Abstract
The introduction of Listeria monocytogenes into the food production chain is a concern, with numerous grouped cases of listeriosis associated with milk-derived or pork-derived products have been documented. Management of this zoonotic pathogen considers all strains as an equal risk. Recently, a new perspective for characterisation of strain virulence was introduced with the discovery of the unaltered sequence of InlA as a determinant of strain virulence; this has also been reported as an infrequent finding among so-called environmental strains, that is, strains isolated from food or from surfaces in food industries. The aim of this study was to differentiate L monocytogenes strains isolated from animal cases versus those from human cases and to differentiate clinical strains from environmental ones using a Caenorhabditis elegans virulence testing model. In Quebec in 2013/2014, the surveillance of L monocytogenes clinical isolates registered a total of 20 strains of animal origin and 16 pulsed-field gel electrophoresis types isolated from human cases. The mixed PCR multiplex agglutination protocol used for geno-serotyping clearly discriminated genogroup IVB strains from bovine and human origins. The presence of a premature stop codon single nucleotide polymorphism in the inlA gene sequence in clinical strains and the identical behaviour of particular strains in the C elegans model are discussed in this paper from the perspective of industrial management of L monocytogenes risk.
Keywords: Listeria monocytogenes, inlA, C elegans, virulence
Introduction
Listeria monocytogenes is a great concern both for industrial and public health stakeholders. These bacteria cause listeriosis, a foodborne disease acquired through the consumption of contaminated ready-to-eat products, including milk or meat products. Pork meat has been associated with large-scale outbreaks in the past (de valk and others 2001) and is considered a risk factor, particularly for sensitive populations such as pregnant women, children and the elderly. In the last few years, the increase in human cases (not associated with intensified surveillance), mainly for the elderly, has increased the concern surrounding management of L monocytogenes in food in Canada and Europe (Lomonaco and others 2015). One critical question to answer is whether the virulence of L monocytogenes has increased. The determinants of virulence have been studied, and there is a large amount of data that allow for discrimination of strains from lineages III or IV from lineages I and II (Ragon and others 2008). Lineages I and II are considered to be the most worrisome because of their potential to cause outbreaks or sporadic listeriosis. Among such strains, discrimination based on pathogenic properties still represents a challenge. A few years ago, the very promising concept of virulence determination through inlA gene sequencing was introduced (Van Stelten and others 2010, Van Stelten and others 2011). Published studies revealed the presence of a premature stop codon for internalin A translation. Because this stop codon inactivated this determinant of the first step of pathogenicity in the digestive tract, a key determinant for virulence differentiation of strains appeared to have been discovered. In Quebec, human listeriosis surveillance works hand-in-hand with food surveillance. Few strains isolated from clinical cases in farms (animal primary productions) are available, so we took the opportunity that arose to test virulent strains isolated from animals or humans with clinical signs of listeriosis. After we performed the serovar determination and comparison according to strain origin, we wanted to confirm if the inlA sequences were complete in these strains and to test their behaviour in a Caenorhabditis elegans infection model, a very attractive but controversial model of pathogenicity testing for L monocytogenes (Thomsen and others 2006, Karthikeyan and others 2015) (see discussion). No relationship between InlA integrity and behaviour of L monocytogenes strain in C elegans has actually been described. Some strains isolated during a previous study involving pig primary production (Larivière-Gauthier and others 2014) were added to the study’s strain collection because only some of them showed stop codon in their inlA sequences and because these strains were (based on pulsotype comparison) frequently or not associated with clinical human cases. These additional strains were expected to show opposite virulence potentials. The aim of this study was to analyse inlA sequences from strains isolated from clinical forms of human or animal listerioses in Quebec and to compare the behaviour of some selected strains in a C elegans virulence model.
