Abstract
Rationale: Idiopathic pulmonary fibrosis (IPF) is an increasingly recognized, often fatal lung disease of unknown etiology.
Objectives: The aim of this study was to use whole-exome sequencing to improve understanding of the genetic architecture of pulmonary fibrosis.
Methods: We performed a case–control exome-wide collapsing analysis including 262 unrelated individuals with pulmonary fibrosis clinically classified as IPF according to American Thoracic Society/European Respiratory Society/Japanese Respiratory Society/Latin American Thoracic Association guidelines (81.3%), usual interstitial pneumonia secondary to autoimmune conditions (11.5%), or fibrosing nonspecific interstitial pneumonia (7.2%). The majority (87%) of case subjects reported no family history of pulmonary fibrosis.
Measurements and Main Results: We searched 18,668 protein-coding genes for an excess of rare deleterious genetic variation using whole-exome sequence data from 262 case subjects with pulmonary fibrosis and 4,141 control subjects drawn from among a set of individuals of European ancestry. Comparing genetic variation across 18,668 protein-coding genes, we found a study-wide significant (P < 4.5 × 10−7) case enrichment of qualifying variants in TERT, RTEL1, and PARN. A model qualifying ultrarare, deleterious, nonsynonymous variants implicated TERT and RTEL1, and a model specifically qualifying loss-of-function variants implicated RTEL1 and PARN. A subanalysis of 186 case subjects with sporadic IPF confirmed TERT, RTEL1, and PARN as study-wide significant contributors to sporadic IPF. Collectively, 11.3% of case subjects with sporadic IPF carried a qualifying variant in one of these three genes compared with the 0.3% carrier rate observed among control subjects (odds ratio, 47.7; 95% confidence interval, 21.5–111.6; P = 5.5 × 10−22).
Conclusions: We identified TERT, RTEL1, and PARN—three telomere-related genes previously implicated in familial pulmonary fibrosis—as significant contributors to sporadic IPF. These results support the idea that telomere dysfunction is involved in IPF pathogenesis.
Keywords: genetics, exome sequencing, pulmonary fibrosis, interstitial lung disease, collapsing analysis
At a Glance Commentary
Scientific Knowledge on the Subject
Idiopathic pulmonary fibrosis (IPF) is an increasingly diagnosed and often fatal lung disease for which no curative treatments exist. Prior genetic studies of pulmonary fibrosis have predominantly been focused on familial forms of the disease and have associated several genes and common risk alleles with disease development. The degree to which these familial genetic associations extend to sporadic IPF remains uncertain.
What This Study Adds to the Field
We performed whole-exome sequencing to identify rare variants of protein-coding genes in a cohort of patients with predominantly sporadic IPF. Our results demonstrate a case enrichment for ultrarare deleterious qualifying variants in three study-wide significant genes: TERT, RTEL1, and PARN. Collectively, these variants, which included dominant loss-of-function alleles in RTEL1 and PARN and ultrarare missense variants predicted to be damaging in TERT and RTEL1, contributed to more than 10% of the sporadic IPF cases. This provides the first evidence that telomere-related genes previously implicated in familial pulmonary fibrosis also make a major contribution to the genetic architecture of sporadic IPF and supports a body of literature implicating telomere dysfunction as a contributor to disease development in this population.
Pulmonary fibrosis describes a group of diseases generally characterized by progressive inflammation and/or fibrosis of the lung parenchyma. Pulmonary fibrosis may be further categorized to known or idiopathic causes on the basis of clinical history, radiographic appearance, and laboratory and/or histological evaluation. Idiopathic pulmonary fibrosis (IPF) is an increasingly diagnosed and often fatal lung disease associated with an estimated survival of 3–5 years (1, 2). Novel therapeutic agents for IPF have recently been approved and have been shown to slow the course of disease progression. Lung transplant, however, remains the only treatment definitively shown to improve survival in select patients with end-stage pulmonary fibrosis, including IPF. In this context, enhancing understanding of the basic pathogenesis of pulmonary fibrosis is critical.
Approximately 10% of pulmonary fibrosis cases are considered familial in origin (3). Genome-wide association studies have revealed associations of common variants in MUC5B, SPPL2C, and TOLLIP with both familial pulmonary fibrosis (FPF) and sporadic IPF (4, 5). Of these, the most promising has been the MUC5B promoter allele (rs35705950G>T) (4); however, these associations generally explain a small proportion of the heritability and have not always resolved to the underlying causal gene variants. By giving researchers the ability to focus specifically on the protein-coding regions of the genome, whole-exome sequencing allows identification of alleles with direct functional consequences on protein products. Indeed, most known human disease–causing variants reside within the exome (6, 7).
Early studies of surfactant proteins A2 (8) and C (9) associated these two proteins with FPF. Yet, the best characterized examples in pulmonary fibrosis have been genes related to telomerase; specifically, variants in TERT and TERC have been found to associate with telomere shortening and increased susceptibility to FPF (10, 11). More recently, exome sequencing of 78 European individuals with FPF identified an excess of deleterious variants in two novel genes important to telomere maintenance: PARN and RTEL1 (12).
Exome analysis provides a high-throughput, cost-effective strategy to discover disease-causing mutations, but to date it has not been extended to the study of sporadic pulmonary fibrosis. The objectives of this study therefore were to (1) gain insight into the genetic architecture of pulmonary fibrosis by applying whole-exome sequencing to identify genes carrying an excess of rare deleterious variants in precisely phenotyped individuals with pulmonary fibrosis, (2) focus specifically on the genetics of a subset of patients with sporadic IPF, and (3) examine the relationship between rare variants associated with pulmonary fibrosis risk as identified through whole-exome sequencing and the previously reported common risk allele in the MUC5B promoter region.
Methods
Study Cohort
The case population comprised 262 unrelated individuals of European ancestry who underwent lung transplant at Duke University Medical Center for pulmonary fibrosis. All patients were clinically well characterized prior to transplant through physician interview for a complete medical history, including for family history of pulmonary fibrosis, chest computed tomography, serological evaluation, and pulmonary function testing. At the time of transplant, explanted native lung tissue was examined by an experienced lung pathologist for histological characterization of pulmonary fibrosis. For this study, a specific pulmonary fibrosis phenotype was adjudicated to each case on the basis of review of the comprehensive medical, radiographic, and histological data by a pulmonologist with expertise in fibrosing lung disease. As illustrated in Table 1, the final case cohort included patients confirmed to have IPF according to the American Thoracic Society/European Respiratory Society/Japanese Respiratory Society/Latin American Thoracic Association (ATS/ERS/JRS/ALAT) guidelines (1) (213 [81.3%] of 262), patients with usual interstitial pneumonia secondary to autoimmune conditions (30 [11.5%] of 262), and patients with fibrosing nonspecific interstitial pneumonia (19 [7.2%] of 262). The majority of the cohort (229 [87%] of 262) reported no family history of pulmonary fibrosis. All case subjects consented to DNA collection and participation in institutional review board–approved genetic studies according to Duke Institutional Review Board Protocols 00009091 and 00056268. The control population comprised 4,141 unrelated individuals of European ancestry selected for control purposes through unrelated studies not focused on pulmonary disorders, severe pediatric disorders, or other clinical phenotypes where pulmonary fibrosis is a recognized comorbidity (see Table E1 in the online supplement).
