Abstract
Rationale: Genetic association studies in chronic obstructive pulmonary disease have primarily tested for association with common variants, the results of which explain only a portion of disease heritability. Because rare variation is also likely to contribute to susceptibility, we used whole-genome sequencing of subjects with clinically extreme phenotypes to identify genomic regions enriched for rare variation contributing to chronic obstructive pulmonary disease susceptibility.
Objectives: To identify regions of rare genetic variation contributing to emphysema with severe airflow obstruction.
Methods: We identified heavy smokers that were resistant (n = 65) or susceptible (n = 64) to emphysema with severe airflow obstruction in the Pittsburgh Specialized Center of Clinically Oriented Research cohort. We filtered whole-genome sequencing results to include only rare variants and conducted single variant tests, region-based tests across the genome, gene-based tests, and exome-wide tests.
Measurements and Main Results: We identified several suggestive associations with emphysema with severe airflow obstruction, including a suggestive association of all rare variation in a region within the gene ZNF816 (19q13.41; P = 4.5 × 10−6), and a suggestive association of nonsynonymous coding rare variation in the gene PTPRO (P = 4.0 × 10−5). Association of rs61754411, a rare nonsynonymous variant in PTPRO, with emphysema and obstruction was demonstrated in all non-Hispanic white individuals in the Pittsburgh Specialized Center of Clinically Oriented Research cohort. We found that cells containing this variant have decreased signaling in cellular pathways necessary for survival and proliferation.
Conclusions: PTPRO is a novel candidate gene in emphysema with severe airflow obstruction, and rs61754411 is a previously unreported rare variant contributing to emphysema susceptibility. Other suggestive candidate genes, such as ZNF816, are of interest for future studies.
Keywords: genetic association studies, whole-genome sequencing, chronic obstructive pulmonary disease, emphysema
At a Glance Commentary
Scientific Knowledge on the Subject
The genetic susceptibility to chronic obstructive pulmonary disease remains incompletely understood, in part because of the contribution of variants that cannot be detected by classic common variant association studies. The well-known contribution of rare variation to emphysema in alpha-1 antitrypsin deficiency and recent whole-exome sequencing studies demonstrate that rare variants contribute to this susceptibility.
What This Study Adds to the Field
Here, we present the first whole-genome sequencing study of chronic obstructive pulmonary disease, identifying PTPRO as a novel candidate gene and rs61754411 as a previously unreported rare, nonsynonymous susceptibility variant contributing to the disease. We offer in vitro evidence that this variant decreases signaling in pathways necessary for cellular survival and proliferation, suggesting how this gene may contribute to emphysema with severe airflow obstruction.
Chronic obstructive pulmonary disease (COPD) is defined as irreversible airflow limitation and is a major cause of worldwide morbidity and mortality (1, 2). The disorder is caused by an interaction of environmental factors, most commonly cigarette smoke, and genetic predisposition (3). This complex etiology results in an equally complex disease presentation, with at least two pathologic entities contributing to airflow limitation (emphysema and small airway obstruction) accompanied by any of numerous comorbidities (4).
Genetic susceptibility to COPD remains incompletely characterized. Common variant association studies have successfully identified genetic loci associated with COPD phenotypes, including several that have been replicated repeatedly, notably loci near the genes HHIP, CHRNA3/5/IREB2, FAM13A, and CYP2A6 (5–7). However, the most frequently replicated single-nucleotide polymorphisms (SNPs) at these loci account for less than 10% of COPD heritability (8). Similar results have been observed in many other complex, polygenic diseases including coronary artery disease, type II diabetes, and schizophrenia (9). This disparity between the heritability explained by common variation in complex disease and what was expected at the advent of the genome-wide association study era is likely explained by multiple factors, including contributions from uncommon and rare genetic variants, terms used to describe variants with minor allele frequency (MAF) of less than 5% and 1% in the population, respectively (10–12). Notably, rare variants in SERPINA1 leading to alpha-1 antitrypsin deficiency suggest a role for rare variation in the genetic susceptibility to COPD (13).
Advances in next generation sequencing technologies have begun to make sequencing studies both technologically and financially feasible (14). Approaches to maximize power in smaller populations have been hypothesized, including studies limited to the extremes of a phenotype and within families (11, 15). Recent whole-exome sequencing studies have taken one of these two approaches by looking at heavy smokers, severe emphysematous phenotypes, or families of severe early onset COPD, identifying candidate genes (16–18). The extreme trait sequencing approach is based on evidence from other diseases that individuals at the extreme of a phenotype are enriched for causal variants (19). This approach is particularly useful outside of the exome, where understanding of the impact of noncoding variants on gene expression remains limited but is clearly important. However, the contribution of such loci has not, to our knowledge, been studied in a sequencing study. Here we report the findings from the first whole-genome sequencing study in COPD. Some of the results of these studies have previously been reported in the form of an abstract (20).
Methods
Study Design
This study was approved by the Institutional Review Board for Human Subject Research at the University of Pittsburgh (IRB0612016). To control for population stratification, only U.S. non-Hispanic white persons (NHW) were included in this study. Subject recruitment and clinical evaluation of subjects in the Pittsburgh COPD Specialized Center of Clinically Oriented Research (SCCOR) has been described previously (21). Briefly, participants were current or former smokers ages 40–79 with a minimum 10-pack-year cigarette smoking history. Each subject completed a chest computed tomography (CT) scan, prebronchodilator and post-bronchodilator spirometry and plethysmographic lung volume measurement, diffusion capacity, and demographic and medical history questionnaires. Emphysema was quantified on CT images using a density mask approach to compute the fraction of voxels depicting the lung with a Hounsfield unit value less than −950 (F-950) (22).
From this population, 102 subjects with emphysema as measured by CT scan (susceptible, F-950 > 0.05) and significant airflow obstruction as quantified by spirometry (% predicted FEV1 < 0.04) and 86 subjects that did not develop emphysema (resistant, F-950 ≤ 0.01) or airflow obstruction (FEV1/FVC > 0.7; % predicted FEV1 > 0.8) were identified. Individuals in each group were matched by age, smoking history, and sex using an algorithm written in Python (see online supplement). After matching, 70 individuals in each subcohort remained for sequencing.
Genotyping of candidate nonsynonymous substitutions was performed in all NHW individuals in the Pittsburgh SCCOR cohort for whom DNA was available. Additional information about these individuals can be found in the online supplement.
Sequencing and Genotyping
Knome Biosciences (Boston, MA) sequenced 40 subjects (20 resistant, 20 susceptible) and Hudson Alpha Biotechnology (Birmingham, AL) sequenced an additional 100 (50 resistant, 50 susceptible). All individuals were sequenced using Illumina HiSeq technology (Illumina, San Diego, CA). Library preparation by both of these centers has previously been described (23, 24). Quality control steps are described in the online supplement.
Genomic DNA was isolated from blood for association testing and from whole-cell lysates for functional studies using the QIAamp DNA isolation kit (QIAgen, Valencia, CA). A nonsynonymous substitution in the gene PTPRO, rs61754411, was genotyped using the TaqMan platform (25) with predesigned primer and probes and 7900 DNA analyzer (ABI, Foster City, CA).
Data Analysis
We followed the Broad Institute’s Genome Analysis Toolkit best practices workflow to align and call variants (26). Briefly, Burrows-Wheeler Alignment (version 0.7.12-r1039) was used to align sequences to GrCh37 and Picard (version 1.126) was used for deduplication (27). Recalibrated variant calls for each individual were generated, followed by joint genotyping and SNP and indel recalibration (28). Final sequencing metrics including depth of coverage, distribution of coverage, and variant calling statistics were calculated after analysis of the entire sample set using Genome Analysis Toolkit (version 3.3–0) and SAM tools (version 1.1) (see Table E1 in the online supplement) (29). Six individuals were removed after sequencing quality control (see online supplement for further details).
