Abstract
The Zetaproteobacteria are ubiquitous in marine environments, yet this class of Proteobacteria is only represented by a few closely-related cultured isolates. In high-iron environments, such as diffuse hydrothermal vents, the Zetaproteobacteria are important members of the community driving its structure. Biogeography of Zetaproteobacteria has shown two ubiquitous operational taxonomic units (OTUs), yet much is unknown about their genomic diversity. Genome-resolved metagenomics allows for the specific binning of microbial genomes based on genomic signatures present in composite metagenome assemblies. This resulted in the recovery of 93 genome bins, of which 34 were classified as Zetaproteobacteria. Form II ribulose 1,5-bisphosphate carboxylase genes were recovered from nearly all the Zetaproteobacteria genome bins. In addition, the Zetaproteobacteria genome bins contain genes for uptake and utilization of bioavailable nitrogen, detoxification of arsenic, and a terminal electron acceptor adapted for low oxygen concentration. Our results also support the hypothesis of a Cyc2-like protein as the site for iron oxidation, now detected across a majority of the Zetaproteobacteria genome bins. Whole genome comparisons showed a high genomic diversity across the Zetaproteobacteria OTUs and genome bins that were previously unidentified by SSU rRNA gene analysis. A single lineage of cosmopolitan Zetaproteobacteria (zOTU 2) was found to be monophyletic, based on cluster analysis of average nucleotide identity and average amino acid identity comparisons. From these data, we can begin to pinpoint genomic adaptations of the more ecologically ubiquitous Zetaproteobacteria, and further understand their environmental constraints and metabolic potential.
Introduction
Microbes are everywhere, and in many ecosystems they are the key drivers of biogeochemical cycles. Iron is the most abundant element in the earth and only microbes are able to utilize it as an energy source. Mineralogical evidence of iron- oxidizers has been found, dating to 1.89 Ga, making iron oxidation a very ancient metabolism (Planavsky et al., 2009). Early Earth hosted a ferruginous ocean where iron oxidation may have been the dominant metabolism (Ilbert and Bonnefoy, 2013; Guilbaud et al., 2015). Microbial iron oxidizers are found suspended in the water column (Field et al., 2016), but extensive microbial growth by iron oxidation is limited to areas of high ferrous iron and low oxygen concentrations, such as hydrothermal vents (Emerson and Moyer, 2010; Scott et al., 2015).
Reduced iron released by hydrothermal vent systems fuels primary production by lithoautotrophic microbes, which in turn support additional trophic levels making hydrothermal vent systems some of the most biologically active regions of the deep-sea (Sievert and Vetriani, 2012). It is estimated that 3 × 1011 mol of Fe(II) is released each year through hydrothermal venting in Earth’s oceans (Holland, 2006), and is transported in the water column thousands of kilometers away from the source (Resing et al., 2015), where it can be utilized by phototrophs in the upper ocean; however, iron is still a limiting factor for phototrophs in the upper ocean (Raven et al., 1999). The abiotic oxidation of Fe(II) by O2 is rapid in fully aerated seawater (Konhauser et al., 2005; Druschel et al., 2008). Therefore, from a microbe’s perspective, Fe(II) is potentially a vast food source, yet it is as ephemeral as it is abundant and bioavailable.
Microbial iron oxidation has been recognized in freshwater systems since the 1890s, whereas microbial iron oxidation in marine systems is just beginning to be recognized (Emerson et al., 2013; Fleming et al., 2013). The isolates of the newest class of Proteobacteria, the Zetaproteobacteria, are described as neutrophilic marine iron-oxidizers (Emerson et al., 2007). Zetaproteobacteria have been identified throughout the Pacific and Atlantic Oceans at hydrothermal vent habitats and estuaries (McAllister et al., 2011; Scott et al., 2015; Field et al., 2016). At sites where the predominant vent effluent is high in ferrous iron, Zetaproteobacteria are the dominant microbial mat community members, with the classes of the Gamma-, Delta- and Epsilon-proteobacteria as well as Nitrospira consistently detected in these habitats (Moyer et al., 1995; Rassa et al., 2009; Fleming et al., 2013). Several Zetaproteobacteria operational taxonomic units have been identified and two are globally ubiquitous in iron-driven microbial mat communities (McAllister et al., 2011).
Zetaproteobacteria are considered ecosystem engineers due to their foundational role in the formation of the microbial mat architecture. This architecture is comprised of exopolysaccharide structures, including twisted helical stalks or tubular sheaths as observed by microscopic analysis of cultures and microbial mats (Chan et al., 2011; Fleming et al., 2013; Chan et al., 2016b). Through the production of stalks or sheaths, the Zetaproteobacteria can alter their physical and chemical environment (Chan et al., 2016a). Furthermore, Zetaproteobacteria are lithoautotrophs and the primary producers in iron-dominated hydrothermal vent systems (Singer et al., 2011; Field et al., 2015). Previous molecular analysis of microbial mats at Lō’ihi Seamount showed that Zetaproteobacteria correlate with the abundance of key functional genes, but that functional gene abundance did not vary across varying mat morphologies; furthermore, vent chemistry was found to be associated with the observed mat morphologies (Jesser et al., 2015), suggesting unrealized genomic diversity within the Zetaproteobacteria.
Zetaproteobacteria were first described at Lō’ihi Seamount, which is located 35 km south-east of the big island of Hawai’i and hosts a plethora of dynamic hydrothermal vents (Moyer et al., 1995). In 1996, a major eruption formed Pele’s Pit, a 300 m wide caldera near the summit, with several active hydrothermal venting sites (Figure 1). Before the 1996 eruption, Lō’ihi was dominated by low-temperature diffuse-flow hydrothermal vents emitting fluids up to ~70 °C and elevated levels of Fe(II), CO2, CH4 and NH4+ (Sedwick et al., 1992; Wheat et al., 2000) and has now returned to these pre-eruption conditions (Glazer and Rouxel, 2009).
