Abstract
Mouse models have been used to provide primary cells to study physiology and pathogenesis of epithelia. However, highly efficient simple approaches to propagate mouse primary epithelial cells remain challenging. Here, we show that pharmacological inhibition of TGF-β signaling enables long-term expansion of p63+ epithelial progenitor cells in low Ca2+ media without the need of progenitor cell-purification steps or support by a feeder cell layer. We find that TGF-β signaling is operative in mouse primary keratinocytes in conventional cultures as determined by the nuclear Smad2/3 localization. Accordingly, TGF-β signaling inhibition in crude preparations of mouse epidermis robustly increases proliferative capacity of p63+ epidermal progenitor cells, while preserving their ability of differentiation in response to Ca2+ stimulation. Notably, inhibition of TGF-β signaling also enriches and expands other p63+ epithelial progenitor cells in primary crude cultures of multiple epithelia, including the cornea, oral and lingual epithelia, salivary gland, esophagus, thymus, and bladder. We anticipate that this simple and efficient approach will facilitate the use of mouse models for studying a wide range of epithelia by providing highly enriched populations of diverse p63+ epithelial progenitor cells in quantity.
Introduction
Homeostasis and regeneration of epithelia are maintained by self-renewal, proliferation, and differentiation of tissue-specific stem cells1–3. The development of 3T3-J2 feeder cell co-culture by Rheinwald and Green4 has allowed expansion of human primary keratinocytes, and later corneal epithelia, and contributed to not only our understanding of epithelial cell biology but also therapeutic strategies in regenerative medicine5–7. 3T3-J2 cells support the proliferative potential of epithelial progenitor cells in a paracrine manner8, 9 and have been used to assess self-renewal capacity of p63+ epithelial stem cells10–12. The transcription factor p63 plays an essential intrinsic role in regulating stem cell self-renewal12, 13. Indeed, mice lacking p63 show loss or severe hypoplasia in all epithelia that normally express p63 in their stem cell compartments, such as the skin, cornea, thymus, salivary gland, esophagus, mammary gland, prostate, and bladder14, 15. Thus, p63 plays a key role in maintaining the proliferative capacity of epithelial progenitor cells of diverse epithelia.
Mouse models have been used in studies of normal and disease conditions of epithelia. However, the growth of p63+ mouse primary epithelial cells (e.g. primary keratinocytes) in culture rapidly declines and the cells become terminally differentiated16, a limit that restricts the use of primary cells for functional analyses of epithelia. As proliferation and differentiation of epithelial cells are tightly coupled through the induction of cyclin-dependent kinase (CDK) inhibitor genes17–19, suppression of growth arrest and of premature differentiation are both potential approaches to improve the lifespan of p63+ mouse primary epithelial progenitor cells17, 20–23. Although the use of low calcium (Ca2+) media has extended proliferation of p63+ mouse epithelial progenitor cells short term16, 24, the most effective protocols to propagate these primary cells rely on modified 3T3-J2 feeder co-culture20, 22 or the use of fluorescence activated cell sorting (FACS)-purified progenitor populations with triple drug inhibitors23. Highly efficient protocols that eliminate the use of undefined factors (e.g. feeder cells), labor-intensive purification steps, and the potentially complex effects of multiple drugs will facilitate the use of primary epithelial cells of mice for studying the physiology and pathogenesis of p63-dependent epithelia.
Transforming growth factor-β (TGF-β) signaling regulates proliferation and differentiation of many different progenitor cells, including those that are controlled by p6325. TGF-β signaling is mediated through the receptor complex consisting of the Type I TGF-β receptor (TGFβR1/ALK5) and the Type II TGF-β receptor (TGFβR2)26. Upon binding of the TGF-β ligands, TGFβR2 phosphorylates and activates TGFβR1/ALK5, resulting in the phosphorylation and nuclear translocation of Smad2/3, downstream effectors of TGF-β signaling27. Consistent with its growth suppressive function, phosphorylated Smad2/3 are barely detectable in basal cells in many different epithelia in mouse, including the skin23. Notably, however, we find that a large fraction of Smad2/3 is localized in the nuclei of mouse primary epidermal keratinocytes in a growth-permissive low Ca2+ condition that would ordinarily support keratinocyte proliferation. We hypothesize that inhibition of TGF-β signaling would suppress Smad2/3 nuclear localization and consequently increase the proliferative capacity of p63+ mouse primary epithelial cells in vitro.
In this report, we show that the use of a single small molecule inhibitor of TGF-β signaling in crude mouse tissue preparations enriches p63+ epithelial progenitor cells and supports their long-term proliferative capacity in a low Ca2+ condition. This methodology can produce a large quantity of diverse p63+ mouse epithelial progenitor cells without the need of a feeder layer or progenitor cell-purification steps. We anticipate that this simple and efficient approach will facilitate the use of mouse models for studying a wide range of p63-dependent epithelia.
Result
Inhibition of TGF-β signaling stimulates the proliferation of mouse primary epidermal cells in vitro
Although mouse epidermal cells can proliferate in low Ca2+ media, their lifespan is relatively short16, 24. As mouse epidermal cells express TGF-β in vitro 28, 29, we sought to determine whether TGF-β signaling is operative in mouse epidermal cells in primary cultures, thus limiting their proliferation. To address this possibility, we investigated the subcellular localization of Smad2/3 in cytokeratin-positive (CK+) mouse primary epidermal cells grown in CnT-PR, one of the best commercially available basal media for low Ca2+ epithelial culture. As epidermal cells in newborn mice are more proliferative than those in postnatal mice, we first focused on the analysis of primary cells from newborn mice in this study. We found that a substantial level of Smad2/3 was localized in the nuclei of virtually all CK+ newborn mouse epidermal cells, including those of potentially proliferative immature keratinocytes with relatively small nuclear-to-cytoplasmic ratios (Fig. 1a). Notably, however, treatment with RepSox, a cell-permeable and selective inhibitor of the TGF-β type 1 receptor/ALK5, substantially reduced the nuclear localization of Smad2/3, while addition of TGF-β ligands in culture promoted it (Fig. 1a,b). These results indicate that TGF-β signaling is active in mouse primary epidermal cells in a growth-permissive condition and that TGF-β signaling activity can be suppressed by the use of small molecule inhibitors.
Figure 1.
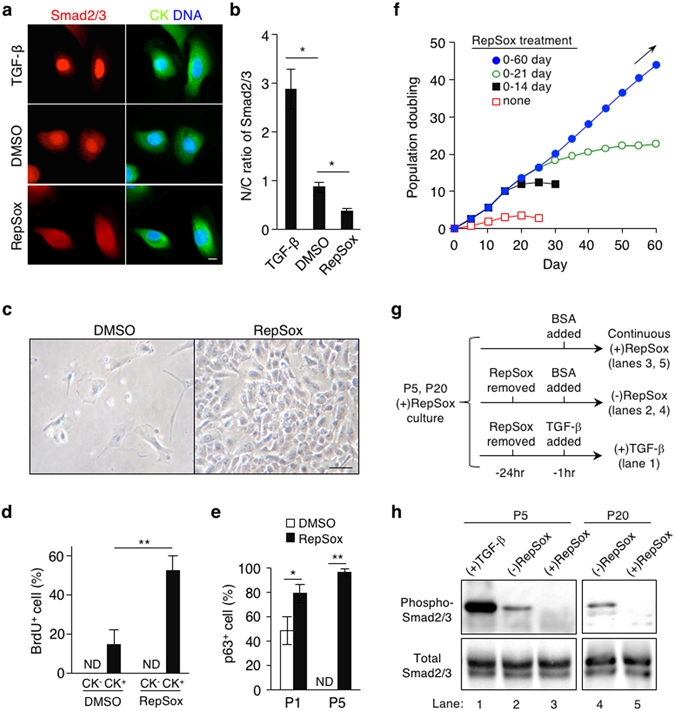
Inhibition of TGF-β signaling enables long-term expansion of p63+ mouse primary epidermal progenitor cells in vitro. (a) Subcellular localization of Smad2/3 in newborn mouse primary epidermal keratinocytes grown in CnT-PR media. The cells were stimulated with 1 ng/ml TGF-β2, 0.1% DMSO (as a control) or 1 μM RepSox for 1 hr. CK, pan-cytokeratin; DNA, nuclear counterstaining with Hoechst 33342; Bar = 10 μm. (b) Nuclear-to-cytoplasmic (N/C) ratios of Smad2/3 as determined by fluorescence intensity. Data shown are mean ± s.e.m. (n = 6). *P < 0.05. (c) Representative images of primary cells of newborn mouse epidermis at day 14 of culture in the presence or absence of 1 μM RepSox. Bar = 25 μm. (d) A short-pulse labeling with BrdU in primary culture of newborn mouse epidermal cells in the presence or absence of 1 μM RepSox. Data shown are % per field, expressed as mean ± s.e.m. (n = 4). ND, not detected; **P < 0.01. (e) Percentages of p63+ cells at passage 1 and 5 (P1 and P5, respectively) grown in the presence (solid) or absence (open) of 1 μM RepSox. Data shown are % per field, expressed as mean ± s.e.m. (n = 3 and 4 for P1 and P5, respectively). ND, not detected; *P < 0.05; **P < 0.01. (f) Population doubling of newborn mouse-derived, RepSox-expanded P2 epidermal cells grown in CnT-PR media in the presence of 1 μM RepSox for 0, 14, 21, and 60 days. Data shown are representative of three independent experiments with similar results. Arrow indicates continuous cell growth. (g,h) Removal of RepSox associates with increased Smad2/3 phosphorylation. (g) Experimental design. Newborn mouse-derived, RepSox-expanded P5 and P20 epidermal keratinocytes were further grown in continuous presence (upper) or absence (middle and lower) of 1 μM RepSox for 24 hrs. Culture of P5 cells stimulated with 1 ng/ml TGF-β for 1 hr prior to lysis (lower) was used as a positive control. (h) Expression of total and phosphorylated Smad2/3 as determined by Western blot. Lane numbers correspond to those in (g). The full-length blots are included in Supplementary Fig. S11.