Population stratification was tested between the two subcohorts by pruning all variants for which there was more than 10% missingness and that were not in Hardy-Weinberg equilibrium using PLINK (version 1.90) (30). Principal components analysis was used on this pruned set of SNPs and Tracy Widom statistics were calculated for each of the top principal components using Eigenstrat software (version 6.0.1) (31). After pruning, three individuals were removed from the susceptible cohort and two from the resistant cohort because of significant variation along the most significant principal components PC2 and PC6 (see Table E2, Figure E1).
Rare Variant Filtering
Variants were annotated using Annovar (version 2014–11–12) and filtered with bcf tools as follows. First, variants were removed if MAF was greater than 0.05 in European populations in 1,000 genomes or if MAF was greater than 0.05 in 6,500 exomes European populations. Then, variants were removed if they lacked AF in European populations but MAF in the entire 1,000 genomes populations greater than 0.05 and finally remaining variants were filtered if the empiric MAF was greater than 0.01.
Association Testing and Statistics
To test for genotype–phenotype association, we dichotomized susceptible and resistant individuals, a phenotype hereafter referred to as “emphysema with severe airflow obstruction.” We tested for association of single rare variants with emphysema with severe airflow obstruction using the efficient mixed-model association expedited (EMMAX) algorithm as implemented in EPACTS (32). Unadjusted P values were also generated using Fisher exact test. Single variants were annotated with Annovar from dbSNP144 and PolyPhen and Sift predictions were downloaded from Ensembl.
We used the optimized sequence kernel association test (SKAT-O) as implemented in EPACTS (version 3.2.6) to test for association of groups of rare SNPs with emphysema with severe airflow obstruction (33). We first tested across standardized 30,000 bp (1,000 bp = kb) windows based on power calculations demonstrating that this window size maximized power to detect in this population (see Figure E7). Then, we tested for all rare variation within genic regions, defined as the intronic, exonic, and 1 kb flanking spaces for each gene in the genome. Finally, we tested for association of nonsynonymous variants (missense and nonsense substitutions) with our phenotype across the exome. Annotations of nonsynonymous variants were made with EPACTS. We tested for association of rs61754411 with FEV1 or F-950 in the entire SCCOR cohort of NHW with a dominant multiple linear regression model in R, treating age, sex, and pack-years as covariates.
Thresholds for genome-wide significance and suggestive significance were defined as Bonferroni-corrected α of 0.05 (0.05/number of tests) and Bonferroni-corrected α of 1 (1/number of tests), respectively. All association tests except for EMMAX and Fisher exact test were adjusted for the following covariates: age, sex, pack-years, and eigenvalues from principal components 2 and 6 identified with Eigenstrat (see Table E2). Because EMMAX directly accounts for population structure, this test was only adjusted for age, sex, and pack-years. Fisher exact test is reported unadjusted. Two-sided Student’s t tests were used to compare population demographic and clinical variables. Tracy Widom statistics were used to compare the principal components generated in Eigenstrat.
Cell Culture
Primary human bronchial epithelial (HBE) cells were cultured from donor lungs obtained from the Center for Organ Recovery and Education that seemed to have no lung disease by high-resolution CT scan but were deemed unacceptable for lung transplant. The cells were isolated and cultured using the method described previously with the following modifications (34). For the in vitro recombinant human epidermal growth factor (EGF) treatment, cells were maintained in monolayer and cultured with bronchial epithelial growth medium (Lonza, Basel, Switzerland) and type VI human placental collagen-treated tissue culture flasks. Cells in 80–90% confluence were used for subsequent seeding and recombinant human EGF treatment. Cigarette smoke extract (CSE) was a gift from Dr. Peter Di (University of Pittsburgh) and has been described previously (35). The HBE cells were starved in culture media without growth factor supplements for 4 hours before the treatment with CSE or 20 ng/ml recombinant human EGF (Thermo-Fisher Scientific, Pittsburgh, PA).
Western Blot
Whole-cell lysates were prepared by homogenization in radioimmunoprecipitation assay buffer. Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membrane, and blocked in 2% milk. Blots were incubated with primary antibodies (p-ErbB1 [Invitrogen #44-784G; ThermoFisher Scientific, Waltham, MA], ErbB1 [Cell Signaling #4267; Cell Signaling Technology, Danvers, MA], p-ErbB2 [Cell Signaling #6942], ErbB2 [Cell Signaling #4290], p-Erk [Cell Signaling #4377], Erk [Cell Signaling #4695], p-STAT3 [Cell Signaling #9145], STAT3 [Cell Signaling #4904], PT-PRO [Santa Cruz Biotech #sc365354; Santa Cruz Biotech, Santa Cruz, CA], GAPDH [Cell Signaling #5174], and β-actin [Sigma #5441; Sigma-Aldrich, St. Louis, MO]) overnight at 4°C and with appropriate secondary antibodies (Santa Cruz Biotech goat antimouse IgG-HRP, #SC2005; goat antirabbit IgG-HRP, #SC2004) for 1 hour at room temperature. Blots were developed using HRP-substrate (Millipore #WBKLS0500; Millipore, Billerica, MA) and imaged using Amersham Imager 600 (GE Health, Chicago, IL). Densitometry was performed using ImageJ software (National Institutes of Health, Bethesda, MD) and values are expressed as mean ± SEM. A paired Student’s t test was performed on densitometry values, and a P less than 0.05 was considered statistically significant.
Results
Cohort Characteristics
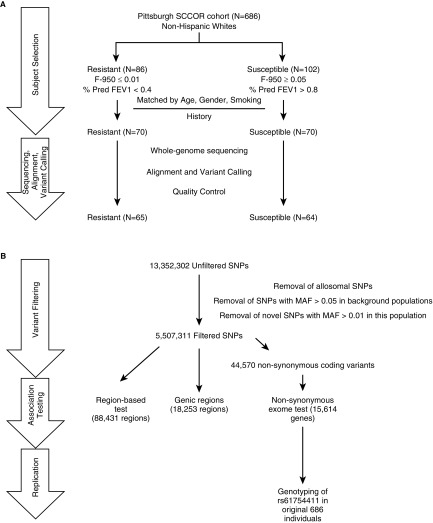
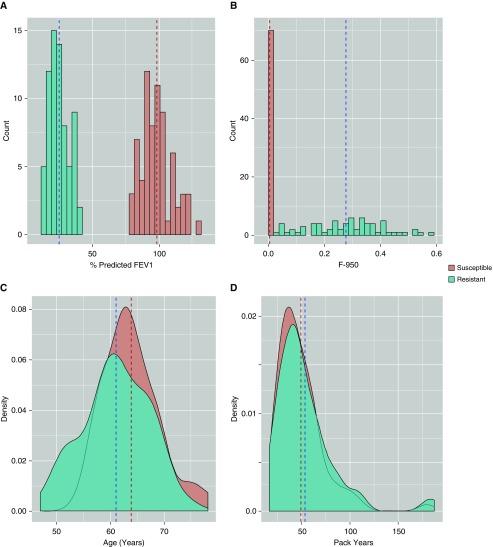
From a population of heavy smokers, we identified 102 NHW subjects with emphysema as measured by CT scan (susceptible, F-950 > 0.05) and significant obstruction (% predicted FEV1 < 0.4) and 86 NHW subjects that did not develop emphysema (resistant, F-950 ≤ 0.01) or obstruction (FEV1/FVC > 0.7 and % predicted FEV1 > 0.8) despite similar smoking histories. Importantly, this approach allowed us to identify individuals susceptible to clinically relevant emphysema, because several studies have shown that emphysema as measured by CT does not always correlate with severity of obstruction, and individuals resistant to emphysema and obstruction (36). We matched individuals in this population to identify 70 susceptible and 70 resistant individuals representative of the larger groups. Six individuals were removed after sequencing quality control and five were removed as population outliers, leaving a final population of 64 resistant and 65 susceptible individuals analyzed here (Figure 1A). Resistant and susceptible individuals shared similar characteristics in terms of sex (% female susceptible, 43.8; resistant, 47.7), age (median years susceptible, 61; resistant, 63), and smoking history (mean pack-years susceptible, 54.4; resistant, 48.5) but were significantly different in spirometric (mean percent predicted FEV1 susceptible, 25.3; resistant, 98.0) and radiologic attributes (mean F-950 susceptible, 0.269; resistant, 0.004) (Figure 2, Table 1).