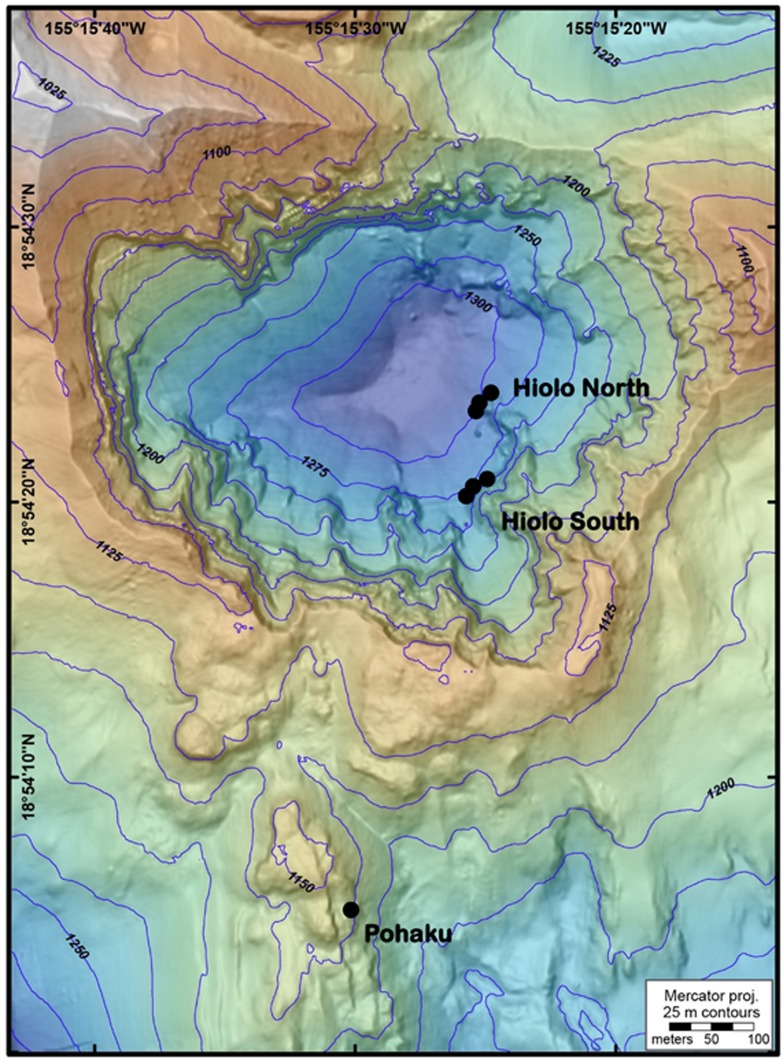
Figure 1.
Bathymetric map (high resolution at <2 m) of sampling sites in and near Pele’s Pit caldera on the summit of Lō’ihi Seamount, Hawai’i. Precise marker locations include Pohaku (Marker 57), Hiolo North (Markers 36, 39 and 31), Hiolo South (Markers 34, 38 and Ku’kulu). Courtesy of Susan Merle, NOAA EOI/OSU.
Biogeographic patterns for marine microbes remain poorly understood in terms of distribution scale and evolutionary divergence rates. To address this, we sequenced six distinct microbial mat communities collected from Lō’ihi Seamount. From this, a shotgun metagenomics approach was used, where we were able to construct a composite assembly for genome binning. We used differential coverage analysis to reconstruct site-specific community composition and compared this to the community structure as determined by taxa specific Quantitative PCR (qPCR) analyses. Here we present genome-resolved metagenomics to further explore patterns of biodiversity and adaptation of Zetaproteobacteria populations, including two ecologically significant Zetaproteobacteria OTUs (zOTUs).
Materials and methods
Sample collection
Microbial mat samples were collected at Lō’ihi Seamount, HI in October 2009 by the remotely operated vehicle (ROV) Jason II onboard the R/V Kilo Moana. Samples were collected from within Pele’s Pit at Hiolo North (Markers 31, 36 and 39), Hiolo South (Markers 34 and 38), and Ku’kulu Base (no marker) or on the caldera rim at Pohaku (Marker 57) (Figure 1). All samples were collected using a single-action Biomat Syringe (BS) sampler (Figure 2c) as described in Fleming et al. (2013).
Figure 2.
Photos illustrating the different mat morphologies. (a) curd-type mat from Marker 34, (b) curd-type mat from Marker 57, (c) veil-type mat from Ku’kulu, (d) veil-type mat from Marker 39, (e) streamers from Marker 31, and (f) streamers from Marker 39. Scale bars are 10 cm.
DNA extraction, T-RFLP analysis, and qPCR
Genomic DNA was extracted from samples using the FastDNA SPIN kit for soil (MP Biomedical, Santa Ana, CA, USA) according to manufacturer’s protocol. Cells were lysed by bead beating twice (stored on ice for 5 min in between) in a FastPrep instrument (MP Biomedical) at a speed setting of 5.5 for 45 s and DNA was eluted with 1 mM Tris at pH 8. Genomic DNA was quantified with a Qubit 2.0 fluorometer (ThermoFisher Scientific, Waltham, MA, USA).
Samples were PCR amplified for use in terminal-restriction fragment length polymorphisms (T-RFLP) as previously described (Davis and Moyer, 2008). PCR was visualized on a 1% agarose gel before restriction digestion. The end-labeled fragments were run on an ABI model 3130XL automated DNA sequencer and the data were analyzed with the BioNumerics v7.6 software (Applied Maths, Austin, TX, USA). SSU rRNA gene clone libraries from five sampled microbial mat communities were constructed as described in McAllister et al. (2011), in order assess putative phylotypes in the T-RFLP dataset (Supplementary Figures 1 and 2).
qPCR conditions along with Bacterial and Zetaproteobacterial primers used were the same as described by Jesser et al. (2015). Zetaproteobacteria abundance was determined using the ratio of Zetaproteobacteria to Bacteria SSU rRNA gene copies per nanogram gDNA. No qPCR data were used unless primers exhibited better than 95% efficiency and yielded single-peak amplicons upon post-PCR melt curve analysis.