Consistent with the inhibition of the nuclear localization of Smad2/3, RepSox treatment enhanced the proliferation of CK+ mouse primary epidermal cells while co-existing CK− cells were not responsive to the RepSox treatment (Fig. 1c,d). Accordingly, RepSox treatment robustly increased the CK+ epidermal cell number in a dose-dependent manner (Supplementary Fig. S1). Other widely used TGF-β signaling inhibitors with unrelated chemical structures also stimulated the growth of CK+ mouse primary epidermal cells in a dose-dependent manner (Supplementary Fig. S1). As the composition of CnT-PR media is not publicly available, we also used a chemically defined alternative, specifically a low Ca2+ keratinocyte basal medium, termed SFM. Our data show that RepSox treatment robustly stimulated the growth of CK+ mouse epidermal cells in SFM media to an extent similar to that in CnT-PR media (Supplementary Fig. S2). These data indicate that inhibition of TGF-β signaling stimulates the proliferation of mouse primary epidermal cells in basal media.
Inhibition of TGF-β signaling enables long-term expansion of p63+ mouse epidermal progenitor cells
As our data show that TGF-β signaling inhibition stimulates the proliferation of CK+ mouse primary epidermal cells but not of CK− cells (Fig. 1d), we next determined whether RepSox treatment enriches p63+ epidermal progenitor cells in crude tissue preparations. Although primary cells harvested from newborn mouse skin included 40% to 50% p63-negative (p63−) cells at passage 1 (P1) in the absence of RepSox (Figs 1e and S3), addition of RepSox in culture yielded approximately 80% of cells with p63 expression (Fig. 1e). As low Ca2+ media preferentially expand epidermal cells16, virtually all cells in both cultures with and without RepSox were CK+p63+ by P2. Notably, however, CK+p63+ epidermal progenitor cells were maintained beyond P5 only in the presence of RepSox, while the absence of RepSox treatment did not allow any cells to survive beyond P4 (Fig. 1e). These results indicate that inhibition of TGF-β signaling enriches p63+ epidermal progenitor cells in crude preparations of mouse epidermis in basal media.
Next, we determined the effect of TGF-β signaling inhibition on long-term proliferation of mouse epidermal progenitor cells that had been expanded by RepSox treatment in CnT-PR media. Notably, p63+ mouse epidermal progenitor cells grew at a constant rate at least for 60 days in the presence of RepSox (Fig. 1f). Proliferation of epidermal progenitor cells was RepSox-dependent, as removal of RepSox from the culture resulted in a rapid decline in the growth of epidermal cells (Fig. 1f), accompanied by elevated expression of phosphorylated Smad2/3 in both P5 and P20 epidermal progenitor cells (Fig. 1g,h). Consistent with these findings, gene expression analysis showed that RepSox treatment suppressed known TGF-β target CDK inhibitor genes30–32, including p21 Waf1/Cip1, p16 Ink4a, p19 Arf, and p15 Ink4b (Supplementary Fig. S4). These results indicate that inhibition of TGF-β signaling enables long-term expansion of p63+ mouse epidermal progenitor cells by promoting cell cycle progression in basal media.
Inhibition of TGF-β signaling supports self-renewal of mouse primary epidermal progenitor cells in 3T3-J2 co-culture
Clonogenic culture with 3T3-J2 cells has been used to assess self-renewal capacity of epithelial progenitor cells of many different species10–12, 33. However, except for some rare cell types (e.g. hair follicle stem cells)34, mouse primary epithelial cells do not form macroscopically visible colonies in serial cultures with 3T3-J2 cells, owing at least in part to their short lifespan in vitro. Given that inhibition of TGF-β signaling enables long-term proliferation of mouse epidermal progenitor cells (Fig. 1f), we next sought to determine whether TGF-β signaling inhibition supports their self-renewal in serial 3T3-J2 co-culture.
The RepSox-expanded newborn mouse epidermal cells were transferred onto 3T3-J2 cells at a clonal density and grown for two weeks in the presence or absence of RepSox. Notably, our data show that inclusion of RepSox in culture drastically enhanced the clonal growth of mouse primary epidermal cells (Fig. 2a,b). Removal of RepSox from the co-culture led to an accumulation of Smad2/3 in the nuclei of epidermal cells (Fig. 2c–f), indicating that TGF-β signaling in epidermal cells inversely correlates with their clonal growth. The majority of large clones grown in the presence of RepSox were composed of cytokeratin 14 (CK14)-positive immature epidermal cells with relatively small nuclear-to-cytoplasmic ratios and high p63 levels (Fig. 2g), resembling those in human epidermal stem cell clones termed holoclones10, 12. In contrast, the absence of RepSox in 3T3-J2 co-culture only produced smaller colonies and, even in the largest group of clones, the majority of epidermal cells were morphologically differentiated. Although indirect effects by feeder cells under TGF-β signaling inhibition need to be elucidated, these results clearly show that RepSox treatment supports self-renewal of p63+ mouse epidermal progenitor cells in 3T3-J2 co-culture.
Figure 2.
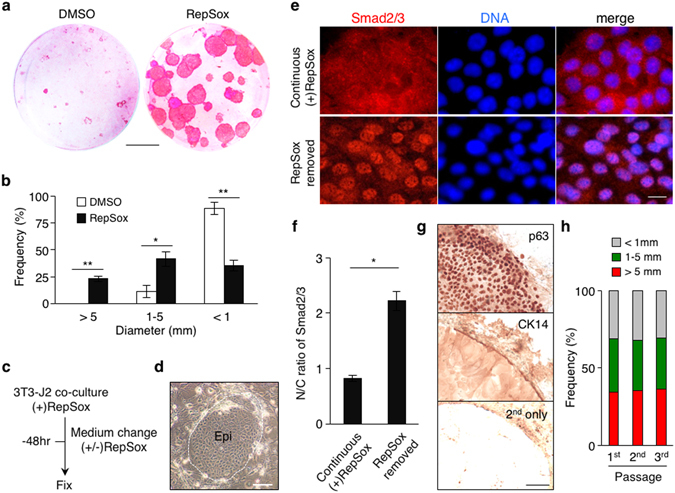
Inhibition of TGF-β signaling supports self-renewal of mouse primary epidermal progenitor cells in serial 3T3-J2 co-culture. (a) Rhodamine B staining of newborn mouse-derived, RepSox-expanded P1 epidermal cells grown in 3T3-J2 co-culture in the presence or absence of 1 μM RepSox for 14 days. Bar = 10 mm. (b) Distribution of epidermal clone sizes at day 14. Data shown are mean ± s.e.m. (n = 3). *P < 0.05; **P < 0.01. (c–f) Removal of RepSox associates with nuclear translocation of Smad2/3 in newborn mouse epidermal cells in 3T3-J2 co-culture. (c) Experimental scheme. Mouse primary epidermal keratinocytes were grown in 3T3-J2 co-culture in the presence of 1 μM RepSox. Subsequently, the cultures were maintained in the presence or absence of 1 μM RepSox for 48 hrs before fixation for immunohistochemistry. (d) A representative image of mouse primary keratinocyte clones in 3T3-J2 co-culture. Note that epidermal cell clones show a cobblestone appearance with a distinct clone border (dotted line). Epi, epidermal cell clone. Bar = 50 μm. (e) Representative immunofluorescence images of mouse epidermal cells without (upper) or with (lower) removal of RepSox, stained with anti-Smad2/3 antibodies (left panels) and counterstained with Hoechst 33342 (middle panels). Right panels show merged images. Bar = 10 μm. (f) Nuclear-to-cytoplasmic (N/C) ratios of Smad2/3 as determined by fluorescence intensity. Data shown are mean ± s.e.m. (n = 10). *P < 0.001. (g) Immunohistochemistry of representative epidermal cell clones grown in 3T3-J2 co-culture for 14 days in the presence of 1 μM RepSox, stained with anti-p63 (upper) and anti-CK14 (middle) antibodies. The lower panel represents a control clone stained with the secondary antibodies alone. Data shown are peripheral regions of three different epidermal clones with similar diameter. Bar = 100 μm. (h) Distribution of epidermal clone sizes in serial co-culture with 3T3-J2 cells in the presence of 1 μM RepSox.
To determine whether continuous suppression of TGF-β signaling enables long-term assessment of self-renewal of mouse epidermal progenitor cells, we harvested epidermal cells from the RepSox-treated 3T3-J2 co-culture and passaged them on fresh 3T3-J2 cells multiple times at a clonal density with two-week intervals in the presence or absence of RepSox. Our results show that, while in the absence of RepSox epidermal cells no longer formed holoclone-like clones in the subsequent passage, the presence of RepSox continuously produced holoclone-like large colonies at similar rates at least for three generations (Fig. 2h). These results indicate that continuous suppression of TGF-β signaling enables long-term assessment of self-renewal of mouse epidermal progenitor cells in serial 3T3-J2 co-culture.