Figure 1.
Extreme trait whole-genome sequencing study design. (A) Patient selection and quality control resulted in (B) final analysis using three different group-wise tests comparing rare variants or rare nonsynonymous variants in 65 individuals resistant to emphysema and 64 individuals susceptible to emphysema. F-950 = Hounsfield units value less than −950; MAF = minor allele frequency; Pred = predicted; SCCOR = Specialized Center of Clinically Oriented Research; SNP = single-nucleotide polymorphisms.
Figure 2.
Analyzed subcohorts represent clinical extremes in terms of obstruction as measured by (A) percent predicted FEV1 and (B) F-950 despite having very similar (D) smoking histories (pack-years) at a similar (C) age. Dashed lines represent subcohort means. F-950 = Hounsfield units value less than −950.
Table 1.
Sex, Age, Smoking History, and Clinical Characteristics of the Analyzed Susceptible and Resistant Subcohorts
| Emphysema Susceptible | Emphysema Resistant | P Value* | |
|---|---|---|---|
| Number of subjects sequenced | 64 | 65 | |
| Females, % | 43.8 | 47.7 | 0.67 |
| Age at PFT, yr, median (IQR) | 61 (57–65) | 63 (60–66) | 0.01 |
| Smoking history, pack-years, mean (IQR) | 54.4 (35.6–61.5) | 48.5 (30.0–60.0) | 0.25 |
| FEV1, % predicted, mean (IQR) | 25.3 (19.8–30.0) | 98.0 (89.0–104.0) | 7.63 × 10−29 |
| F-950, mean (IQR) | 0.269 (0.172–0.351) | 0.004 (0.002–0.006) | 2.00 × 10−24 |
Definition of abbreviations: F-950 = Hounsfield units value less than −950; IQR = interquartile range; PFT = pulmonary function testing.
Two-tailed Student’s t test.
Sequencing Results
The average depth of sequencing in all analyzed samples was ×30.2 across the genome, with 83.5% coverage of the genome at ×20 depth of coverage (see Table E1). We identified 13,352,302 SNPs, 2,682,220 of which were novel (compared with dbSNP144) in 135 individuals that passed sequencing quality control.
We used a stepwise approach to filter our variant calls for rare variation, which we defined as any variant with a MAF less than 0.05 in background European populations or MAF less than 0.01 empirically if no MAF existed in any of these populations (see Methods section). Using this approach, we identified 5,673,659 rare SNPs, with an average of 79,207 rare SNPs per subject (see Table E1). We tested across a final 5,507,311 rare autosomal SNPs after excluding sex-linked variants.
Genome-Wide Single Rare Variant Association Tests
We tested each rare autosomal SNP individually for its association with emphysema with severe airflow obstruction using the EMMAX algorithm and found no significant associations after Bonferroni correction (0.05/5,507,311 tests; P ≤ 8.8 × 10−9). The association with the lowest P value was with a set of four noncoding SNPs at 9p13 (rs117400947, rs80121798, rs77945177, rs75985055; P = 7.1 × 10−5). The nearest genes to these four SNPs are PAX5 and EBLN3P. The nonsynonymous substitution with the lowest P value was rs75683534 (P = 1.4 × 10−3), a C to A transversion resulting in a premature stop codon in the gene PIF1 that occurred in nine susceptible individuals and no resistant individuals (see Table E3).
Genome-Wide Region-based Rare Variant Association Tests
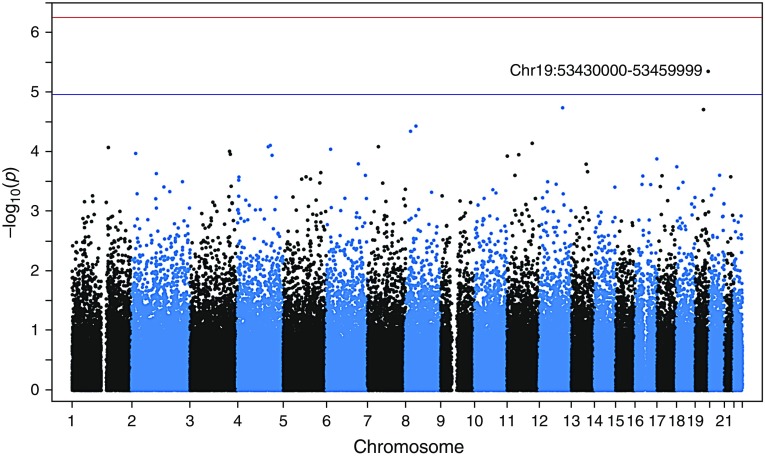
We grouped rare autosomal SNPs in 30-kb windows across the genome and tested for association using SKAT-O (33). The association with the lowest P value in this genome-wide scan was a locus at 19q13.41 (Figure 3, Table 2) (chr19:53430000–53459999; P = 4.5 × 10−6). Although not reaching genome-wide significance after Bonferroni multiple-testing correction (0.05/88,431 tests; P ≤ 5.4 × 10−7), this locus reached suggestive significance (1/88,431 tests; P ≤ 1.1 × 10−5). Fifty-six percent of individuals in this study harbored at least one rare variant in a total of 79 different loci within this region (Table 2). Rare variants were preferentially harbored by individuals with susceptibility to emphysema, with 75% of all alternate alleles in this region occurring in the susceptible population (129/172 alleles) (see Table E4). The 10 most significantly associated regions can be seen in Table 2, and quantile–quantile plots are available in the online supplement.
Figure 3.
Manhattan plot of 30-kb regions spanning the whole genome. Each dot represents a single 30-kb region and the strength of its association with emphysema is plotted on the y-axis. The top association at 19q31 is labeled. Other top associations are reported in Table 2. The top threshold represents genome-wide significant association (P ≤ 5.4 × 10−7), and the bottom threshold represents suggestive association (P ≤ 1.1 × 10−5) after Bonferroni correction.
Table 2.
Top Associations with Emphysema in 30-kb Region–based Whole-Genome Scan and Any Genes in the Regions Identified
| Chromosome | Start | End | Fraction of Population with Rare* | Unique Rare† | Unique Singletons‡ | P Value | Gene(s) in Region |
|---|---|---|---|---|---|---|---|
| 19 | 53430000 | 53459999 | 0.55814 | 79 | 62 | 4.51 × 10−6 | ZNF816, ZNF321P |
| 12 | 96300000 | 96329999 | 0.37209 | 33 | 18 | 1.84 × 10−5 | CCDC38 |
| 19 | 33810000 | 33839999 | 0.36434 | 43 | 34 | 1.97 × 10−5 | — |
| 8 | 42060000 | 42089999 | 0.45736 | 67 | 48 | 3.74 × 10−5 | PLAT |
| 8 | 19560000 | 19589999 | 0.62791 | 77 | 51 | 4.56 × 10−5 | CSGALNACT1 |
| 11 | 104400000 | 104429999 | 0.51938 | 66 | 47 | 7.26 × 10−5 | — |
| 4 | 136500000 | 136529999 | 0.57364 | 62 | 44 | 7.88 × 10−5 | — |
| 7 | 45630000 | 45659999 | 0.51163 | 66 | 41 | 8.28 × 10−5 | ADCY1 |
| 4 | 128040000 | 128069999 | 0.44961 | 50 | 40 | 8.30 × 10−5 | — |
| 1 | 151350000 | 151379999 | 0.3876 | 44 | 32 | 8.53 × 10−5 | PSMB4, POGZ |
Fraction of entire population (129 individuals) that harbors at least one rare variant.
