Abstract
Swimming motility is considered a beneficial trait among bacterial species as it enables movement across fluid environments and augments invasion of tissues within the host. However, non-swimming bacteria also flourish in fluid habitats, but how they effectively spread and colonize distant ecological niches remains unclear. We show that non-motile staphylococci can gain motility by hitchhiking on swimming bacteria, leading to extended and directed motion with increased velocity. This phoretic interaction was observed between Staphylococcus aureus and Pseudomonas aeruginosa, Staphylococcus epidermidis and P. aeruginosa, as well as S. aureus and Escherichia coli, suggesting hitchhiking as a general translocation mechanism for non-motile staphylococcal species. By leveraging the motility of swimming bacteria, it was observed that staphylococci can colonize new niches that are less available in the absence of swimming carriers. This work highlights the importance of considering interactions between species within polymicrobial communities, in which bacteria can utilize each other as resources.
Bacteria often exist as polymicrobial communities in a multitude of environments (Fernandez et al., 2000; Hosni et al., 2011; Burmølle et al., 2014). Here we investigate the ways in which bacteria in the same community may affect each other’s motility in liquid environments. Swimming motility offers a considerable advantage for bacteria by enabling movement toward environments of favorable conditions (Stocker et al., 2008; Dennis et al., 2013), and movement away from toxins or predators (Adler, 1966; Berg, 1975). Non-flagellated bacteria do not have the capacity to independently translocate with this mechanism. The genus Staphylococcus, for example, is classically considered non-motile in fluid environments due to the lack of flagella (Kloos and Bannerman, 1994; Freney et al., 1999). Despite their limitations in motility, staphylococcal species effectively reach and thrive in their preferred ecological niches.
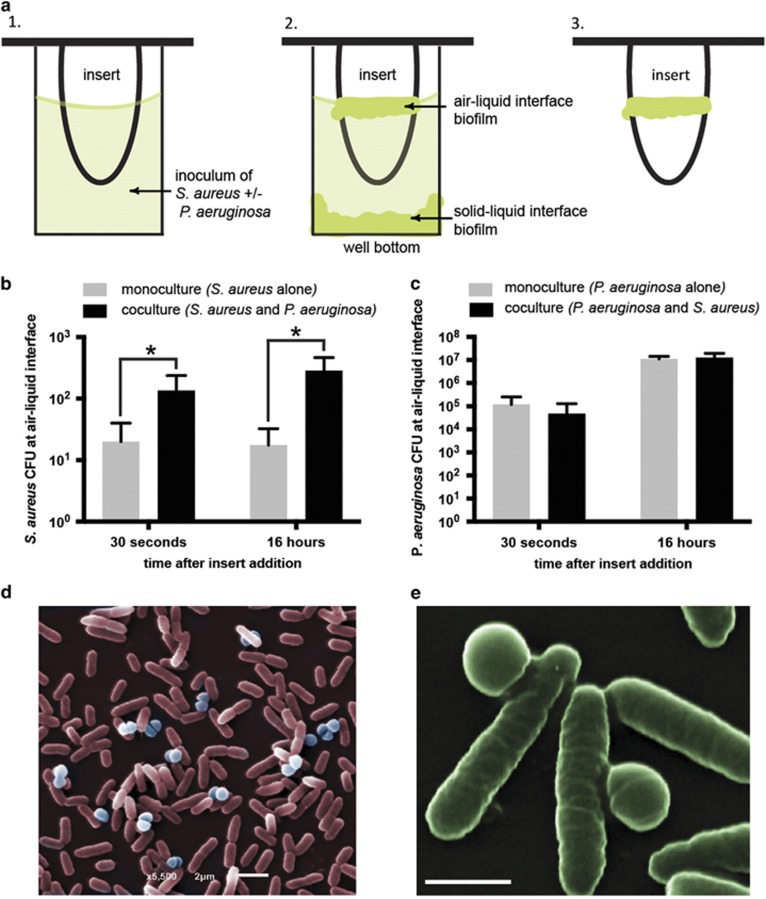
We tested if non-motile species may benefit from the swimming motility of flagellated bacteria. To address this, we studied two human pathogens that are found in the same ecological habitats, but rely on different mechanisms for translocation. Staphylococcus aureus is a Gram-positive, non-motile cocci species and Pseudomonas aeruginosa is a Gram-negative, flagellated rod species capable of swimming motility. Using a standardized biofilm assay (Ceri et al., 1999), a vertical insert was placed into a microwell containing bacterial inoculum. The insert does not touch the bottom or sides of the microwell, thereby creating two distinct niches for potential colonization: the first at the bottom of the well, and the second at the top of the inoculum (air–liquid interface) on the lateral surface of the insert (Figure 1a). We expected non-motile S. aureus to settle and form a biofilm at the bottom of the microwell and motile P. aeruginosa to build a biofilm at the air-liquid interface, which requires upward swimming. Figures 1b and c show that indeed, P. aeruginosa readily colonized the air-liquid interface while S. aureus was largely absent from