Materials and methods
Strain collections
Strains were collected from L monocytogenes surveillance in 2013/2014: 20 strains were obtained from animal disease surveillance (laboratory of the Ministry of Agriculture, Fisheries and Food, Quebec (MAPAQ)), among which 13 were included in this study because they satisfied the required ‘clinical’ criteria (ie, isolated from organs after animal necropsy, excluding isolation from milk and silage) and 16 strains isolated from clinical cases in humans were provided by the Provincial Public Health Laboratory (public health surveillance). The collection was completed with four strains of pig origin isolated in a previous project (Larivière-Gauthier and others 2014). These strains showed a single nucleotide polymorphism (SNP) premature stop codon in the inlA sequence (Pulso1 and Pulso9 strains) or an integrated inlA sequence (Pulso4 and Pulso6 strains). Strains Pulso6 and Pulso9 showed 100% similarity between pulsed-field gel electrophoresis (PFGE) banding patterns with strains recovered from clinical human cases; strains Pulso1 and Pulso4 have not previously been identified in clinical human surveillance. These added strains were expected to show opposite virulence potentials.
Serotyping
L monocytogenes strains were serotyped based on a combination of multiplex PCR-based geno-serogrouping, completed by detection of flaA (Kérouanton and others 2010) and agglutination against discriminatory O serum (OI, OVII, OVIII; Oxoid Thermo Fisher Scientific, Nepean, ON, Canada) (Burall and others 2011).
InlA sequencing
The inlA gene sequences were determined after overlapping amplification (Ragon and others 2008) using the Sanger method at the Centre d’Innovation Génome Québec (Applied Biosystems 3730xl DNA analyzer). InlA gene sequencing was extensively analysed, and the occurrence and position of SNP before in silico translation of the sequences were compared. Sequences were aligned and screened for mutations causing a premature stop codon or amino acid deletion using ClustalX 2.1 software, with inlA L monocytogenes EGD-e (NCBI: NC_003210.1) as reference.
C elegans virulence model
Bacterial strains (Fig 1) were cultured 24 hours at 37°C in BHI (Brain Heart Infusion), acid shocked (0.1 N HCl pH 4.5, one hour) then washed and suspended in phosphate-buffered saline (PBS) (concentration, see Fig 1) before being used in trials. Strain N2 worms, maintained for five days on nematode-growing medium plates and inoculated with Escherichia coli OP50 feeding strain at 25°C, were harvested (non-synchronised culture), washed three times in PBS, then inoculated in 96-well plates in PBS and maintained for five more days at 30°C with daily addition of fresh bacteria.
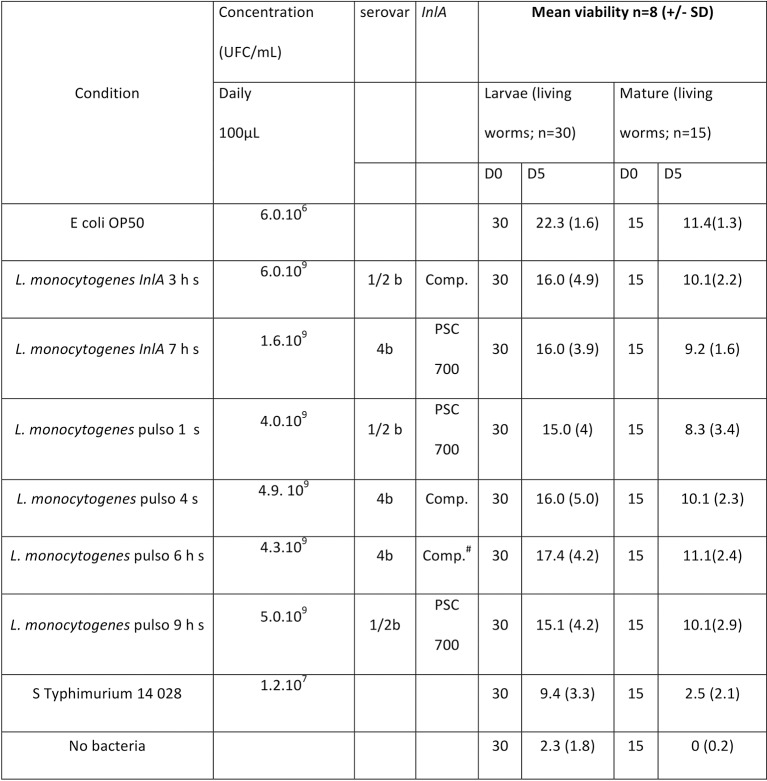
FIG 1:
Listeria monocytogenes virulence on Caenorhabditis elegans model isolated in human (h) or pig (s). h s: strains from animal origin related to human cases (identical pulsed-field gel electrophoresis profile). Comp: complete InlA; PSC 700: truncated InlA due to the presence of a 700 aa position premature stop codon. Comp.#: complete but with substitution of first amino acids. aa, amino acid.