Metagenomic sequencing, assembly and annotation
Extracted DNA was cleaned and concentrated using an Aurora (Boreal Genomics, Vancouver, BC, Canada) prior to sequencing; libraries were prepared with the Nextera DNA Library Kit (Illumina, San Diego, CA, USA). Sample J2-479-BS3 was run on an Illumina HiSeq 2000 using paired-end sequencing with reads of 101 bp from each end. Sample J2-483-BS63 was run using paired-end sequencing with reads of 84 bp on an Illumina MiSeq and was a combination of two samples collected from the same microbial mat. The remainder of the samples were run using an Illumina MiSeq with paired-end sequencing of 308 bp reads (Supplementary Table 1).
Sequenced reads were quality checked using FastQC (Andrews, 2010) and were trimmed of adaptors, and pairs were matched using cutadapt (Martin, 2011). Trimmed reads were normalized using BBnorm (target depth: 18). The resulting reads were assembled with IDBA-UD (Peng et al., 2012) (k-mer sizes: 50–240 in steps of 10 without correction). Reads from each sample were mapped, with bowtie2 (Langmead and Salzberg, 2012) to the composite assembly to get coverage information. This was then used to construct genome bins with MaxBin 2.0 (Wu et al., 2016) using default parameters. The resulting genome bins were evaluated with CheckM (Parks et al., 2015). The assembled composite metagenome was uploaded to Integrated Microbial Genomics (IMG) for annotation. Genome bins were separated from bulk data after annotation.
Genes annotated as specific proteins identified in M. ferrooxydans PV-1 were identified by BLASTp searches of the composite metagenome with an e-value cutoff of 10−5 (Supplementary Tables 2). Cyc1PV-1 (DAA64808.1), and Cyc2PV-1 (AKN35166.1) were identified via proteomics (Barco et al., 2015) and Mob (SPV1_03948) identified via fosmid library genome analysis (Singer et al., 2011).
Average nucleotide and average amino acid identities
Genome bins identified as Zetaproteobacteria by CheckM were compared to genome sequences of Zetaproteobacteria single amplified genomes (SAGs) and Zetaproteobacteria isolate genomes. The average nucleotide identity (ANI) was calculated using the BLAST-based algorithm tool in JSpecies v1.2.1 (Richter and Rosselló-Móra, 2009). The average amino acid identity was calculated using the enveomics toolbox (Rodriguez-R and Konstantinidis, 2016). Hierarchical cluster analysis was calculated in R using the gplots package.
Phylogenetic analysis
All genes annotated as a ribulose 1,5-bisphosphate carboxylase (RubisCO) were further analyzed for binning and taxonomic placement. IMG phylogeny was used for the unbinned genes, whereas CheckM was used for the genes within a genome bin. The identified RubisCO Form II amino acid sequences were then aligned using the Geneious v9.1 aligner (Kearse et al., 2012). The resulting alignments were manually screened, and all sequences less than 110 amino acids were removed from analysis. The resulting alignment was then used to create a phylogenic consensus tree with RAxML v7.2.8 using the gamma GTR protein model with 1000 bootstrap iterations, again with Geneious (Kearse et al., 2012).
Accession numbers
Representative sequences from each operational taxonomic unit identified in the clone libraries were submitted to GenBank. Accession numbers are JQ287646−JQ287657, JX468894 (Fleming et al., 2013) and KY417831−KY417866 (this study). All metagenomic contigs have been made available in the IMG Database (IMG Genome ID 3300009408). All sequence data are also available from NCBI SRA (Biosample accessions SAMN06226859-SAMN06226864).
Results and Discussion
Site description and community structure
Microbial mats vary in and around Pele’s Pit in gross morphology and color, from white-yellow to burnt orange (Figures 2a–f). In addition to variation in color, the mats had variable textures that were assigned to three specific mat morphological groups associated with variable fluid flow regimes. These were described as curds in the presence of direct flow (Figures 2a and b), veils associated with diffuse flow (Figures 2c and d), and streamers also found in direct flow (Figures 2e and f). Pohaku is the only sample site located on the outside of Pele’s Pit on the southern rim of this caldera (Figure 1), and has been characterized as highest in reduced iron, at nearly 1 mM (Glazer and Rouxel, 2009). Microscopic analysis of the curd-type mat shows the predominance of helical stalks (Chan et al., 2016b), whereas analysis of veil-type mats showed a prevalence of the sheathed morphology (Fleming et al., 2013).
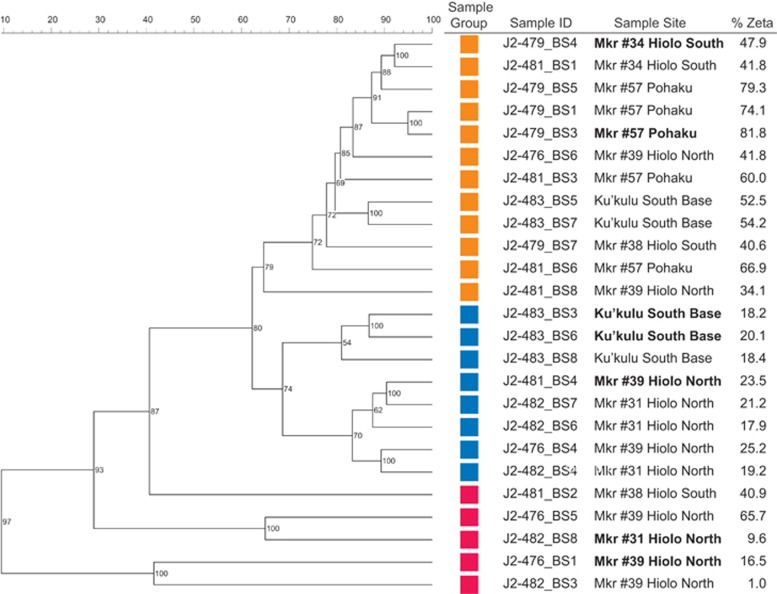
A comprehensive community fingerprint analysis by T-RFLP of 25 mat communities from seven vent sites showed three distinct groups, which corresponded to the gross mat morphology of curds, veils and streamers (Figure 3); however, these groups did not correlate with location or site temperature. All of the microbial mats were collected with a Biomat Syringe sampler, allowing for precision sampling of the topmost active layer of the mat. The morphology of Group I mats are characterized as light yellow to light orange curds, Group II are yellow veiled-type mats and Group III are comprised of white to dark orange streamers attached to the vent orifice. Group I mats had the greatest abundance (56.3%±15.5%) of Zetaproteobacteria within the bacterial community, whereas Group II had significantly less Zetaproteobacteria (20.5%±2.7%) and Group III had the lowest (17.7%±13.7%) as determined by qPCR.