this location in monoculture. However, when the two species were co-cultured in the same microwell, significantly more S. aureus cells were isolated from the air-liquid interface. On average, there were 6-fold more S. aureus cells isolated 30 seconds after initiation of biofilm formation, and 16-fold more after 16 hours (Figure 1b). Together, these results indicate that colonization of this niche was strongly enhanced by the presence of P. aeruginosa. A similar trend was observed with a 100 times higher inoculation density of S. aureus cells, with 30-fold more S. aureus cells at the air-liquid interface after 30 seconds, and 3-fold more cells at the same location 16 hours after inoculation (Supplementary Figure 1A). For comparison, P. aeruginosa cell numbers at the air-liquid interface were largely unaffected by the presence of S. aureus (Figure 1c; Supplementary Figure 1B). Scanning electron micrographs of cells from vertical inserts confirmed co-localization of P. aeruginosa and S. aureus in the biofilm at the air-liquid interface after 2 h (Figures 1d and e). Taken together, these data suggest S. aureus has acquired, through P. aeruginosa, an increased capacity to travel longer distances, allowing it to colonize niches that are relatively inaccessible in the absence of swimming carrier bacteria.
Figure 1.
P. aeruginosa augments S. aureus localization to air-liquid interface. (a) Inoculum containing S. aureus with or without P. aeruginosa is added to a microwell and allowed to settle for one hour before a polystyrene insert is gently placed into the inoculum (a, panel 1). This set up provides two niches where the bacteria can colonize over time, the bottom of the well and the lateral surface of the insert at the air–liquid interface (a, panel 2). After 30 seconds or 16 hours, the insert is removed to isolate cells that colonize the air–liquid interface (a, panel 3) (b) S. aureus CFUs at the air-liquid interface at 30 seconds and 16 hours after addition of the insert. An initial inoculation density of ~106 cells per ml for S. aureus and ~108 cells per ml for P. aeruginosa (100:1 P. aeruginosa to S. aureus ratio) was used. The presence of P. aeruginosa increases colonization of the air-liquid interface by S. aureus at both time points. Mean±s.d. (n=4). *P<0.05 (30 seconds P=0.0098; 16 hours P=0.0016). (c) P. aeruginosa CFUs are not affected by the presence of S. aureus. (d, e) False colored scanning electron micrographs of P. aeruginosa (rod cells) and S. aureus (spherical cells) at the air-liquid interface after 2 h revealed onset of co-localization during the initial attachment stage of biofilm development. Scale bars in d and e represent 2 and 1 μm, respectively.
We hypothesized that the increased colonization of the air-liquid interface by S. aureus may be due to hitchhiking of S. aureus on P. aeruginosa. To evaluate this possibility, fluorescently labeled P. aeruginosa and S. aureus were combined at equal numbers and placed in a microchamber (Supplementary Figure 2) for observation by live confocal microscopy. When imaged concurrently, S. aureus cells were observed associated to P. aeruginosa cells (Figure 2a) and moving together for a period of time (Supplementary Video 1).
