegetation of burned soils. For this objective, we use, for the first time, a strategy based on the use of high-throughput technologies for the identification of the key microorganisms found in the tree rhizosphere three years after a wildfire, when the holm-oaks were naturally re-growing. This identification was followed by the isolation of the most abundant genera and their characterization as PGP rhizobacteria to select single isolates that could be used as bacterial inoculants to promote forest recovery after a wildfire.
Results
Effects of fire on soil physicochemical properties
Both the control and burned soil samples belong to haplic phaeozem type, classified as loam with regard to their texture (Table S1). Due to the wildfire, there was an increase in soil pH, but in contrast, there was a decrease in soil organic matter and iron concentration. No other shifts due to fire were observed in the physicochemical characteristics of the soil samples assessed (Table S1).
Bacterial community composition
Firstly, a total of 62,072 raw reads from BOF (Burned Oak Forest) and 56,620 raw reads from UOF (Undisturbed Oak Forest) were obtained. After the trimming, a total of 22,370 and 14,394 quality sequences were extracted from BOF and UOF, respectively. The sequences were normalized to 4,631 sequences per sample, which was the minimum number at UOF1 sample, thus yielding 13,893 sequences at each rhizospheric situation (Table S2). We were able to classify 92.76% of these sequences. The dominant phyla (or class in the case of Proteobacteria) in both samples were Actinobacteria, Alphaproteobacteria, Betaproteobacteria, Acidobacteria, Bacteroidetes, Gammaproteobacteria and Planctomycetes, accounting for more than 83% of the bacterial sequences from each of the soils (Fig. 1). In addition, Gemmatimonadetes, Verrucomicrobia and Deltaproteobacteria were present in both soils but at relatively low proportion, and other rare phyla were identified (Table S3).
Figure 1.
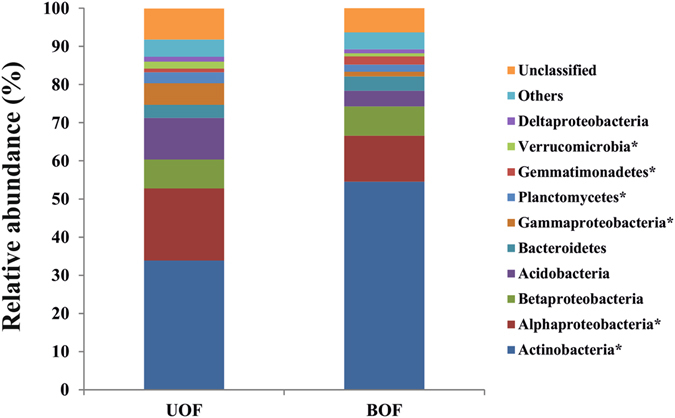
Relative abundances of the dominant bacterial phyla in soils. Relative (%) abundances are based on the proportional frequencies of 16S rRNA gene sequences that could be classified at the phylum level. Asterisks represent a statistically significant difference between UOF and BOF, measured by Fisher’s exact test (p < 0.05). UOF: Undisturbed Oak Forest; BOF: Burned Oak Forest.
Fire significantly (p < 0.05) shifted the relative abundance of some dominant phyla (Fig. 1). BOF has more relative abundance of Actinobacteria and Gemmatimonadetes meanwhile UOF is over-represented in Alphaproteobacteria, Gammaproteobacteria, Planctomycetes and Verrucomicrobia the greatest observed differences being in the Gram-positive phyla (Fig. 1). Furthermore, the comparison of the two communities at genus level showed statistically significant differences (p < 0.05) in 26 genera. However, only 10 genera showed biologically relevant differences (Fig. 2). The genera with a significant increase in the BOF rhizosphere were: Arthrobacter and Blastococcus (phylum Actinobacteria), Adhaeribacter (phylum Bacteroidetes), Gemmatimonas (phylum Gemmatimonadetes) and Microvirga (class Alphaproteobacteria). However, other genera from the same phyla were significantly more abundant in the UOF rhizosphere, like: Solirubrobacter, Mycobacterium and Nakamurella (phylum Actinobacteria), Spartobacteria (phylum Verrucomicrobia) and Rhizomicrobium (class Alphaproteobacteria).
Figure 2.
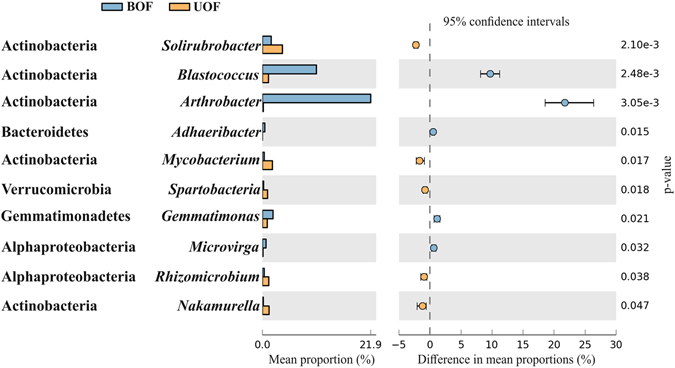
Genera that exhibited significant and biologically relevant changes in abundance. Significant differences were calculated using White’s non-parametric t-test at a 95% confidence interval. Biologic relevance was determined choosing genera with a difference between proportions >0.5 and a ratio of proportions >2. UOF: Undisturbed oak forest; BOF: Burned oak forest.
Bacterial diversity
In terms of both phylotype (i.e. number of OTUs, Operational Taxonomic Units) and phylogenetic diversity (Table S2), which were analysed at a depth of 13,893 randomly selected sequences per sample, the diversity of bacterial communities was lower in BOF. Phylotype diversity measured with the Shannon index had statistically significant differences, being higher in UOF (6.05) than in BOF (5.07). Good’s coverage was lower in BOF (91.28%) than in UOF (92.50%, Table S2).
Strains isolation and RAPD fingerprinting
Since the genus Arthrobacter had the highest proportion (over 21% of total sequences) and showed a statistically significant difference between BOF and UOF (Fig. 2), we isolated single colonies from rhizospheric BOF soil samples. An average of 100 CFU (Colony-Forming Units) were obtained from 10−2 dilution petri dishes, and 231 CFU were selected growing in selective broth media. After filtered selection (see experimental procedures), 55 CFU from the genus Arthrobacter were selected.
High-resolution Random Amplification of Polymorphic DNA (RAPD) fingerprints were obtained for the isolated strains (Fig. S1). The amplified fragments ranged from 0.3 to 2.9 kb. We clustered our strains at a 60% similarity level, which defined 12 groups and revealed a high genetic diversity in our isolates. Fifty-one strains were distributed in the clusters containing 2–9 strains per cluster; the remaining 4 isolates had a unique profile. Moreover, no clones were found amongst the isolates.
Phylogenetic analysis of selected Arthrobacter strains
Near complete 16S rRNA gene sequences (1460 bp) were obtained for the 55 selected strains. Sequence similarities between the new isolates and currently described Arthrobacter species ranged from 98.99 to 99.93%. A significant number of the isolates sequenced (approx. 62%) showed a 16S rRNA sequence similarity of over 99.5% with the already described Arthrobacter species. However, none of them showed a 100% sequence similarity to any type strain in the databases (Table S4).