Observation of worm viability (based on worm locomotion and/or pharyngeal pumping and assessed under an inverted phase contrast microscope according to Thomsen and others (2006) allowed determination of a living worm count of 15 mature worms and 30 L1/L2 larvae in each of the tested conditions. The distribution of number of living worms counts (n=8) were compared with control conditions (Mann-Whitney U test; SPSS Statistics V.17, Licenced U de Montreal, α=0.05)
Results
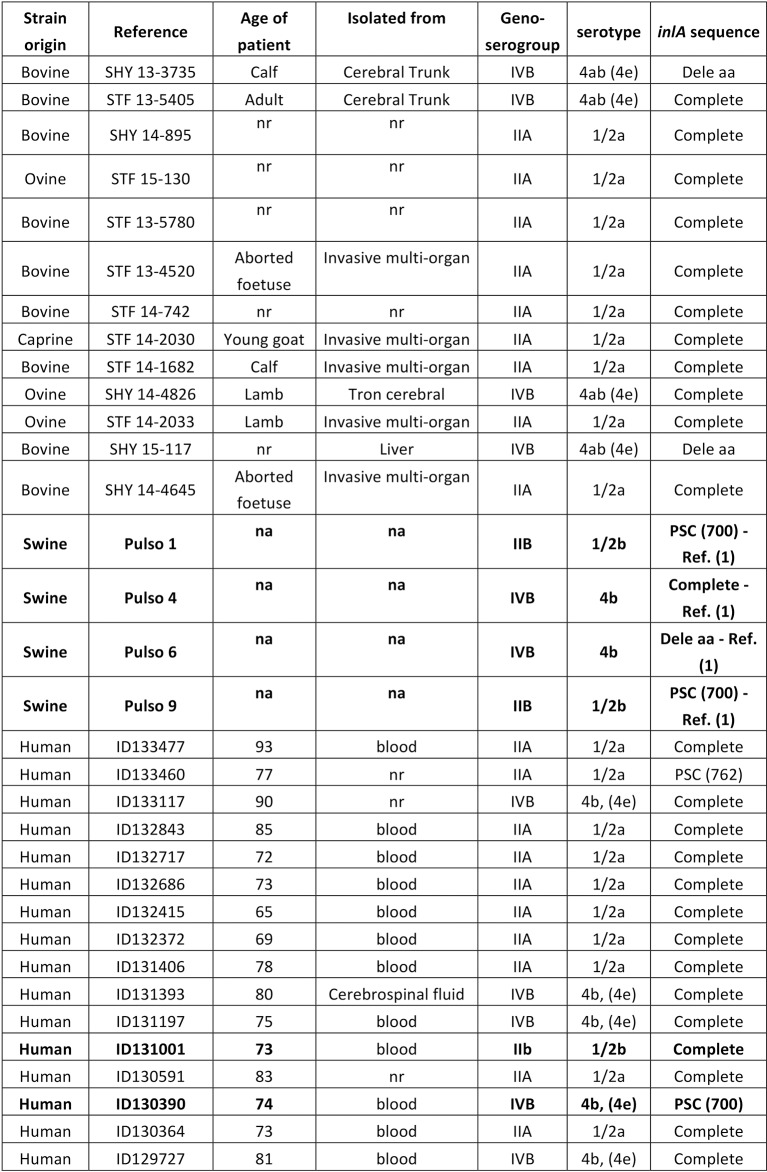
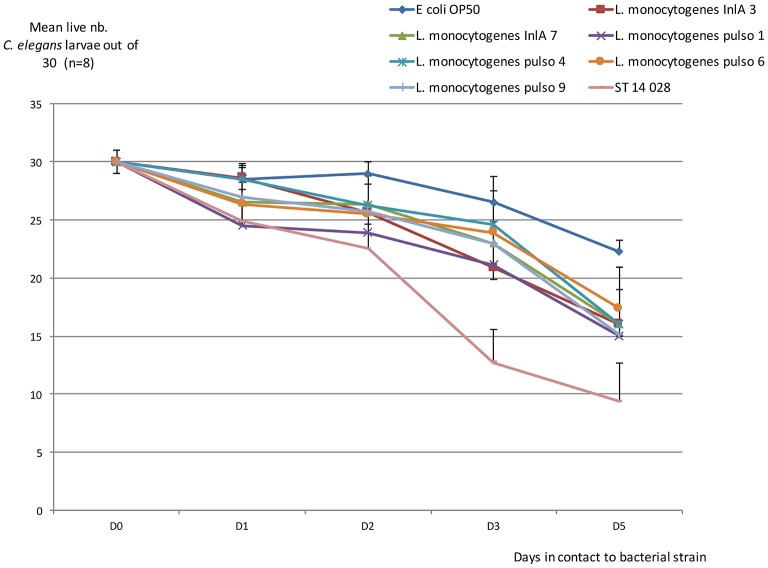
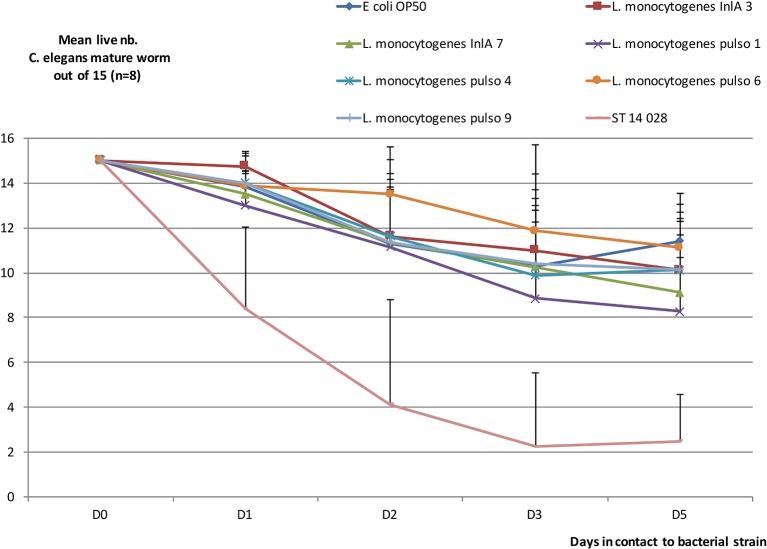
All 16 of the strains of human origin were considered clinical because they were isolated from hospitalised patients. From the 20 strains provided by MAPAQ, only 13 strains strictly associated with clinical listeriosis in animals were included. It should be noted that we excluded strains isolated from milk tanks and silage, all belonging to the 1/2a serovar. Although serotyping revealed that the greater proportion of clinical strains (regardless of origin) belonged to the serovar 1/2a (18/29, 62%), there were still some strains (nine out of 29 with similar proportions from humans and animals, that is, 5/16 and 4/13, respectively) that belonged to the IVB serogroup (a serogroup that contains 4b and 4a/ab serovars). Focusing on this IVB group, a clear distinction appeared where only 4b serovar strains were found in human surveillance, whereas strains shown in ruminant clinical cases all belonged to 4a/4ab serovars (table presented in Fig 2). No SNP occurred on the sequence coding for the InlA LPXTG domain. Moreover, inlA sequencing showed that a majority of strains had a complete sequence (table presented in Fig 2). But it should be noted that two out of the 16 strains isolated from clinical listeriosis cases had a premature stop codon; the mutations were expected to induce truncations of InlA at 700 and 762 amino acid (aa) positions, respectively. One