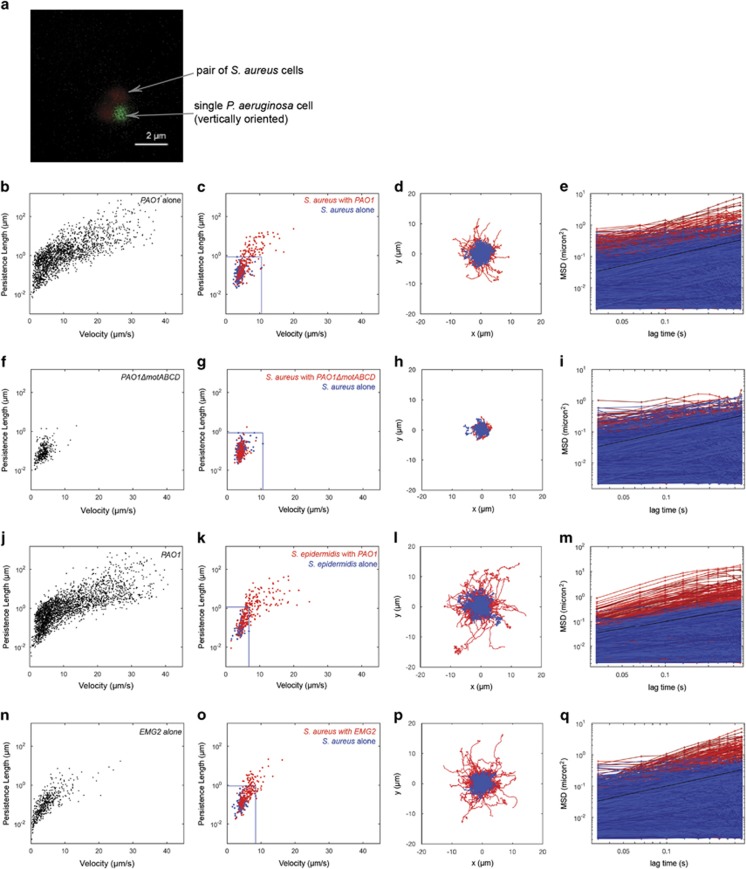
Figure 2.
Swimming bacteria alter the motility of non-motile Staphylococci. (a) Confocal image of S. aureus attached to P. aeruginosa taken from a time-lapse confocal microscopy series. Scale bar represents 2 μm. (b) Individual trajectories of P. aeruginosa alone in culture plotted on axes of persistence length vs velocity. (c) The trajectories of S. aureus alone (blue), and of S. aureus in the presence of P. aeruginosa (co-culture) (red) were also plotted on axes of persistence length vs velocity. The blue box indicates the upper bound of persistence length and velocity for S. aureus alone. There is a cluster of trajectories in the co-culture condition with persistence lengths several orders of magnitude higher than those in the central cluster for the S. aureus alone. (d) The actual traces represented in (c) are re-centered to begin at coordinate (0,0), using the same color code as c. There is a set of trajectories in the co-culture condition that are further reaching and more linear than those of S. aureus alone. (e) The MSD of all individual trajectories, using the same color code as c. There is an increase in motility of the S. aureus in co-culture, as seen by the higher MSDs. (f) Individual trajectories of PAO1ΔmotABCD, a non-motile P. aeruginosa mutant, alone in culture, exhibiting smaller persistence lengths and velocities than P. aeruginosa. (g) The trajectories of S. aureus alone (blue), and of S. aureus in the presence of PAO1ΔmotABCD (red). The motility pattern of the S. aureus is preserved in the co-culture with PAO1ΔmotABCD, confirming that the altered S. aureus motility is dependent on the swimming motility of P. aeruginosa. The blue box indicates the upper bound of persistence length and velocity for the S. aureus alone (h) The actual traces represented in g are re-centered to begin at coordinate (0,0), using the same color code as g. S. aureus trajectories in the presence of P. aeruginosa do not look different then trajectories of S. aureus alone in terms of extent and linearity (i) MSDs of S. aureus in co-culture with PAO1ΔmotABCD compared to S. aureus alone are not appreciably different. (j–m) Repeating the analysis in a–d for another non-motile staphylococci cargo, S epidermidis, alone (blue) and in the presence of P. aeruginosa (red). S. epidermidis trajectories were more linear and further reaching in the presence of P. aeruginosa, suggesting that the hitchhiking observed is generalizable to members of genus Staphylococcus. (n–q) Repeating the analysis in a–d with S. aureus as the cargo, but with a different motile carrier, E. coli EMG2. S. aureus alone is shown in blue, while S. aureus in the presence of E. coli is shown in red.