The inferred phylogenetic tree based on 16S rRNA gene sequences performed using maximum likelihood (Fig. 3) and neighbor-joining methods (Figure S2) showed several groups containing both type strains and our isolates. Other groups contained only sequences from our strains, which indicates the diversity of the isolates from this study compared to the Arthrobacter type strains used. A large number of our isolates are closely related to four Arthrobacter species: A. globiformis (20), A. siccitolerans (8), A. nitroguajacolicus (7) and A. oxydans (5). It should also be noted that the 20 strains related to A. globiformis belong to 10 different RAPD groups (Fig. S1) and the same applies to the other strains grouped close to the aforementioned Arthrobacter species (Table S4). The tree topology generated by both maximum likelihood and neighbor-joining methods was strongly supported by bootstrap values, which were similar for both methods (Fig. 3 and Fig. S2).
Figure 3.
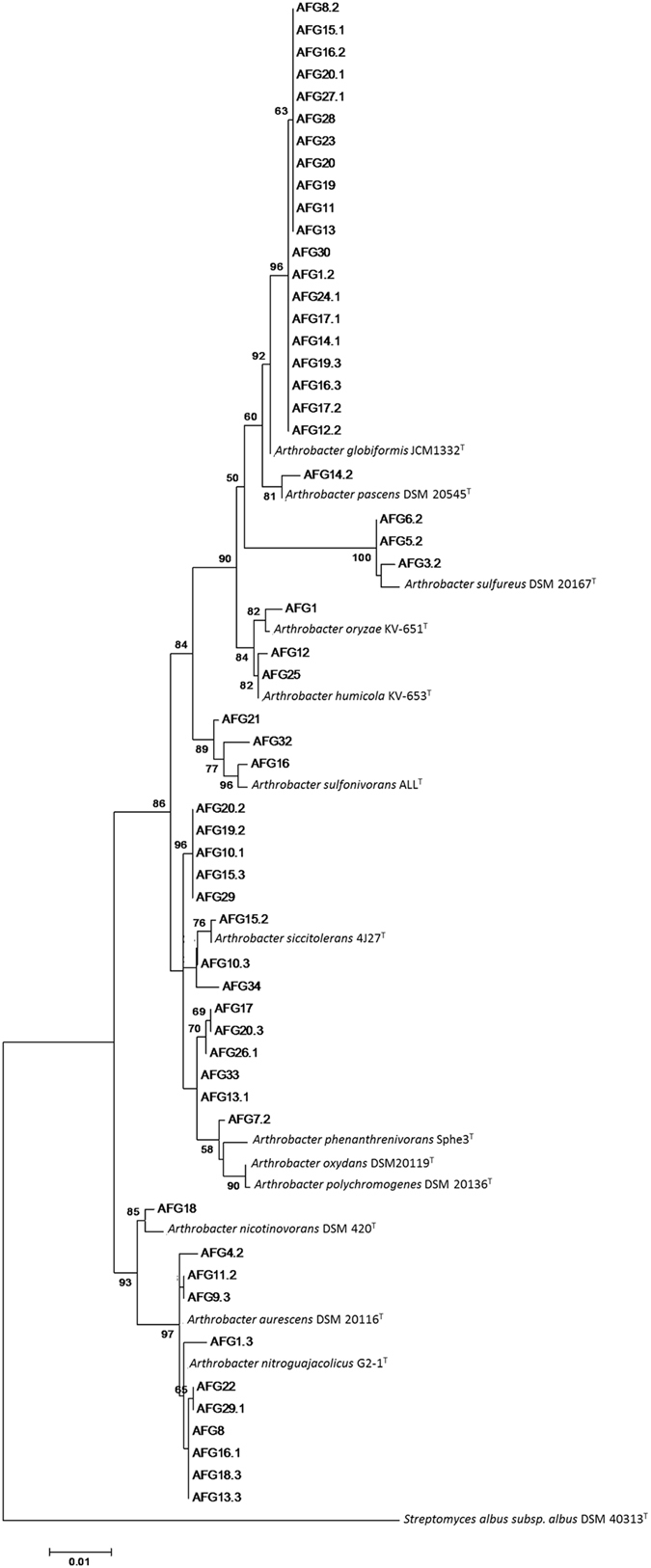
Maximum likelihood phylogenetic tree based on 16S rRNA gene sequences showing the relationship between the 55 isolated Arthrobacter strains and the closest recognized Arthrobacter species. Bar, 0.01 substitutions per nucleotide position. Bootstrap percentages (1000 replicates) above 50% are shown at nodes.
The 16S rRNA gene sequences from the 55 isolates were compared to the pyrotags clustered in the largest OTU from the BOF rhizosphere, all of them with similarity to the Arthrobacter 16S rRNA gene. The alignment of the V6-V8 regions from the sequences generated one OTU at 97%, but 46 OTUs at 100% similarity. These 46 OTUs were ordered from major to minor according to the number of pyrotags contained in them (Table 1). The first OTU had 991 pyrotags and it clustered 41 out of 55 sequences from the Arthrobacter isolates (Table 1). No isolates were obtained from the second OTU which clustered 468 pyrotags. Three isolates were clustered to the third OTU, which contained 14 pyrotags. The fourth OTU with 7 pyrotags was clustered with 11 isolates (Table 1).
Table 1.
OTUs clustered at 100% similarity belonging to the genus Arthrobacter. Clustering was performed with aligned sequences from 16S rRNA gene region obtained by both pyrosequencing and Sanger method of isolates from rhizospheric BOF soil samples.
| OTU | Number of Pyrotags Sequences | Number of Isolates | Species* |
|---|---|---|---|
| 1 | 991 | 41 | A. globiformis 20 A. oxydans 13 A. sulfonivorans 3 A. humicola 2 A. oryzae 1 A. pascens 1 A. scleromae 1 |
| 2 | 468 | 0 | — |
| 3 | 14 | 3 | A. sulfureus 3 |
| 4 | 7 | 11 | A. nitroguajacolicus 10 A. nicotinovorans 1 |
| 5 | 6 | 0 | — |
| 6 | 6 | 0 | — |
| 7 | 3 | 0 | — |
| 8 | 2 | 0 | — |
| 9 | 2 | 0 | — |
| 10 | 2 | 0 | — |
| 11 | 2 | 0 | — |
Asterisk indicates species most closely related to the isolates according to near complete 16S rRNA gene BLASTn results, identity ≥ 99%.
Effect of Arthrobacter strains isolated from BOF soil on plant growth
The role of the 55 Arthrobacter isolates as putative PGPR was tested in an inoculation experiment with Medicago sativa (alfalfa) and Capsicum annuum (pepper) plants because plant growth promotion (PGP) experiments on a hardwood like oak are impractical due to time limitations. The reasons for choosing these crops were two-fold. Firstly, we valued a fast confirmation of in vivo PGP capabilities of our strains, hence, the use of two crops with a fast growing rate. The second reason for choosing alfalfa and pepper is that these two species are commonly used in such PGP studies and thus are very well characterized. The results on plant growth showed that a large number of Arthrobacter strains clearly increased the shoot weight of alfalfa (36, Fig. S3a) or pepper (34, Fig. S3b) and only two of our isolates negatively affected the growth of pepper plants (Fig. S3b). After this preliminary experiment, the best five strains were selected for their ability to significantly increase the weight of the shoot of both inoculated plants (alfalfa and pepper) when compared with control (non-inoculated) plants. Using these five selected strains, a mesocosm experiment was conducted using only alfalfa as the plant of choice (Table 2). The inoculation with any of the five Arthrobacter strains selected produced a statistically (p < 0.05) significant higher shoot dry weight than the control treatment. The mean shoot dry weight of plants inoculated with Arthrobacter AFG15.2, AFG3.2, AFG16.1, AFG7.2 or AFG20 were, respectively, 53%, 44% 48% 46% and 43% greater than that of control plants (Table 2).