Figure 3.
T-RFLP analysis of BioMat Samples collected from Lō’ihi microbial mats. Colored boxes represent the three different groups that resulted from the cluster analysis: Group I, orange; Group II, blue; Group III, red. The sampled mat communities in bold were selected for Illumina metagenomic sequencing.
SSU rRNA gene clone libraries were constructed from representative mat communities in an attempt to identify the microbial community members driving the T-RFLP clustering. Group I mats had a lower bacterial diversity compared to the other two groups, and exhibited high levels of zOTUs 1 and 2. Group II mats contained a higher abundance of zOTUs 4, 6 and 10 along with Gammaproteobacteria. Group III mats were dominated by sulfur- and hydrogen-metabolizing Epsilonproteobacteria, with a smaller contribution from zOTUs. These results highlight a clear difference between Fe-rich (Groups I and II) and S-rich (Group III) habitats, and between direct flow (Groups I and III) and indirect/diffuse-flow (Group II) environments (Figures 2 and 3; Supplementary Figures 1 and 2).
Assembly and annotation
Six samples, two representatives from each morphotype group, were chosen for metagenomic sequencing. The resulting composite assembly had 162 376 contigs comprised of 289 114 522 bases with an overall GC% of 51.1% and an n50 of 3483. This composite assembly was separated into genome bins based on coverage and tetranucleotide frequencies of the scaffolds with MaxBin 2.0. These bins contained 77.9% of the total composite metagenome bases and 37.4% of the scaffolds. There were no sequences in multiple bins. Genome binning of the composite metagenome resulted in 93 total bins. These genome bins were assessed for completeness and taxonomic classification using CheckM (Parks et al., 2015). Two of the bins were identified as Archaea, which is consistent with previous analysis showing Archaea were either below the detection limit or less than 5% of the community, and generally derived from deep-sea archaeoplankton retention in the mats (Moyer et al., 1998; Rassa et al., 2009). Nine genomes were unresolved to the class level; however, one of these bins contained a full-length SSU rRNA gene identified as a Deferribacteres (LoihiBin_014). Unclassified bins were removed from further analysis. The most numerous genome bins identified belonged to the Zetaproteobacteria (Table 1). Overall, the bins had an average n50 of 12 376 in an average of 707 scaffolds. The genome bins range in completeness from 6.55 to 100%, with an average of 70.8% (±30.8%). Contamination ranged from 0.0 to 83%, with an average of 12.6% (±15.0%). On average, the Zetaproteobacteria genome bins were 62.9% (±34.1%) complete, with an average contamination level of 11.6% (±13.4%).
Table 1. Summary statistics of 77 population genomes, which have been assigned to a phylogenetic class.
| Bin Id | Class | GC Content (%) | Genome size (Mbp) | Gene count | Compl. (%) | Cont. (%) | Scaffolds (no.) | Longest scaffold (bp) |
|---|---|---|---|---|---|---|---|---|
| ZetaBin011 | Zetaproteobacteria | 58.51 | 2.14 | 2472 | 96.64 | 2.43 | 305 | 50 762 |
| ZetaBin022 | Zetaproteobacteria | 55.75 | 2.27 | 3048 | 83.51 | 12.68 | 1107 | 17 841 |
| ZetaBin030 | Zetaproteobacteria | 49.62 | 0.14 | 202 | 9.40 | 0.00 | 57 | 8654 |
| ZetaBin035 | Zetaproteobacteria | 60.80 | 2.92 | 3306 | 94.26 | 22.37 | 525 | 99 453 |
| ZetaBin037 | Zetaproteobacteria | 58.78 | 1.98 | 2929 | 33.29 | 9.66 | 1254 | 16 849 |
| ZetaBin040 | Zetaproteobacteria | 47.98 | 2.91 | 2812 | 98.74 | 0.84 | 145 | 144 433 |
| ZetaBin041 | Zetaproteobacteria | 50.51 | 2.19 | 2391 | 96.80 | 12.77 | 321 | 43 694 |
| ZetaBin042 | Zetaproteobacteria | 51.09 | 2.64 | 2707 | 97.06 | 3.21 | 188 | 129 034 |
| ZetaBin043 | Zetaproteobacteria | 50.12 | 1.90 | 2339 | 63.98 | 5.43 | 538 | 25 129 |
| ZetaBin047 | Zetaproteobacteria | 51.36 | 3.18 | 4581 | 73.66 | 33.61 | 1364 | 25 119 |
| ZetaBin049 | Zetaproteobacteria | 52.24 | 2.65 | 3747 | 72.41 | 26.54 | 955 | 18 206 |