Epidermal progenitor cells expanded by TGF-β signaling inhibition are capable of differentiation in vitro
To determine whether mouse epidermal progenitor cells expanded by TGF-β signaling inhibition are capable of differentiation in vitro, we performed Ca2+-mediated differentiation35 of epidermal progenitor cells that had been expanded by RepSox treatment in primary culture of newborn mouse epidermis. Treatment of the cells with Ca2+ for 3 days in the absence of RepSox led to an enlarged and flattened morphology with an increase in nuclear-to-cytoplasmic ratios, suggesting that they are differentiating (Fig. 3a). This is supported by changes in expression of epidermal cell differentiation markers (Fig. 3b,c). For example, treatment with Ca2+ decreased p63 expression at both mRNA and protein levels although a higher concentration of Ca2+ (1.3 mM) was required for this effect on mRNA levels than protein levels (0.3 mM Ca2+). These results are consistent with a previous report showing that p63 is subject to proteasome-mediated degradation during keratinocyte differentiation36. Expression of CK1, CK10, and loricrin, early-to-intermediate differentiation markers37–39, increased in response to 0.3 mM Ca2+ treatment (Fig. 3b,c). Likewise, expression of filaggrin and involucrin, markers for terminal differentiation40, 41, also increased by 0.3 mM Ca2+ treatment although the transcription of the Ivl gene remained relatively unchanged (Fig. 3b,c). These results indicate that mouse epidermal progenitor cells expanded by TGF-β signaling inhibition are capable of differentiation in response to Ca2+ stimulation in vitro.
Figure 3.
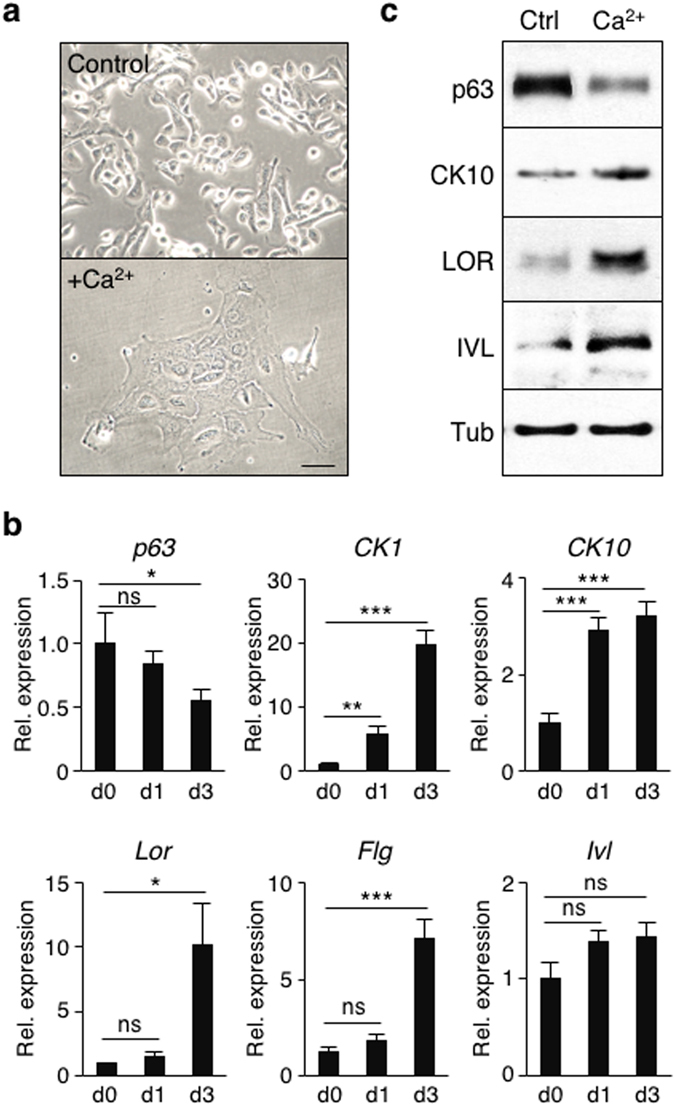
Mouse epidermal progenitor cells expanded by TGF-β inhibition are capable of differentiation in response to Ca2+ stimulation in vitro. (a) Representative images of newborn mouse-derived, RepSox-expanded P18 epidermal cells before (upper) and after (lower) treatment with 0.3 mM Ca2+ for 3 days. Bar = 25 μm. (b) Quantitative RT-PCR analysis of epidermal cell differentiation marker genes. Newborn mouse-derived, RepSox-expanded P17 epidermal cells were induced to differentiate by treatment with 0.3 mM Ca2+ (for CK1, CK10, Lor, Flg, and Ivl) or 1.3 mM Ca2+ (for p63) for 0–3 days as indicated. Data shown are normalized to the housekeeping gene Gapdh and expressed as mean ± s.e.m. (n = 3). *P < 0.05; **P < 0.01; ***P < 0.005; ns, not significant. (c) Western blot analysis using antibodies against p63, CK10, loricrin (LOR), and involucrin (IVL). Newborn mouse-derived, RepSox-expanded P17 epidermal cells were induced to differentiate by the treatment with 0.3 mM Ca2+ for 0 (Ctrl) and 3 (Ca2+) days. Tubulin-α (Tub) was used as a loading control. The full-length blots are included in Supplementary Figure S12.
Inhibition of TGF-β signaling enables long-term expansion of p63+ epidermal progenitor cells of postnatal mice
Our studies thus far utilized newborn mice to demonstrate that inhibition of TGF-β signaling enables long-term expansion of p63+ epidermal progenitor cells. To determine whether TGF-β signaling inhibition also expands p63+ epidermal progenitor cells of postnatal mice, we first cultivated crude primary cells from newborn, 4-week-old, and 10-week-old mouse epidermis in CnT-PR media (Fig. 4a,b). Our data show that, albeit at a lower level compared to the primary culture of newborn mouse epidermis, TGF-β signaling inhibition produced significant numbers of p63+ epidermal cells in cultures of 4-week-old mouse epidermis, and somewhat less in cultures of 10-week-old mouse epidermis (Fig. 4a,b). Similar to newborn mouse epidermis (Fig. 1e), RepSox treatment in crude culture of 4-week-old mouse epidermis yielded over 90% p63+ progenitor cells at P1 (Fig. 4c,d). Subsequently, RepSox-expanded P2 epidermal cells showed high clonogenic potential in 3T3-J2 co-culture (Fig. 4e,f) and were capable of differentiation in response to Ca2+ stimulation (Fig. 4g). We also show that p63+ epidermal progenitor cells derived from 4-week-old mice grew at a constant rate for at least 60 days in a RepSox-dependent manner (Fig. 4h). Together, these results indicate that inhibition of TGF-β signaling in crude primary cell culture of postnatal mouse epidermis selectively expands p63+ epidermal progenitor cells and supports their proliferative potential long term.
Figure 4.
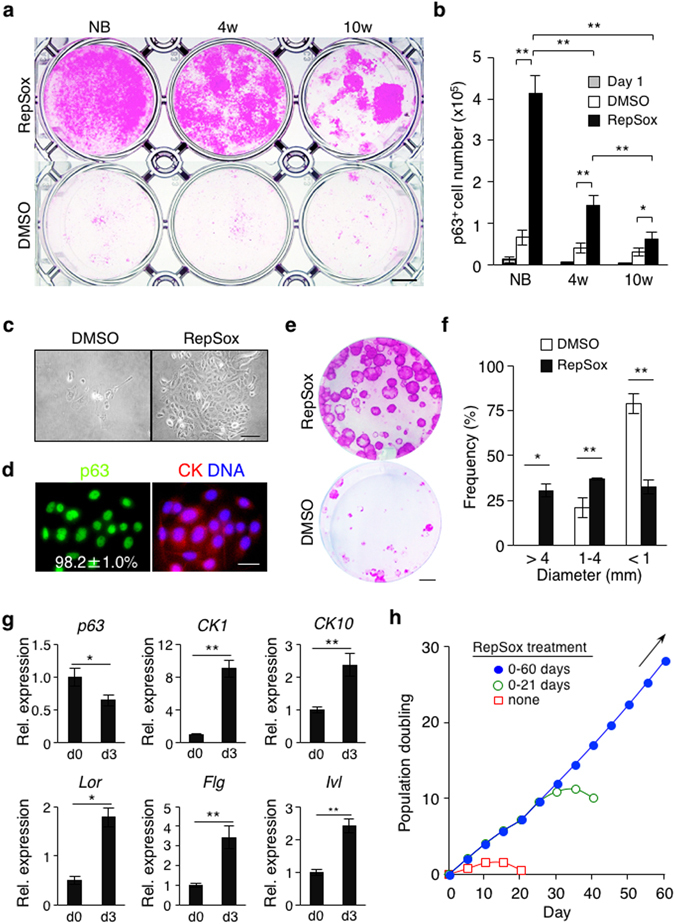
Inhibition of TGF-β signaling supports high proliferative potential of p63+ epidermal progenitor cells of postnatal mice. (a) Representative images of Rhodamine B staining of crude culture of epidermal cells harvested from newborn (NB), 4-week-old (4w), and 10-week-old (10w) mice and grown in CnT-PR media in the presence (upper) or absence (lower) of 1 μM RepSox for 14 days. One million primary cells were seeded. Data shown are representative of two independent experiments with similar results. Bar = 5 mm. (b) Total number of p63+ epidermal cells. One million primary cells were seeded and adherent cells were counted at day1 in replicative wells (grey bars) as the majority of the cells remained in suspension. Solid bars and open bars represent total numbers of p63+ epidermal cells at day 14 of culture in the subsequent presence of 1 μM RepSox or 0.1% DMSO, respectively (n = 3). *P < 0.05; **P < 0.01. (c) Representative images of 4-week-old mouse-derived P1 epidermal cells, grown in the presence (right) or absence (left) of 1 μM RepSox for 6 days. Bar = 50 μm. (d) Representative immunofluorescence images of 4-week-old mouse-derived, RepSox-expanded P1 epidermal cells stained with anti-p63 and anti-pan-CK antibodies and counterstained with Hoechst 33342 (DNA). The number shown in the left panel represents percentages of p63+ cells per field, expressed as mean ± s.e.m. (n = 4). Bar = 25 μm. (e) Representative images of Rhodamine B staining of epidermal clones grown for 14 days in the presence (upper) or absence (lower) of 1 μM RepSox. Four-week-old mouse-derived, RepSox-expanded P2 epidermal cells were used. Bar = 5 mm. (f) Distribution of epidermal clone sizes at day 14. Data shown are mean ± s.e.m (n = 3). *P < 0.05; **P < 0.01. (g) Quantitative RT-PCR analysis of epidermal cell differentiation marker genes. Four-week-old mouse-derived, RepSox-expanded P7 epidermal cells were induced to differentiate by treatment with 0.3 mM Ca2+ (for CK1, CK10, Lor, Flg, and Ivl) or 1.3 mM Ca2+ (for p63) for 3 days. Data shown are normalized to the housekeeping gene Gapdh and expressed as mean ± s.e.m (n = 3). *P < 0.05; **P < 0.01. (h) Population doubling of 4-week-old mouse-derived, RepSox-expanded P2 epidermal cells grown in CnT-PR media in the presence of 1 μM RepSox for 0, 21, and 60 days. Data shown are representative of three independent experiments with similar results. Arrow indicates continuous cell growth.