Table 2.
Growth parameters of alfalfa plants inoculated with selected Arthrobacter strains.
| Strain | Length shoot (cm) | Dry weight shoot (mg) | ||
|---|---|---|---|---|
| Mean | Std. error | Mean | Std. error | |
| Control | 34.75 | 1.75 | 279.83*** | 14.73 |
| AFG15.2 | 38.63 | 2.32 | 428.92*** | 24.24 |
| AFG3.2 | 37.58 | 1.66 | 404.33*** | 27.26 |
| AFG16.1 | 38.83 | 2.24 | 414.80*** | 28.71 |
| AFG7.2 | 39.33 | 2.24 | 408.42*** | 32.29 |
| AFG20 | 37.29 | 1.38 | 400.17** | 16.42 |
Means and standard error (N = 12) are shown. Within columns, treatment means showing significant differences with control plants are marked with asterisks according to Tukey tests at P ≤ 0.1 (*), P ≤ 0.05 (**) and P ≤ 0.01 (***). Uninoculated alfalfa plants were used as control.
Functional characterization of the selected Arthrobacter strains
All of the Arthrobacter strains selected showed the ability to grow using Galactose, Glucose and Mannose as the sole carbon source. Furthermore, most of them demonstrated the ability to degrade two or more plant cell wall components, namely cellulose, pectin and xylan (Table 3). Moreover, all of the tested strains showed lipase activity; with them being able to degrade Tween 80, Tween 20 or both. Additionally, all of the isolates showed the ability to produce cellulose with the exception of strains AFG15.2 and AFG7.2 all the Arthrobacter strains were able to produce and excrete siderophores. Three isolates (AFG15.2, AFG16.1 and AFG20) were able to solubilize phosphate and only two (AFG16.1 and AFG7.2) were able to grow in N-free medium. Finally, all selected strains were able to produce Indole Acetic Acid (IAA), with the strain AFG7.2 showed production levels at one order of magnitude higher than the rest of the strains (Table 3).
Table 3.
Ecological, plant growth promoting related enzymatic activities and indolacetic acid production in selected Arthrobacter strains.
| PPB traits | Cellulase | Pectinase | Xylanase | Tween 20 | Tween 80 | Cellulose production | Growtn in N free medium | C source Galactose | C source Glucose | C source Manose | C source Xylose | Phosphate solubilization | IAA production | Siderophore production |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arthrobacter strains | ||||||||||||||
| AFG 15.2 | − | + | + | − | + | + | − | + | + | + | + | + | 0.034 | − |
| AFG 3.2 | + | − | + | + | + | + | − | + | + | w | − | − | 0.034 | + |
| AFG 16.1 | − | + | + | + | + | + | + | + | + | + | + | + | 0.02 | + |
| AFG 7.2 | + | + | + | w | − | + | + | + | + | + | + | − | 0.308 | − |
| AFG 20 | − | + | + | − | + | + | − | + | + | + | + | + | 0.042 | + |
(+) positive, (−) negative and (w) weak. Indolacetic acid production is expressed in µg ml−1.
Discussion
Three years after wildfire, the physico-chemical analysis of UOF and BOF soils showed minimal differences in mineral composition which should not be relevant for the structure of the microbial community. The main differences between UOF and BOF soils are the increase in pH and the decrease in N and C due to the fire, which according to previous results11 are the most important soil factors that influenced the holm-oak rhizospheric communities after wildfire in these sampling sites. The role of pH as an important factor for the structure of the microbial communities is well established23, 24. Even after a wildfire, pH had the strongest effect on bacterial community composition and diversity over soil moisture, ammonium content and total nitrogen9. In our case, the increase of pH was not an indication of an increase in richness; in fact we report a significant reduction of richness and diversity because of the wildfire (Table S2). This contradiction could be due to the fact that UOF is a slightly acid soil and BOF is a slightly basic soil (Table S1).
The sharpest effect of wildfire in soil microbial community after 3 years was the predominance of phylum Actinobacteria mainly due to the high proportion of genus Arthrobacter (Figs 1 and 2). The increase of this phylum after the wildfire coincides with the findings of other studies on wildfires by analysis of microbial communities7, 8, 10 and metagenomics11. It should be pointed out that previous results11 were obtained at the same sampling sites but with a metagenomic, functional approach. Thus, our current results obtained by pyrosequencing of 16S rRNA amplicons show that two different molecular approaches offer quite a good comparison, and confirm the main role of the phylum Actinobacteria and the genus Arthrobacter in microbial communities following Mediterranean oak forest fires. This high proportion of Actinobacteria is probably a reflection of its capacity to withstand high temperatures and proliferate on partially sterile burned soils in the form of spores; and in the same way, specific genera like Arthrobacter are adapted to oligotrophic conditions. Moreover, the presence of Arthrobacter in burned soils could be related to its capacity to form “cyst-like” resting cells and its described physiology to resist starvation, desiccation and oxidative stress8, 21, 22, 25.
Therefore, our objective was to obtain and to characterize the dominant bacterial genus from burned soil. Our results showed that the free-living Arthrobacter strains isolated in this study were physiologically (Table 3) and genetically diverse (Fig. S1), since a high degree of genetic variation was observed among the 55 isolates when analysed by RAPD fingerprinting. The RAPD grouping provided a useful background for determining the relationship of the isolated strains. It should also be noted that none of them were clones of any other, supporting the idea of the existence of high genetic diversity among isolated Arthrobacter strains. The RAPD groups also served to detect strain diversity among those grouped very closely in the phylogenetic tree based on 16S rRNA gene sequences (Fig. 3). RAPD fingerprinting has been shown to be a useful tool to discriminate highly related strains and has been applied to study the genetic diversity of different bacterial taxa26–28.
It also should be pointed out that while all our isolates belong to the same genus, they show a very different ability to be cultivated in vitro (cultivability, Table 1). With a clustering of OTUs at 100% similarity, we got a ratio of 1 isolate for each 24 sequences of the OTU with highest proportion, which was 1 per each 4 sequences of the third most abundant and not even a single isolate of the second OTU. Others authors have observed a similar discrepancy between cultured microorganisms and the results of clone libraries and pyrosequencing on Thymus zygis rhizosphere29. The absence of a lineal ratio in the Arthrobacter OTUs cultivability could be related to different ecosystemic requirements of each species of the same genus or with the defined composition of the selective media used in this isolation.
Most of the strains in this study have more than 99% 16S rRNA gene similarity with the described Arthrobacter species but none of them showed a 100% sequence similarity to any strain type mentioned (Table S4). Despite nearly complete 16S rRNA genes having been sequenced and the high similarity shown, it should not be inferred that our isolates belong to a particular Arthrobacter species because of the lack of resolution at the species level of 16S rRNA gene sequence comparisons30, 31 (Figs 3 and S2).
The inferred phylogenetic trees based on 16S rRNA gene sequences using maximum likelihood (Fig. 3) and neighbor-joining methods (Fig. S2) were very similar. The topologies of both trees were strongly supported by the high bootstrap values at many branching points which suggest that is very stable (Figs 3 and S2). Taking into account the distribution of our strains in the phylogenetic trees it can be observed that many of these strains are grouped independently from the type strains and could indicate that they were not related to any of the already known Arthrobacter species. A more extensive characterization work would be needed to elucidate the taxonomic status of the strains used for this study and the description of new species if needed.