| ZetaBin050 | Zetaproteobacteria | 51.42 | 3.33 | 4118 | 95.66 | 18.26 | 600 | 76 915 |
| ZetaBin052 | Zetaproteobacteria | 43.64 | 0.60 | 682 | 22.86 | 0.05 | 146 | 16 046 |
| ZetaBin055 | Zetaproteobacteria | 43.43 | 0.75 | 909 | 40.87 | 0.14 | 221 | 11 516 |
| ZetaBin056 | Zetaproteobacteria | 43.77 | 0.64 | 837 | 26.42 | 3.09 | 247 | 12 705 |
| ZetaBin057 | Zetaproteobacteria | 43.16 | 0.45 | 625 | 19.75 | 1.72 | 202 | 10 079 |
| ZetaBin058 | Zetaproteobacteria | 43.59 | 0.33 | 438 | 14.11 | 0.19 | 153 | 11 627 |
| ZetaBin059 | Zetaproteobacteria | 42.76 | 0.71 | 956 | 30.09 | 9.80 | 275 | 11 729 |
| ZetaBin060 | Zetaproteobacteria | 42.09 | 0.41 | 587 | 6.55 | 0.34 | 180 | 7595 |
| ZetaBin062 | Zetaproteobacteria | 43.75 | 0.62 | 783 | 34.87 | 0.00 | 216 | 11 379 |
| ZetaBin064 | Zetaproteobacteria | 43.02 | 0.73 | 953 | 18.26 | 0.00 | 304 | 16 336 |
| ZetaBin065 | Zetaproteobacteria | 41.82 | 4.32 | 5575 | 78.50 | 34.27 | 1556 | 27 653 |
| ZetaBin066 | Zetaproteobacteria | 48.52 | 3.69 | 4265 | 99.58 | 5.46 | 541 | 87 185 |
| ZetaBin069 | Zetaproteobacteria | 42.71 | 0.79 | 1109 | 10.27 | 0.69 | 342 | 10 884 |
| ZetaBin077 | Zetaproteobacteria | 50.37 | 2.17 | 2648 | 93.63 | 16.43 | 738 | 17 579 |
| ZetaBin078 | Zetaproteobacteria | 43.08 | 0.50 | 702 | 19.48 | 0.00 | 259 | 9548 |
| ZetaBin079 | Zetaproteobacteria | 51.17 | 2.24 | 2521 | 86.27 | 10.85 | 365 | 24 239 |
| ZetaBin080 | Zetaproteobacteria | 47.96 | 2.96 | 3001 | 96.22 | 15.64 | 280 | 89 556 |
| ZetaBin084 | Zetaproteobacteria | 49.43 | 4.46 | 5555 | 95.77 | 36.35 | 2073 | 22 352 |
| ZetaBin088 | Zetaproteobacteria | 52.51 | 2.94 | 3356 | 90.99 | 22.27 | 827 | 30 533 |
| ZetaBin089 | Zetaproteobacteria | 46.84 | 7.73 | 8861 | 98.59 | 55.28 | 2338 | 145 501 |
| ZetaBin090 | Zetaproteobacteria | 44.39 | 2.66 | 3080 | 91.22 | 22.57 | 836 | 26 177 |
| ZetaBin091 | Zetaproteobacteria | 43.81 | 1.60 | 2257 | 59.55 | 3.32 | 900 | 7813 |
| ZetaBin092 | Zetaproteobacteria | 48.98 | 2.19 | 2486 | 90.17 | 8.05 | 579 | 33 624 |
| PlanctoBin028 | Planctomycetia | 70.38 | 2.53 | 3027 | 66.15 | 18.11 | 1153 | 15 570 |
| PlanctoBin046 | Planctomycetia | 56.90 | 3.93 | 5336 | 59.95 | 29.64 | 2392 | 62 755 |
| NitroBin001 | Nitrospira | 42.82 | 2.70 | 2876 | 100.00 | 2.73 | 145 | 102 379 |
| NitroBin004 | Nitrospira | 49.60 | 2.94 | 3178 | 91.13 | 3.52 | 423 | 72 390 |
| NitroBin006 | Nitrospira | 55.00 | 2.97 | 2728 | 97.41 | 1.72 | 107 | 231 086 |
| NitroBin008 | Nitrospira | 54.70 | 3.51 | 3231 | 99.08 | 2.44 | 130 | 308 593 |
| NitroBin010 | Nitrospira | 48.18 | 1.27 | 1628 | 10.85 | 0.16 | 525 | 19 238 |
| NitroBin051 | Nitrospira | 66.96 | 1.91 | 2303 | 59.18 | 1.72 | 873 | 9924 |
| IgnaviBin015 | Ignavibacteria | 34.92 | 4.28 | 4351 | 100.00 | 22.15 | 792 | 62 071 |
| GemmaBin005 | Gemmatimonadetes | 65.73 | 3.33 | 2732 | 97.80 | 1.10 | 93 | 433 639 |
| GemmaBin009 | Gemmatimonadetes | 70.00 | 3.08 | 2652 | 100.00 | 1.10 | 139 | 186 026 |
| GammaBin013 | Gammaproteobacteria | 62.84 | 3.47 | 3308 | 97.46 | 4.56 | 366 | 92 648 |
| GammaBin018 | Gammaproteobacteria | 64.56 | 3.40 | 3474 | 98.28 | 10.53 | 449 | 84 061 |
| GammaBin021 | Gammaproteobacteria | 64.89 | 1.72 | 2122 | 61.34 | 2.37 | 470 | 21 092 |
| GammaBin025 | Gammaproteobacteria | 63.75 | 2.91 | 3074 | 97.56 | 7.71 | 383 | 39 220 |
| GammaBin034 | Gammaproteobacteria | 45.52 | 3.17 | 4017 | 97.93 | 21.32 | 1080 | 37 727 |
| GammaBin036 | Gammaproteobacteria | 55.82 | 3.34 | 4213 | 68.97 | 18.28 | 1152 | 33 248 |
| GammaBin038 | Gammaproteobacteria | 60.57 | 5.80 | 6109 | 93.89 | 83.17 | 1221 | 60 894 |
| GammaBin045 | Gammaproteobacteria | 60.12 | 1.96 | 2816 | 72.49 | 22.73 | 1122 | 9308 |
| GammaBin063 | Gammaproteobacteria | 43.85 | 1.44 | 2097 | 22.48 | 0.79 | 507 | 197 530 |
| GammaBin076 | Gammaproteobacteria | 50.35 | 3.06 | 3378 | 95.77 | 8.63 | 588 | 44 851 |
| GammaBin082 | Gammaproteobacteria | 42.31 | 3.44 | 3647 | 88.17 | 5.25 | 565 | 52 724 |
| GammaBin093 | Gammaproteobacteria | 38.54 | 4.19 | 4496 | 97.06 | 3.98 | 476 | 90 958 |
| FlavoBin054 | Flavobacteriia | 40.70 | 1.45 | 2303 | 17.41 | 2.47 | 812 | 27 641 |