Inhibition of TGF-β signaling supports high proliferative potential of p63+ epithelial progenitor cells of diverse mouse epithelia
As p63 is expressed in the stem cell compartment of diverse epithelia in mice42, we next determined whether TGF-β signaling inhibition would also enrich and expand p63+ progenitor cells of epithelia in mouse tissues other than epidermis. To begin with, we prepared crude primary cells of various newborn mouse epithelia that contain p63+ progenitor cells, including the cornea, tongue, esophagus, salivary gland, bladder, oral epithelium, and thymus (Supplementary Fig. S5) and placed them in CnT-PR media in the presence or absence of RepSox. To validate the feasibility of TGF-β signaling inhibition in primary cultures of postnatal mouse epithelia, we also included some representative 4-week-old mouse epithelia, including the esophagus, salivary gland, bladder, and thymus. Our data show that RepSox treatment robustly enhanced the growth of primary cells of all newborn and 4-week-old mouse epithelia we examined (Figs 5a and S6a). In contrast, RepSox treatment in CnT-PR culture failed to stimulate the growth of primary cells of p63-independent epithelia, such as the small intestine and colon14, 15, (Supplementary Fig. S7). Although crude preparations of primary cultures of all p63-dependent epithelia contained non-epithelial cells and terminally differentiated p63− epithelial cells, just as in epidermis, >90% of cells from all epithelial cultures showed high p63 expression by P1 to P2 (Figs 5b and S6b), and virtually all growing cells were p63+ thereafter in all cultures derived from both newborn and 4-week-old mice. These results indicate that inhibition of TGF-β signaling in CnT-PR media selectively expands p63+ epithelial progenitor cells in crude tissue preparations of diverse mouse epithelia.
Figure 5.
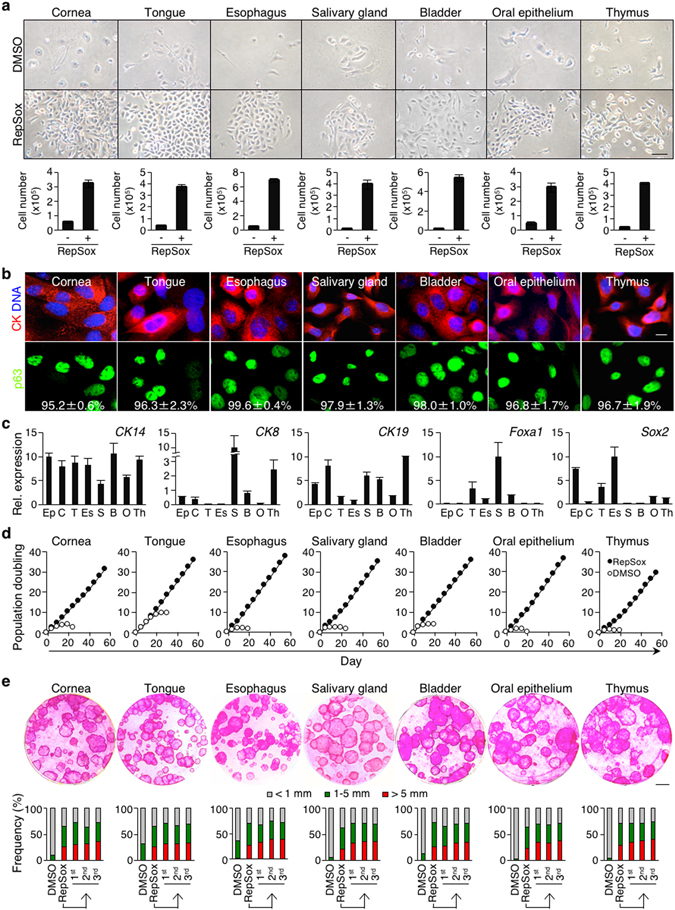
Inhibition of TGF-β signaling enables long-term expansion of p63+ epithelial progenitor cells of diverse mouse epithelia. (a) Representative images of primary cells of various newborn mouse epithelia, grown in the presence (lower) or absence (upper) of 1 μM RepSox for 6 days. Bar = 50 μm. Bar graphs below indicate numbers of primary cells at day 7 of culture. CnT-PR-expanded CK+ mouse primary epithelial cells (2 × 104) were seeded. Data shown are mean ± s.e.m. (n = 3). (b) Representative immunofluorescence images of newborn mouse-derived, RepSox-expanded P2 epithelial cells stained with anti-p63 and anti-pan-CK antibodies and counterstained with Hoechst 33342 (DNA). Numbers shown in lower panels represent percentages of p63+ cells per field, expressed as mean ± s.e.m. (n ≥ 4). Bar = 10 μm. (c) Quantitative RT-PCR analysis of epithelial progenitor cell-associated genes using newborn mouse-derived, RepSox-expanded P9 (thymus and salivary gland), P10 (epidermis), and P11 (cornea, oral epithelium, tongue, esophagus and bladder) cells. For each gene, the highest expression was set to 10. Data shown are normalized to the housekeeping gene Rps18 and expressed as mean ± s.e.m. (n = 3). Ep, epidermis; C, cornea; T, tongue; Es, esophagus; S, salivary gland; B, bladder; O, oral epithelium; and Th, thymus. (d) Population doubling of newborn mouse-derived, RepSox-expanded P2 epithelial cells grown in CnT-PR media in the presence (solid) or absence (open) of 1 μM RepSox. (e) 3T3-J2 co-culture. Representative images of Rhodamine B staining of epithelial clones grown for 14 days in the presence of 1 μM RepSox. Newborn mouse-derived, RepSox-expanded P1 epithelial cells were used. Bar = 5 mm. Data shown below indicate the distribution of epithelial clone sizes during serial 3T3-J2 co-cultures. Epithelial cells harvested from RepSox-treated primary 3T3-J2 co-cultures (lane 2) were serially cultivated with two-week intervals with fresh 3T3-J2 cells in the presence of 1 μM RepSox (lanes 3–5). DMSO, primary cultures without RepSox (lane 1).
To determine whether epithelial progenitor cells express markers for the corresponding progenitor cells in each epithelium after prolonged exposure to TGF-β inhibition, we examined relative expression of five representative, well-characterized genes, including CK14, CK8, CK19, Foxa1, and Sox2, using RepSox-expanded P9 to P11 newborn mouse epithelial progenitor cells (Fig. 5c). Consistent with uniformly high expression of p63, CK14, a common basal epithelial cell marker43, was expressed in all progenitor cells to a similar extent (Fig. 5c). In contrast, expression of tissue-specific CK markers44–49, CK8 and CK19, was relatively higher in progenitor cells of the salivary gland, thymus, bladder, and cornea than in the other progenitor cells (Fig. 5c). Expression of the transcription factor Foxa1 50 was substantially higher in the progenitor cells of the salivary gland, tongue, bladder, and esophagus than in the other progenitor cells, while Sox2 transcription factor51, 52 was expressed at considerably higher levels in the progenitor cells of the esophagus, epidermis, tongue, oral epithelium, and thymus than in the other progenitor populations (Fig. 5c). These results are consistent with previous reports on marker expression in tissue-specific progenitor cells43–52, suggesting that the gene expression signature in each progenitor population is preserved after prolonged exposure to TGF-β signaling inhibition.