Arthrobacter was the bacterial genus that had the highest relative increase in the prokaryotic soil community following the forest fire (Fig. 2). The diversity of Arthrobacter strains recovered from the burned soil sample (BOF) (Figs 3, S1 and S2) suggests that members of this genus in the phylum Actinobacteria are resistant to drastic changes in environmental conditions associated with wildfire, and could play an important ecological role probably related to the natural recovery of the burned area. One possible ecological role of Arthrobacter could be facilitating the recovery of the vegetation cover of the burned areas. Arthrobacter spp. can utilize a wide variety of aromatic compounds like those that appear in the soil after a wildfire32 and also play an important role in the transformation of organic matter in the natural environment20. Moreover, Arthrobacter has a main role in the nitrogen cycle of rhizospheric burned soils11; therefore, it could play a key role facilitating the change from oligotrophic to copiotrophic conditions. In addition, several Arthrobacter species have been described as PGPR and have been used in field revegetation33–35, and some of these microorganisms are also tolerant to desiccation21, 22.
The role of the 55 Arthrobacter isolates as putative PGPR was tested in an inoculation experiment with Medicago sativa (alfalfa) and Capsicum annuum (pepper) plants. Our goal was to select the microorganisms with PGP activity in both crops. With agronomical characteristics of these plants being very different, positive results in PGP capabilities for both plants would mean that the beneficial effect was not plant-specific. Consequently we could extrapolate that our strains were beneficial to a range of plants, which could include oak. Most of the strains isolated performed well in planta, and their PGP effects were significantly different than the control except for two isolates (Fig. S3). This result is very interesting because of its agreement with previous literature in which a high number of beneficial bacteria are systematically found in the rhizosphere36, compared to the low numbers of detrimental strains found. This suggests that the plant and/or the bacterial activities favour mutualism. Interestingly in this particular case, the bacterial ecosystem analysed is a burned rhizosphere sample (BOF) and, however, the results are very similar to those described for wild rhizospheric samples. Considering the difficulties of working with oak trees, we decided to scale down the experiments with other plants of interest. Medicago sativa is a widely used leguminous plant and Capsicum annum has also been stated to have properties that make it a very good model plant for several types of studies such as the study of genetic diversity to develop new cultivars37. The PGP results obtained with the two plants are very similar, so we suggest that the observed effect on growth is not dependent on the host and because of the lack of specificity, Arthrobacter could help a more rapid recovery of vegetation cover in burned areas, accelerating the growth of plants that would prevent the loss of soil. However, more studies in field conditions in burnt soil would be needed to confirm the role of Arthrobacter in vegetation recovery. The five selected strains that were used in the more in-depth study resulted in a 40% increase in growth when compared to the un-inoculated plants. The increase in growth of M. sativa in the second experiment (Table 2) is not surprising, given that several studies demonstrate Actinobacteria, including strains of Arthrobacter, promote both shoot growth in alfalfa as well as in the actinorhizal plant species when co-inoculated with the corresponding nitrogen-fixing micro-symbiont, Ensifer or Frankia.
As mentioned previously, the increase in shoot growth could be caused by the diverse molecules produced by the Arthrobacter strains considered responsible for the PGP action38, 39. Siderophore production is commonly found in Actinobacteria and is regarded as an important PGPR trait used to overcome low iron availability40, 41. The fact that three of our five selected strains produce siderophores is probably related to their capability as PGPR and to the low iron concentration in the BOF sample. Furthermore, IAA production has been directly related with plant growth42, 43 and all the Arthrobacter strains selected showed IAA production based on the Salkowsky test. Thus, both traits should be considered as general, nonspecific PGP.
Most Actinobacteria are saprophytes able to produce a wide range of extracellular hydrolytic enzymes, and all our studied strains synthesize hydrolytic enzymes able to cleave complex polymeric substrates (Table 3), strongly suggesting that Arthrobacter can favour plant nutrition by mineralization of soil organic matter. Nonetheless, further research is needed to fully explain the rationale for improved plant growth in plants inoculated with Arthrobacter. Phosphorous (P) is also a very limited nutrient which makes the strains able to solubilize phosphate very interesting PGPRs. Three Arthrobacter strains showed phosphate solubilisation activity (Table 3), which can thus make soil P more available to plants. Indeed, several Arthrobacter isolates have been recently reported as Phosphate-solubilizing rhizobacteria35, 44.
The ability to form biofilms plays a fundamental role in the bacterial colonization of the root as well as in the regulation of plant beneficial properties45. It has been reported that the production of cellulose is important for biofilm formation46, 47. All selected strains are capable of producing cellulose, which will probably aid colonization of plant roots. The ability to fix atmospheric nitrogen is often associated with growth in nitrogen-free media. Our results are preliminary, as it has been suggested before that these methods are not accurate or can lead to falsely positive results48 but it is certain that these bacteria can grow in an environment with very limited nitrogen availability, rendering them as excellent candidates for recovering soils, where they could be inoculated to enhance afforestation efforts.
In this study, we obtained and characterized the dominant bacterial genus from a burned rhizospheric soil. Thus, 55 Arthrobacter strains were isolated and characterized using RAPDs, 16S rRNA gene sequencing and their interaction when used as an inoculant in pepper and alfalfa plants. It is important to note that Arthrobacter is a bacterium that can endure harsh environmental conditions and can naturally be found in soil. But our results indicate that Arthrobacter strains isolated from burned soil samples have a very high genetic diversity. We have shown that Arthrobacter could play an important ecological role in interacting with the host plant by enhancing aerial growth as a general phenomenon. The selected strains exhibited in vitro a great ability to degrade organic polymers as well as possibly presenting a direct mechanism for plant growth promotion (IAA production). It remains to be elucidated whether these positive effects also occur in other plant species. All the above data suggest that, in general, Arthrobacter can be considered as an excellent PGPR, although a correct selection of strains is of capital importance because of the detrimental effect that some of them may have for plant growth. However, even though most of the 55 isolates showed a trend towards plant growth enhancement, the five strains examined in detail excel at plant growth promotion. These findings could lead to the formulation of bioinoculants for the recovery of reserves, endangered or endemic plant species.
Methods
Experimental site
The study area is located in the Sierra Nevada Natural and National Park (SE Spain); in which, in September 2005, a wildfire burned 3426.74 ha, included 412 ha of evergreen holm oaks (Quercus ilex subsp. ballota). Soil samples were collected at the valley of the Lanjarón River, where two sites were selected: one was a holm oak forest affected by the wildfire (burned oak forest, BOF) and another one was a control site in the evergreen oak forest not affected by wildfire (undisturbed oak forest, UOF). These sites are the same sites described previously11 and the sampled trees, processes and time of sampling were also the same. The sampling was done three years after the fire, when the holm-oaks were re-growing and a new microbial community, possibly involved in ecosystem recovery, was established. The BOF and UOF sites were on a steep south-facing slope. Three sampling plots were randomly chosen within each study site along transects of 1.0 km length. At the BOF and UOF sites, we sampled the rhizosphere of three trees per plot, each with a diameter of at least 15 cm at breast height and separated by at least 5 m. The specific sampled trees were marked, and the positions of sites were registered with the Global Positioning System (GPS, Fig. S4).