Table 1.
Characteristics of the Pulmonary Fibrosis Cohort
| Characteristics | Pulmonary Fibrosis Cohort |
|---|---|
| Number of individuals | 262 |
| Sex, n (%) | |
| Male | 204 (78%) |
| Female | 58 (22%) |
| Transplant age, yr, mean ± SD | 63.2 ± 8.2 |
| Clinical pulmonary fibrosis phenotype, n (%) | |
| IPF | 213 (81.3%) |
| CTD UIP | 30 (11.5%) |
| Fibrosing NSIP | 19 (7.2%) |
| Self-reported family history of pulmonary fibrosis, n (%) | |
| No | 229 (87.4%) |
| Yes | 33 (12.6%) |
Definition of abbreviations: CTD UIP = usual interstitial pneumonia associated with connective tissue disease; IPF = idiopathic pulmonary fibrosis; NSIP = nonspecific interstitial pneumonia.
Exome Sequencing and Bioinformatic Processing
Exome sequencing of blood-extracted DNA was performed at the Institute for Genomic Medicine at Columbia University using the SureSelect Human All Exon (65 MB; Agilent Technologies, Santa Clara, CA) or the NimbleGen SeqCap EZ version 2.0 or 3.0 exome enrichment kit (Roche NimbleGen, Madison, WI) on HiSeq 2000 or 2500 sequencers (Illumina, San Diego, CA) according to standard protocols. Whole-exome sequence data from the 262 case subjects with sporadic IPF and 4,141 control subjects were processed using the same bioinformatic pipeline (see Methods section in online supplement).
On average, at least 10-fold sequencing read coverage was achieved for 96.9% and 95.7% of the 33.27 megabase pairs (Mbp) of the Consensus Coding Sequence (CCDS; release 14) for case and control subjects, respectively. To alleviate confounding attributable to differential coverage, for all of the 33.27-Mbp positions in the CCDS sequence, we determined both the percentage of case subjects and the percentage of control subjects who had at least 10-fold coverage at the site (see Methods section in online supplement). An individual CCDS site was excluded from analysis if the absolute difference in percentages of case subjects compared with control subjects who achieved at least 10-fold coverage at the site was greater than 6.0% (Figure E2). This site-based pruning resulted in 7.8% of the CCDS sites being excluded. All collapsing tests were then performed on the pruned 30.67 Mbp of CCDS sites (i.e., 92.2% of the CCDS) where case and control subjects had a similar opportunity to call variants. For the remaining 30.67 Mbp, on average, case and control subjects had at least 10-fold coverage for 98.1% and 97.9% of CCDS sites, respectively. To further confirm no preferential inflation of background variation, we assessed the exome-wide tally of rare autosomal synonymous (i.e., presumed neutral) variants per individual and did not find a significant difference between the case and control groups (P = 0.68) (Figure E3, Table E3). Autosomal read depth (i.e., sequencing coverage) was also consistent between case and control subjects, with a case average of 96.35 ± 25.78 reads and a control average of 97.88 ± 24.22 reads (P = 0.35 by two-sample t test) (Table E3).
Statistical Analysis
To search for genes conferring pulmonary fibrosis risk, we implemented a genetic collapsing test (13, 14). After site-based pruning, we focused our analyses on CCDS protein-coding sites with minimal variability in coverage between the case and control populations. As initially introduced in our earlier work (13), we use the term qualifying variant to refer to the subset of genetic variations within the sequence data that meets specific population allele frequency and predicted variant effect criteria. We defined seven different qualifying variant models (Table E4). Our primary model was focused on searching for “ultrarare” nonsynonymous variants to capture the category of genetic variation expected to be most enriched for variants of high effect. To identify ultrarare variants, we use internal (test cohort) and external (Exome Variant Server and Exome Aggregation Consortium release 0.3 [15]) sequence data to find variants with a minor allele frequency (MAF) of less than 0.05% among our combined case and control test populations and absent (MAF of 0%) among the two external reference control cohorts. For the primary model, qualifying variants were restricted to indels and single-nucleotide variants annotated as having either a loss-of-function (LoF) effect, an in-frame indel, or a “probably damaging” missense prediction by Polymorphism Phenotyping version 2 (PolyPhen, HumDiv; http://genetics.bwh.harvard.edu/pph2/) (16). These analyses relied on the predicted effects of the LoF and missense annotated variants whose functions have not been individually confirmed in the laboratory. We subsequently performed analyses of CCDS genes using six alternative qualifying variant models as defined in Table E4, including an autosomal recessive model and a synonymous variant negative control model.
For each of the seven models, we tested the list of 18,668 CCDS genes. For each gene, an indicator variable (1/0 states) was assigned to each individual on the basis of presence of at least one qualifying variant in the gene (state 1) or no qualifying variants in that gene (state 0). A two-tailed Fisher’s exact test (FET) was then performed for each gene to compare the rate of case subjects carrying a qualifying variant compared with the rate of control subjects. For our study-wide significance threshold, after Bonferroni correction for the number of genes tested across the six nonsynonymous models, the study-wide multiplicity-adjusted significance threshold was calculated as α = (0.05/[6 × 18,668]) = 4.46 × 10−7 (Table E4). We did not correct for the synonymous (negative control) model.
Because of the discordance in sex-sampling rates between the case (78% male) and control (48% male) cohorts, for genes on the X chromosome, we randomly sampled 565 control female subjects from among the original female control group and ran a separate X chromosome assessment using matched male/female ratios. Thus, despite following the same qualifying criteria as the autosomes, the X chromosome tests are reported separately.
To investigate the genetics of sporadic IPF, we subsequently examined only the 186 (71.0%) case subjects confirmed to have IPF on the basis of ATS/ERS/JRS/ALAT guidelines (1) and without a family history of pulmonary fibrosis. We repeated the primary and LoF collapsing analyses to compare just these 186 case subjects with sporadic IPF with the 4,141 control subjects.
Collapsing analyses were performed using an in-house package, Analysis Tool for Annotated Variants (https://redmine.igm.cumc.columbia.edu/projects/atav). Additional binomial analyses, logistic regression analyses, and FETs were completed using the ‘stats’ package in R version 3.2.2 (R Foundation for Statistical Computing, Vienna, Austria).
MUC5B Risk Allele Genotyping
Genotyping of rs35705950 within the promoter region of the MUC5B gene (chr11:g.1241221G>T, NCBI Build 37) was performed with the Applied Biosystems TaqMan SNP Genotyping Assay on a 7900HT Fast Real-Time PCR System (Thermo Fisher Scientific, Foster City, CA). The context sequence is CCTTCCTTTATCTTCTGTTTTCAGC[G/T]CCTTCAACTGTGAAGAGGTGAACTC. Amplification was performed according to the TaqMan Universal PCR protocol in a 5-μl reaction volume using 2× TaqMan Universal PCR Master Mix (Life Technologies, Carlsbad, CA). Of the 262 case subjects with pulmonary fibrosis, 258 were successfully genotyped at this locus. We also genotyped 342 European control subjects to generate in-house control frequency estimates for the rs35705950 variant.