Next, we determined whether inhibition of TGF-β signaling supports long-term proliferation and self-renewal of epithelial progenitor cells. Our data show that RepSox treatment robustly stimulated the expansion of all newborn and 4-week-old mouse epithelial progenitor cells long term, for at least 60 days when the experiment was terminated (Figs 5d and S6c). Our data also show that RepSox-expanded progenitor cells of all newborn and 4-week-old mouse epithelia produced holoclone-like large clones in 3T3-J2 co-cultures (Figs 5e and S6d), and in the case of newborn mouse epithelial progenitor cells, they all produced large clones at similar rates in three consecutive subcultures in the presence of RepSox (Fig. S5e, lower panels). The absence of RepSox in subculture produced very small or abortive clones that did not survive in the subsequent passage. These results indicate that inhibition of TGF-β signaling supports long-term proliferation and self-renewal of epithelial progenitor cells of diverse mouse epithelia.
Finally, we determined whether mouse epithelial progenitor cells expanded by TGF-β signaling inhibition are capable of differentiation in vitro using the esophagus, salivary gland, bladder, and thymus of both newborn and 4-week-old mice. To induce differentiation of epithelial progenitor cells, RepSox-expanded p63+ progenitor cells derived from each epithelium were cultivated in cFAD media10 in the absence of RepSox. Although cFAD has been developed for stimulating the growth of human epidermal keratinocytes in 3T3-J2 feeder cell co-culture, it allows differentiation of mouse epidermal progenitor cells in monolayer culture without 3T3-J2 cell support (Supplementary Fig. S8). Similar to epidermal progenitor cells, our data show that all epithelial progenitor cells of both newborn and 4-week-old mice tested decreased p63 expression in culture with cFAD as determined by qPCR (Figs 6a–d and S6e). Decrease in p63 protein levels was verified by Western blot using newborn mouse epithelial progenitor cells (Fig. 6a–d). We then analyzed expression of epithelial cell differentiation markers that are relatively well characterized and unique to each epithelial cell type. Our data show that expression of cytokeratin 4 (CK4), a marker of suprabasal cells in the esophagus53, increased by 28-fold and 10-fold in newborn and 4-week-old mouse epithelial progenitor cells, respectively, in response to cFAD treatment (Figs 6a and S6e). Expression of cytokeratin 7 (CK7) and aquaporin 5 (Aqp5), lineage-specific markers of ductal epithelia and acinar epithelia in the salivary glands, respectively54, significantly increased in progenitor cells derived from both newborn and 4-week-old mice in response to cFAD treatment (Figs 6b and S6e). Likewise, expression of cytokeratin 18 (CK18), a marker of the superficial layer of the bladder55, substantially increased in both newborn and 4-week-old mouse-derived epithelial progenitor cells in response to cFAD treatment (Figs 6c and S6e). Lastly, expression of CK18 and Ivl, markers of cortical epithelia and medullary epithelia of the thymus, respectively56, 57, increased significantly in both newborn and 4-week-old mouse epithelial progenitor cells upon cultivation in cFAD (Figs 6d and S6e). Increase of all differentiation markers at the protein levels was verified by Western blot using newborn mouse epithelial progenitor cells (Fig. 6a–d). Although the mechanisms of how cFAD induces differentiation of each epithelial cell type need to be determined, these results indicate that p63-dependent mouse epithelial progenitor cells expanded by TGF-β signaling inhibition are capable of differentiation in vitro.
Figure 6.
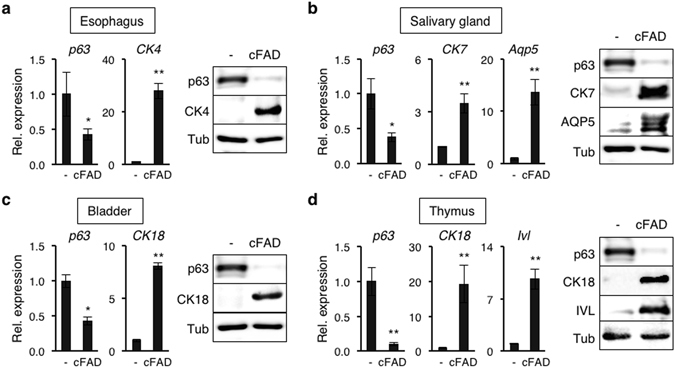
Mouse epithelial progenitor cells expanded by TGF-β inhibition are capable of differentiation in vitro. (a–d) Newborn mouse-derived epithelial progenitor cells expanded by RepSox treatment were grown in CnT-PR media in the presence of 1 μM RepSox to sub-confluence, followed by cultivation in cFAD for 7 days in the absence of RepSox. Epithelial progenitor cells used were derived from the (a) esophagus, (b) salivary gland, (c) bladder, and (d) thymus. Bar graphs on the left, quantitative RT-PCR analysis of epithelial cell differentiation marker genes, p63, CK4, CK7, CK18, Ivl, and Aqp5. Data shown are normalized to the housekeeping gene Gapdh and expressed as mean ± s.e.m. (n = 3). *P < 0.05; **P < 0.01. Right, Western blot analysis with antibodies against p63, CK4, CK7, CK18, aquaporin 5 (AQP5), and involucrin (IVL). Tubulin-α (Tub) was used as a loading control. The full-length blots are included in Supplementary Figure S13.
Discussion
In this report, we describe a simple and effective method of expanding p63+ epithelial progenitor cells from crude tissue preparations of diverse mouse epithelia using a single small molecule inhibitor of TGF-β signaling in a low Ca2+, feeder-free condition. Although dedifferentiation of terminally differentiated p63− epithelial cells into p63+ progenitor cells and/or transdifferentiation of p63− non-epithelial cells into a p63+ epithelial lineage could contribute to the observed increase of p63+ epithelial progenitor cell numbers, our data described below suggest that these two possibilities are unlikely. First, terminally differentiated mouse keratinocytes induced by Ca2+ stimuli remained non-proliferative upon exposure to TGF-β signaling inhibition (Supplementary Fig. S9). Second, inhibition of TGF-β signaling did not allow any cells from tissues supported by p63-independent progenitor cells (i.e. small intestine and colon) to expand in CnT-PR media (Supplementary Fig. S7), likely due to a lack of growth supporting factors for these epithelial cell types58, 59. Thus, we conclude that inhibition of TGF-β signaling in CnT-PR or other low Ca2+ basal media selectively expands p63+ epithelial progenitor cells without involving dedifferentiation of terminally differentiated p63− epithelial cells and/or transdifferentiation of p63− non-epithelial cells. This is particularly important as our methodology can significantly reduce the labor and cost associated with progenitor cell pre-purification steps, while efficiently expanding highly enriched populations of p63+ epithelial progenitor cells with high proliferative capacity.
Our data show that the numbers of p63+ epidermal progenitor cells expanded by RepSox treatment in culture declined in an age-dependent manner (Fig. 4b, solid bars). As cultures of newborn, 4-week-old, and 10-week-old mouse epidermal cells produced similar numbers of p63+ progenitor cells without RepSox treatment (Fig. 4b, open bars), these results suggest that the relative insensitivity to TGF-β signaling inhibition, rather than a decrease in proliferative cell numbers, was responsible for the age-associated decline in the expansion of p63+ epidermal progenitor cells. Although less profound, similar age-associated decrease in RepSox-mediated expansion of p63+ progenitor cells was observed in the other epithelial tissues, such as the esophagus, salivary gland, bladder, and thymus (Figs 5d and S6c). Despite intensive studies, however, the mechanisms that regulate age-associated alterations in quantity and/or quality of progenitor cells are controversial60, likely due to the involvement of complex signaling acting in these processes in both cell-autonomous and extrinsic manner. In this regard, it is important to note that our present work will provide a unique opportunity to assess age-dependent alterations in TGF-β signaling that may control versatile cell-intrinsic properties61 of a diverse array of p63+ epithelial progenitor cells.
Recently, it has been shown that several different mouse epithelial progenitor cells can be expanded from p63-dependent epithelia, such as the skin, prostate, mammary gland, and thymus, in 3T3-J2 co-culture in the presence of Rho-associated protein kinase (Rock) inhibitors20, 22, 62–64. Although the molecular mechanisms underlying Rock inhibition-mediated expansion of epithelial progenitor cells are unclear, TGF-β signaling has been shown to regulate the Rho signaling pathway in normal mouse mammary gland epithelial cells65. In addition, TGF-β signaling phosphorylates p63, which in turn leads to decreased proliferative capacity of epithelial progenitor cells66, 67. Elucidating potential mechanisms by which TGF-β and Rho signaling pathways cooperate with p63 may help to better understand the molecular mechanisms controlling the proliferative potential of epithelial progenitor cells. Nonetheless, the development of feeder-free culture of p63+ mouse primary epithelial progenitor cells described in this study will allow us to better perform many downstream applications of epithelial analysis, in which high numbers of p63+ epithelial progenitor cells are required. Some examples of such applications would include gene expression studies, molecular and cellular analyses in vitro, and manipulation of stem cells ex vivo, followed by their transplantation and characterization in vivo.
Smad-dependent growth control and differentiation are mediated by both TGF-β and bone morphogenetic protein (BMP) signaling pathways in a context-dependent manner25. Indeed, dual inhibition of both pathways has been shown to synergistically promote neural conversion of embryonic stem (ES) cells and induced pluripotent stem (iPS) cells68, 69. A recent report by Mou et al. has described an analogous approach in which dual inhibition of TGF-β and BMP signaling, in addition to Rock inhibition, can expand mouse airway epithelial progenitor cells23. However, it is not clear if inhibition of BMP signaling is required for promoting proliferation of other mouse epithelial progenitor cells. Indeed, our data show that inhibition of BMP signaling did not improve the enhanced proliferation of mouse primary epidermal progenitor cells mediated by TGF-β signaling inhibition (Supplementary Figs S1 and S10). Nevertheless, we showed that inhibition of TGF-β signaling selectively expands diverse p63+ mouse epithelial progenitor cells and increases their cellularity by 109-fold (equivalent to PD 30) to 1012-fold (PD 40) within 60 days (Figs 1f, 4h and 5d and S6c). Mou et al. achieved similar results by dual TGF-β/BMP inhibition in mouse airway epithelial progenitor cells23. These data strongly suggest that inhibition of TGF-β signaling alone is sufficient for the expansion of p63+ epithelial progenitor cells of many different mouse epithelia.