Sample collection and soil chemical analysis
The rhizospheric samples were collected from a previous work11, by following the tree’s main roots until young cork-free roots were found at a distance of less than 50 cm from the trunk. The soil attached to the roots was manually removed and the roots with rhizospheric soil were put into 50 ml Falcon tubes filled with 20 ml of sterilized NaCl 0.8%. After shaking 25 min at 150 rpm and room temperature, the roots were removed and the tubes were centrifuged at 12,850 g for 5 min. The pelleted soil was used to extract environmental DNA. On the other hand, we processed up to 2 kg of soil from each site, sieved through a 2 mm mesh, for physicochemical analysis including soil type, pH, available water, total nitrogen, organic matter, electrical conductivity, etc. All these analyses were carried out with standardized procedures (Table S1) at the Food and Agriculture Laboratory of the Andalusian regional government at Atarfe (Granada, Spain).
DNA extraction, PCR amplification and pyrosequencing
Soil DNA was extracted from each individual soil sample using the PowerSoil™ DNA Isolation Kit (MoBio, Laboratories Inc., CA), following the manufacturer’s recommendations within 24 hours of sample collection. DNA yields and quality were checked after electrophoresis in 0.8% (w/v) agarose gel stained with ethidium bromide under UV light and with a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). An Equimolecular amount of DNA from the three rhizospheres of the same plot was pooled prior to pyrosequencing.
Partial bacterial 16S rRNA gene sequences were obtained from the analysis of each plot sample as described previously49 with the amplification of the hypervariable V6-V8 regions with primers 926F (5′-AAACTYAAAKGAATTGRCGG-3′) and 1392R (5′-ACGGGCGGTGTGTRC-3′). The PCR mixtures (25 μl) contained 10 pmol of each primer, 1.8 mM MgCl2, 0.4 mM dNTPs, 1 x the corresponding Taq buffer and Taq enhancer buffer, 1 U of Taq Master (5 Prime, USA) and 10 ng of the same DNA template used above. The PCR program consisted of an initial denaturation step at 94 °C for 4 min, 25 cycles of denaturation at 94 °C for 15 s, primer annealing at 55 °C for 45 s and extension at 72 °C for 1 min, followed by a final step of heating at 72 °C for 10 min. For each sample, amplicons were generated in three replicated PCRs. All the amplicons generated from the PCR of each individual sample (9 per treatment) were pooled in equimolar amounts50–52 in two composite samples (one composite sample per treatment) that were subjected to pyrosequencing with the Genome Sequencer Titanium FLX system (454| Life Sciences, Branford, CT, USA) at LifeSequencing S.L. (Valencia, Spain). Sequence files were submitted to the NCBI Sequence Read Archive (www.ncbi.nlm.nih.gov/sra) and are available with BioProject accession number PRJNA 291009.
Pyrosequencing data analysis
Raw sequences were processed using MOTHUR version 1.34.053. Briefly, sequencing errors were reduced using the AmpliconNoise algorithm and low-quality sequences were removed [minimum length of 150 base pairs (bp), allowing 2 mismatch in the barcode, 2 mismatches in the primer, and homopolymers no longer than 8 bp]. Sequences were then aligned using align.seqs function and the SILVA database as template54. The chimera.uchime function was then used to identify potentially chimeric sequences which were subsequently removed55. Subsequently, the high-quality bacterial sequences were clustered into operational taxonomic units (OTUs) with a similarity of at least 97%. The number of sequences per sample was rarefied before OTU definition. OTU (phylotype richness) distribution among samples was used to calculate rarefaction curves, the phylotype Shannon diversity index (H’), phylotype evenness (Pielou), Chao 1 (abundance-based coverage estimation) richness estimator index, Good’s coverage as well as phylogenetic diversity (phylodiversity)56 by the use of MOTHUR software.
Significant differences in Shannon diversity indices between undisturbed and burned samples were assessed using the diversity t test, with p < 0.05 being regarded as statistically significant57.
Finally, to examine changes in the relative abundance of the different microbial groups mediated by fire, the normalized bacterial sequences were classified with an 80% bootstrap cutoff to the Ribosomal Database Project (RDP-II) 16S rRNA reference database Release 1058 using the classify.seqs function from MOTHUR. Furthermore, significant and biologically relevant differences in the proportions between undisturbed and burned samples at genus level were assessed in STAMP v2.0.959, using White’s non-parametric t-test at a 95% confidence interval. Biologic relevance was determined choosing genera with a difference between proportions >0.5 and a ratio of proportions >260.
Strains isolation and classification
With the purpose of obtaining cultures of the most abundant genus from burned rhizosphere (BOF), we isolated some bacteria from the first BOF plot (BOF1, located in UTM coordinates, 30S 0458909, 4090493, elevation 1557 m above sea level). From sampling till bacterial isolation, BOF1 soil was stored at −80 °C for three years. The isolation was as follows: Firstly, a pre-enrichment with tryptone 0.5%, 1 g of BOF1 soil and 30 ml dH2O (Peptone solution) was gently shaken for one hour at room temperature61. Subsequently, after 15 min waiting for the biggest soil particles to decant, 100 μl of Peptone solution was spread over the surface of a sterile HH’ medium in petri dishes and incubated for one week at 30 °C. This process was repeated with serial dilutions up to 10–7. The HH’ medium was a modification of a previous medium62, and consisted of 0.4% Trypticase soy broth, 0.2% Yeast extract, 3% NaCl, 0.01% Cycloheximide and 1.5% agar.
Following a 7–8 days incubation period, 231 CFU (Colony-Forming Units) were selected. The genomic DNA was obtained by heating 100 μl colony suspensions in dH2O at 96 °C for 10 min in a QBD2 dry block heater (Grant, Cambridge, England). This DNA from isolates was amplified with 9bfm forward63 and 1512UR reverse64 universal primers to almost reach the complete 16S rRNA sequence, with the following PCR specifications: an initial denaturation step at 94 °C for 4 min, 25 cycles of denaturation at 94 °C for 1 min, primer annealing at 52 °C for 1 min and extension at 72 °C for 1.5 min, followed by a final step of heating at 72 °C for 10 min. Partial sequencing of the 16S rRNA gene was performed from forward primer (9bfm) and BLASTed against nt/nr database (https://blast.ncbi.nlm.nih.gov). Finally, 55 CFU from the genus Arthrobacter were selected.
Fingerprinting and phylogenetic analysis of Arthrobacter strains
RAPD fingerprinting profiles from bacterial genomic DNA were obtained as described previously65. Strain clusters were defined at 60% similarity. The phylogenetic tree was performed once with 55 sequences from 16S rRNA gene from isolates (GenBank accession numbers KT314102-KT314156) and with 63 Arthrobacter type strains into RDP-II database. Sequences of reference strains were obtained from the NCBI (http://ncbi.nlm.nih.gov/) and Eztaxon (http://www.ezbiocloud.net/eztaxon) public databases.
For a second tree, Streptomyces albus subsp. albus DSM 40313T (GenBank accession number AJ621602.2) was selected as an outgroup and the 12 most closely related type strains were added for the alignment. In addition, the alignment was produced by Infernal Aligner tool66 from RDP-II and curated with Geneious R8.1.2. (Biomatters Ltd, Auckland, New Zealand).
Finally, the phylogenetic tree was inferred using the neighbor-joining67 and maximum likelihood68 methods. Bootstrap analysis was based on 1,000 replicates. MEGA569 was used for neighbor-joining and maximum likelihood analysis.