Number of unique variants that passed filtering in this region.
Number of unique variants that only occurred once in either subcohort of this population.
Gene-based and Nonsynonymous Rare Variant Association Tests
We tested for an association of all rare variation with emphysema with severe airflow obstruction in coding and noncoding regions of genic regions (introns, exons, and 1-kb flanking region in each direction) across the exome. The association with the lowest P value in this test was the gene ZNF816 (P = 7.2 × 10−6), located at 19q13 and partially covered by the 30-kb locus with the lowest P value in the region-based test (see Figure E4). The 129 individuals in the population harbored a total of 120 different rare variants in this gene (Table 3). Similar to the region covered in the gene-based test, rare variation was more abundant in the susceptible population (see Table E4). Most of the rare variation located in this gene was intronic, although there were six nonsynonymous variants that also occurred preferentially in the susceptible population (see Table E4).
Table 3.
Top Associations with Emphysema in Gene-based Tests
| Gene | Chromosome | Start | End | Fraction of Population with Rare* | Unique Rare† | Unique Singletons‡ | P Value |
|---|---|---|---|---|---|---|---|
| ZNF816 | 19 | 53429414 | 53466404 | 0.60465 | 120 | 96 | 4.52 × 10−6 |
| BCAT2 | 19 | 49298537 | 49314089 | 0.17829 | 20 | 15 | 0.000245 |
| TMPRSS2 | 21 | 42835502 | 42903906 | 0.84496 | 181 | 126 | 0.000263 |
| DAGLA | 11 | 61447355 | 61515326 | 0.74419 | 124 | 83 | 0.00042 |
| COL5A3 | 19 | 10070204 | 10121507 | 0.71318 | 103 | 67 | 0.00045 |
| BBIP1 | 10 | 112661005 | 112680031 | 0.30233 | 25 | 16 | 0.000561 |
| ATP6V1B2 | 8 | 20054059 | 20084860 | 0.55039 | 63 | 36 | 0.000581 |
| KRTAP24 | 21 | 31652835 | 31655692 | 0.062016 | 5 | 3 | 0.000665 |
| CDC34 | 19 | 531546 | 543025 | 0.37209 | 39 | 20 | 0.000711 |
| C11orf53 | 11 | 111125842 | 111157151 | 0.72093 | 56 | 25 | 0.000759 |
Fraction of entire population (129 individuals) that harbors at least one rare variant.
Number of unique variants that passed filtering in this region.
Number of unique variants that only occurred once in either subcohort of this population.
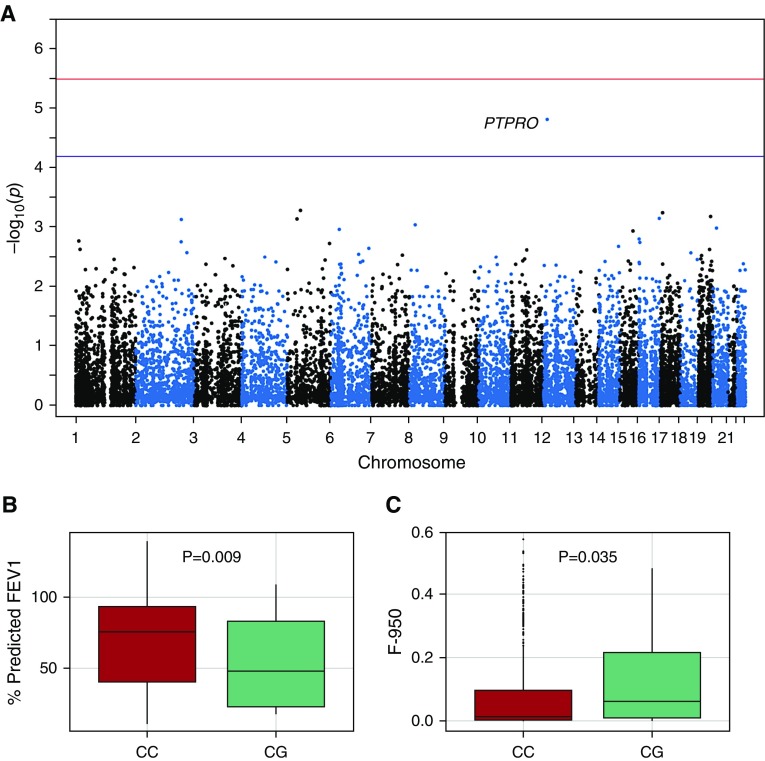
In our final test we tested for association with nonsynonymous coding variants grouped by gene across the exome (37). The association with the lowest P value was with the gene PTPRO located on chromosome 12 (Figure 4A, Table 4) (P = 4.0 × 10−5). We identified four separate rare, nonsynonymous substitutions in the gene, with the alternate allele of each substitution occurring only in the susceptible population (Table 5). One of these substitutions, rs61754411, occurred in eight individuals in the susceptible group but not at all in the resistant group and was predicted to be deleterious by both PolyPhen and Sift (Table 5). We genotyped this SNP in all 686 NHW individuals in the Pittsburgh SCCOR cohort and found that it was significantly associated with F-950 (P = 0.035) and % predicted FEV1 (P = 0.009) under a dominant model (Figures 4B and 4C, Table 6). Interestingly, we did not identify any individuals homozygous for the alternate allele, nor could we find an individual with this genotype in reported populations.
Figure 4.
Nonsynonymous variants in PTPRO are associated with emphysema. (A) Manhattan plot of exome-wide test taking into consideration only nonsynonymous variation. The top association is the gene PTPRO on chromosome 12, and other top associations are reported in Table 4. The top threshold represents genome-wide significant association, and the bottom threshold represents genome-wide suggestive association after Bonferroni correction. The most prevalent nonsynonymous single-nucleotide polymorphism in this gene was significantly associated with (B) % predicted FEV1 and (C) F-950 in the entire Pittsburgh Specialized Center of Clinically Oriented Research cohort. F-950 = Hounsfield units value less than −950; PTPRO = protein tyrosine phosphatase, type O.
Table 4.
Top Associations with Emphysema Including Only Nonsynonymous Variants across the Exome
| Gene | Chromosome | Start | End | Fraction of Population with Rare* | Unique Rare NS† | Unique NS Singletons‡ | P Value |
|---|---|---|---|---|---|---|---|
| PTPRO | 12 | 15654574 | 15656846 | 0.085271 | 4 | 2 | 1.57 × 10−5 |
| IL6ST | 5 | 55237014 | 55272085 | 0.054264 | 4 | 3 | 5.26 × 10−4 |
| ALOX15B | 17 | 7942928 | 7950394 | 0.062016 | 6 | 3 | 5.75 × 10−4 |
| TMEM143 | 19 | 48845944 | 48866796 | 0.062016 | 4 | 1 | 6.65 × 10−4 |
| ADAD2 | 16 | 84227605 | 84230332 | 0.069767 | 6 | 4 | 7.17 × 10−4 |
| CARD6 | 5 | 40841579 | 40860097 | 0.14729 | 7 | 2 | 7.32 × 10−4 |
| TFPI | 2 | 188331704 | 188348943 | 0.085271 | 2 | 0 | 7.47 × 10−4 |
| EPB49 | 8 | 21917015 | 21929924 | 0.069767 | 5 | 2 | 9.18 × 10−4 |
| FLRT3 | 20 | 14306868 | 14307107 | 0.062016 | 4 | 3 | 1.04 × 10−3 |
| ETV7 | 6 | 36336764 | 36343720 | 0.14729 | 8 | 5 | 1.10 × 10−3 |
Definition of abbreviation: NS = nonsynonymous variants.
Fraction of entire population (129 individuals) that harbors at least one rare variant.
Number of unique variants that passed filtering in this region.
Number of unique variants that only occurred once in either subcohort of this population.
Table 5.