In conclusion, we have shown that inhibition of TGF-β signaling in basal media enables the expansion of p63+ epithelial progenitor cells without the need of progenitor cell-purification steps or a feeder cell layer. We anticipate that this simple and effective approach to enrich and propagate p63+ primary epithelial progenitor cells will facilitate the use of normal as well as genetically engineered mouse models for downstream analyses of diverse p63-dependent epithelia.
Methods
Mice
All experimental procedures were carried out in accordance with protocols approved by the Institutional Animal Care and Use Committees (IACUC) at the University of Pennsylvania and Boston University. Mice used in this study were on a C57BL/6 background (Jackson Laboratories, Bar Harbor, ME; Charles River, Wilmington, MA). The day of the vaginal plug was designated embryonic day 0.5 (E0.5).
Tissue isolation and cell culture
All tissues harvested were sterilized in Isojin (Meiji Seika Pharma Co., Ltd., Tokyo, Japan), followed by three washes in phosphate buffered saline (PBS). Primary cells of mouse epidermis were prepared as described16, 70 with some modifications. Briefly, newborn mouse skin was harvested from E19.0 to postnatal day 1 mice and incubated in 0.25% trypsin for 2 hrs at room temperature (RT), followed by collection of epidermis using forceps and incubation in 0.25% trypsin for an additional 5 min at RT. Postnatal mouse skin was harvested from 4-week-old and 10-week-old mice and processed as described above. Newborn mouse tissues including the salivary gland, thymus, esophagus, colon, small intestine, and bladder were harvested from E19.0 to postnatal day1 mice, minced into small pieces by scissors, and incubated in 0.25% trypsin for 30 min at 37 °C with vigorous pipetting every 10 min. To isolate primary cells from corneal, lingual, and oral epithelia of newborn mice, eyeballs71, tongue72, and palatal shelves73, respectively, were harvested from E19.0 to postnatal day 1 mice and incubated in 0.25% trypsin for 2 hrs at RT. Loosened epithelia were removed using forceps and incubated in 0.25% trypsin for an additional 5 min at 37 °C. Postnatal mouse tissues including the salivary gland, thymus, esophagus, and bladder were harvested from 4-week-old mice and minced into small pieces by scissors. Subsequently, the salivary gland and thymus were incubated in DMEM containing a mixture of Dispase II (0.5 mg/ml, Roche Diagnostics, Indianapolis, IN), Collagenase (0.5 mg/ml, Worthington, Lakewood, NJ), DNase (0.01 mg/ml, Worthington), and 0.125% trypsin for 20 min at 37 °C with vigorous pipetting every 10 min. The esophagus and bladder were incubated in DMEM containing Dispase II (1 mg/ml), Collagenase (1 mg/ml), and DNase (0.02 mg/ml) for 40 min at 37 °C with vigorous pipetting every 20 min. Dissociated cells from all newborn and postnatal mouse tissues were neutralized with fetal bovine serum (FBS, Hyclone, Logan, UT) and filtered through a 70 μm cell strainer. After centrifugation at 1,500 rpm (400 g) for 5 min, the cells were washed twice in PBS and suspended in CnT-PR epithelial culture basal media (Cellntec, Bern, Switzerland). In some experiments, crude primary cells were plated at a high density and serially passaged 2–3 times in CnT-PR media to deplete all non-epithelial cells. By passage 3, virtually all growing cells in CnT-PR media were cytokeratin (CK)-positive epithelial cells as determined by immunohistochemistry. Other basal media used in this study include Keratinocyte-SFM basal media with growth supplement (Invitrogen). The cells were grown in the presence or absence of 1 ng/ml TGF-β2 (Invitrogen, Carlsbad, CA) or 0.1–10 μM inhibitors (RepSox, A83-01, LY364947, SB525334, SB431542, and DMH-1) (all from Cayman Chemical, Ann Arbor, MI) as indicated. All primary cell cultures were performed in a humidified chamber at 37 °C with 5% CO2.
To assess cell numbers, the cells were incubated in 0.25% trypsin for 5–10 min, neutralized with 10% FBS in DMEM, and washed in PBS, followed by cell counting in a glass hemocytometer chamber (Hausser Scientific, Horsham, PA) under a CKX41 Inverted Microscope (Olympus, Center Valley, PA) with a 10X objective. To measure population doubling (PD), the cells were seeded at a density of 2 × 104 cells per well in 12-well plates, grown in CnT-PR media in the presence or absence of 1 μM RepSox, and serially passaged every 5 days. PD was calculated as PD = 3.32 (log [cell number yielded/cell number inoculated]).
To induce differentiation of epidermal keratinocytes, sub-confluent cells in 12-well plates were washed twice in PBS and cultivated in CnT-PR media containing 0.3 or 1.3 mM CaCl2 for 1–5 days as indicated. To induce differentiation of other epithelial progenitor cells of the esophagus, salivary gland, bladder, and thymus, RepSox (1 μM)-expanded cells were grown to sub-confluence in CnT-PR media in 12-well plates, followed by cultivation in cFAD media for 7 days in the absence of RepSox. In both differentiation assays, culture media were replaced every 24 to 48 hrs and cells were harvested at the time points indicated for subsequent analyses.
3T3-J2 co-culture
Clonogenic culture with 3T3-J2 cells was performed as described10. Briefly, mouse epithelial cells were seeded at a clonal density on lethally γ-irradiated 3T3-J2 cells (gift from H. Green, Harvard Medical School) in complete FAD (cFAD) media and grown in the presence or absence of 1 μM RepSox in a humidified chamber at 37 °C with 5% CO2. To perform serial co-culture with 3T3-J2 cells, epithelial cells were harvested from the initial 3T3-J2 co-cultures by trypsinization in 0.25% trypsin for 5–10 min and equal numbers of epithelial cells were serially cultured with γ-irradiated fresh 3T3-J2 cells with two-week intervals. To visualize epithelial colonies, culture plates were fixed in 10% buffered formalin for 10 min and stained with 1% Rhodamine B (Sigma-Aldrich, St. Louis, MO). To determine clone sizes, the whole plates were photographed with a Nikon digital camera Coolpix S10 (Nikon, Melville, NY) and the diameter of each clone was measured with the aid of Adobe Photoshop (Adobe, San Jose, CA) software.
Immunohistochemistry and Immunofluorescence
Immunofluorescence and immunohistochemistry were performed as described67. Briefly, cultured cells were fixed in 10% buffered formalin for 10 min and permeabilized in PBS/0.1% Triton X-100 for 5 min, followed by blocking with 10% FBS for 30 min and incubation with primary antibodies overnight at 4 °C. After three washes in PBS containing 0.1% Tween-20 (PBST), the cells were incubated with either fluorescence-labeled secondary antibodies for 1 hr followed by counterstaining with 0.1 μg/ml Hoechst 33342 (Invitrogen) or HRP-conjugated secondary antibodies followed by detection of the antigens using a DAB reagent kit (KPL, Gaithersburg, MD).
Mouse tissues were fixed in 10% buffered formalin overnight at 4 °C. Paraffin-embedded sections were cut into 6 μm sections and antigen retrieval was performed by incubating the slides in 0.01 M citric acid buffer (pH6.0) at 95 °C for 20 min. Sections were then blocked with 10% FBS at RT for 30 min and incubated with primary antibodies overnight at 4 °C. After three washes in PBST, the slides were incubated with HRP-conjugated secondary antibodies and the antigens were detected using a DAB reagent kit (KPL).
BrdU labeling and detection
A short-pulse with 5-bromo-2′-deoxyuridine (BrdU) was performed as described9. Briefly, cultured cells were incubated for 30 min with 10 μM BrdU (Roche Diagnostics), fixed in 10% buffered formalin at RT for 10 min, and permeabilized in PBS/0.1% Triton X-100 for 5 min, followed by denaturation in 2 N HCl for 15 min at 37 °C. After blocking with 10% FBS, the cells were incubated with mouse anti-pan-CK and rat anti-BrdU antibodies, and then stained with pre-adsorbed secondary antibodies. The frequency of CK+BrdU+ cell populations was determined using a Nikon Eclipse 80i microscope and NIS Elements software (Nikon).
Western blot
Cells were lysed in RIPA buffer containing 50 mM Tris-HCl (pH6.8), 5% 2-mercaptoethanol, 2% sodium dodecyl sulfate, 10% glycerol and 1 mM phenylmethanesulfonyl fluoride (all from Sigma-Aldrich). Equal amounts of proteins were separated by 8% SDS-PAGE and transferred to Immobilon-P membranes (Millipore, Billerica, MA). Blots were developed using the ECL detection reagent (GE Healthcare Bio-Sciences, Pittsburgh, PA).