of these—which showed the longest InlA size but lost LPXTG domain—was used in the C elegans virulence model, as well as another strain presenting a complete sequence (tables presented in Figs 1,2). Four more strains (different PFGE type) from porcine origin were added; some had been recurrently detected in production (and food) but had never been associated with human cases (table presented in Fig 1; Pulso1 and Pulso4 strains). The two others were kept because of their 100% homology with strains involved in clinical human cases (sporadically for strain Pulso9 or recurrently for strain Pulso6; table presented in Fig 1). After five days, unfed C elegans worms were almost all dead. Using E coli OP 50 as a control, viability was maintained at 74% after five days, both in larvae and in mature worm forms. By comparison, L monocytogenes lowered the viability of the worms and significantly so for the larvae forms (Mann-Whitney U test, P<0.05). No differences in virulence behaviour were revealed using this model to compare the L monocytogenes strains (Fig 3 and Fig 4). The strain that was expected to be the least virulent (Pulso1) showed the largest decrease in value for worm viability for both forms (50% and 55%, respectively, for larvae and mature worms) (table presented in Fig 1).
FIG 2.
: Characterisation of Listeria monocytogenes strains of clinical origin. PSC, premature stop codon (aa position). Dele aa: deletion of first amino acids in the protein. na: not applicable; nr: not recorded. Bold: retained for C elegans testing.
FIG 3:
Evolution of mean live number Caenorhabditis elegans larvae out of 30 (n=8). Mean (n=8) number of live C elegans larvae out of 30 after one to five days in contact with bacterial strain (see Fig 1). Bars: SD.
FIG 4:
Evolution of mean number of live mature Caenorhabditis elegans out of 15 (n=8). Mean (n=8) number of live C elegans mature out of 15 after one to five days in contact with bacterial strain (see Fig 1). Bars: SD.