To further quantify the motility of S. aureus in the absence and presence of P. aeruginosa, single cell tracking was employed on videos obtained of the cells in single and dual species cultures. For tracking experiments, we used P. aeruginosa PAO1-eGFP and S. aureus stained with hexidium iodide. From the resultant cell trajectories, we calculated persistence length (defined as the length scale of decay for angular autocorrelation of a trajectory, which provides a measurement of how linear a trajectory is), velocity and mean squared displacement (MSD). S. aureus showed random motion patterns (Supplementary Video 2) with MSDs similar to a sphere undergoing Brownian motion in liquid medium (Supplementary Figure 3). For comparison, P. aeruginosa exhibited a run-reverse motility pattern consistent with previous accounts of swimming motility among this species (Supplementary Video 3). P. aeruginosa also exhibited greater velocities and persistence lengths than non-motile S. aureus (Figure 2b; Supplementary Figure 4). S. aureus trajectories changed distinctly when P. aeruginosa was present, with persistence lengths increased by an order of magnitude compared to values obtained for S. aureus in monoculture (Figures 2c and d; Supplementary Video 4). Cell trajectories set to start at coordinate (0,0), plotted for S. aureus alone (Figure 2d; blue) and S. aureus mixed with P. aeruginosa (Figure 2d; red) illustrate the extended and directed motion of S. aureus when P. aeruginosa was present. Comparing the MSD of S. aureus in the absence (Figure 2e; blue) and presence (Figure 2e; red) of P. aeruginosa, a shift was observed toward increased directed motility for S. aureus when P. aeruginosa was present. These S. aureus trajectories are superdiffusive (Supplementary Figure 5), which has previously been suggested for bacteria undergoing flagellar motility (Matthäus et al., 2009). For comparison, the trajectories of S. aureus in the presence of non-motile P. aeruginosa mutants, PAO1ΔmotABCD and PAO1ΔflgE, did not measurably deviate from those of S. aureus alone (Figures 2f–i; Supplementary Figure 6; Supplementary Videos 5 and 6), further suggesting that the extended motility of S. aureus is dependent on the swimming motility of P. aeruginosa.
It was also observed that P. aeruginosa can carry another staphylococcal cargo, Staphylococcus epidermidis. S. epidermidis trajectories in the presence of P. aeruginosa were characterized by longer persistence lengths and increased velocity compared with S. epidermidis alone (Figures 2j–m; Supplementary Figures 5B and E; Supplementary Video 7). Moreover, P. aeruginosa was capable of transporting carboxylated polystyrene beads (Supplementary Videos 8–11). Together, these results suggest that P. aeruginosa can engage with a variety of cargos with distinct biochemistries, presumably through different types of chemical interactions. Another motile carrier species, E. coli EMG2 also has the ability to carry staphylococci as cargo (Figures 2n–q; Supplementary Figures 5C and F; Supplementary Video 12). Collectively, these data may support a generalized mechanism for translocation among staphylococcal species via interaction with flagellated bacteria.
The findings presented here quantitatively indicate that staphylococcal species, classically defined as non-motile, have altered motility patterns in the presence of flagellated P. aeruginosa and E. coli. Specifically, P. aeruginosa and E. coli may function as microbial carriers for staphylococcal species and result in enhanced dispersal range in fluid environments. The carrier-dependent movement described here appears mechanistically distinct from previously described spreading of S. aureus on surfaces, which occurs independent of a second, swimming bacterium (Kaito and Sekimizu, 2007; Pollitt et al., 2015).
While there exists a wealth of information regarding flagella-mediated bacterial self-propulsion (Lauga and Powers, 2009), in most natural environments, bacteria are part of polymicrobial communities (Sibley et al., 2008; Consortium, 2012) and the contributions of interspecies phoretic interactions to bacterial dispersal in aqueous environments are not well understood. The ability of non-motile bacterial species to leverage motility from other bacteria has been observed to occur between microbes on the surface of plants, and in soil (Hagai et al., 2014; Finkelshtein et al., 2015). The primary motivation of this work is to highlight the impact of microbial hitchhiking on translocation and distribution of non-motile bacteria in liquid. We found that this phoretic mobility could directly change the localization patterns of staphylococci and open new niches for colonization, as observed in biofilm formation at the air-liquid interface. Looking forward, such behavior could influence community diversity, microbial dispersal, and perhaps enhance transmission of non-motile pathogenic strains. From a clinical perspective, our observations may have important implications on how non-motile pathogens disseminate.