Results
Insights into the Genetic Architecture of Pulmonary Fibrosis Using Exome Sequencing
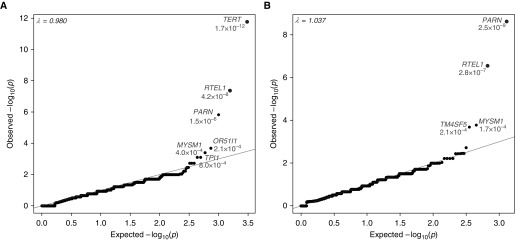
In our primary analysis of the pulmonary fibrosis cohort (n = 262), we identified two genes that achieved study-wide significance (Figure 1, Tables E5 and E6). Five percent of case subjects with pulmonary fibrosis had a qualifying variant in TERT, a well-known FPF gene, compared with 0.1% of control subjects (odds ratio [OR], 35.9; 95% confidence interval [CI], 12.6–116.2; P = 1.7 × 10−12 by two-tailed FET). The second gene to achieve study-wide significance was the recently implicated FPF gene RTEL1 (12, 17, 18), with 2.3% of case subjects with pulmonary fibrosis carrying a qualifying variant, as compared with 0% of control subjects (OR, >96.8; 95% CI, 18.9 to >4,335; P = 4.2 × 10−8 by two-tailed FET). The third ranked gene, just under the prespecified level of statistical significance, was another recently implicated FPF gene, PARN (12). We found PARN qualifying variants in 2.7% of case subjects compared with 0.1% of control subjects (OR, 22.7; 95% CI, 6.1–91.0; P = 1.5 × 10−6 by two-tailed FET) (Figure 1). These results reflect the 95.7% (TERT), 84.6% (RTEL1), and 98.9% (PARN) of the protein-coding sequence of these genes that had reliable sequence coverage in both the case and control samples (Table E5). No individual case subject carried a qualifying variant in more than one of these three genes, and a higher-resolution cryptic relatedness screen confirmed that no two case subjects carrying one of four recurring TERT, RTEL1, or PARN qualifying variants (Table 2) shared more than 1% of their exome-wide rare protein-coding variants (Table E10).
Figure 1.
Quantile–quantile plot of pulmonary fibrosis collapsing analyses. Results are shown for the analysis of 262 pulmonary fibrosis case and 4,141 control subjects. (A) A total of 15,393 genes had at least one case or control carrier. Qualifying variants have a minor allele frequency less than 0.05% in the test cohort and are absent among external reference cohorts. Variants are annotated as loss-of-function, in-frame indel, or missense predicted to be “probably damaging” by Polymorphism Phenotyping version 2 (HumDiv). Two genes, TERT and RTEL1, achieved study-wide significance (adjusted α = [0.05/(6 × 18668)] = 4.46 × 10−7). (B) A total of 10,710 genes had at least one loss-of-function case or control carrier. Qualifying variants are variants with a population minor allele frequency less than or equal to 0.1% and are annotated as loss-of-function single-nucleotide variants or indels. PARN and RTEL1 achieved study-wide significance (P < 4.46 × 10−7).
Table 2.
Qualifying Variants among the Top Three Pulmonary Fibrosis Genes
| Patient | Sex | Family History | Fibrosis Phenotype | Gene | Chromosomal Coordinates GRCh37/hg19 (rs refsnp) | HGVS Identifier (Protein) | Effect | ExAC Genotype Distribution | Literature Phenotype | OMIM Variant Identifier | Qualifying Model* | GERP | CADD | PolyPhen-2 HumDiv |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pf316 | M | N | IPF | TERT | chr5:g.1279426G>A (rs199422297) | NP_937983.2 p.Pro704Ser | Missense | 29,430/0/0 | Dyskeratosis congenita AD | 0.0014 | Primary | 2.47 | 5.96 | 0.998 |
| pf350 | M | Y | IPF | |||||||||||
| pf1132 | F | N | IPF | |||||||||||
| pf1180 | F | Y | IPF | |||||||||||
| pf65 | M | N | IPF | TERT | chr5:g.1278817C>T (rs727503468) | NP_937983.2 p.Arg742His | Missense | 60,706/0/0 | — | — | Primary | 3.74 | 24.3 | 0.987 |
| pf1318 | M | N | IPF | TERT | chr5:g.1279547G>C | NP_937983.2 p.Ser663Arg | Missense | 10,193/0/0 | — | — | Primary | −5.1 | 10.59 | 0.998 |
| pf132 | F | Y | CTD UIP | TERT | chr5:g.1254522G>A | NP_937983.2 p.Arg1086Cys | Missense | 59,219/0/0 | — | — | Primary | −6.21 | 17.94 | 0.999 |
| pf238 | M | N | IPF | TERT | chr5:g.1279455A>T | NP_937983.2 p.Val694Glu | Missense | 28,149/0/0 | — | — | Primary | 0.97 | 23.5 | 1 |
| pf514 | M | N | IPF | TERT | chr5:g.1264550G>A | NP_937983.2 p.Arg938Trp | Missense | 58,867/0/0 | — | — | Primary | 3.42 | 33 | 1 |
| pf681 | M | N | IPF | TERT | chr5:g.1279467C>G | NP_937983.2 p.Trp690Ser | Missense | 25,430/0/0 | — | — | Primary | 4.67 | 25.9 | 1 |
| pf717 | M | N | IPF | TERT | chr5:g.1280338C>G | NP_937983.2 p.Gly629Arg | Missense | 60,609/0/0 | — | — | Primary | 4.48 | 25.4 | 1 |
| pf1110 | F | Y | IPF | TERT | chr5:g.1266613T>C | NP_937983.2 p.Thr874Ala | Missense | 56,038/0/0 | — | — | Primary | 3.99 | 23.8 | 1 |
| pfB103 | F | N | IPF | TERT | chr5:g.1280455T>C | NM_198253.2 c.1770-2A>G | Splice site acceptor | 60,075/0/0 | — | — | Primary and LoF 0.1% | — | 16.51 | — |
| pf166 | M | Y | IPF | RTEL1 | chr20:g.62324564C>T (rs398123017) | NP_116575.3 p.Arg998Ter | Stop gain | 59,955/2/0 | Dyskeratosis congenita AR | 0.0004 | LoF 0.1% | — | 37 | — |
| pf941 | M | N | IPF | |||||||||||
| pf1532 | M | N | IPF | |||||||||||
| pf1226 | F | Y | IPF | RTEL1 | chr20:g.62324600C>T (rs373740199) | NP_116575.3 p.Arg1010Ter | Stop gain | 59,848/10/0 | Dyskeratosis congenita AD | 0.0012 | LoF 0.1% | — | 35 | — |
| pf53 | F | N | IPF | RTEL1 | chr20:g.62320919A>AC | NP_116575.3 p.Arg675ThrfsTer15 | Frameshift | 58,946/0/0 | — | — | Primary and LoF 0.1% | — | 28.4 | — |
| pf191 | M | N | IPF | RTEL1 | chr20:g.62321438A>G | NM_032957.4 c.2214-2A>G | Splice site acceptor | 59,613/0/0 | — | — | Primary and LoF 0.1% | — | 22.2 | — |
| pf249 | M | Y | IPF | RTEL1 | chr20:g.62321112G>A | NP_116575.3 p.Gly703Arg | Missense | 58,909/0/0 | — | — | Primary | 4.61 | 27.5 | 1 |
| pf304 | M | N | IPF | RTEL1 | chr20:g.62321483C>T | NP_116575.3 p.Arg753Cys | Missense | 59,716/0/0 | — | — | Primary | 4.57 | 29.7 | 1 |
| pf1191 | M | N | IPF | |||||||||||