Discussion
L monocytogenes virulence determination is a current research concern, and many authors have contributed to the advancement of this endeavour. A quite recent proposition (relevant when considering the capital role of InlA in pathogenicity) is that the truncation of InlA, induced by the presence of a premature stop codon in inlA, is associated with lower virulence properties (Nightingale and others 2008). Moreover, a great proportion of truncated forms were shown in strains isolated from food or food-related environments but not in L monocytogenes strains involved in clinical human cases (Van Stelten and others 2010). Discussion of the relevance of this virulence determinant arose when it was demonstrated that some truncated forms can express virulence in animal models (Van Stelten and others 2011, Holch and others 2013). Our analyses confirm that truncated forms can be found in clinical human cases in Quebec. The truncation positions, induced by SNP stop codon (700 and 762 aa) were previously reported (Kovacevic and others 2013). For the 700 aa truncation position, the entire anchorage region (between 700 and 800 aa), including the determinant proline in the LPXTG motif, was not translated. So, at least for the strain harbouring the 700 aa truncated InlA, pathogenicity was expected to be extremely low. The clinical origin of such strains led us to question the dose of ingested inoculum. In studies that demonstrated, in experimental conditions, a maintenance of virulence for strains harbouring truncated InlA, high doses of inoculum were used (Van Stelten and others 2011, Holch and others 2013). High-dose ingestion of L monocytogenes could be observed in outbreaks, giving an opportunity for a strain harbouring InlA truncated form to cause disease. We suggest that inlA analysis alone is not sufficient to characterise the virulence potential of L monocytogenes strains.