Four Nonsynonymous Variants Identified in PTPRO Are Harbored in the Emphysema-Susceptible Population
| Chromosome | BP | Variation Name | Ref | Alt | MAF* | Resistant Count† | Susceptible Count† | AA change | PolyPhen Prediction | SIFT Prediction |
|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 15654574 | rs117540301 | A | G | 0.0078 | 65/0/0 | 62/2/0 | Ile228Val | Probably damaging | Tolerated |
| 12 | 15654578 | C | G | 0.0039 | 65/0/0 | 63/1/0 | Ser229Cys | |||
| 12 | 15654977 | rs141042273 | A | G | 0.0039 | 65/0/0 | 63/1/0 | His362Arg | Benign | Deleterious |
| 12 | 15656846 | rs61754411 | C | G | 0.0310 | 65/0/0 | 56/8/0 | Asn370Lys | Probably damaging | Deleterious |
Definition of abbreviations: Alt = alternate; BP = base pair; MAF = minor allele frequency; PolyPhen = predictions of functional effects of nonsynonymous SNPs by Polymorphism Phenotyping V2 (PolyPhen-2) algorithm; PTPRO = protein tyrosine phosphatase, type O; Ref = reference; SIFT Prediction = predictions of functional effects of nonsynonymous by Sorting Intolerant from Tolerant (SIFT) algorithm; SNP = single-nucleotide polymorphism.
Minor allele frequency in the entire population (129 individuals).
Counts are given as number of individuals homozygous for the reference allele/number of individuals heterozygous/number of individuals homozygous for the alternate allele.
Table 6.
Top Nonsynonymous Variant Identified in PTPRO Is Also Significantly Associated with Obstruction and Emphysema in a Larger Cohort
| rs61754411 Genotype | n | Age (yr) [Median (IQR)] | Females (%) | Smoking History (Pack-Years) [Mean (IQR)] | F-950 [Mean (IQR)] | FEV1 (% Predicted) [Mean (IQR)] |
|---|---|---|---|---|---|---|
| CC | 652 | 64 (60–69) | 46.6 | 54.8 (30–60) | 0.076 (0.004–0.096) | 0.692 (0.403–0.931) |
| CG | 34 | 64 (59–66) | 41.2 | 56.1 (35–70) | 0.126 (0.010–0.216) | 0.542 (0.232–0.828) |
| P value* | 0.217 | 0.539 | 0.843 | 0.035 | 0.009 |
Definition of abbreviations: F-950 = Hounsfield units value less than −950; IQR = interquartile range; PTPRO = protein tyrosine phosphatase, type O.
Two-tailed Student’s t test.
Protein Tyrosine Phosphatase, Type O Functional Assays
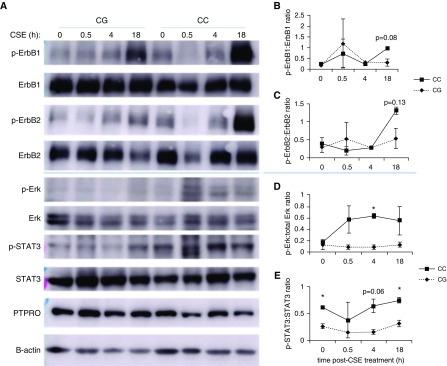
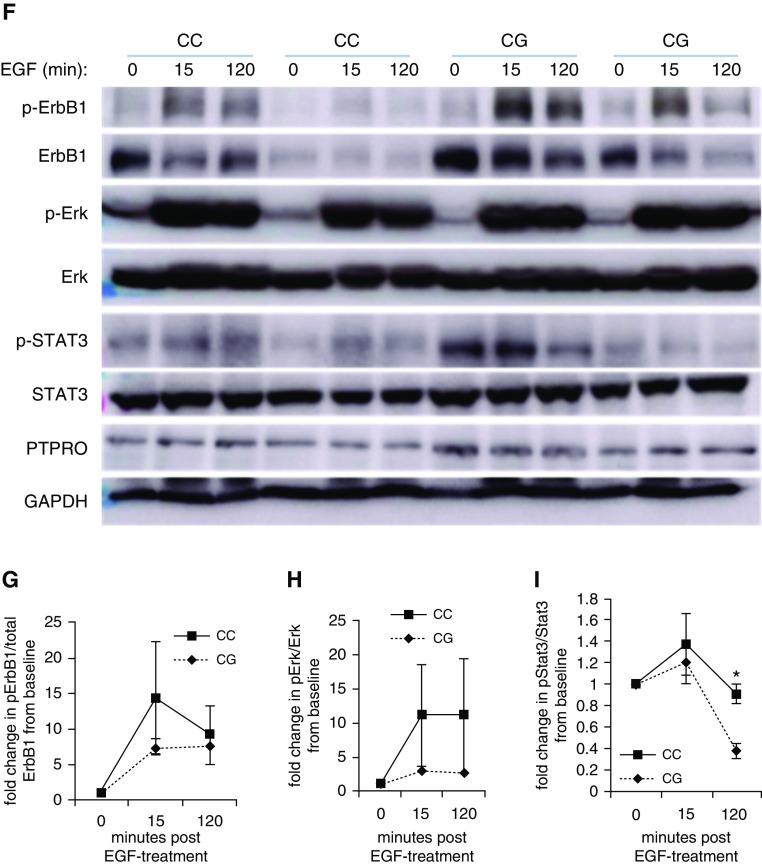
We obtained human primary HBE cells from donor lungs with CC or CG genotype of rs61754411 and assayed genotype-specific signaling characteristics in vitro. Because protein tyrosine phosphatase, type O (PTPRO) is a phosphatase that indirectly regulates the phosphorylation status of the intracellular domain of subunits of the EGF receptor, including the most prevalent, ErbB1 and ErbB2, we chose to focus on these pathways. In response to treatment with EGF ligand, individuals with CC genotype demonstrated a trend toward increased phosphorylation of ErbB1 (Figure 5G), which was associated with sustained phosphorylation of downstream mediators Erk (trend, Figure 5H), and STAT3 (P < 0.05 at 120 min post-treatment; n = 2) (Figure 5I). Similar findings were observed when HBEs were cultured in air-liquid interface and treated with 12.5% CSE (Figures 5A–5E). A trend toward increased phosphorylation of ErbB1 and ErbB2 was observed at 18 hours post-CSE-exposure. Importantly, PTPRO rs61754411-CC cells demonstrated significantly increased Erk and STAT3 phosphorylation at several time points post-CSE. There were no genotype-specific differences for PTPRO levels with any treatment, consistent with a coding sequence change that alters function rather than expression levels.
Figure 5.
PTPRO rs61754411 genotype-specific intracellular signaling necessary for cell survival and proliferation after CSE and EGF treatments of human primary human bronchial epithelial cells. Human bronchial epithelial cells from individuals with no lung diseases bearing the PTPRO rs61754411-CG or -CC genotype (n = 2 of each) were cultured in air–liquid interface and treated with CSE for the times indicated (A–E). (A) Whole-cell lysates from treated cells were blotted for the EGFR subunits ErbB1 and ErbB2 and for the downstream signaling molecules Erk and STAT3. Blots above the blue line are representative of one unique individual, and blots below are representative of the other unique individual. As controls, PTPRO and β-actin were also analyzed. (B–E) Densitometry analysis (ratio of p-Akt to total Akt) was performed on these results. Data are presented as mean ± SEM. These same cell lines were cultured in monolayer and treated with recombinant human EGF for the times indicated (F–I). (F) Whole-cell lysates from treated cells were blotted for the EGFR subunit ErbB1 and for the downstream signaling molecules Erk and STAT3. Similarly, PTPRO and GAPDH were also analyzed as controls. (G–I) Densitometry analysis was performed on the blots shown in F for two unique individuals with PTPRO rs61754411-CC and -CG genotypes. Values are presented as a relative ratio of p-Akt to total Akt band intensity compared with those observed in basal, untreated conditions. *P < 0.05. For both treatments, the cells were starved with growth media without growth factor supplements for 4 hours before the treatment in growth media with reduced growth factor supplements (10% of normal culture conditions). Akt = protein kinase B; CSE = cigarette smoke extract; EGF = epidermal growth factor; EGFR = EGF receptor; ErbB1/2 = EGFR/human epidermal growth factor receptor (HER2); Erk = extracellular signal regulated kinase; GAPDH = glyceraldehyde 3-phosphate dehydrogenase; p-Akt = phosphorylated Akt; p-Erk = phosphorylated Erk; p-STAT3 = phosphorylated STAT3; PTPRO = protein tyrosine phosphatase, type O; STAT3 = signal transducer and activator of transcription 3.