| pf1576 | M | N | IPF | RTEL1 | chr20:g.62321477T>C | NP_116575.3 p.Trp751Arg | Missense | 59,716/0/0 | — | — | Primary | 4.57 | 24.9 | 1 |
| pf143 | F | Y | Fibrosing NSIP | PARN | chr16:g.14698077G>A (rs760506977) | NP_002573.1 p.Arg237Ter | Stop gain | 27,816/1/0 | — | — | LoF 0.1% | — | 38 | — |
| pf244 | M | N | IPF | PARN | chr16:g.14704526G>A (rs876661305) | NP_002573.1 p.Gln177Ter | Stop gain | 59,377/0/0 | Pulmonary fibrosis AD | 0.0006 | Primary and LoF 0.1% | — | 37 | — |
| pfB467 | M | N | IPF | |||||||||||
| pf1099 | M | N | IPF | |||||||||||
| pf1311 | M | N | IPF | |||||||||||
| pf566 | M | N | IPF | PARN | chr16:g.14540858CCT>C | NP_002573.1 p.Glu585AspfsTer5 | Frameshift | 5,9377/0/0 | — | — | Primary and LoF 0.1% | — | 28.6 | — |
| pf1495 | M | N | IPF | PARN | chr16:g.14676110C>A | NP_002573.1 p.Glu374Ter | Stop gain | 59,377/0/0 | — | — | Primary and LoF 0.1% | — | 43 | — |
| pf1466 | M | N | IPF | PARN | chr16:g.14647946G>C | NP_002573.1 p.Leu461Val | Missense | 58,345/0/0 | — | — | Primary | 5.93 | 27.7 | 0.994 |
Definition of abbreviations: AD = autosomal dominant; AR = autosomal recessive; CADD = Combined Annotation Dependent Depletion score; CTD UIP = usual interstitial pneumonia associated with connective tissue disease; ExAC = Exome Aggregation Consortium; GERP = Genomic Evolutionary Rate Profiling score; HGVS = Human Genome Variation Society; IPF = idiopathic pulmonary fibrosis; LoF = loss of function; N = no; NSIP = nonspecific interstitial pneumonia; OMIM = Online Mendelian Inheritance in Man database; PolyPhen-2 = Polymorphism Phenotyping version 2; Y = yes.
The ExAC genotype distribution reflects the corresponding homozygous reference, heterozygous variant, and homozygous variant counts found in the ExAC browser of up to 60,706 samples. Annotations for qualifying variants derived from models other than the primary and LoF model are available in Table E9 in the online supplement. Refsnp (rs) single-nucleotide polymorphism identifiers are provided where available. A higher-resolution relatedness screen confirmed that no two cases carrying one of the recurring TERT, RTEL1, or PARN qualifying variants shared more than 1% of their protein-coding variants at a minor allele frequency less than or equal to 1% (Table E10).
See Table E4 in the online supplement for details.
The results derived from the LoF model suggested haploinsufficiency as a leading disease mechanism for both RTEL1 and PARN (Tables E5 and E6), consistent with earlier literature (12). Among our case subjects, 2.7% had an LoF qualifying variant in PARN, as compared with no control carriers, enabling PARN to achieve study-wide significance under the LoF model (OR, >113.3; 95% CI, 23.2 to >4,593; P = 2.4 × 10−9 by two-tailed FET). RTEL1 was also significantly enriched for LoF alleles in the case subjects (2.3% vs. 0.02%; OR, 96.7, 95% CI, 11.7–4,333; P = 2.8 × 10−7 by two-tailed FET). Although TERT LoF alleles have been reported to segregate with disease in familial pulmonary fibrosis pedigrees (10, 11), our present sample of 262 case subjects with pulmonary fibrosis is not significantly enriched for putative LoF TERT alleles (1 of 262 case subjects vs. 0 of 4,141 control subjects; uncorrected P = 0.06) (Table E5). No additional genes achieved study-wide significance across the six nonsynonymous models.
The cumulative findings for the three study-wide significant genes (TERT, RTEL1, and PARN) indicate that 11.8% of case subjects (31 of 262) and 0.3% of control subjects (12 of 4,141) carry a qualifying variant (Figure 2, Table 2). This suggests that approximately 11.5% of our case subjects with pulmonary fibrosis could be partially explained by the identified qualifying variants (OR, 46.1; 95% CI, 22.6–99.5; P = 1.5 × 10−29 by two-tailed FET). Indeed, given the 0.3% rate of qualifying variation among control subjects, we expected to see 0.76 (95% CI, 0.41–1.37) carriers among 262 case subjects rather than the observed 31 carriers. Comparison of the 31 TERT, RTEL1, or PARN qualifying variant carriers with the remaining 231 noncarriers did not reveal distinguishing clinical features. No significant difference was found for the average age at transplant among the 31 carriers (62.2 ± 7.8 yr) compared with 231 noncarriers (63.3 ± 8.3 yr) (P = 0.5 by two-sample t test). No significant difference was found when we assessed the proportion of case subjects where the clinical pulmonary fibrosis diagnosis was strictly IPF and not usual interstitial pneumonia associated with connective tissue disease or fibrosing nonspecific interstitial pneumonia (29 of 31 carriers and 184 of 231 noncarriers; P = 0.08 by FET), and no significant difference was observed for sex, with the male proportion among carriers being 74.2% (23 of 31) compared with that among noncarriers at 78.4% (181 of 231) (P = 0.6 by FET). A negative control analysis was also performed, which confirmed no enrichment of synonymous genetic variation across the three study-wide significant genes (1.1% of case subjects vs. 0.9% of control subjects; P = 0.74 by two-tailed FET).
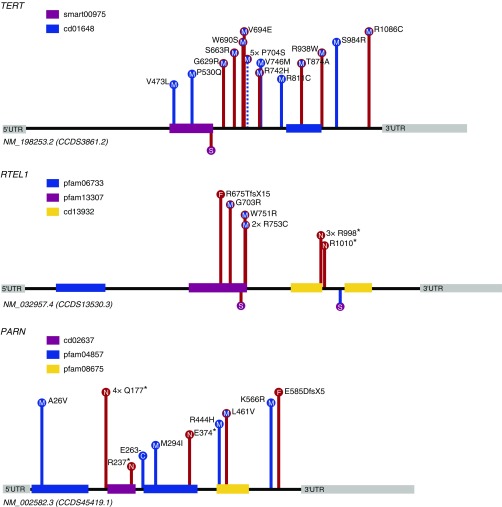
Figure 2.
Distribution of case and control qualifying variants for TERT, RTEL1, and PARN from the primary and loss-of-function model analyses. Case and control variants are plotted on the canonical transcript of the corresponding gene. Frameshift (F) and nonsense (N) variants are filled in red. Missense (M) and in-frame indel (C) variants are filled in blue. Case variants are shown with red lines, and control variants are shown with blue lines. Dashed lines indicate a qualifying variant found in both case and control subjects. Variants reported below the protein line indicate canonical splice (S) variants. Known conserved domains from the Conserved Domains Database are listed below the gene names. UTR = untranslated region.