Functional characterization of selected Arthrobacter strains
Siderophore synthesis was investigated as described previously70. Selected strains of Arthrobacter were spotted on Chrome Azurol S (CAS) plates; development of deep blue to yellow or orange halo was indicative of siderophore production. Xylan and Pectin were measured as carbon sources using a Yeast Nitrogen Base (Difco) following the manufacturer’s instructions and complemented with xylan or pectin at a 1% concentration. Tween 80 and 20 were added in a concentration of 10 g l−1 to a medium containing bacto-peptone (10 g l−1), Calcium chloride (5 g l−1) and Sodium chloride (0.1 g l−1). Degradation halos can be seen in this medium for positive strains.
Cellulose and cellulase production were tested following a procedure stated previously71. Degradation of carbon sources was assayed in a TSA medium supplemented with Starch, Gelatin and Casein. Degradation halos appeared in the medium of positive strains.
Phosphate solubilisation activity of bacterial strains was determined by plating onto Pikovskaya’s agar medium72 with 0.5% tricalcium phosphate [Ca3(PO4)2] as the inorganic phosphate source. The plates were incubated at 28 °C for 72 h. The formation of a clear halo around the bacterial colonies indicated phosphate solubilisation.
The indole acetic acid (IAA) production was determined as reported previously73. Flasks with GPAM medium supplemented with filter-sterilized L- tryptophan (100 μg ml−1) were inoculated with our selected Arthrobacter strains (at a concentration of 109 cells per ml) and incubated at 28 ± 2 °C for 4 days. Fully grown cultures were centrifuged at 5,600 g for 5 min. 1 ml of the supernatant was mixed with 1 ml of the Salkowski reagent (50 ml, 35% of perchloric acid, 1 ml 0.5 M FeCl3 solution). The mixture was incubated at room temperature for 25 min, and the absorbance of pink colour developed was read at 530 nm by the spectrophotometer Unicam 8625 (Unicam Limited). The concentration of IAA produced by cultures was calculated by using a standard graph of IAA concentrations (range of 10–100 μg ml−1).
Growth in nitrogen-free media
The ability to fix N2 was tested on an N-free semisolid SM medium74 supplemented with the carbon sources used in the isolation medium. 55 Arthrobacter strains were grown in nitrogen-free semisolid agar to determine their potential nitrogen-fixing activity. Strains grown on modified HH’ medium were point inoculated in the bottom of test tubes with 10 ml semi-solid agar, which contained 1% Yeast Carbon Base (Difco, Sparks, MD, USA) and 1% Noble Agar (Difco). The strains were incubated in the dark at 28 °C for 8 days. After the first incubation period, the strains were transferred to fresh media using the first tubes as inoculating source and incubated for an additional 8 days period. The above medium supplemented with (NH4)2SO4 (2 g l−1) was used as a positive control.
Plant assays
Medicago sativa var. Aragón seeds were surface-sterilized with HgCl2 2.5% for 5 min and then washed ten times with sterile dH2O. Seeds were axenically germinated in petri dishes with filter paper soaked in water for 48 hours. The seedlings were then aseptically transferred to pots with a 1litre capacity, filled with tyndallized agricultural soil. One seedling was aseptically transferred to each pot. Capsicum annuum (Pepper) plant experiments were carried out sterilizing the seeds with sodium hypochlorite for 10 min and rinsing them three times in sterile distilled water. Seeds were placed on sterile vermiculite, grown until the first true leaf appeared and then transplanted to pots with commercial substrate. Alfalfa and pepper plantlets were inoculated with 1 ml of bacterial suspensions (109 cells per ml) of each microbial strain. The bacterial suspensions were carefully spotted onto the soil near the roots of the seedling using a micropipette. Ten replicates per treatment were used. Greenhouse conditions were set as follows: 50–60% humidity, with a photoperiod of 16/8 hours. Temperature ranged between 25–15 °C day/night. The plants were harvested 6 weeks after transplantation. The parameters measured were shoot and root dry weight.
In a second experiment, only Medicago sativa was used with selected Arthrobacter strains. Alfalfa plants were individually grown in pots (1 L volume) containing tyndallized soil in a greenhouse under controlled environmental conditions. The experimental design included 5 treatments with 10 replicates per treatment. Plants were harvested 14 weeks after inoculation.
Statistical analysis
Plant growth data was studied with univariate (ANOVA) analyses with SPSS v21.0 software for Windows (IBM Corp., Armonk, NY). Dunnett’s one-tailed t-tests were used to identify inoculation treatments with means significantly different from the control at P ≤ 0.05.
Electronic supplementary material
Acknowledgements
This work was funded by the following grants: P08-CVI-03549 from The Department of Innovation, Science and Enterprise of the Autonomous Government of Andalusia; OAPN 021/2007 from The National Parks Autonomous Body (Ministry of the Environment) and 20134R069, RECUPERA 2020 from the Spanish Ministry of Economy and Competitiveness and CSIC, including ERDF (European Regional Development Fund). The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. AJFG was awarded a pre-doctoral fellowship (FPU) from the Spanish Ministry of Education; JFCD was awarded a pre-doctoral fellowship from the Junta de Andalucía, and both with a postdoctoral contract from RECUPERA 2020. PMH was awarded a postdoctoral fellowship from Ramón Areces foundation.
Author Contributions
M.F.L. and N.T. designed the research and obtained the financial support. A.J.F.G., J.F.C.D. and P.J.V. made the soil sample collection and strains isolation. A.J.F.G. and S.G.T. made the data analysis. P.M.H. and E.M.M. made the P.G.P.R. and plant experiments. A.J.F.G., P.M.H. and M.F.L. wrote the original draft. N.T., E.M.M., S.G.T. and M.F.L. review and edit the final version.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Antonio J. Fernández-González and Pilar Martínez-Hidalgo contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-06112-3
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pausas JG, Fernández-Muñoz S. Fire regime changes in the Western Mediterranean Basin: from fuel-limited to drought-driven fire regime. Climatic Change. 2012;110:215–222. doi: 10.1007/s10584-011-0060-6. [DOI] [Google Scholar]
- 2.IPCC, Intergovernmental Panel for Climate Change. Working Group I Contribution to the IPCC Fifth Assessment Report Climate Change 2013: The Physical Science Basis Summary for Policymakers (2013).