Discussion
We aimed to use whole-genome sequencing of two populations of heavy smokers with extreme phenotypes to identify genetic associations using group-wise rare variant tests. The selection of individuals with extreme traits has been shown to enrich for causal variants and this approach has been used successfully to identify candidate genes in sequencing studies (19, 38). Our cohort included two groups of heavy smokers that had extremely different outcomes in terms of both obstruction and emphysema, a phenotype we have referred to as emphysema with severe airflow obstruction (Figure 2, Table 1).
A primary goal of this study was to test for association of nongenic variants with emphysema with severe airflow obstruction. However, our knowledge of the effect of variants in noncoding regions remains limited, making it extremely difficult to prioritize the effects of variants as is possible in coding regions (39). Based on this, we tested for association with all filtered rare variants in standardized windows across the genome. Using physical distance rather than genetic distance to define the windows tested with SKAT-O was a simple approach to identify clusters of rare variation between two extreme populations. Although more complex approaches have been described, using SKAT in this manner remains an ideal approach when it is unknown whether variants in a region will have similar or opposing directions of effect (40).
We identified a single suggestive association using this approach, located on 19q13.41. This region is enriched for rare variants within the gene ZNF816 in the emphysema susceptible subcohort. Notably, when we tested for association of all rare variation with emphysema across gene-based regions, ZNF816 was also the result with the lowest P value (P = 7.2 × 10−6) (Table 3). Based on its sequence, this gene, which encodes zinc finger protein 816 (ZNF816), contains 15 zinc finger domains and is likely to be a DNA-binding protein, but its function is otherwise unknown. It is notable, however, that early linkage studies of COPD identified linkage between 19q and FEV1 in smokers (41, 42).
Several associations with the lowest P values from our whole-genome scan, although not reaching genome-wide significance, are of interest because they offer knowledge-based validation of our approach (Table 2). The association with the second lowest P value in this test included variants of the coiled-coil domain containing 38 (CCDC38) gene (P = 1.84 × 10−5). This gene was identified as a candidate in the first whole-exome sequencing study of heavy smokers, looking at resistance to airflow obstruction (18). In that study, the authors identified the nonsynonymous SNP rs10859974 as nominally associated with their phenotype. This SNP was filtered in our study suggesting that we identified this gene in an independent manner. Although the function of CCDC38 is not well-understood, it has been suggested to play a role in ciliary function (18).
In addition, several regions with the lowest P values in this study that do not contain any coding material are in close proximity to previously identified common variant associations with a phenotype of COPD. One of these is at 11q22.3, 1.6 Mb from an association with severe COPD between the genes MMP3 and MMP12 (43). Mice deficient in MMP12, which encodes the gene macrophage elastase, have been shown to be protected from cigarette-smoke-induced emphysema (44). Recent exome array studies have identified common coding variants in both MMP12 and MMP3 that are associated with COPD risk, suggesting that the rare variant association identified in this study is more likely caused by a novel functional effect on these or other nearby genes that contribute to disease (45, 46). Another region with a low P value in this study is located 8.9 Mb from a locus on 4q31 near the gene HHIP that is perhaps the most replicated association of phenotypes of pulmonary function and COPD, including FEV1 in smokers (43). Recent whole-exome sequencing studies have had similar results, with top associations within relatively short genomic distances of previously identified common variant associations, such as the results of a study of lung cancer cases with emphysema in which the authors identified a variant at 4q22 approximately 1 Mb from a frequently replicated locus near FAM13A (47).
Although whole-genome sequencing allows interrogation of the noncoding regions of the genome, it also offers more efficient coverage of exonic SNPs than whole-exome sequencing. By testing for association of rare nonsynonymous variants collapsed by gene with emphysema, we identified a suggestive association with the gene PTPRO on 12p12.3. We replicated the association between the most prevalent rare nonsynonymous SNP in this gene, rs61754411, and emphysema with airflow obstruction in a larger cohort. Similar to the top association from our region-based test, the location of this gene is notable because it is located less than 1 Mb from the short tandem repeat D12S1715, which was shown to demonstrate the most significant linkage with moderate airflow obstruction in smokers and FEV1 in smokers in two early linkage studies of COPD (42, 48). A candidate association study in this region showed association with COPD susceptibility near the gene SOX5; however, it remains possible that more than one gene contributed to this early linkage result (49).
In this study, only individuals susceptible to emphysema with severe obstruction harbored any of the four predicted deleterious substitutions in this gene, which encodes the receptor PTPRO. PTPRO is a phosphatase that targets residue Y416 of Src kinase for dephosphorylation, thus down-regulating its activity (50). Because Src is responsible for the activation of EGFR via phosphorylation of the cytoplasmic domain of its subunits, including ErbB1 and ErbB2, PTPRO serves as an indirect negative regulator of EGF signaling. Many studies have implicated the importance of EGF signaling in lung epithelial cell function during lung development, acute lung injury, and notably in response to cigarette smoke exposure (51, 52). Dysregulated EGFR phosphorylation or expression in patients with COPD compared with normal control subjects has been documented in several studies (53, 54). A functional effect on PTPRO, then, may play an important role in regulating the EGF pathway and account for the increased emphysema severity observed in patients with the CG genotype of rs61754411. We screened several dozen primary lung epithelial cell lines from donor lungs to identify cells from two donor lungs with the CG genotype of rs61754411. Cells expressing this allelic variant demonstrated reduced phosphorylation of the EGFR subunits ErbB1 and ErbB2, and reductions in downstream signaling activity in response to both EGF and CSE treatment. These observed reductions in signaling suggest that the CG variant in may in fact be hypermorphic, because we would predict that increased dephosphorylation of Src would result in less EGFR activation. Additional studies are required to refine and to better understand the mechanisms underlying these findings.
Hence, we have identified a suggestive association of rare variation with a gene previously associated in an exome sequencing study with protection from heavy smoking, two associated regions at loci previously shown to be in linkage with phenotypes of COPD, and two regions near associations of common variation with numerous phenotypes of COPD. It is likely that coding regions can be affected by both common and rare variation, as seen with CCDC38. However, regions that are in relatively close proximity but too distant to be plausible regulatory regions for the same genes, such as the association in this study identified near 4q31, may represent larger “disease susceptibility regions” in which multiple nearby loci may contribute to COPD susceptibility independently, as has been seen in other complex diseases (55).
Our study has limitations. As the first whole-genome sequencing study of emphysema, our sample sizes were limited; even with the extremes of a well-defined phenotype, we were unable to demonstrate significant association at the genome-wide level. Our filtering approach used an arbitrary MAF, which is more inclusive than has been used in some sequencing studies but still lower than what is measured in a common variant study where markers are specifically chosen for MAF greater than 0.05. Finally, our exome-based tests do not correct for gene size and thus favor the detection of variants in larger genes, although our most significant gene is actually quite small in size (13 kb).