Looking for a complementary test that avoids labour intensive and ethically questioned experimentations on mammal models, we considered the C elegans model for comparing the virulence of our characterised strains. The C elegans model was first proposed in 2006 and confirmed in 2007 (Thomsen and others 2006, Forrester and others 2007) when studies showed that the worm is as sensitive to pathogenic L monocytogenes as humans. But Guha and others questioned the model in 2013 (Guha and others 2013) after failing to obtain loss of viability of worms inoculated by L monocytogenes. That same year, Neuhaus and others (2013) confirmed that primed strains (acid shocked) increase the lethality to the worms and ultimately confirmed the worm model as able to compare virulence for food-derived strains (Karthikeyan and others 2015). Our results, when incubating worms at 30°C with acid-shocked L monocytogenes (to promote prfA-driven virulence factors in the pathogen), confirmed a decrease of viability of worms in contact with L monocytogenes (in a lesser proportion than Salmonella Typhimurium). Contrary to the work of Karthikeyan and others (2015), the worms did not show any morphological changes when they were submitted to our L monocytogenes strain inoculations. The loss of viability was solely based on immobility and loss of pharyngeal pumping activity, in accordance with Neuhaus and others (2013). In our conditions, neither on both the mature form nor on the young larvae, L monocytogenes showed differences in virulence regardless of origin (environmental or clinical) and in their inlA integrity.
Conclusion
Using complete serotyping, differences in IVB geno-serogroup between clinical strains from bovine and human origins were shown, with serovar 4b found only in human clinical case strains. The virulence analysis tools used in this study (inlA sequencing and C elegans model) on both confirmed virulent and presumed less virulent strains could not univocally established the public health risk associated with L monocytogenes strains. No pig clinical case strains were provided by the MAPAQ laboratory, but serovar 4b strains were previously found in the collection of strains from healthy pigs in primary production (Larivière-Gauthier and others 2014). As calls for farm to fork strain traceability continue to be made, accurate surveillance is needed to enlarge the collection of strains in animal surveillance, particularly from pig production. It should be mentioned that we did receive all the collected L monocytogenes strains isolated in the MAPAQ laboratory, and we observed that they mainly originated from bovine clinical surveillance (where L monocytogenes present economical relevance).
Acknowledgments
C elegans were kindly provided by Caenorhabditis Genetics Center, University of Minnesota, Minneapolis, Minnesota, USA.
Footnotes
Contributors: All authors equally contributed to the conception of the study; data acquisition was equally distributed among Philippe Fravalo, Tamazight Cherifi and Kersti Neira laboratory work. The paper was written by Philippe Fravalo after analyses and discussions of the results done by all authors.
Funding: This work was financially supported by CDEVQ.
Competing interests: None declared.
References
- Burall L. S., Simpson A. C., Datta A. R (2011) Evaluation of a serotyping scheme using a combination of an antibody-based serogrouping method and a multiplex PCR assay for identifying the major serotypes of Listeria monocytogenes. Journal of Food Protection 74, 403–409 doi:10.4315/0362-028X.JFP-10-355 [DOI] [PubMed] [Google Scholar]
- de Valk H., Vaillant V., Jacquet C., Rocourt J., Le Querrec F., Stainer F., Quelquejeu N., Pierre O., Pierre V., Desenclos J. C., Goulet V (2001) Two consecutive nationwide outbreaks of listeriosis in France, October 1999-February 2000. American Journal of Epidemiology 154, 944–950 doi:10.1093/aje/154.10.944 [DOI] [PubMed] [Google Scholar]
- Forrester S., Milillo S. R., Hoose W. A., Wiedmann M., Schwab U (2007) Evaluation of the pathogenicity of Listeria spp. in Caenorhabditis elegans. Foodborne Pathogens and Disease 4, 67–73 doi:10.1089/fpd.2006.64 [DOI] [PubMed] [Google Scholar]
