Abstract
We describe here the partial purification of a novel Escherichia coli endoribonuclease, RNase K. This protein catalyses site-specific cleavages in the 5' region of in vitro transcribed ompA and bla transcripts. Some of the resulting cleavage products are also found in cellular ompA mRNA, defining the in vivo activity of RNase K. The following evidence suggests that RNase K initiates mRNA degradation. First, RNase K cleavages are suppressed in the ams mutant, which has a generally prolonged mRNA half-life. Secondly, RNase K cleavage products seem to have very short half-lives in vivo, indicating that they are decay intermediates rather than processing products. Thirdly, the differences in in vivo half-life between the ompA and bla mRNAs are mimicked in in vitro decay reactions with purified RNase K. The relationship between RNase K and the ams locus might point to a more general role of RNase K in mRNA degradation. We discuss the influence of mRNA secondary structure on RNase K cleavage specificity.
Full text
PDF
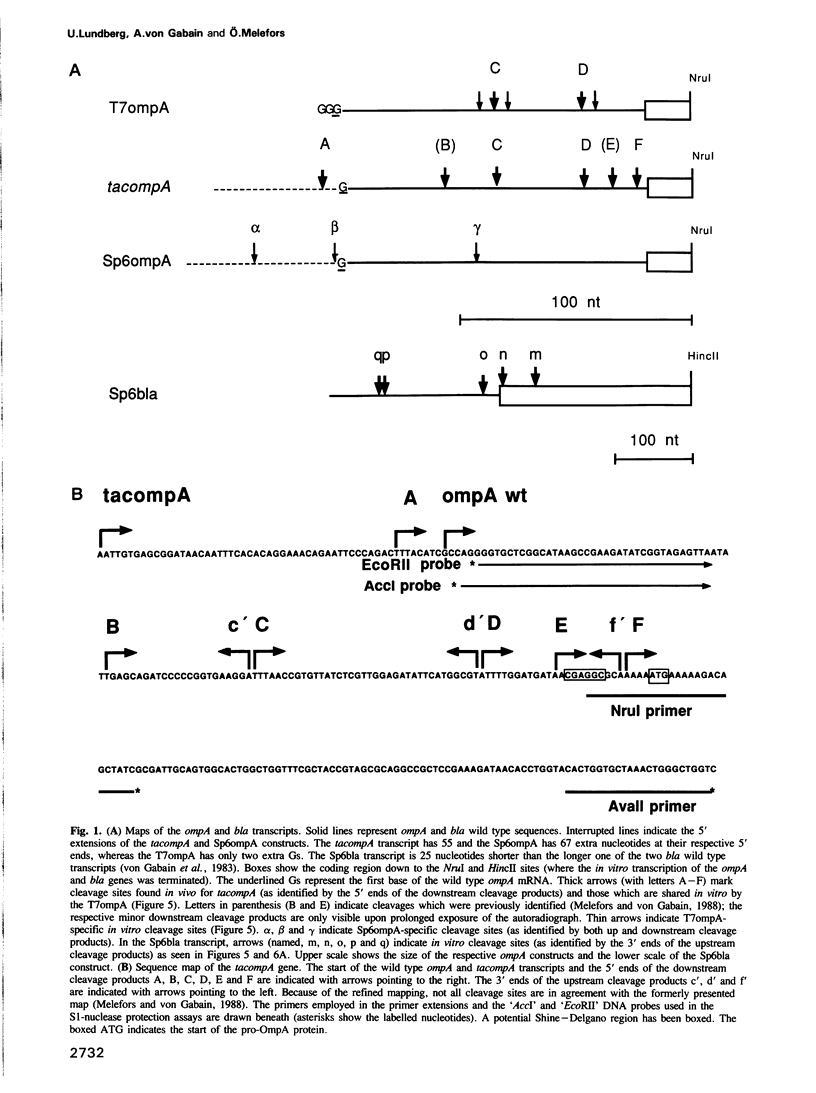



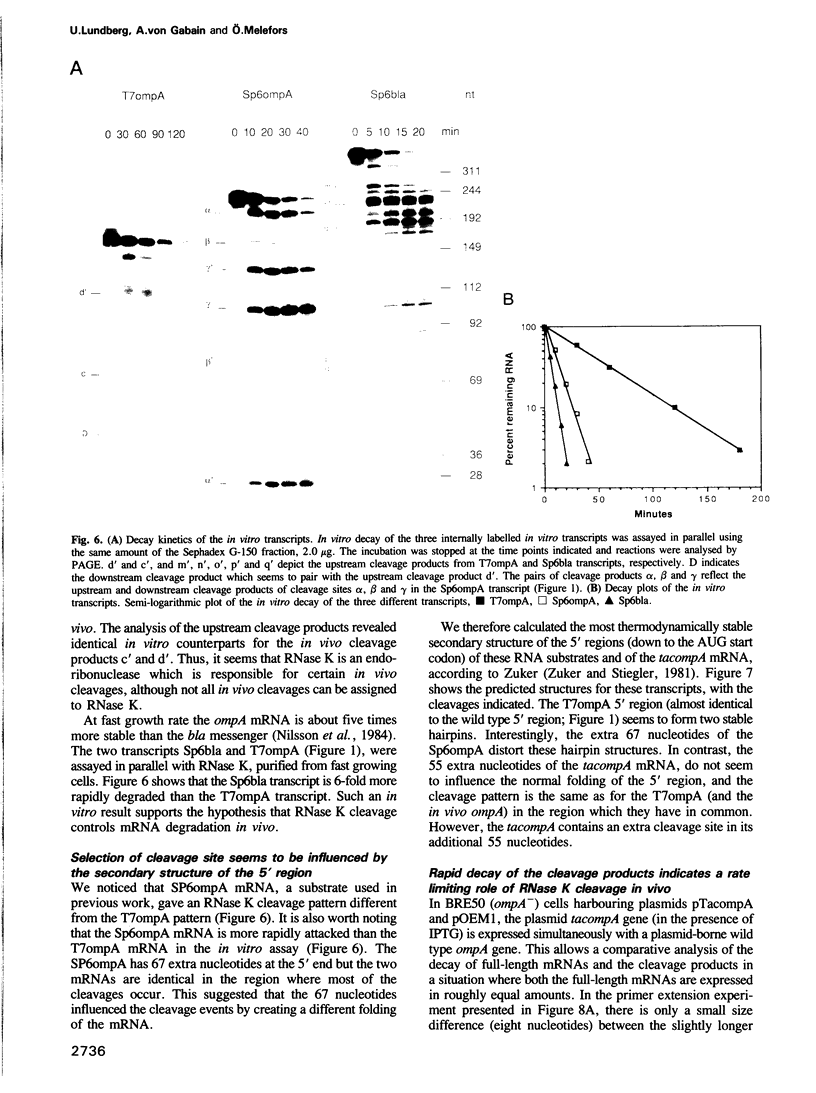

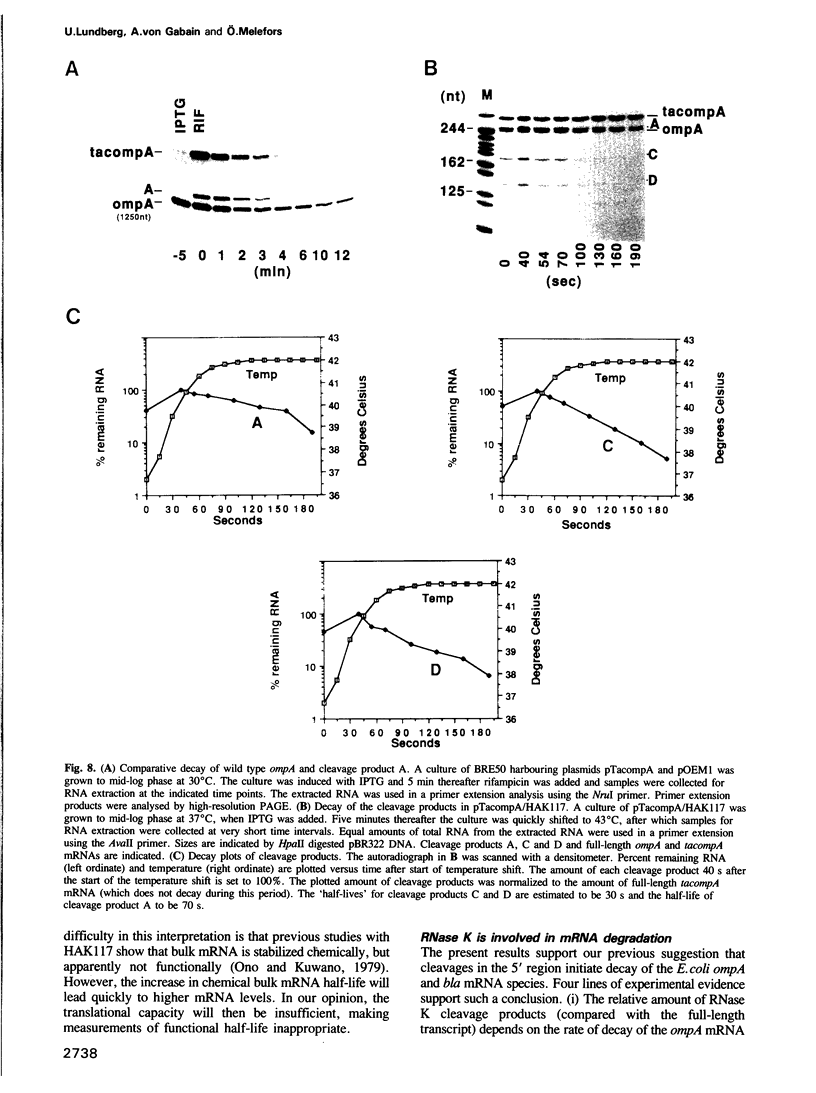



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arraiano C. M., Yancey S. D., Kushner S. R. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J Bacteriol. 1988 Oct;170(10):4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell J. C., Régnier P., Chen S. M., Nakamura Y., Grunberg-Manago M., Court D. L. Autoregulation of RNase III operon by mRNA processing. EMBO J. 1989 Nov;8(11):3401–3407. doi: 10.1002/j.1460-2075.1989.tb08504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G., Squires C., Squires C. L. Attenuation and processing of RNA from the rplJL--rpoBC transcription unit of Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3331–3335. doi: 10.1073/pnas.77.6.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer D. H., Dubnau D. Induced mRNA stability in Bacillus subtilis. Proc Natl Acad Sci U S A. 1987 Jan;84(2):498–502. doi: 10.1073/pnas.84.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Beatty J. T., Adams C. W., von Gabain A., Cohen S. N. Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell. 1985 Jan;40(1):171–181. doi: 10.1016/0092-8674(85)90320-4. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Higgins C. F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988 Dec 10;72(1-2):15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Nilsson G., von Gabain A., Cohen S. N. The stability of E. coli gene transcripts is dependent on determinants localized to specific mRNA segments. Cell. 1986 Jul 18;46(2):245–251. doi: 10.1016/0092-8674(86)90741-5. [DOI] [PubMed] [Google Scholar]