This is the first study in which individuals with a phenotype of COPD were whole-genome sequenced and the first investigation of the contribution of genome-wide rare variation to a phenotype of COPD. Although we did not identify any associations reaching genome-wide significance, we identified associations reaching suggestive significance with the genes ZNF816 and PTPRO. In addition, we identified a rare susceptibility variant in PTPRO, rs61754411, which is significantly associated with emphysema and airway obstruction in an expanded population and alters the response to CSE and EGF treatments in primary human HBE cells in vitro. The replication of an association with the gene CCDC38 by exome sequencing using an alternate rare variant analysis approach gives further evidence that rare variation in this gene contributes to emphysema susceptibility. The nominal associations of numerous loci near previous human genetic evidence for linkage or association with COPD supports the hypothesis that rare variation contributes to this disease in a complex manner. However, as also seen in recent exome studies, the effect is likely to be caused by contributions from multiple genes, and potentially as seen here, from multiple forms of variation affecting multiple genes in nearby disease susceptibility regions.
Acknowledgments
Acknowledgment
The authors thank the patients who participated in the Pittsburgh Specialized Center of Clinically Oriented Research study. The authors also acknowledge Dr. Annerose Berndt of University of Pittsburgh Medical Center’s Enterprise Analytics and Jacob Yundt of University of Pittsburgh Medical Center’s Internet Services Division for technical assistance. The authors appreciate Chad Karoleski’s and Melissa Saul’s assistance with patient data. The authors thank Dr. Peter Di for sharing cigarette smoke extract and Dr. Joseph Pilewski and his laboratory for sharing human bronchial epithelial cells. Finally, the authors thank Stefanie Brown, Yanxia Chu, and Quanwei Yang for support in the preparation of human bronchial epithelial cells and biologic samples.
Footnotes
Supported by NHLBI grants P01 HL103455 and R01 HL107883 (S.D.S.), P50 HL084948 (F.C.S.), R21 HL129917 (F.C.S., Y.Z.), and T32 HL094295 (N.J.K.); Commonwealth of Pennsylvania, Department of Health, CURE (Commonwealth Universal Research Enhancement) Program SAP 4100062224 (F.C.S. and Y.Z.); National Institute of General Medical Sciences grant 5T32 GM008208 (N.J.K.); the Flight Attendant Medical Research Institute (A.D.G.); and the University of Pittsburgh Medical Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NHLBI or the National Institutes of Health.
Author Contributions: Study design, J.E.R., Y.Z., A.D.G., S.Y., J.K.L., N.K., F.C.S., and S.D.S. Clinical phenotyping and sample collection, F.C.S. and Y.Z. Data collection, J.E.R., Y.Z., A.D.G., S.Y., N.J.K., J.K.L., N.K., F.C.S., and S.D.S. Data quality control and analysis, J.E.R. Western blotting, A.D.G., S.Y., and Y.Z. Manuscript writing, J.E.R. Manuscript revision, J.E.R., Y.Z., A.D.G., S.Y., N.J.K., J.K.L., N.K., F.C.S., and S.D.S.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201606-1147OC on February 15, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Rabe KF, Hurd S, Anzueto A, Barnes PJ, Buist SA, Calverley P, Fukuchi Y, Jenkins C, Rodriguez-Roisin R, van Weel C, et al. Global Initiative for Chronic Obstructive Lung Disease. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176:532–555. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 2. National Heart, Lung, and Blood Institute. Morbidity and mortality: 2012 chart book on cardiovascular, lung and blood diseases. Bethesda, MD: National Institutes of Health; 2012 [updated 2013 May; accessed 2016 May 21]. Available from: https://www.nhlbi.nih.gov/research/reports/2012-mortality-chart-book.
- 3.Shapiro SD, Ingenito EP. The pathogenesis of chronic obstructive pulmonary disease: advances in the past 100 years. Am J Respir Cell Mol Biol. 2005;32:367–372. doi: 10.1165/rcmb.F296. [DOI] [PubMed] [Google Scholar]
- 4.Barr RG, Celli BR, Mannino DM, Petty T, Rennard SI, Sciurba FC, Stoller JK, Thomashow BM, Turino GM. Comorbidities, patient knowledge, and disease management in a national sample of patients with COPD. Am J Med. 2009;122:348–355. doi: 10.1016/j.amjmed.2008.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho MH, Castaldi PJ, Wan ES, Siedlinski M, Hersh CP, Demeo DL, Himes BE, Sylvia JS, Klanderman BJ, Ziniti JP, et al. ICGN Investigators; ECLIPSE Investigators; COPDGene Investigators. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet. 2012;21:947–957. doi: 10.1093/hmg/ddr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castaldi PJ, Cho MH, San José Estépar R, McDonald ML, Laird N, Beaty TH, Washko G, Crapo JD, Silverman EK COPDGene Investigators. Genome-wide association identifies regulatory loci associated with distinct local histogram emphysema patterns. Am J Respir Crit Care Med. 2014;190:399–409. doi: 10.1164/rccm.201403-0569OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardin MS, Silverman EK. Chronic obstructive pulmonary disease genetics: a review of the past and a look into the future. Chronic Obstr Pulm Dis. 2014;1:33–46. doi: 10.15326/jcopdf.1.1.2014.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou JJ, Cho MH, Castaldi PJ, Hersh CP, Silverman EK, Laird NM. Heritability of chronic obstructive pulmonary disease and related phenotypes in smokers. Am J Respir Crit Care Med. 2013;188:941–947. doi: 10.1164/rccm.201302-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.So HC, Gui AH, Cherny SS, Sham PC. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet Epidemiol. 2011;35:310–317. doi: 10.1002/gepi.20579. [DOI] [PubMed] [Google Scholar]
- 10.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, Kathiresan S, Daly MJ, Neale BM, Sunyaev SR, Lander ES. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci USA. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laurell CB. Electrophoretic microheterogeneity of serum alpha-1-antitrypsin. Scand J Clin Lab Invest. 1965;17:271–274. doi: 10.1080/00365516509075347. [DOI] [PubMed] [Google Scholar]
- 14.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 15.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 16.Bruse S, Moreau M, Bromberg Y, Jang JH, Wang N, Ha H, Picchi M, Lin Y, Langley RJ, Qualls C, et al. Whole exome sequencing identifies novel candidate genes that modify chronic obstructive pulmonary disease susceptibility. Hum Genomics. 2016;10:1. doi: 10.1186/s40246-015-0058-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiao D, Lange C, Beaty TH, Crapo JD, Barnes KC, Bamshad M, Hersh CP, Morrow J, Pinto-Plata VM, Marchetti N, et al. Lung GO; NHLBI Exome Sequencing Project; COPDGene Investigators. Exome sequencing analysis in severe, early-onset chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2016;193:1353–1363. doi: 10.1164/rccm.201506-1223OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wain LV, Sayers I, Soler Artigas M, Portelli MA, Zeggini E, Obeidat ME, Sin DD, Bossé Y, Nickle D, Brandsma C-A, et al. Whole exome re-sequencing implicates CCDC38 and cilia structure and function in resistance to smoking related airflow obstruction. PLoS Genet. 2014;10:e1004314. doi: 10.1371/journal.pgen.1004314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnett IJ, Lee S, Lin X. Detecting rare variant effects using extreme phenotype sampling in sequencing association studies. Genet Epidemiol. 2013;37:142–151. doi: 10.1002/gepi.21699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radder JE, Zhang Y, Sciurba FC, Shapiro SD. Whole genome rare variant association study identifies a non-genic region and several genes associated with severe emphysema [abstract] Am J Resp Crit Care Med. 2016;193:A7477. [Google Scholar]