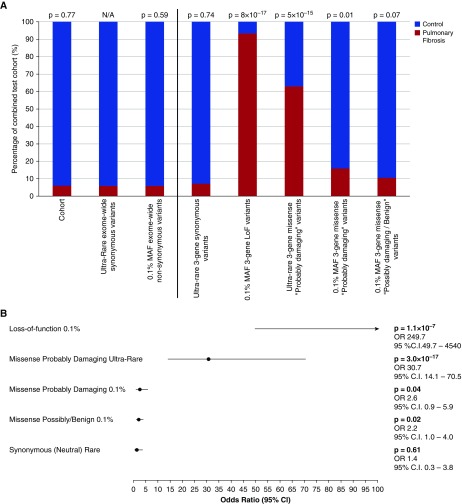
The 262 case subjects accounted for 5.95% of the overall test cohort. Of the autosomal synonymous qualifying variants (neutral model), 5.85% were found to belong to case subjects, which indicated a close match between the proportion of individuals in the test set who were case subjects and the proportion of synonymous (neutral) qualifying variants in the test set that were found in case subjects (P = 0.77 by binomial exact test). After establishing the lack of case enrichment for synonymous variation, we binned the collection of variants found among the three study-wide significant genes into various frequency and effect bins to identify categories that significantly departed from the synonymous variation background rate. Among these three genes, the two classes that stood out were LoF annotated variants (P = 8 × 10−17) and ultrarare missense variants predicted to be “probably damaging” by PolyPhen-2 (P = 5 × 10−15). There was little additional signal contributed by increasing the MAF to 0.1% or relaxing the in silico PolyPhen-2 criteria (Figure 3A, Table E7).
Figure 3.
Enrichment levels of different qualifying variant categories among the three study-wide significant pulmonary fibrosis genes. We compared enrichment of qualifying variants across increasing minor allele frequency (MAF) and varying in silico effects to assess which variation type was most attributable to pulmonary fibrosis disease risk. (A) Six paired comparisons were done to a single background variation estimate: the percentage of exome-wide ultrarare autosomal synonymous variants that belong to the case subjects (8,279 of 141,449; Ps = 0.0583). (B) Forest plot derived from a multivariate logistic regression assessing the relative contribution of different qualifying variant categories. All categories are mutually exclusive (see Methods section in online supplement). The loss-of-function (LoF) category represents LoF variants with an MAF less than or equal to 0.1%. The LoF category is prematurely truncated for illustrative purposes; however, the effect size and 95% confidence interval (CI) are provided. The missense probably damaging ultrarare category includes variants predicted to be “probably damaging” by Polymorphism Phenotyping version 2 with an MAF less than 0.05% in the test population and absent in Exome Variant Server (EVS) and Exome Aggregation Consortium (ExAC release 0.3.1). The missense probably damaging MAF less than or equal to 0.1% represents variants not in the ultrarare category and that are predicted to be “probably damaging” with a MAF less than or equal to 0.1% across the test population, EVS, and ExAC cohorts. The missense “possibly/benign” category includes missense variants predicted to be “possibly damaging” or “benign” with a MAF less than or equal to 0.1% across the test population, EVS, and ExAC cohorts (including ultrarare possibly/benign missense variants). The “neutral” category represents ultrarare putatively neutral (synonymous) variants across the three pulmonary fibrosis genes (TERT, RTEL1, and PARN). N/A = not applicable; OR = odds ratio.
We also used a multivariate logistic regression model to specifically assess the relative contribution that variant effects and allele frequency bins have on pulmonary fibrosis risk among these three genes (see Methods section in online supplement; Figure 3B). This approach ensured that each qualifying category is relative to the same baseline category and is naturally adjusted for variation in the other categories (see Methods section in online supplement). For missense variants, in comparison with the risk contribution from PolyPhen-2 “probably damaging” variants that are ultrarare in the population (OR, 30.7; 95% CI, 14.1–70.5; P = 3.0 × 10−17), neither the “probably damaging” missense variants that are more common nor those predicted to be nondamaging by PolyPhen-2 contributed substantial additional disease risk (Figure 3).
Subanalysis of Familial Pulmonary Fibrosis and Sporadic Idiopathic Pulmonary Fibrosis
Our cohort of 262 case subjects with pulmonary fibrosis included 33 subjects with FPF. We found that 8 (24.2%) of our 33 case subjects with FPF have a qualifying variant in one of these three pulmonary fibrosis genes, as compared with 0.3% of our control cohort. The contribution of these three genes to the genetics of this FPF group is striking (24.2% vs. 0.3%; OR, 108.6; 95% CI, 35.4–323.3; P = 7.2 × 10−13 by two-tailed FET).
To assess the genetic signal in a strictly homogeneous sporadic IPF cohort, we restricted our case cohort to the 186 individuals with no reported family history of pulmonary fibrosis and who were clinically confirmed to have IPF on the basis of ATS/ERS/JRS/ALAT guidelines (1). Comparison of these 186 IPF case subjects with the 4,141 control subjects showed that the three genes remained study-wide significant in sporadic IPF (Figure E4, Table E8). TERT achieved a P value of 1.7 × 10−9 on the basis of 4.8% of case subjects with sporadic IPF carrying a qualifying variant, as compared with 0.14% of control subjects (OR, 34.9; 95% CI, 11.0–120.7). RTEL1 achieved a P value of 1.4 × 10−7 (2.7% case subjects vs. 0% of control subjects; OR, >114; 95% CI, 20.7 to >5,208). PARN achieved a P value of 1.6 × 10−7 (3.8% of case subjects vs. 0.12% of control subjects; OR, 32.2; 95% CI, 8.7–130.1). Remarkably, we found that, among the sporadic IPF collection, 11.3% of samples carried a qualifying variant in one of these three IPF genes, as compared with the 0.3% rate seen among control subjects (OR, 47.7; 95% CI, 21.5–111.6; P = 5.5 × 10−22 by two-tailed FET).
Relationship between Identified Rare Variants and the Known MUC5B Common Risk Allele
Compared with both the 1000 Genomes Project European control subjects (19) and in-house European control data, our collection of European ancestry case subjects with pulmonary fibrosis showed an elevated rate of the MUC5B promoter risk allele rs35705950G>T (MAF, 31.6% of case subjects vs. 11.0% of control subjects; OR, 3.7; 95% CI, 2.7–5.1; P = 8.8 × 10−19) (Table 3). Although this elevated rate is consistent with prior literature (4), it is remarkable that the MUC5B risk allele remained significantly elevated among individuals with a qualifying variant in the three study-wide significant IPF genes (Table 3), suggesting the possibility of an oligogenic model contributing to pulmonary fibrosis disease risk.
Table 3.