- Blumer K. J., Steege D. A. mRNA processing in Escherichia coli: an activity encoded by the host processes bacteriophage f1 mRNAs. Nucleic Acids Res. 1984 Feb 24;12(4):1847–1861. doi: 10.1093/nar/12.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawerman G. mRNA decay: finding the right targets. Cell. 1989 Apr 7;57(1):9–10. doi: 10.1016/0092-8674(89)90166-9. [DOI] [PubMed] [Google Scholar]
- Bremer E., Beck E., Hindennach I., Sonntag I., Henning U. Cloned structural gene (ompA) for an integral outer membrane protein of Escherichia coli K-12: localization on hybrid plasmid pTU100 and expression of a fragment of the gene. Mol Gen Genet. 1980;179(1):13–20. doi: 10.1007/BF00268440. [DOI] [PubMed] [Google Scholar]
- Båga M., Göransson M., Normark S., Uhlin B. E. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell. 1988 Jan 29;52(2):197–206. doi: 10.1016/0092-8674(88)90508-9. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Kennell D. Escherichia coli lac operator mRNA affects translation initiation of beta-galactosidase mRNA. Nature. 1979 Feb 1;277(5695):407–409. doi: 10.1038/277407a0. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Kennell D. Evidence that the 5' end of lac mRNA starts to decay as soon as it is synthesized. J Bacteriol. 1985 Feb;161(2):820–822. doi: 10.1128/jb.161.2.820-822.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannistraro V. J., Kennell D. Purification and characterization of ribonuclease M and mRNA degradation in Escherichia coli. Eur J Biochem. 1989 May 1;181(2):363–370. doi: 10.1111/j.1432-1033.1989.tb14733.x. [DOI] [PubMed] [Google Scholar]
- Carpousis A. J., Mudd E. A., Krisch H. M. Transcription and messenger RNA processing upstream of bacteriophage T4 gene 32. Mol Gen Genet. 1989 Oct;219(1-2):39–48. doi: 10.1007/BF00261155. [DOI] [PubMed] [Google Scholar]
- Cho K. O., Yanofsky C. Sequence changes preceding a Shine-Dalgarno region influence trpE mRNA translation and decay. J Mol Biol. 1988 Nov 5;204(1):51–60. doi: 10.1016/0022-2836(88)90598-0. [DOI] [PubMed] [Google Scholar]