- 3.Wang Q, Zhong M, Wang S. A meta-analysis on the response of microbial biomass, dissolved organic matter, respiration, and N mineralization in mineral soil to fire in forest ecosystems. For. Ecol. Manage. 2012;271:91–97. doi: 10.1016/j.foreco.2012.02.006. [DOI] [Google Scholar]
- 4.San-Miguel-Ayanz J, Moreno JM, Camia A. Analysis of large fires in European Mediterranean landscapes: Lessons learned and perspectives. For. Ecol. Manage. 2013;294:11–22. doi: 10.1016/j.foreco.2012.10.050. [DOI] [Google Scholar]
- 5.Hart SC, DeLuca TH, Newman GS, MacKenzie MD, Boyle SI. Post-fire vegetative dynamics as drivers of microbial community structure and function in forest soils. For. Ecol. Manage. 2005;220:166–184. doi: 10.1016/j.foreco.2005.08.012. [DOI] [Google Scholar]
- 6.Álvarez R, et al. Spatial and temporal patterns in structure and diversity of Mediterranean forest of Quercus pyrenaica in relation to fire. For. Ecol. Manage. 2009;257:1596–1602. doi: 10.1016/j.foreco.2009.01.016. [DOI] [Google Scholar]
- 7.Bárcenas-Moreno G, García-Orenes F, Mataix-Solera J, Mataix-Beneyto J, Bååth E. Soil microbial recolonisation after a fire in a Mediterranean forest. Biol. Fert. Soils. 2011;47:261–272. doi: 10.1007/s00374-010-0532-2. [DOI] [Google Scholar]
- 8.Weber CF, Lockhart JS, Charaska E, Aho K, Lohse KA. Bacterial composition of soils in ponderosa pine and mixed conifer forests exposed to different wildfire burn severity. Soil Biol. Biochem. 2014;69:242–250. doi: 10.1016/j.soilbio.2013.11.010. [DOI] [Google Scholar]
- 9.Xiang X, et al. Rapid recovery of soil bacterial communities after wildfire in a Chinese boreal forest. Sci. Rep. 2014;4:3829. doi: 10.1038/srep03829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeager CM, Northup DE, Grow CC, Barns SM, Kuske CR. Changes in nitrogen-fixing and Ammonia-oxidizing bacterial communities in soil of a mixed conifer forest after wildfire. Appl. Environ. Microbiol. 2005;71:2713–2722. doi: 10.1128/AEM.71.5.2713-2722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobo-Díaz JF, et al. Metagenomic assessment of the potential microbial nitrogen pathways in the rhizosphere of a Mediterranean forest after a wildfire. Microb. Ecol. 2015;69:895–904. doi: 10.1007/s00248-015-0586-7. [DOI] [PubMed] [Google Scholar]
- 12.Isobe K, Otsuka S, Sudiana I, Nurkanto A, Senoo K. Community composition of soil bacteria nearly a decade after a fire in a tropical rainforest in East Kalimantan, Indonesia. J. Gen. Appl. Microbiol. 2009;55:329–337. doi: 10.2323/jgam.55.329. [DOI] [PubMed] [Google Scholar]
- 13.Velázquez, E. et al. Nodular endophytes: an untapped diversity. In Beneficial Plant-Microbial Interactions: Ecology and Applications. (eds Rodelas, M. B. & González-López, J.). 215-235 (CRC press, 2013).
- 14.El-Tarabily KA, Nassar AH, Sivasithamparam K. Promotion of growth of bean (Phaseolus vulgaris L.) in a calcareous soil by a phosphate-solubilizing, rhizosphere-competent isolate of Micromonospora endolithica. Appl. Soil Ecol. 2008;39:161–171. doi: 10.1016/j.apsoil.2007.12.005. [DOI] [Google Scholar]
- 15.Franco C, et al. Actinobacterial endophytes for improved crop performance. Austral. Plant Pathol. 2007;36:524–531. doi: 10.1071/AP07067. [DOI] [Google Scholar]
- 16.Martínez-Hidalgo P, Galindo-Villardón P, Trujillo ME, Igual JM, Martínez-Molina E. Micromonospora from nitrogen fixing nodules of alfalfa (Medicago sativa L.). A new promising plant probiotic bacteria. Sci. Rep. 2014;4:6389. doi: 10.1038/srep06389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cacciari I, Giovannozzi-Sermanni G, Grappelli A, Lippi D. Nitrogen fixation by Arthrobacter sp. I. Taxonomic study and evidence of nitrogenase activity of two new strains. Ann. Microbiol. Enzimol. 1971;21:97–105. [Google Scholar]
- 18.Knapp, R. & Jurtshuk, P. Characterization of free-living nitrogen-fixing Streptomyces species and factors which affect their rates of acetylene redpuction. In Abstr Annu Meet Am Soc Microbiol (Vol. 88, p. 219) (1988).
- 19.Baranova NA, Gogotov IN. Molecular nitrogen fixation by propionic acid bacteria. Mikrobiologiya. 1974;43:791. [PubMed] [Google Scholar]
- 20.Gkorezis P, et al. Draft genome sequence of Arthrobacter sp. strain SPG23, a hydrocarbon-degrading and plant growth-promoting soil bacterium. Genome announcements. 2015;3:e01517–15. doi: 10.1128/genomeA.01517-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.SantaCruz-Calvo L, González-López J, Manzanera M. Arthrobacter siccitolerans sp. nov., a highly desiccation-tolerant, xeroprotectant-producing strain isolated from dry soil. Int. J. Syst. Evol. Microbiol. 2013;63:4174–4180. doi: 10.1099/ijs.0.052902-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manzanera M, Narváez-Reinaldo JJ, García-Fontana C, Vílchez JI, González-López J. Genome sequence of Arthrobacter koreensis 5J12A, a plant growth-promoting and desiccation-tolerant strain. Genome Announcements. 2015;3:e00648–15. doi: 10.1128/genomeA.00648-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen C, et al. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol. Biochem. 2013;57:204–211. doi: 10.1016/j.soilbio.2012.07.013. [DOI] [Google Scholar]
- 24.Griffiths RI, et al. The bacterial biogeography of British soils. Environ. Microbiol. 2011;13:1642–1654. doi: 10.1111/j.1462-2920.2011.02480.x. [DOI] [PubMed] [Google Scholar]
- 25.Mongodin EF, et al. Secrets of soil survival revealed by the genome sequence of Arthrobacter aurescens TC1. PLoS Genet. 2006;2:e214. doi: 10.1371/journal.pgen.0020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gillespie BE, Oliver SP. Comparison of an automated ribotyping system, pulsed-field gel electrophoresis and randomly amplified polymorphic DNA fingerprinting for differentiation of Streptococcus uberis strains. Biotechnology. 2004;3:165–172. doi: 10.3923/biotech.2004.165.172. [DOI] [Google Scholar]
- 27.Zhang H, Lee YK, Zhang W, Lee HK. Culturable Actinobacteria from the marine sponge Hymeniacidon perleve: isolation and phylogenetic diversity by 16S rRNA gene-RFLP analysis. Anton Leeuw. 2006;90:159–169. doi: 10.1007/s10482-006-9070-1. [DOI] [PubMed] [Google Scholar]
- 28.Armas-Capote N, et al. Core and symbiotic genes reveal nine Mesorhizobium genospecies and three symbiotic lineages among the rhizobia nodulating Cicer canariense in its natural habitat (La Palma, Canary Islands) Syst. Appl. Microbiol. 2014;37:140–148. doi: 10.1016/j.syapm.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Pascual J, et al. Assessing bacterial diversity in the rhizosphere of Thymus zygis growing in the Sierra Nevada National Park (Spain) through culture-dependent and independent approaches. PLoS ONE. 2016;11:e0146558. doi: 10.1371/journal.pone.0146558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fox GE, Wisotzkey JD, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Evol. Microbiol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 31.Gevers D, et al. Re-evaluating prokaryotic species. Nature Rev. Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 32.Westerberg K, Elvang AM, Stackebrandt E, Jansson JK. Arthrobacter chlorophenolicus sp. nov., a new species capable of degrading high concentrations of 4-chlorophenol. Int. J. Syst. Evol. Microbiol. 2000;50:2083–2092. doi: 10.1099/00207713-50-6-2083. [DOI] [PubMed] [Google Scholar]
- 33.Kim KM, Song HG. Revegetation of barren lakeside land through growth enhancement of Xanthium italicum by rhizobacteria. Paddy Water Environ. 2014;12:125–131. doi: 10.1007/s10333-014-0428-0. [DOI] [Google Scholar]
- 34.Pereira SIA, Castro PML. Phosphate-solubilizing rhizobacteria enhance Zea mays growth in agricultural P-deficient soils. Ecological Engineering. 2014;73:526–535. doi: 10.1016/j.ecoleng.2014.09.060. [DOI] [Google Scholar]
- 35.Gusain YS, Kamal R, Mehta CM, Singh US, Sharma AK. Phosphate solubilizing and indole-3-acetic acid producing bacteria from the soil of Garhwal Himalaya aimed to improve the growth of rice. J. Environ. Biol. 2015;36:301–307. [PubMed] [Google Scholar]
- 36.Antoun, H. & Prévost, D. Ecology of plant growth promoting rhizobacteria. In PGPR: Biocontrol and Biofertilization (ed. Siddiqui, Z. A.) 1–38 (Springer, 2005).