Acknowledgments
We thank Gerardo Cárcamo-Oyarce, Erica Shapiro Frenkel and Jacob Witten for helpful feedback on the manuscript. TS was supported by the National Science Foundation Graduate Research Fellowship under Grant 1122374. NB was supported by NIH-NIEHS Training Grant in Toxicology T32 ES7020-37. AB was supported by Hugh Hampton Young Memorial Fellowship and NIH-NIAID F30 Fellowship 1F30AI110053. TC was supported by The Marie Curie International Outgoing Fellowship (BIOMUC). PSD acknowledged support from Singapore-MIT Alliance for Research and Technology (SMART). This work is funded in part by the MRSEC Program of the National Science Foundation under award number DMR-0819762, the CAREER program of the National Science Foundation under award number PHY-1454673, NIH-NIBIB Grant R01EB017755 and P30ES002109 NIH-NIEHS Core Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Adler J. (1966). Chemotaxis in bacteria. Science 153: 708–716. [DOI] [PubMed] [Google Scholar]
- Berg HC. (1975). Chemotaxis in bacteria. Annu Rev Biophys Bioeng 4: 119–136. [DOI] [PubMed] [Google Scholar]
- Burmølle M, Ren D, Bjarnsholt T, Sørensen SJ. (2014). Interactions in multispecies biofilms: do they actually matter? Trends Microbiol 22: 84–91. [DOI] [PubMed] [Google Scholar]
- Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. (1999). The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 37: 1771–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium THMP. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PG, Seymour J, Kumbun K, Tyson GW. (2013). Diverse populations of lake water bacteria exhibit chemotaxis towards inorganic nutrients. ISME J 7: 1661–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AS, Hashsham SA, Dollhopf SL, Raskin L, Glagoleva O, Dazzo FB et al. (2000). Flexible community structure correlates with stable community function in methanogenic bioreactor communities perturbed by glucose. Appl Environ Microbiol 66: 4058–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelshtein A, Roth D, Jacob EB, Ingham CJ. (2015). Bacterial swarms recruit cargo bacteria to pave the way in toxic environments. mBio 6: e00074–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freney J, Kloos WE, Hajek V, Webster JA, Bes M, Brun Y et al. (1999). Recommended minimal standards for description of new staphylococcal species. Subcommittee on the taxonomy of staphylococci and streptococci of the International Committee on Systematic Bacteriology. Int J Syst Bacteriol 49: 489–502. [DOI] [PubMed] [Google Scholar]
- Hagai E, Dvora R, Havkin-Blank T, Zelinger E, Porat Z, Schulz S et al. (2014). Surface-motility induction, attraction and hitchhiking between bacterial species promote dispersal on solid surfaces. ISME J 8: 1147–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosni T, Moretti C, Devescovi G, Suarez-Moreno ZR, Fatmi MB, Guarnaccia C et al. (2011). Sharing of quorum-sensing signals and role of interspecies communities in a bacterial plant disease. ISME J 5: 1857–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaito C, Sekimizu K. (2007). Colony spreading in Staphylococcus aureus. J Bacteriol 189: 2553–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloos WE, Bannerman TL. (1994). Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev 7: 117–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauga E, Powers TR. (2009). The hydrodynamics of swimming microorganisms. Rep Prog Phys 72: 96601. [Google Scholar]
- Matthäus F, Jagodic M, Dobnikar J. (2009). E. coli superdiffusion and chemotaxis-search strategy, precision, and motility. Biophys J 97: 946–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitt EJG, Crusz SA, Diggle SP. (2015). Staphylococcus aureus forms spreading dendrites that have characteristics of active motility. Sci Rep 5: 17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CD, Parkins MD, Rabin HR, Duan K, Norgaard JC, Surette MG. (2008). A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc Natl Acad Sci USA 105: 15070–15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker R, Seymour JR, Samadani A, Hunt DE, Polz MF. (2008). Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci USA 105: 4209–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.