- 21.Bon J, Fuhrman CR, Weissfeld JL, Duncan SR, Branch RA, Chang CC, Zhang Y, Leader JK, Gur D, Greenspan SL, et al. Radiographic emphysema predicts low bone mineral density in a tobacco-exposed cohort. Am J Respir Crit Care Med. 2011;183:885–890. doi: 10.1164/rccm.201004-0666OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller NL, Staples CA, Miller RR, Abboud RT. “Density mask”. An objective method to quantitate emphysema using computed tomography. Chest. 1988;94:782–787. doi: 10.1378/chest.94.4.782. [DOI] [PubMed] [Google Scholar]
- 23.Allen EG, Grus WE, Narayan S, Espinel W, Sherman SL. Approaches to identify genetic variants that influence the risk for onset of fragile X-associated primary ovarian insufficiency (FXPOI): a preliminary study. Front Genet. 2014;5:260. doi: 10.3389/fgene.2014.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuen RK, Thiruvahindrapuram B, Merico D, Walker S, Tammimies K, Hoang N, Chrysler C, Nalpathamkalam T, Pellecchia G, Liu Y, et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med. 2015;21:185–191. doi: 10.1038/nm.3792. [DOI] [PubMed] [Google Scholar]
- 25.De Gregori M, Diatchenko L, Belfer I, Allegri M. OPRM1 receptor as new biomarker to help the prediction of post mastectomy pain and recurrence in breast cancer. Minerva Anestesiol. 2014 [PubMed] [Google Scholar]
- 26.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;11:11.10.11–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 32.Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, Christiani DC, Wurfel MM, Lin X NHLBI GO Exome Sequencing Project—ESP Lung Project Team. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91:224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myerburg MM, Harvey PR, Heidrich EM, Pilewski JM, Butterworth MB. Acute regulation of the epithelial sodium channel in airway epithelia by proteases and trafficking. Am J Respir Cell Mol Biol. 2010;43:712–719. doi: 10.1165/rcmb.2009-0348OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao J, Harper R, Barchowsky A, Di YPP. Identification of multiple MAPK-mediated transcription factors regulated by tobacco smoke in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2007;293:L480–L490. doi: 10.1152/ajplung.00345.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez FJ, Foster G, Curtis JL, Criner G, Weinmann G, Fishman A, DeCamp MM, Benditt J, Sciurba F, Make B, et al. NETT Research Group. Predictors of mortality in patients with emphysema and severe airflow obstruction. Am J Respir Crit Care Med. 2006;173:1326–1334. doi: 10.1164/rccm.200510-1677OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belkadi A, Bolze A, Itan Y, Cobat A, Vincent QB, Antipenko A, Shang L, Boisson B, Casanova JL, Abel L. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci USA. 2015;112:5473–5478. doi: 10.1073/pnas.1418631112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emond MJ, Louie T, Emerson J, Zhao W, Mathias RA, Knowles MR, Wright FA, Rieder MJ, Tabor HK, Nickerson DA, et al. National Heart, Lung, and Blood Institute (NHLBI) GO Exome Sequencing Project; Lung GO. Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat Genet. 2012;44:886–889. doi: 10.1038/ng.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang F, Lupski JR. Non-coding genetic variants in human disease. Hum Mol Genet. 2015;24:R102–R110. doi: 10.1093/hmg/ddv259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin W-Y. Association testing of clustered rare causal variants in case-control studies. PLoS One. 2014;9:e94337. doi: 10.1371/journal.pone.0094337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Celedón JC, Lange C, Raby BA, Litonjua AA, Palmer LJ, DeMeo DL, Reilly JJ, Kwiatkowski DJ, Chapman HA, Laird N, et al. The transforming growth factor-β1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD) Hum Mol Genet. 2004;13:1649–1656. doi: 10.1093/hmg/ddh171. [DOI] [PubMed] [Google Scholar]
- 42.Silverman EK, Mosley JD, Palmer LJ, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ, et al. Genome-wide linkage analysis of severe, early-onset chronic obstructive pulmonary disease: airflow obstruction and chronic bronchitis phenotypes. Hum Mol Genet. 2002;11:623–632. doi: 10.1093/hmg/11.6.623. [DOI] [PubMed] [Google Scholar]
- 43.Cho MH, McDonald ML, Zhou X, Mattheisen M, Castaldi PJ, Hersh CP, Demeo DL, Sylvia JS, Ziniti J, Laird NM, et al. NETT Genetics, ICGN, ECLIPSE and COPDGene Investigators. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2:214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hautamaki RD, Kobayashi DK, Senior RM, Shapiro SD. Requirement for macrophage elastase for cigarette smoke-induced emphysema in mice. Science. 1997;277:2002–2004. doi: 10.1126/science.277.5334.2002. [DOI] [PubMed] [Google Scholar]
- 45.Hobbs BD, Parker MM, Chen H, Lao T, Hardin M, Qiao D, Hawrylkiewicz I, Sliwinski P, Yim JJ, Kim WJ, et al. NETT Genetics Investigators; ECLIPSE Investigators; COPDGene Investigators; International COPD Genetics Network Investigators. Exome array analysis identifies a common variant in IL27 associated with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2016;194:48–57. doi: 10.1164/rccm.201510-2053OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackson VE, Ntalla I, Sayers I, Morris R, Whincup P, Casas J-P, Amuzu A, Choi M, Dale C, Kumari M, et al. Exome-wide analysis of rare coding variation identifies novel associations with COPD and airflow limitation in MOCS3, IFIT3 and SERPINA12. Thorax. 2016;71:501–509. doi: 10.1136/thoraxjnl-2015-207876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lusk CM, Wenzlaff AS, Dyson G, Purrington KS, Watza D, Land S, Soubani AO, Gadgeel SM, Schwartz AG. Whole-exome sequencing reveals genetic variability among lung cancer cases subphenotyped for emphysema. Carcinogenesis. 2016;37:139–144. doi: 10.1093/carcin/bgv248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silverman EK, Palmer LJ, Mosley JD, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ, et al. Genomewide linkage analysis of quantitative spirometric phenotypes in severe early-onset chronic obstructive pulmonary disease. Am J Hum Genet. 2002;70:1229–1239. doi: 10.1086/340316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hersh CP, Silverman EK, Gascon J, Bhattacharya S, Klanderman BJ, Litonjua AA, Lefebvre V, Sparrow D, Reilly JJ, Anderson WH, et al. SOX5 is a candidate gene for chronic obstructive pulmonary disease susceptibility and is necessary for lung development. Am J Respir Crit Care Med. 2011;183:1482–1489. doi: 10.1164/rccm.201010-1751OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asbagh LA, Vazquez I, Vecchione L, Budinska E, De Vriendt V, Baietti MF, Steklov M, Jacobs B, Hoe N, Singh S, et al. The tyrosine phosphatase PTPRO sensitizes colon cancer cells to anti-EGFR therapy through activation of SRC-mediated EGFR signaling. Oncotarget. 2014;5:10070–10083. doi: 10.18632/oncotarget.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geraghty P, Hardigan A, Foronjy RF. Cigarette smoke activates the proto-oncogene c-src to promote airway inflammation and lung tissue destruction. Am J Respir Cell Mol Biol. 2014;50:559–570. doi: 10.1165/rcmb.2013-0258OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finigan JH, Downey GP, Kern JA. Human epidermal growth factor receptor signaling in acute lung injury. Am J Respir Cell Mol Biol. 2012;47:395–404. doi: 10.1165/rcmb.2012-0100TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ganesan S, Unger BL, Comstock AT, Angel KA, Mancuso P, Martinez FJ, Sajjan US. Aberrantly activated EGFR contributes to enhanced IL-8 expression in COPD airways epithelial cells via regulation of nuclear FoxO3A. Thorax. 2013;68:131–141. doi: 10.1136/thoraxjnl-2012-201719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Filosto S, Becker CR, Goldkorn T. Cigarette smoke induces aberrant EGF receptor activation that mediates lung cancer development and resistance to tyrosine kinase inhibitors. Mol Cancer Ther. 2012;11:795–804. doi: 10.1158/1535-7163.MCT-11-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hughes T, Coit P, Adler A, Yilmaz V, Aksu K, Düzgün N, Keser G, Cefle A, Yazici A, Ergen A, et al. Identification of multiple independent susceptibility loci in the HLA region in Behçet’s disease. Nat Genet. 2013;45:319–324. doi: 10.1038/ng.2551. [DOI] [PubMed] [Google Scholar]