| FlavoBin072 | Flavobacteriia | 30.74 | 2.76 | 3716 | 67.07 | 22.79 | 1138 | 21 266 |
| FlavoBin087 | Flavobacteriia | 29.74 | 3.28 | 3737 | 78.86 | 37.63 | 543 | 33 162 |
| EpsilonBin027 | Epsilonproteobacteria | 31.35 | 3.20 | 4464 | 91.12 | 40.00 | 1399 | 17 806 |
| EpsilonBin032 | Epsilonproteobacteria | 38.78 | 1.83 | 2035 | 96.67 | 6.35 | 226 | 62 498 |
| EpsilonBin033 | Epsilonproteobacteria | 35.69 | 1.33 | 1883 | 33.03 | 5.34 | 669 | 22 159 |
| EpsilonBin053 | Epsilonproteobacteria | 38.02 | 1.49 | 1694 | 95.90 | 4.17 | 243 | 33 985 |
| EpsilonBin071 | Epsilonproteobacteria | 38.65 | 2.82 | 3865 | 51.40 | 18.20 | 1184 | 37 928 |
| DeltaBin002 | Deltaproteobacteria | 62.33 | 3.07 | 2716 | 98.21 | 1.21 | 268 | 89 342 |
| DeltaBin003 | Deltaproteobacteria | 56.12 | 2.31 | 2081 | 94.19 | 2.80 | 105 | 128 268 |
| DeltaBin016 | Deltaproteobacteria | 72.27 | 6.62 | 4801 | 89.52 | 4.19 | 561 | 102 686 |
| DeltaBin031 | Deltaproteobacteria | 54.46 | 2.27 | 2924 | 64.58 | 11.80 | 1102 | 10 516 |
| DeltaBin044 | Deltaproteobacteria | 51.08 | 2.66 | 2843 | 95.24 | 11.92 | 441 | 54 886 |
| DeltaBin048 | Deltaproteobacteria | 50.60 | 2.90 | 3774 | 77.60 | 38.52 | 1238 | 24 934 |
| DeferriBin019 | Deferribacteres | 46.16 | 5.66 | 5252 | 100.00 | 59.31 | 1600 | 40 272 |
| CaldiBin024 | Caldilineae | 58.61 | 5.11 | 4978 | 99.09 | 8.58 | 766 | 132 167 |
| AnaeroBin020 | Anaerolineae | 61.54 | 4.25 | 4986 | 96.55 | 26.33 | 1477 | 26 958 |
| AlphaBin023 | Alphaproteobacteria | 61.16 | 2.39 | 3062 | 86.31 | 9.04 | 950 | 16 719 |
| AlphaBin068 | Alphaproteobacteria | 48.13 | 1.31 | 1697 | 16.25 | 0.86 | 512 | 23 917 |
| ActinoBin026 | Actinobacteria | 71.81 | 2.90 | 3229 | 94.02 | 12.54 | 566 | 35 545 |
Abbreviations: Compl., completeness; Cont., contamination; OTU, operational taxonomic unit.
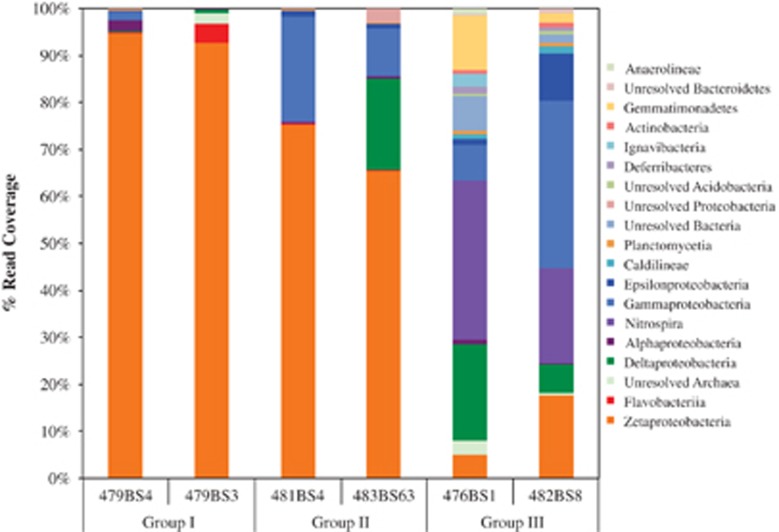
T-RFLP and qPCR results both indicate that Group I mat communities were less diverse than Group II or Group III. This is again corroborated by coverage analysis of the genome bins, where communities from Group I have the lowest diversity and Group III had the highest diversity. Zetaproteobacteria genome bins were still present, though as minor community members, in the representative Group III communities (Figure 4). Group III also had higher coverage estimates within the Nitrospira, Gamma-, Epsilon- and Alphaproteobacteria. Zetaproteobacteria genome bins had the highest coverage in the Group I mat communities. The bacterial taxa distribution observed in the clone libraries is consistent with that estimated by genome binning from metagenomics (Figure 4; Supplementary Figure 1).
Figure 4.
Relative abundance estimated by read coverage of taxonomically-classified genome bins from the sampled microbial communities. Genome bins were assessed for taxonomic classification using CheckM.
Carbon utilization
All isolates of Zetaproteobacteria grow via lithoautotrophy and encode for the RubisCO protein for carbon fixation from CO2. Mariprofundus ferrooxydans PV-1, M. ferrooxydans JV-1, M. ferrooxydans M34 and Mariprofundus DIS-1 encode for both Form I and Form II large subunit RubisCO gene, whereas Zetaproteobacterium TAG-1 and Mariprofundus sp. EFK-M39 only encodes a Form II RubisCO (Field et al., 2015). In total, 87 genes were identified as the large subunit of RubisCO. Of these, 67 were Form II and 11 were Form I. Of the Form I genes, only one was binned into a Zetaproteobacteria genome bin (ZetaBin022). The majority of the RubisCO Form II genes belonged to Zetaproteobacteria and 28 of the Zetaproteobacteria genome bins encoded a Form II gene, including the bin with the Form I gene (ZetaBin022). The Gammaproteobacteria had the second highest abundance of RubisCO genes, with four Form I and sixteen Form II genes detected. Twenty-three of the RubisCO genes were not placed into genome bins, but nine of these had the highest similarity to Zetaproteobacteria genes and six were most similar to Gammaproteobacteria (Supplementary Table 5).