MUC5B Promoter Variant (rs35705950) Genotypes and Minor Allele Frequencies across the Collections
| All PF* | Familial PF | Sporadic IPF | All PF QV Carriers | In-House Control Subjects | 1KGP EUR | |
|---|---|---|---|---|---|---|
| Number of samples genotyped | 258 | 33 | 183 | 31 | 342 | 503 |
| G/G, n (%) | 115 (44.6) | 12 (36.4) | 79 (43.2) | 16 (51.6) | 272 (79.5) | 397 (78.9) |
| G/T, n (%) | 123 (47.7) | 18 (54.5) | 88 (48.1) | 15 (48.4) | 65 (19.0) | 104 (20.7) |
| T/T, n (%) | 20 (7.7) | 3 (9.1) | 16 (8.7) | 0 (0) | 5 (1.5) | 2 (0.4) |
| MAF (T allele), % | 31.59 | 36.36 | 32.79 | 24.19 | 10.96 | 10.74 |
| HWE exact test†P value | 0.11 | 0.46 | 0.25 | 0.15 | 0.58 | 0.10 |
| Allelic Fisher’s exact test P value (OR; 95% CI) compared with in-house control subjects | 8.8 × 10−19 (3.7; 2.7–5.1) | 3.8 × 10−7 (4.6; 2.5–8.3) | 3.1 × 10−17 (4.0; 2.8–5.6) | 0.007 (2.6; 1.3–5.0) | N/A | 0.87 (1.0; 0.7–1.4) |
| Cochran-Armitage test P value for trend compared with in-house control subjects | 1.3 × 10−18 | 1.0 × 10−8 | 1.7 × 10−17 | 0.002 | N/A | 0.88 |
Definition of abbreviations: CI = confidence interval; HWE = Hardy-Weinberg equilibrium; IPF = idiopathic pulmonary fibrosis; 1KGP EUR = 1000 Genomes Project European samples; MAF = minor allele frequency; N/A = not applicable; OR = odds ratio; PF = pulmonary fibrosis; QV = qualifying variant.
All allelic and trend tests were performed by comparison with a set of 342 in-house rs35705950 genotyped European control subjects. “All PF” column reports results derived from the full set of European case subjects with pulmonary fibrosis successfully genotyped for rs35705950. “Familial PF” column reports results for the subset of case subjects with a self-reported family history of pulmonary fibrosis. “Sporadic IPF” presents results for a homogeneous pulmonary fibrosis cohort with confirmed no reported family history of pulmonary fibrosis and clinically confirmed to have IPF on the basis of American Thoracic Society/European Respiratory Society/Japanese Respiratory Society/Latin American Thoracic Association guidelines. “All PF QV carriers” column reports results for the set of 31 case subjects with pulmonary fibrosis with a qualifying variant in one of the three study-wide significant genes. “In-House Control Subjects” column displays a sampling of 342 European control subjects genotyped using the same assay employed for the case genotyping. 1KGP EUR column gives the genotyping results derived from the 1000 Genomes Project (phase 3 European samples).
No significance (allelic or additive model) was observed when we compared the 31 PF QV carriers with the remaining 227 PF QV noncarriers (P > 0.1).
Adopting the recursion equations in function HWExact of R package HardyWeinberg.
Discussion
This study provides new evidence that three telomere-related genes (TERT, RTEL1, and PARN) previously implicated in FPF also confer risk for sporadic IPF. The relevance of this study is far reaching. It not only provides insight into the ultrarare variant genetic architecture of IPF, which should encourage the ongoing sequencing of larger case populations, but also, by correcting for background variation, provides a catalog of genetic variation in these three pulmonary fibrosis genes that is enriched for IPF risk alleles. This work, like our previous studies in other disorders (13, 14), also provides a powerful example of an analytic approach that can be employed to manage the volume of rare variant data generated through exome analysis of disease populations. Our results suggest that IPF could be characterized by modest locus heterogeneity, perhaps in part owing to the careful clinical phenotyping in this cohort, including the highly unique aspect of lung tissue assessment for histological characterization of fibrosis.
Although patients with IPF and other idiopathic fibrosing lung disorders have been shown to have significantly shorter telomeres in leukocytes or alveolar epithelial cells than control subjects, these observations have been incompletely explained by mutations in TERT or TERC, suggesting that other genes could be involved (20, 21). Our results provide further support for recent studies implicating deleterious variants in RTEL1 and PARN (12, 17) as additional genes critical to telomere maintenance and thus provide possible explanations for the shorter telomere lengths observed in the pulmonary fibrosis population. The PARN gene encodes a poly(A)-specific RNase protein that has been shown to regulate gene expression through deadenylation and thus shortening of mRNA poly(A) tail length (22). With regard to pulmonary fibrosis, by using induced pluripotent stem cells from patients, Moon and colleagues showed that patients with a disrupted PARN gene have decreased levels of TERC, thus highlighting the important role for PARN in the biogenesis of TERC and further showing that depleted TERC levels could be returned to normal after restoring PARN (23). The RTEL1 gene encodes the regulator of telomere elongation helicase 1 protein. As the name suggests, the RTEL1 protein is an ATP-dependent DNA helicase that has been shown to be crucial both for regulating telomere length and also for preventing genetic instability through its role in DNA repair mechanisms (24). Growing evidence suggests that defects in telomere maintenance predispose affected individuals to epithelial cell dysfunction. In particular, telomere defects have been associated with epithelial cell senescence, alveolar epithelial stem cell failure, and an impaired epithelial response to injury (25, 26).
None of the patients in our cohort had the classic multisystem clinical manifestations of dyskeratosis congenita (DC), a disease hallmarked by skin and nail changes, bone marrow failure, and organ fibrosis. Whereas the prevalence of pulmonary fibrosis is low in the general population (0.01 to 0.06%) (27), it is a common comorbidity with DC (28), and 8 of our 31 TERT, RTEL1, or PARN case variant observations (Table 2) have been reported previously in DC (24, 29, 30). This is consistent with IPF involving a disease mechanism driven by telomere dysfunction, a mechanism recognized to be responsible for various genetic disorders with a spectrum of clinical manifestations, including lung fibrosis (31). This overlap also suggests that variable expressivity could be more common among TERT, RTEL1, or PARN risk allele carriers than previously appreciated.
Across the various qualifying variant criteria, including an LoF-specific analysis (Table E4), we did not find significant enrichment of rare qualifying variants among other reported DC genes (DKC1, NHP2, NOP10, TINF2, and WRAP53) (Tables E5, E6, and E8). We did not identify any NAF1 LoF qualifying variant carriers among this collection of 262 cases (32). We did not find an enrichment of rare qualifying variants among genes that have been associated with pulmonary fibrosis risk via genome-wide association study loci (MUC5B, TOLLIP, SPPL2C, DSP, MAPT, and DPP9) (5, 33). Given that our study was focused on the protein-coding exome, its design is not amenable to assessing the RNA-encoding TERC gene, a gene previously implicated in both DC and pulmonary fibrosis risk, and thus some of our TERT, RTEL1, or PARN qualifying variant noncarriers may carry risk alleles in this noncoding gene. The knowledge of IPF risk associated with variants in the TERC noncoding gene also raises an interesting opportunity to investigate in future genome sequencing studies the contribution to IPF disease risk of ultrarare noncoding variants in noncoding genes such as TERC and also the noncoding sequences of implicated protein-coding genes such as TERT, RTEL1, and PARN.
This study does not rule out the possible influence of more complex patterns contributing to IPF risk among the TERT, RTEL1, or PARN qualifying variant noncarriers. However, given the remarkable signals identified using 262 case subjects, we also anticipate that it is likely that additional IPF risk genes, each explaining smaller fractions of the patient population, will be identified by analyzing larger case samples using this analytical framework focused on rare deleterious variations.