- Churchward G., Belin D., Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984 Nov;31(1-3):165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- Claverie-Martin F., Diaz-Torres M. R., Yancey S. D., Kushner S. R. Cloning of the altered mRNA stability (ams) gene of Escherichia coli K-12. J Bacteriol. 1989 Oct;171(10):5479–5486. doi: 10.1128/jb.171.10.5479-5486.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. J., Roberts G. P., Brill W. J. Posttranscriptional control of Klebsiella pneumoniae nif mRNA stability by the nifL product. J Bacteriol. 1986 Oct;168(1):173–178. doi: 10.1128/jb.168.1.173-178.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D. L., Subbarao M. N., Blattner F. R., Lozeron H. A. Q-mediated late gene transcription of bacteriophage lambda: RNA start point and RNase III processing sites in vivo. Virology. 1988 Dec;167(2):568–577. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan W. P., Kushner S. R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1986 Jan;83(1):120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski K., Roch J. M., Prentki P., Krisch H. M. The stability of bacteriophage T4 gene 32 mRNA: a 5' leader sequence that can stabilize mRNA transcripts. Cell. 1985 Dec;43(2 Pt 1):461–469. doi: 10.1016/0092-8674(85)90176-x. [DOI] [PubMed] [Google Scholar]
- Guarneros G., Montañez C., Hernandez T., Court D. Posttranscriptional control of bacteriophage lambda gene expression from a site distal to the gene. Proc Natl Acad Sci U S A. 1982 Jan;79(2):238–242. doi: 10.1073/pnas.79.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M. N., Hayashi M. Cloned DNA sequences that determine mRNA stability of bacteriophage phi X174 in vivo are functional. Nucleic Acids Res. 1985 Aug 26;13(16):5937–5948. doi: 10.1093/nar/13.16.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koraimann G., Högenauer G. A stable core region of the tra operon mRNA of plasmid R1-19. Nucleic Acids Res. 1989 Feb 25;17(4):1283–1298. doi: 10.1093/nar/17.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mackie G. A. Posttranscriptional regulation of ribosomal protein S20 and stability of the S20 mRNA species. J Bacteriol. 1987 Jun;169(6):2697–2701. doi: 10.1128/jb.169.6.2697-2701.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melefors O., von Gabain A. Site-specific endonucleolytic cleavages and the regulation of stability of E. coli ompA mRNA. Cell. 1988 Mar 25;52(6):893–901. doi: 10.1016/0092-8674(88)90431-x. [DOI] [PubMed] [Google Scholar]