In comparison, only seven ATP citrate lyase (encoded by aclB) genes were identified. This is a key gene in the reductive tricarboxylic acid cycle and is found in autotrophic Epsilonproteobacteria and Aquifacales (Hügler and Sievert, 2011). Five of the seven aclB genes were binned into Epsilonproteobacteria genome bins (Supplementary Table 6). The closest taxonomic hits were to Sulfurovum sp AR, Sulfurimonas autotrophica OK10 and Nitratiruptor sp SB155-2. Two of these organisms, S. autotrophica OK10 and Nitratiruptor sp. SB155-2, were isolated from Iheya North hydrothermal field sediments and chimneys, respectively (Sikorski et al., 2010; Inoue et al., 2016). Sulfurovum sp. AR was isolated from deep marine sediments collected near Svalbard, within the Arctic Circle (Park et al., 2012).
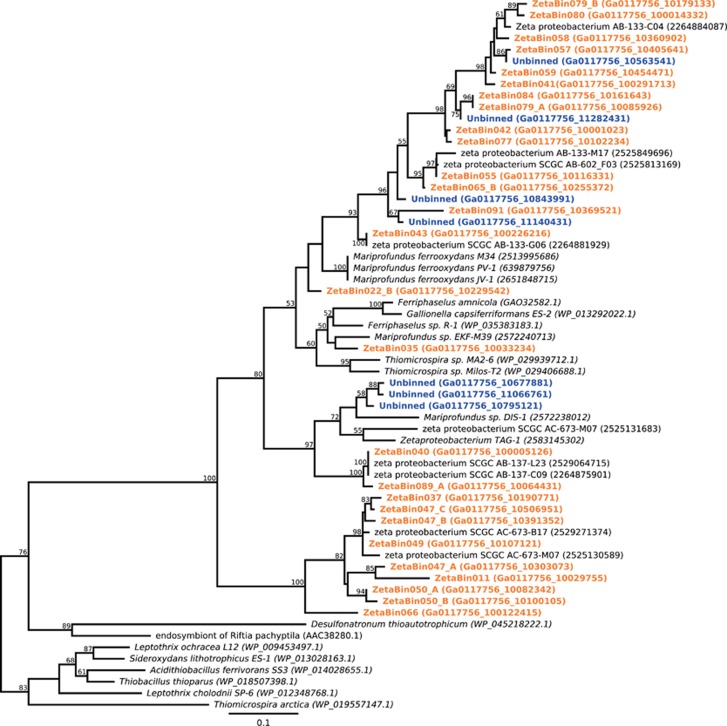
There was a high diversity of Form II RubisCO proteins recovered from Zetaproteobacteria genome bins and unbinned proteins identified as Zetaproteobacteria by IMG (Figure 5; Supplementary Table 5). Many of these RubisCO proteins were most similar to RubisCO proteins from the Zetaproteobacteria SAGs belonging to zOTU 2. This zOTU was one of the two considered as cosmopolitan because it is found throughout the Pacific Ocean (McAllister et al., 2011).
Figure 5.
RAxML phylogenetic tree of the Form II RubisCO proteins from Zetaproteobacteria genome bins (orange) and unbinned proteins identified as Zetaproteobacteria by IMG (blue). Numbers within parenthesis are gene identification numbers. Bootstrap values (⩾50) are representative of 1000 iterations.
Targeted qPCR on RubisCO Form II (cbbM) showed high abundance of the gene correlated strongly with a high abundance of Zetaproteobacteria (Jesser et al., 2015). The abundance of Form II RubisCO genes in comparison to Form I is indicative of adaptations to high CO2 and very low O2 environments (Hernandez et al., 1996; Tabita et al., 2008). The prevalence of Form II RubisCO in the genome bins of the Zetaproteobacteria (Table 2) shows an adaptation to growth in very low O2 environments similar to what is found in and around Pele’s Pit (Glazer and Rouxel, 2009). Zetaproteobacteria SAGs showed a similar pattern, in that Form I RubisCO was undetected (Field et al., 2015). Only a single Zetaproteobacteria genome bin contained both forms of RubisCO, suggesting that genotypes containing only Form II are the most prevalent.
Table 2. Number of genes per Zetaproteobacteria genome bins. Bins were sorted by their closest zOTU as determined by both ANI and AAI.
| OTU by Closest SAG | GenomeBin | cbbM | ccoN | ccoO | coxA | cyc2 (PV-1) | cyc1 (PV-1) | arsC | nirK/nirS | napA | narG | nasAB | nirB | nirD | nifH | amt |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ZetaBin042 | 1 | 4 | 3 | 1 | 1 | 3 | 3 | 1 | 1 | 1 | 2 | ||||
| 1 | ZetaBin066 | 1 | 4 | 2 | 2 | 2 | 3 | 2 | 1 | 1 | 1 | 2 | ||||
| 1 | ZetaBin077 | 1 | 6 | 4 | 1 | 2 | 3 | 1 | 2 | 1 | 2 | |||||
| 1 | ZetaBin079 | 2 | 4 | 2 | 1 | 3 | 1 | 1 | 2 | 1 | 2 | |||||
| 1 | ZetaBin080 | 1 | 4 | 2 | 1 | 1 | 3 | 1 | 1 | 1 | 2 | |||||
| 1 | ZetaBin084 | 1 | 6 | 2 | 1 | 5 | 2 | 4 | 5 | 1 | 2 | 2 | 5 | 2 | 7 | |
| 1 | ZetaBin088 | 5 | 4 | 1 | 2 | 2 | 6 | 2 | 3 | 2 | 2 | |||||
| 1 | ZetaBin092 | 3 | 3 | 2 | 3 | 2 | 1 | 1 | 2 | |||||||
| 1 | ZetaBin041 | 1 | 3 | 1 | 1 | 1 | 3 | 5 | 1 | 1 | 1 | 2 | ||||
| 1 | ZetaBin043 | 1 | 4 | 2 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| 2 | ZetaBin052 | 1 | 1 | |||||||||||||
| 2 | ZetaBin055 | 1 | 1 | 1 | 3 | |||||||||||
| 2 | ZetaBin056 | 1 | 1 | 1 | 1 | 1 | ||||||||||
| 2 | ZetaBin057 | 1 | 1 | 1 | ||||||||||||
| 2 | ZetaBin058 | 1 | 1 | 1 | 1 | 1 | ||||||||||
| 2 | ZetaBin059 | 1 | 4 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | |||||
| 2 | ZetaBin060 | 1 | ||||||||||||||
| 2 | ZetaBin062 | 3 | 3 | 1 | 1 | 3 | ||||||||||
| 2 | ZetaBin064 | 3 | 1 | 1 | 4 | 1 | 1 | |||||||||
| 2 | ZetaBin065 | 2 | 4 | 2 | 2 | 2 | 5 | 5 | ||||||||
| 2 | ZetaBin069 | 1 | 1 | |||||||||||||
| 2 | ZetaBin078 | 1 | ||||||||||||||
| 2 | ZetaBin090 | 3 | 3 | 3 | 3 | 2 | 3 | 1 | 1 | |||||||
| 4 | ZetaBin030 | |||||||||||||||
| 4 | ZetaBin047 | 3 | 3 | 3 | 1 | 2 | 1 | 2 | 1 | 6 | ||||||
| 4 | ZetaBin037 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | ||||||||
| 4 | ZetaBin050 | 2 | 3 | 2 | 2 | 2 | 1 | 4 | 2 | 4 | ||||||
| 6 | ZetaBin040 | 1 | 3 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 3 | |||
| 9 | ZetaBin089 | 2 | 5 | 1 | 2 | 2 | 1 | 2 | 1 | 1 | 4 | 2 | 1 | 12 | ||
| 9 | ZetaBin091 | 1 | 3 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | |||||