Our conditional analysis emphasizes that a large genetic component of pulmonary fibrosis risk is mediated by dominant LoF alleles in RTEL1 and PARN, and for TERT and RTEL1, missense variants predicted to be damaging and below the lowest currently detectable frequencies in the general population (i.e., absent in Exome Aggregate Consortium and Exome Variant Server; MAF, <0.001%). For some of the LoF variants in PARN and RTEL1, there is evidence derived from the external reference cohorts to support the possibility of incomplete penetrance, as reported previously in FPF pedigrees (12) and also similarly described in our earlier findings derived using this same framework to associate LoF alleles in TBK1 with amyotrophic lateral sclerosis risk (13).
We also found that deleterious variants in IPF genes as well as the presence of genetic risk factors might collectively cause or modify the disease phenotype, as reflected by a possible oligogenic model. This could in part explain the incomplete penetrance of some of the PARN, RTEL1, and TERT deleterious variants reported in earlier familial studies (12). In our data, the MUC5B promoter risk allele frequency remained significantly enriched among case subjects carrying a qualifying variant in one of the three study-wide significant pulmonary fibrosis disease genes, albeit at a lower rate than among noncarrier case subjects. A similar observation was previously reported (34). Certainly, such a proposed oligogenic model requires further studies with screening of larger cohorts of familial and sporadic pulmonary fibrosis case subjects to better elucidate the nature and extent of the possible oligogenic model.
Because lung transplant is currently the only intervention demonstrated to improve survival in select candidates with advanced pulmonary fibrosis, our results may provide a new opportunity to better understand post-transplant outcomes in this population. Previously published case series have demonstrated that although lung transplant is feasible in patients with mutations in the telomerase genes TERT or TERC, these patients experience a higher rate of post-transplant complications, including leukopenia, thrombocytopenia, and renal failure (35–37). Broadening the assessment of potential lung transplant recipients with pulmonary fibrosis to include evaluation of established genetic variations in TERT, RTEL1, and PARN may help clinicians better inform patients of potential risks and tailor treatment interventions, such as choice of immunosuppressive agents, in anticipation of these complications.
In conclusion, we have demonstrated that three telomere-related genes previously implicated in FPF also confer risk for sporadic IPF, contributing to more than 10% of the genetic risk in this population. This work provides genetic evidence that telomere dysfunction also plays an important role in sporadic pulmonary fibrosis. An additional novelty is the enrichment of the MUC5B promoter allele among individuals with ultrarare PARN, RTEL1, and TERT qualifying variants, suggesting the possibility of a more complex oligogenic model contributing to the development of pulmonary fibrosis. Further genetic stratification of pulmonary fibrosis may aid in understanding or improving lung transplant outcomes in these patients. It is also plausible that genetic stratification could provide an effective approach to identifying individuals most likely to benefit from existing or newly developed treatments. We anticipate that the catalog of risk alleles, enriched for disease-causing variants, in the three pulmonary fibrosis genes we have defined could assist in delineating meaningful genetic subphenotypes.
Acknowledgments
Acknowledgment
The authors thank members of the Institute for Genomic Medicine, Columbia University (B. Copeland, S. Kamalakaran, B. Krueger, and R. Padmanabhan), and Matt McKevitt of Gilead Sciences, Inc., for ongoing commitment that enables this work. S.P. is a National Health and Medical Research Council Career Development Fellowship fellow. The authors also thank the NHLBI GO Exome Sequencing Project and its ongoing studies, which produced and provided exome variant calls for comparison: the Lung Grand Opportunity (GO) Sequencing Project (HL-102923), the Women’s Health Initiative Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926), and the Heart GO Sequencing Project (HL-103010). In addition, the authors thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about.
Footnotes
Supported by internal funding from the Duke University School of Medicine and Department of Medicine, National Institutes of Health (NIH)/NHLBI K24 grant 1K24 HL091140-01A1 (S.M.P.), R. D. Wright Career Development fellowship 1126877 (S.P.), and a grant from Gilead Sciences (D.B.G.). J.W.C., T.O’R., and J.G.M. are employees of Gilead Sciences, Inc. The Duke and NIH funds supported sample collection and clinical phenotyping, and Gilead funding supported exome analysis.
Author Contributions: D.B.G. and S.M.P. conceived of and designed the study; J.L.T., M.T.D., F.L.K., C.F., A.F.C., C.B., and S.M.P. acquired and processed the clinical samples; M.T.D. performed the clinical phenotyping with support from J.L.T. and S.M.P.; S.P., Q.W., Z.R., and J.B. performed the bioinformatic processing; C.M.M. and C.D.M. performed the TaqMan genotyping; S.P. analyzed the data with support from Q.W., A.S.A., and D.B.G.; S.P., J.L.T., A.S.A., S.M.P., and D.B.G. interpreted the data; S.P., J.L.T., M.T.D., S.M.P., and D.B.G. drafted the manuscript; and all authors critically revised the manuscript for important intellectual content.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201610-2088OC on January 18, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, Colby TV, Cordier JF, Flaherty KR, Lasky JA, et al. ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raghu G, Chen SY, Yeh WS, Maroni B, Li Q, Lee YC, Collard HR.Idiopathic pulmonary fibrosis in US Medicare beneficiaries aged 65 years and older: incidence, prevalence, and survival, 2001–11 Lancet Respir Med 20142566–572.[Published erratum appears in Lancet Respir Med 2014;2:e12.] [DOI] [PubMed] [Google Scholar]
- 3.Steele MP, Schwartz DA. Molecular mechanisms in progressive idiopathic pulmonary fibrosis. Annu Rev Med. 2013;64:265–276. doi: 10.1146/annurev-med-042711-142004. [DOI] [PubMed] [Google Scholar]
- 4.Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, Loyd JE, Fingerlin TE, Zhang W, Gudmundsson G, Groshong SD, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. 2011;364:1503–1512. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noth I, Zhang Y, Ma SF, Flores C, Barber M, Huang Y, Broderick SM, Wade MS, Hysi P, Scuirba J, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med. 2013;1:309–317. doi: 10.1016/S2213-2600(13)70045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein DB, Allen A, Keebler J, Margulies EH, Petrou S, Petrovski S, Sunyaev S. Sequencing studies in human genetics: design and interpretation. Nat Rev Genet. 2013;14:460–470. doi: 10.1038/nrg3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Kuan PJ, Xing C, Cronkhite JT, Torres F, Rosenblatt RL, DiMaio JM, Kinch LN, Grishin NV, Garcia CK. Genetic defects in surfactant protein A2 are associated with pulmonary fibrosis and lung cancer. Am J Hum Genet. 2009;84:52–59. doi: 10.1016/j.ajhg.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nogee LM, Dunbar AE, III, Wert SE, Askin F, Hamvas A, Whitsett JA. A mutation in the surfactant protein C gene associated with familial interstitial lung disease. N Engl J Med. 2001;344:573–579. doi: 10.1056/NEJM200102223440805. [DOI] [PubMed] [Google Scholar]