- Melin L., Rutberg L., von Gabain A. Transcriptional and posttranscriptional control of the Bacillus subtilis succinate dehydrogenase operon. J Bacteriol. 1989 Apr;171(4):2110–2115. doi: 10.1128/jb.171.4.2110-2115.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra T. K., Apirion D. Characterization of an endoribonuclease, RNase N, from Escherichia coli. J Biol Chem. 1978 Aug 25;253(16):5594–5599. [PubMed] [Google Scholar]
- Misra T. K., Apirion D. RNase E, an RNA processing enzyme from Escherichia coli. J Biol Chem. 1979 Nov 10;254(21):11154–11159. [PubMed] [Google Scholar]
- Morikawa N., Imamoto F. Degradation of tryptophan messenger. On the degradation of messenger RNA for the tryptophan operon in Escherichia coli. Nature. 1969 Jul 5;223(5201):37–40. doi: 10.1038/223037a0. [DOI] [PubMed] [Google Scholar]
- Morse D. E., Mosteller R., Baker R. F., Yanofsky C. Direction of in vivo degradation of tryptophan messenger RNA--a correction. Nature. 1969 Jul 5;223(5201):40–43. doi: 10.1038/223040a0. [DOI] [PubMed] [Google Scholar]
- Mott J. E., Galloway J. L., Platt T. Maturation of Escherichia coli tryptophan operon mRNA: evidence for 3' exonucleolytic processing after rho-dependent termination. EMBO J. 1985 Jul;4(7):1887–1891. doi: 10.1002/j.1460-2075.1985.tb03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd E. A., Prentki P., Belin D., Krisch H. M. Processing of unstable bacteriophage T4 gene 32 mRNAs into a stable species requires Escherichia coli ribonuclease E. EMBO J. 1988 Nov;7(11):3601–3607. doi: 10.1002/j.1460-2075.1988.tb03238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Higgins C. F. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell. 1987 Dec 24;51(6):1131–1143. doi: 10.1016/0092-8674(87)90599-x. [DOI] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Robinson E. C., Hiles I. D., Higgins C. F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987 Jan 30;48(2):297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- Nilsson G., Belasco J. G., Cohen S. N., von Gabain A. Growth-rate dependent regulation of mRNA stability in Escherichia coli. Nature. 1984 Nov 1;312(5989):75–77. doi: 10.1038/312075a0. [DOI] [PubMed] [Google Scholar]
- Nilsson G., Lundberg U., von Gabain A. In vivo and in vitro identity of site specific cleavages in the 5' non-coding region of ompA and bla mRNA in Escherichia coli. EMBO J. 1988 Jul;7(7):2269–2275. doi: 10.1002/j.1460-2075.1988.tb03067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M., Kuwano M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of messenger RNA. J Mol Biol. 1979 Apr 15;129(3):343–357. doi: 10.1016/0022-2836(79)90500-x. [DOI] [PubMed] [Google Scholar]