- 37.Taranto F, D’Agostino N, Greco B, Cardi T, Tripodi P. Genome-wide SNP discovery and population structure analysis in pepper (Capsicum annuum) using genotyping by sequencing. BMC Genomics. 2016;17:943. doi: 10.1186/s12864-016-3297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Souza RD, Ambrosini A, Passaglia LM. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 2015;38:401–419. doi: 10.1590/S1415-475738420150053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo JH, Jiang CH, Xie P, Huang ZY, Fa ZH. The plant healthy and safety guards plant growth promoting rhizo bacteria (PGPR) Transcriptomics. 2015;3:2. [Google Scholar]
- 40.Bossier P, Verstraete W. Ecology of Arthrobacter JG-9-detectable hydroxamate siderophores in soils. Soil Biol. Biochem. 1986;18:487–492. doi: 10.1016/0038-0717(86)90005-2. [DOI] [Google Scholar]
- 41.Lee J, Postmaster A, Soon HP, Keast D, Carson KC. Siderophore production by actinomycetes isolates from two soil sites in Western Australia. Biometals. 2012;25:285–296. doi: 10.1007/s10534-011-9503-9. [DOI] [PubMed] [Google Scholar]
- 42.Radwan ESED, Mohamed ZK, Massena-Reis V. Production of indole-3-acetic acid by different strains of Azospirillum and Herbaspirillum spp. Symbiosis. 2002;32:39–53. [Google Scholar]
- 43.Merzaeva OV, Shirokikh IG. The production of auxins by the endophytic bacteria of winter rye. Appl. Biochem. Microbiol. 2010;46:44–50. doi: 10.1134/S0003683810010072. [DOI] [PubMed] [Google Scholar]
- 44.Pereira SIA, Barbosa L, Castro PML. Rhizobacteria isolated from a metal-polluted area enhance plant growth in zinc and cadmium-contaminated soil. Int. J. Environ. Sci. Technol. 2015;12:2127–2142. doi: 10.1007/s13762-014-0614-z. [DOI] [Google Scholar]
- 45.Drogue B, Doré H, Borland S, Wisniewski-Dyé F, Prigent-Combaret C. Which specificity in cooperation between phytostimulating rhizobacteria and plants? Res. Microbiol. 2012;163:500–510. doi: 10.1016/j.resmic.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 46.Robledo M, et al. Role of Rhizobium endoglucanase CelC2 in cellulose biosynthesis and biofilm formation on plant roots and abiotic surfaces. Microb. Cell Fact. 2012;11:125. doi: 10.1186/1475-2859-11-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robledo, M. et al. Role of Rhizobium cellulase CelC2 in host root colonization and infection. In Biological Nitrogen Fixation. (ed. de Bruijn, F. J.) 525–532 (Wiley-Blackwell, 2015).
- 48.Martínez-Hidalgo P, Olivares J, Delgado A, Bedmar E, Martínez-Molina E. Endophytic Micromonospora from Medicago sativa are apparently not able to fix atmospheric nitrogen. Soil Biol. Biochem. 2014;74:201–203. doi: 10.1016/j.soilbio.2014.03.011. [DOI] [Google Scholar]
- 49.Curiel-Yuste J, et al. Changes in soil bacterial community triggered by drought-induced gap succession preceded changes in soil C stocks and quality. Ecol. Evol. 2012;2:3016–3031. doi: 10.1002/ece3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheik CS, et al. Effect of warming and drought on grassland microbial communities. The ISME journal. 2011;5:1692–1700. doi: 10.1038/ismej.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yergeau E, et al. Shifts in soil microorganisms in response to warming are consistent across a range of Antarctic environments. The ISME journal. 2012;6:692–702. doi: 10.1038/ismej.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Curiel-Yuste J, et al. Strong functional stability of soil microbial communities under semiarid Mediterranean conditions and subjected to long-term shifts in baseline precipitation. Soil Biol. Biochem. 2014;69:223–233. doi: 10.1016/j.soilbio.2013.10.045. [DOI] [Google Scholar]
- 53.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microb. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marcon E, Hérault B. Decomposing phylodiversity. Meth. Ecol. Evol. 2015;6:333–339. doi: 10.1111/2041-210X.12323. [DOI] [Google Scholar]
- 57.Magurran, A. Ecological diversity and its measurement. (University Press, 1988).
- 58.Cole JR, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucl Acids Res. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics. 2010;26:715–721. doi: 10.1093/bioinformatics/btq041. [DOI] [PubMed] [Google Scholar]
- 61.Jones, D. & Keddie, R. M. The genus Arthrobacter. In The Prokaryotes, a Handbook on the Biology of Bacteria, 3rd edition. (eds Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K. H. & Stackebrandt, E.). 945–960 (Springer, 2006).
- 62.Hagedorn C, Holt JG. Ecology of soil arthrobacters in Clarion-Webster toposequences of Iowa. Appl. Microbiol. 1975;29:211–218. doi: 10.1128/am.29.2.211-218.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mühling M, Woolven-Allen J, Murrel JC, Joint I. Improved group-specific PCR primers for denaturing gradient gel electrophoresis analysis of the genetic diversity of complex microbial communities. The ISME journal. 2008;2:379–392. doi: 10.1038/ismej.2007.97. [DOI] [PubMed] [Google Scholar]
- 64.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:687–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rivas R, et al. Biodiversity of populations of phosphate solubilizing rhizobia that nodulate chickpea in different Spanish soils. Plant Soil. 2006;287:23–33. doi: 10.1007/s11104-006-9062-y. [DOI] [Google Scholar]
- 66.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 68.Rogers JS, Swofford DL. A fast method for approximating maximum likelihoods of phylogenetic trees from nucleotide sequences. Systematic Biology. 1998;47:77–89. doi: 10.1080/106351598261049. [DOI] [PubMed] [Google Scholar]
- 69.Tamura K, et al. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 71.Mateos PF, et al. Cell-associated pectinolytic and cellulolytic enzymes in Rhizobium leguminosarum biovar trifolii. Appl. Environ. Microbiol. 1992;58:1816–1822. doi: 10.1128/aem.58.6.1816-1822.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pikovskaya RI. Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya. 1948;17:362–370. [Google Scholar]
- 73.Khalid A, Arshad M, Zahir ZA. Screening plant growth-promoting rhizobacteria for improving growth and yield of wheat. J. Appl. Microbiol. 2004;96:473–480. doi: 10.1046/j.1365-2672.2003.02161.x. [DOI] [PubMed] [Google Scholar]
- 74.Reinhold B, Hurek T, Niemann EG, Fendrik I. Close association of Azospirillum and diazotrophic rods with different root zones of Kallar grass. Appl. Environ. Microbiol. 1986;52:520–526. doi: 10.1128/aem.52.3.520-526.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