- 10.Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, Markin C, Lawson WE, Xie M, Vulto I, Phillips JA, III, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med. 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 11.Tsakiri KD, Cronkhite JT, Kuan PJ, Xing C, Raghu G, Weissler JC, Rosenblatt RL, Shay JW, Garcia CK. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci USA. 2007;104:7552–7557. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stuart BD, Choi J, Zaidi S, Xing C, Holohan B, Chen R, Choi M, Dharwadkar P, Torres F, Girod CE, et al. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat Genet. 2015;47:512–517. doi: 10.1038/ng.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, Couthouis J, Lu YF, Wang Q, Krueger BJ, et al. FALS Sequencing Consortium. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagnall RD, Crompton DE, Petrovski S, Lam L, Cutmore C, Garry SI, Sadleir LG, Dibbens LM, Cairns A, Kivity S, et al. Exome-based analysis of cardiac arrhythmia, respiratory control, and epilepsy genes in sudden unexpected death in epilepsy. Ann Neurol. 2016;79:522–534. doi: 10.1002/ana.24596. [DOI] [PubMed] [Google Scholar]
- 15.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cogan JD, Kropski JA, Zhao M, Mitchell DB, Rives L, Markin C, Garnett ET, Montgomery KH, Mason WR, McKean DF, et al. Rare variants in RTEL1 are associated with familial interstitial pneumonia. Am J Respir Crit Care Med. 2015;191:646–655. doi: 10.1164/rccm.201408-1510OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kannengiesser C, Borie R, Ménard C, Réocreux M, Nitschké P, Gazal S, Mal H, Taillé C, Cadranel J, Nunes H, et al. Heterozygous RTEL1 mutations are associated with familial pulmonary fibrosis. Eur Respir J. 2015;46:474–485. doi: 10.1183/09031936.00040115. [DOI] [PubMed] [Google Scholar]
- 19.1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alder JK, Chen JJ, Lancaster L, Danoff S, Su SC, Cogan JD, Vulto I, Xie M, Qi X, Tuder RM, et al. Short telomeres are a risk factor for idiopathic pulmonary fibrosis. Proc Natl Acad Sci USA. 2008;105:13051–13056. doi: 10.1073/pnas.0804280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cronkhite JT, Xing C, Raghu G, Chin KM, Torres F, Rosenblatt RL, Garcia CK. Telomere shortening in familial and sporadic pulmonary fibrosis. Am J Respir Crit Care Med. 2008;178:729–737. doi: 10.1164/rccm.200804-550OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tummala H, Walne A, Collopy L, Cardoso S, de la Fuente J, Lawson S, Powell J, Cooper N, Foster A, Mohammed S, et al. Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J Clin Invest. 2015;125:2151–2160. doi: 10.1172/JCI78963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moon DH, Segal M, Boyraz B, Guinan E, Hofmann I, Cahan P, Tai AK, Agarwal S. Poly(A)-specific ribonuclease (PARN) mediates 3′-end maturation of the telomerase RNA component. Nat Genet. 2015;47:1482–1488. doi: 10.1038/ng.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walne AJ, Vulliamy T, Kirwan M, Plagnol V, Dokal I. Constitutional mutations in RTEL1 cause severe dyskeratosis congenita. Am J Hum Genet. 2013;92:448–453. doi: 10.1016/j.ajhg.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alder JK, Barkauskas CE, Limjunyawong N, Stanley SE, Kembou F, Tuder RM, Hogan BL, Mitzner W, Armanios M. Telomere dysfunction causes alveolar stem cell failure. Proc Natl Acad Sci USA. 2015;112:5099–5104. doi: 10.1073/pnas.1504780112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kropski JA, Lawson WE, Young LR, Blackwell TS. Genetic studies provide clues on the pathogenesis of idiopathic pulmonary fibrosis. Dis Model Mech. 2013;6:9–17. doi: 10.1242/dmm.010736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nalysnyk L, Cid-Ruzafa J, Rotella P, Esser D. Incidence and prevalence of idiopathic pulmonary fibrosis: review of the literature. Eur Respir Rev. 2012;21:355–361. doi: 10.1183/09059180.00002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dokal I. Dyskeratosis congenita. Hematology Am Soc Hematol Educ Program. 2011;2011:480–486. doi: 10.1182/asheducation-2011.1.480. [DOI] [PubMed] [Google Scholar]
- 29.Du HY, Pumbo E, Manley P, Field JJ, Bayliss SJ, Wilson DB, Mason PJ, Bessler M. Complex inheritance pattern of dyskeratosis congenita in two families with 2 different mutations in the telomerase reverse transcriptase gene. Blood. 2008;111:1128–1130. doi: 10.1182/blood-2007-10-120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ballew BJ, Yeager M, Jacobs K, Giri N, Boland J, Burdett L, Alter BP, Savage SA. Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in dyskeratosis congenita. Hum Genet. 2013;132:473–480. doi: 10.1007/s00439-013-1265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarek G, Marzec P, Margalef P, Boulton SJ. Molecular basis of telomere dysfunction in human genetic diseases. Nat Struct Mol Biol. 2015;22:867–874. doi: 10.1038/nsmb.3093. [DOI] [PubMed] [Google Scholar]
- 32.Stanley SE, Gable DL, Wagner CL, Carlile TM, Hanumanthu VS, Podlevsky JD, Khalil SE, DeZern AE, Rojas-Duran MF, Applegate CD, et al. Loss-of-function mutations in the RNA biogenesis factor NAF1 predispose to pulmonary fibrosis-emphysema. Sci Transl Med. 2016;8:351ra107. doi: 10.1126/scitranslmed.aaf7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fingerlin TE, Murphy E, Zhang W, Peljto AL, Brown KK, Steele MP, Loyd JE, Cosgrove GP, Lynch D, Groshong S, et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet. 2013;45:613–620. doi: 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coghlan MA, Shifren A, Huang HJ, Russell TD, Mitra RD, Zhang Q, Wegner DJ, Cole FS, Hamvas A. Sequencing of idiopathic pulmonary fibrosis-related genes reveals independent single gene associations. BMJ Open Respir Res. 2014;1:e000057. doi: 10.1136/bmjresp-2014-000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silhan LL, Shah PD, Chambers DC, Snyder LD, Riise GC, Wagner CL, Hellström-Lindberg E, Orens JB, Mewton JF, Danoff SK, et al. Lung transplantation in telomerase mutation carriers with pulmonary fibrosis. Eur Respir J. 2014;44:178–187. doi: 10.1183/09031936.00060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tokman S, Singer JP, Devine MS, Westall GP, Aubert JD, Tamm M, Snell GI, Lee JS, Goldberg HJ, Kukreja J, et al. Clinical outcomes of lung transplant recipients with telomerase mutations. J Heart Lung Transplant. 2015;34:1318–1324. doi: 10.1016/j.healun.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borie R, Kannengiesser C, Hirschi S, Le Pavec J, Mal H, Bergot E, Jouneau S, Naccache JM, Revy P, Boutboul D, et al. Groupe d’Etudes et de Recherche sur les Maladies “Orphelines Pulmonaires (GERM“O”P) Severe hematologic complications after lung transplantation in patients with telomerase complex mutations. J Heart Lung Transplant. 2015;34:538–546. doi: 10.1016/j.healun.2014.11.010. [DOI] [PubMed] [Google Scholar]