- Plamann M. D., Stauffer G. V. Escherichia coli glyA mRNA decay: the role of 3' secondary structure and the effects of the pnp and rnb mutations. Mol Gen Genet. 1990 Jan;220(2):301–306. doi: 10.1007/BF00260498. [DOI] [PubMed] [Google Scholar]
- Plunkett G., 3rd, Echols H. Retroregulation of the bacteriophage lambda int gene: limited secondary degradation of the RNase III-processed transcript. J Bacteriol. 1989 Jan;171(1):588–592. doi: 10.1128/jb.171.1.588-592.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier C., Dondon L., Grunberg-Manago M., Régnier P. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5' end. EMBO J. 1987 Jul;6(7):2165–2170. doi: 10.1002/j.1460-2075.1987.tb02484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. D., Webster R. E., Zinder N. D. Purification and properties of ribonuclease III from Escherichia coli. J Biol Chem. 1968 Jan 10;243(1):82–91. [PubMed] [Google Scholar]
- Roy M. K., Apirion D. Purification and properties of ribonuclease E, an RNA-processing enzyme from Escherichia coli. Biochim Biophys Acta. 1983 Sep 28;747(3):200–208. doi: 10.1016/0167-4838(83)90098-5. [DOI] [PubMed] [Google Scholar]
- Sandler P., Weisblum B. Erythromycin-induced stabilization of ermA messenger RNA in Staphylococcus aureus and Bacillus subtilis. J Mol Biol. 1988 Oct 20;203(4):905–915. doi: 10.1016/0022-2836(88)90116-7. [DOI] [PubMed] [Google Scholar]
- Takata R., Mukai T., Hori K. RNA processing by RNase III is involved in the synthesis of Escherichia coli polynucleotide phosphorylase. Mol Gen Genet. 1987 Aug;209(1):28–32. doi: 10.1007/BF00329832. [DOI] [PubMed] [Google Scholar]
- Uzan M., Favre R., Brody E. A nuclease that cuts specifically in the ribosome binding site of some T4 mRNAs. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8895–8899. doi: 10.1073/pnas.85.23.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H. C., Chang S. Identification of a positive retroregulator that stabilizes mRNAs in bacteria. Proc Natl Acad Sci U S A. 1986 May;83(10):3233–3237. doi: 10.1073/pnas.83.10.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Imamoto F. Differential stability of trp messenger RNA synthesized originating at the trp promoter and pL promoter of lambda trp phage. J Mol Biol. 1975 Feb 25;92(2):289–304. doi: 10.1016/0022-2836(75)90228-4. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]