Antibodies
Primary antibodies used in this study were mouse anti-p63 (4A4, Santa Cruz Biotechnology, Santa Cruz, CA, sc-8431, 1:1,000 IF/IHC/WB), rabbit anti-p63 (H137, Santa Cruz Biotechnology, sc-8343, 1:100 IF), mouse anti-pan-cytokeratin (CK) (AE1/AE3, Thermo Fisher Scientific, Waltham, MA, MS-343, 1:100 IF), mouse anti-CK4 (6B10, Santa Cruz Biotechnology, sc-52321, 1:100 WB), mouse anti-CK7 (RCK105, Santa Cruz Biotechnology, sc-23876, 1:500 WB), mouse anti-CK10 (RSKE60, Abcam, Cambridge, MA, ab9025, 1:1,000 WB), mouse anti-CK14 (LL001, Santa Cruz Biotechnology, sc-53253, 1:100 IHC), mouse anti-CK18 (RGE53, Santa Cruz Biotechnology, sc-32329, 1:200 WB), rabbit anti-loricrin (BioLegend, San Diego, CA, PRB-145P, 1:4,000 WB), rabbit anti-involucrin (M-116, Santa Cruz Biotechnology, sc-28558, 1:2,000 IF and 1:100 WB), mouse anti-aquaporin 5 (D-7, Santa Cruz Biotechnology, sc-514022, 1:50 WB), mouse anti-tubulin-α (12G10, Developmental Studies Hybridoma Bank, University of Iowa, 1:500 WB), rat anti-BrdU (ICR1, Abcam, ab6326, 1:100 IF), rabbit anti-phosphorylated Smad2/3 (D27F4, Cell Signaling Technology, Danvers, MA, #8628, 1:1,000 WB), and rabbit anti-Smad2/3 (D7G7, Cell Signaling Technology, #8685, 1:1,000 IF/WB).
Secondary antibodies used for immunofluorescence were Alexa 488-goat anti-mouse IgG, Alexa 488-goat anti-rabbit IgG, Alexa 594-goat anti-mouse IgG, Alexa 594-goat anti-rabbit IgG, (Molecular Probes, Grand Island, NY, 1:1,000 IF), pre-adsorbed DyLight488-goat anti-rat IgG, and pre-adsorbed DyLight594-goat anti-mouse IgG (Abcam, 1:1,000 IF). Secondary antibodies used for immunohistochemistry and Western blot were horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (KPL, 1:250 IHC and 1:5,000 WB), and HRP-conjugated goat anti-rabbit IgG (Cell Signaling Technology, 1:500 WB).
RNA isolation and quantitative RT-PCR
Total RNA was isolated using Trizol reagent according to the manufacturer’s instructions (Invitrogen) and 1 μg total RNA was reverse-transcribed using the ProtoScript M-MuLV First Strand cDNA Synthesis Kit (New England Biolabs, Ipswich, MA). Quantitative PCR was performed using the SYBR Green PCR Master Mix on an ABI 7900 HT or an ABI StepOnePlus machine according to the manufacturer’s instructions (Invitrogen). Relative expression of each gene was determined using the ∆∆Ct method and normalized to the housekeeping gene Gapdh or Rps18 as indicated. Primer sequences used in this study are listed in Supplementary Table S1.
Image analysis
Imaging was done on a Nikon Eclipse 80i microscope with a Nikon DS-Qi1Mc digital camera (Nikon) or a BZ-X710 microscope (Keyence Corp., Osaka, Japan) and analyzed with ImageJ (NIH, Bethesda, MD) and Adobe Photoshop software. To determine the nuclear-to-cytoplasmic ratios, signal intensities of Smad2/3 staining in the nucleus (NSmad2/3) and cytoplasm (CSmad2/3) of individual cells were measured using ImageJ and applied to the equation Ratio = (N Smad2/3 − BG)/(C Smad2/3 − BG), where BG indicates a background signal. To determine BrdU+, p63+, and CK+ cell numbers, randomly selected areas were photographed (n = 3–5 fields) and the captured immunofluorescence images were processed for manual counting of positive cells using Adobe Photoshop software. Counterstaining with Hoechst 33342 was used to determine the total cell numbers.
Statistical analysis
Values are reported as mean ± standard error of the mean (s.e.m.). Student’s t-tests were performed where P < 0.05 was considered statistically significant.
Data Availability
All data generated or analyzed during this study are included in this published article and its Supplementary Information files.
Electronic supplementary material
Acknowledgements
We thank Jeff Holcombe for proofreading of the manuscript. This study was supported by an R01AR066755 grant from the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health to M.S.
Author Contributions
D.S. designed and performed the experiments, collected and analyzed data, and drafted the manuscript; F.P. designed and performed the experiments, and collected and analyzed data; M.S. conceived and designed the work, interpreted data, and wrote the manuscript. All authors approved the final version of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-06470-y
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Blanpain, C. & Fuchs, E. Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science344, 1242281(1–10) (2014). [DOI] [PMC free article] [PubMed]
- 2.Donati G, Watt FM. Stem cell heterogeneity and plasticity in epithelia. Cell Stem Cell. 2015;16:465–476. doi: 10.1016/j.stem.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Van Keymeulen A, Blanpain C. Tracing epithelial stem cells during development, homeostasis, and repair. J. Cell Biol. 2012;197:575–584. doi: 10.1083/jcb.201201041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rheinwald JG, Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 1975;6:331–343. doi: 10.1016/S0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- 5.Green H. The birth of therapy with cultured cells. Bioessays. 2008;30:897–903. doi: 10.1002/bies.20797. [DOI] [PubMed] [Google Scholar]
- 6.Pellegrini G, Rama P, Mavilio F, De Luca M. Epithelial stem cells in corneal regeneration and epidermal gene therapy. J. Pathol. 2009;217:217–228. doi: 10.1002/path.2441. [DOI] [PubMed] [Google Scholar]
- 7.Rama P, et al. Limbal stem-cell therapy and long-term corneal regeneration. N. Engl. J. Med. 2010;363:147–155. doi: 10.1056/NEJMoa0905955. [DOI] [PubMed] [Google Scholar]
- 8.Barreca A, et al. In vitro paracrine regulation of human keratinocyte growth by fibroblast-derived insulin-like growth factors. J. Cell Physiol. 1992;151:262–268. doi: 10.1002/jcp.1041510207. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki D, Senoo M. Dact1 regulates the ability of 3T3-J2 cells to support proliferation of human epidermal keratinocytes. J. Invest. Dermatol. 2015;135:2894–2897. doi: 10.1038/jid.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl. Acad. Sci. USA. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oshima H, Rochat A, Kedzia C, Kobayashi K, Barrandon Y. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell. 2001;104:233–245. doi: 10.1016/S0092-8674(01)00208-2. [DOI] [PubMed] [Google Scholar]
- 12.Senoo M, Pinto F, Crum CP, McKeon F. p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 13.Truong AB, Kretz M, Ridkey TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mills AA, et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 15.Yang A, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 16.Lichti U, Anders J, Yuspa SH. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat. Protoc. 2008;3:799–810. doi: 10.1038/nprot.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ha L, Ponnamperuma RM, Jay S, Ricci MS, Weinberg WC. Dysregulated ΔNp63α inhibits expression of Ink4a/arf, blocks senescence, and promotes malignant conversion of keratinocytes. PLoS One. 2011;6:e21877. doi: 10.1371/journal.pone.0021877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Missero C, Di Cunto F, Kiyokawa H, Koff A, Dotto GP. The absence of p21Cip/WAF1 alters keratinocyte growth and differentiation and promotes ras-tumor progression. Genes Dev. 1996;10:3065–3075. doi: 10.1101/gad.10.23.3065. [DOI] [PubMed] [Google Scholar]
- 19.Paramio JM, et al. The ink4a/arf tumor suppressors cooperate with p21cip1/waf in the processes of mouse epidermal differentiation, senescence, and carcinogenesis. J. Biol. Chem. 2001;276:44203–44211. doi: 10.1074/jbc.M105650200. [DOI] [PubMed] [Google Scholar]
- 20.Chapman S, McDermott DH, Shen K, Jang MK, McBride AA. The effect of Rho kinase inhibition on long-term keratinocyte proliferation is rapid and conditional. Stem Cell Res. Ther. 2014;5:60. doi: 10.1186/scrt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King KE, et al. ΔNp63α functions as both a positive and a negative transcriptional regulator and blocks in vitro differentiation of murine keratinocytes. Oncogene. 2003;22:3635–3644. doi: 10.1038/sj.onc.1206536. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mou H, et al. Dual SMAD signaling inhibition enables long-term expansion of diverse epithelial basal cells. Cell Stem Cell. 2016;19:217–231. doi: 10.1016/j.stem.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Well-differentiated human airway epithelial cell cultures. Methods Mol. Med. 2005;107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- 25.Watabe T, Miyazono K. Roles of TGF-β family signaling in stem cell renewal and differentiation. Cell Res. 2009;19:103–115. doi: 10.1038/cr.2008.323. [DOI] [PubMed] [Google Scholar]
- 26.Schmierer B, Hill CS. TGFβ-SMAD signal transduction: molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 2007;8:970–982. doi: 10.1038/nrm2297. [DOI] [PubMed] [Google Scholar]