- Guha S., Klees M., Wang X., Li J., Dong Y., Cao M (2013) Influence of planktonic and sessile Listeria monocytogenes on Caenorhabditis elegans. Archives of Microbiology 195, 19–26 doi:10.1007/s00203-012-0841-y [DOI] [PubMed] [Google Scholar]
- Holch A., Ingmer H., Licht T. R., Gram L (2013) Listeria monocytogenes strains encoding premature stop codons in inlA invade mice and guinea pig fetuses in orally dosed dams. Journal of Medical Microbiology 62(Pt 12), 1799–1806 doi:10.1099/jmm.0.057505-0 [DOI] [PubMed] [Google Scholar]
- Karthikeyan R., Gunasekaran P., Rajendhran J., Serotyping M (2015) Molecular serotyping and pathogenic potential of Listeria monocytogenes isolated from milk and milk products in Tamil Nadu, India. Foodborne Pathogens and Disease 12, 522–528 doi:10.1089/fpd.2014.1872 [DOI] [PubMed] [Google Scholar]
- Kérouanton A., Marault M., Petit L., Grout J., Dao T. T., Brisabois A (2010) Evaluation of a multiplex PCR assay as an alternative method for Listeria monocytogenes serotyping. Journal of Microbiological Methods 80, 134–137 doi:10.1016/j.mimet.2009.11.008 [DOI] [PubMed] [Google Scholar]
- Kovacevic J., Arguedas-Villa C., Wozniak A., Tasara T., Allen K. J (2013) Examination of food chain-derived Listeria monocytogenes strains of different serotypes reveals considerable diversity in inlA genotypes, mutability, and adaptation to cold temperatures. Applied and Environmental Microbiology 79, 1915–1922 doi:10.1128/AEM.03341-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larivière-Gauthier G., Letellier A., Kérouanton A., Bekal S., Quessy S., Fournaise S., Fravalo P (2014) Analysis of Listeria monocytogenes strain distribution in a pork slaughter and cutting plant in the province of Quebec. Journal of Food Protection 77, 2121–2128 doi:10.4315/0362-028X.JFP-14-192 [DOI] [PubMed] [Google Scholar]
- Lomonaco S., Nucera D., Filipello V (2015) The evolution and epidemiology of Listeria monocytogenes in Europe and the United States. Infection, Genetics and Evolution 35, 172–183 doi:10.1016/j.meegid.2015.08.008 [DOI] [PubMed] [Google Scholar]
- Neuhaus K., Satorhelyi P., Schauer K., Scherer S., Fuchs T. M (2013) Acid shock of Listeria monocytogenes at low environmental temperatures induces prfA, epithelial cell invasion, and lethality towards Caenorhabditis elegans. BMC Genomics 14, 285 doi:10.1186/1471-2164-14-285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale K. K., Ivy R. A., Ho A. J., Fortes E. D., Njaa B. L., Peters R. M., Wiedmann M (2008) inlA premature stop codons are common among Listeria monocytogenes isolates from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Applied and Environmental Microbiology 74, 6570–6583 doi:10.1128/AEM.00997-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragon M., Wirth T., Hollandt F., Lavenir R., Lecuit M., Le Monnier A., Brisse S (2008) A new perspective on Listeria monocytogenes evolution. PLoS Pathogens 4, e1000146 doi:10.1371/journal.ppat.1000146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen L. E., Slutz S. S., Tan M. W., Ingmer H (2006) Caenorhabditis elegans is a model host for Listeria monocytogenes. Applied and Environmental Microbiology 72, 1700–1701 doi:10.1128/AEM.72.2.1700-1701.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Stelten A., Simpson J. M., Chen Y., Scott V. N., Whiting R. C., Ross W. H., Nightingale K. K (2011) Significant shift in median guinea pig infectious dose shown by an outbreak-associated Listeria monocytogenes epidemic clone strain and a strain carrying a premature stop codon mutation in inlA. Applied and Environmental Microbiology 77, 2479–2487 doi:10.1128/AEM.02626-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Stelten A., Simpson J. M., Ward T. J., Nightingale K. K (2010) Revelation by single-nucleotide polymorphism genotyping that mutations leading to a premature stop codon in inlA are common among Listeria monocytogenes isolates from ready-to-eat foods but not human listeriosis cases. Applied and Environmental Microbiology 76, 2783–2790 doi:10.1128/AEM.02651-09 [DOI] [PMC free article] [PubMed] [Google Scholar]