- 27.Ikushima H, Miyazono K. TGFβ signalling: a complex web in cancer progression. Nat. Rev. Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 28.Bascom CC, et al. Complex regulation of transforming growth factor β1, β2, and β3 mRNA expression in mouse fibroblasts and keratinocytes by transforming growth factors β1 and β2. Mol. Cell. Biol. 1989;9:5508–5515. doi: 10.1128/MCB.9.12.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glick AB, et al. Loss of expression of transforming growth factor β in skin and skin tumors is associated with hyperproliferation and a high risk for malignant conversion. Proc. Natl. Acad. Sci. USA. 1993;90:6076–6080. doi: 10.1073/pnas.90.13.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reynisdóttir I, Polyak K, Iavarone A, Massagué J. Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-β. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 31.Tremain R, et al. Defects in TGFβ signaling overcome senescence of mouse keratinocytes expressing v-rasHa. Oncogene. 2000;19:1698–1709. doi: 10.1038/sj.onc.1203471. [DOI] [PubMed] [Google Scholar]
- 32.Vijayachandra K, Higgins W, Lee J, Glick A. Induction of p16ink4a and p19ARF by TGFβ1 contributes to growth arrest and senescence response in mouse keratinocytes. Mol. Carcinog. 2009;48:181–186. doi: 10.1002/mc.20472. [DOI] [PubMed] [Google Scholar]
- 33.Majo F, Rochat A, Nicolas M, Jaoudé GA, Barrandon Y. Oligopotent stem cells are distributed throughout the mammalian ocular surface. Nature. 2008;456:250–254. doi: 10.1038/nature07406. [DOI] [PubMed] [Google Scholar]
- 34.Nowak JA, Fuchs E. Isolation and culture of epithelial stem cells. Methods Mol. Biol. 2009;482:215–232. doi: 10.1007/978-1-59745-060-7_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L. Mouse epidermal keratinocyte culture. Methods Mol. Biol. 2013;945:177–191. doi: 10.1007/978-1-62703-125-7_12. [DOI] [PubMed] [Google Scholar]
- 36.Rossi M, et al. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc. Natl. Acad. Sci. USA. 2006;103:12753–12758. doi: 10.1073/pnas.0603449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freedberg IM, Tomic-Canic M, Komine M, Blumenberg M. Keratins and the keratinocyte activation cycle. J. Invest. Dermatol. 2001;116:633–640. doi: 10.1046/j.1523-1747.2001.01327.x. [DOI] [PubMed] [Google Scholar]
- 38.Ivanyi D, et al. New monoclonal antibodies recognizing epidermal differentiation-associated keratins in formalin-fixed, paraffin-embedded tissue. Keratin 10 expression in carcinoma of the vulva. J. Pathol. 1989;159:7–12. doi: 10.1002/path.1711590105. [DOI] [PubMed] [Google Scholar]
- 39.Hohl D, et al. Characterization of human loricrin. Structure and function of a new class of epidermal cell envelope proteins. J. Biol. Chem. 1991;266:6626–6636. [PubMed] [Google Scholar]
- 40.Fleckman P, Dale BA, Holbrook KA. Profilaggrin, a high- molecular-weight precursor of filaggrin in human epidermis and cultured keratinocytes. J. Invest. Dermatol. 1985;85:507–512. doi: 10.1111/1523-1747.ep12277306. [DOI] [PubMed] [Google Scholar]
- 41.Eckert RL, Green H. Structure and evolution of the human involucrin gene. Cell. 1986;46:583–589. doi: 10.1016/0092-8674(86)90884-6. [DOI] [PubMed] [Google Scholar]
- 42.Yang A, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998;2:305–316. doi: 10.1016/S1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 43.Vassar R, Rosenberg M, Ross S, Tyner A, Fuchs E. Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice. Proc. Natl. Acad. Sci. USA. 1989;86:1563–1567. doi: 10.1073/pnas.86.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennett AR, et al. Identification and characterization of thymic epithelial progenitor cells. Immunity. 2002;16:803–814. doi: 10.1016/S1074-7613(02)00321-7. [DOI] [PubMed] [Google Scholar]
- 45.Kawashima M, Kawakita T, Yoshida S, Shimmura S, Tsubota K. Nucleostemin as a possible progenitor marker of corneal epithelial cells. Mol. Vis. 2009;15:1162–1168. [PMC free article] [PubMed] [Google Scholar]
- 46.Kobayashi T, Owczarek TB, McKiernan JM, Abate-Shen C. Modelling bladder cancer in mice: opportunities and challenges. Nat. Rev. Cancer. 2015;15:42–54. doi: 10.1038/nrc3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsumoto S, et al. Isolation of tissue progenitor cells from duct-ligated salivary glands of swine. Cloning Stem Cells. 2007;9:176–190. doi: 10.1089/clo.2006.0022. [DOI] [PubMed] [Google Scholar]
- 48.Nanduri LS, et al. Salisphere derived c-Kit+ cell transplantation restores tissue homeostasis in irradiated salivary gland. Radiother. Oncol. 2013;108:458–463. doi: 10.1016/j.radonc.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 49.Zhang W, Zhao J, Chen L, Urbanowicz MM, Nagasaki T. Abnormal epithelial homeostasis in the cornea of mice with a destrin deletion. Mol. Vis. 2008;14:1929–1939. [PMC free article] [PubMed] [Google Scholar]
- 50.Besnard V, Wert SE, Hull WM, Whitsett JA. Immunohistochemical localization of Foxa1 and Foxa2 in mouse embryos and adult tissues. Gene Expr. Patterns. 2004;5:193–208. doi: 10.1016/j.modgep.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 51.Arnold K, et al. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu K, et al. Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell Stem Cell. 2013;12:304–315. doi: 10.1016/j.stem.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeWard AD, Cramer J, Lagasse E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 2014;9:701–711. doi: 10.1016/j.celrep.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nanduri LS, et al. Purification and ex vivo expansion of fully functional salivary gland stem cells. Stem Cell Reports. 2014;3:957–964. doi: 10.1016/j.stemcr.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karni-Schmidt O, et al. Distinct expression profiles of p63 variants during urothelial development and bladder cancer progression. Am. J. Pathol. 2011;178:1350–1360. doi: 10.1016/j.ajpath.2010.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klug DB, et al. Interdependence of cortical thymic epithelial cell differentiation and T-lineage commitment. Proc. Natl. Acad. Sci. USA. 1998;95:11822–11827. doi: 10.1073/pnas.95.20.11822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yano M, et al. Aire controls the differentiation program of thymic epithelial cells in the medulla for the establishment of self-tolerance. J. Exp. Med. 2008;205:2827–2838. doi: 10.1084/jem.20080046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 59.Wang X, et al. Cloning and variation of ground state intestinal stem cells. Nature. 2015;522:173–178. doi: 10.1038/nature14484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gundermann, S., Stark, H. J. & Boukamp, P. Aging of stem cells in skin: What is driving the aging process? in Stem cell aging: Mechanisms, consequences, Rejuvenation (ed. Geiger, H., Jasper, H., Florian, M. C.) 149–174 (Springer, 2015).
- 61.Budi, E. H., Duan, D. & Derynck, R. Transforming growth factor-β receptors and Smads: Regulatory complexity and functional versatility. Trends Cell Biol. pii:S0962-8924(17)30063-6, doi:10.1016/j.tcb.2017.04.005 (2017). [DOI] [PubMed]
- 62.Kim MJ, Miller CM, Shadrach JL, Wagers AJ, Serwold T. Young, proliferative thymic epithelial cells engraft and function in aging thymuses. J. Immunol. 2015;194:4784–4795. doi: 10.4049/jimmunol.1403158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saenz FR, et al. Conditionally reprogrammed normal and transformed mouse mammary epithelial cells display a progenitor-cell-like phenotype. PLoS One. 2014;9:e97666. doi: 10.1371/journal.pone.0097666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang L, et al. ROCK inhibitor Y-27632 suppresses dissociation-induced apoptosis of murine prostate stem/progenitor cells and increases their cloning efficiency. PLoS One. 2011;6:e18271. doi: 10.1371/journal.pone.0018271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bhowmick NA, et al. TGF-β-induced RhoA and p160ROCK activation is involved in the inhibition of Cdc25A with resultant cell-cycle arrest. Proc. Natl. Acad. Sci. USA. 2003;100:15548–15553. doi: 10.1073/pnas.2536483100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cherukuri P, et al. Phosphorylation of ΔNp63α via a novel TGFβ/ALK5 signaling mechanism mediates the anti-clonogenic effects of TGFβ. PLoS One. 2012;7:e50066. doi: 10.1371/journal.pone.0050066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suzuki D, Senoo M. Increased p63 phosphorylation marks early transition of epidermal stem cells to progenitors. J. Invest. Dermatol. 2012;132:2461–2464. doi: 10.1038/jid.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou J, et al. High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor beta superfamily receptors. Stem Cells. 2010;28:1741–1750. doi: 10.1002/stem.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dlugosz AA, Glick AB, Tennenbaum T, Weinberg WC, Yuspa SH. Isolation and utilization of epidermal keratinocytes for oncogene research. Methods Enzymol. 1995;254:3–20. doi: 10.1016/0076-6879(95)54003-2. [DOI] [PubMed] [Google Scholar]
- 71.Kobayashi T, Yoshioka R, Shiraishi A, Ohashi Y. New technique for culturing corneal epithelial cells of normal mice. Mol. Vis. 2009;15:1589–1593. [PMC free article] [PubMed] [Google Scholar]
- 72.Michlig S, Damak S, Le Coutre J. Claudin-based permeability barriers in taste buds. J. Comp. Neurol. 2007;502:1003–1011. doi: 10.1002/cne.21354. [DOI] [PubMed] [Google Scholar]
- 73.Nawshad A, Medici D, Liu CC, Hay ED. TGFβ3 inhibits E-cadherin gene expression in palate medial-edge epithelial cells through a Smad2-Smad4-LEF1 transcription complex. J. Cell Sci. 2007;120:1646–1653. doi: 10.1242/jcs.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Supplementary Information files.