| 10 | ZetaBin049 | 1 | 2 | 1 | 3 | 5 | 1 | 1 | 1 | 1 | ||||||
| 11 | ZetaBin011 | 1 | 3 | 1 | 2 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | ||||
| 11 | ZetaBin022 | 2 | 2 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | ||||||
| 11 | ZetaBin035 | 1 | 3 | 1 | 4 | 2 | 1 | 3 | 1 | 2 | ||||||
| Bin Total | 34 | 23 | 28 | 22 | 10 | 22 | 19 | 28 | 15 | 8 | 1 | 19 | 23 | 19 | 2 | 25 |
| Gene totals | 30 | 91 | 42 | 18 | 41 | 24 | 71 | 32 | 11 | 2 | 30 | 37 | 20 | 2 | 71 |
Abbreviations: AAI, average amino acid identity; ANI, average nucleotide identity; OTU, operational taxonomic unit; SAG, single amplified genomes; zOTU, zetaproteobacteria OTU.
Nitrogen cycling
Biological nitrogen fixation is a key process in any ecosystem. The gene nifH encodes the nitrogenase reductase subunit, and is commonly used to track abundance and diversity among nitrogen-fixing organisms (Gaby and Buckley, 2012). Of the Zetaproteobacteria, Mariprofundus sp. EKF-M39, DIS-1 and M. ferrooxydans M34 encode a nifH gene, and qPCR estimates showed very low occurrence of nifH in microbial mat communities from Lō’ihi Seamount (Jesser et al., 2015). Consistent with this notion, only eleven nifH genes were identified and only two of these were within Zetaproteobacteria genome bins (ZetaBin035 & ZetaBin089). ZetaBin089 also encodes for nifD and nifK, the nitrogenase alpha and beta subunits, respectively. These genes are encoded on the same contig and are syntenous with the other identified nifH-containing Zetaproteobacteria isolates (Supplementary Figure 3). ZetaBin035 is lacking the alpha and beta subunits, but encodes the dinitrogenase iron-molybdenum cofactor, which is involved in the synthesis of the iron-molybdenum cofactor that binds the active site of the nitrogenase enzyme. Based on these annotations, it appears that these two Zetaproteobacteria bins (ZetaBin035 and ZetaBin089) are potentially capable of nitrogen fixation.
Diverse nifH genes have been identified at Axial Seamount, located along the Juan de Fuca Ridge (Mehta et al., 2003) and interestingly, ammonium has been detected to similar levels as found at Lō’ihi microbial mats, where nifH genes were either below detection or at very low abundance (Jesser et al., 2015). Ammonium transport proteins (amt) were found in 26 of the Zetaproteobacteria genome bins, including the genome bins that encode a nifH (Table 2). Use of nitrate and/or nitrite as a nitrogen source appears to be the most common across the Zetaproteobacteria genome bins. The majority of the Zetaproteobacteria genome bins contained genes for nitrate reduction (nasAB) and/or nitrite reductase (nirBD) for the assimilation of nitrogen. The dissimilatory nitrate reductase (napAB) and nitrite reductase (nirK/nirS) were also identified in 18 of the Zetaproteobacteria genome bins (Table 2) showing that denitrification is also possible. The prevalence of the ammonium transport proteins, presence of assimilatory nitrogen pathways, and the low recovery of nifH suggest that Zetaproteobacteria rely more on the presence of bioavailable nitrogen compounds accessed from the environment, rather than by dinitrogen fixation.
Arsenic cycling
Arsenic has been found at hydrothermal vents and the arsenic detoxification gene, arsenate reductase (encoded by arsC) has been identified in abundance in microbial mats from Lō’ihi hydrothermal habitats (Jesser et al., 2015). ArsC reduces arsenate to arsenite, which can then be exported from the cell via an arsenite specific transporter. In the composite assembly, there were 195 identified arsC genes in 67 of the genome bins representing every taxonomic class. The majority of the binned arsC genes were contained within either the Zetaproteobacteria or Gammaproteobacteria genome bins, with 71 and 28 gene copies, respectively. Of the identified arsC genes that were unbinned, taxonomic placement by IMG shows these genes to again be similar to genes from Zetaproteobacteria and Gammaproteobacteria. Arsenite transport proteins were identified in 23 of the Zetaproteobacteria genome bins. All of the Zetaproteobacteria genome bins with an arsenite transport protein contained an arsenate reductase as well.
At Tutum Bay, a shallow water hydrothermal vent system, ~1.5 kg of arsenic per day is released into the environment (Meyer-Dombard et al., 2013). This system also releases reduced iron, and Zetaproteobacteria were shown to heavily colonize slides incubated in situ. Although arsenic geochemistry has yet to be recorded at Lō’ihi vents, the abundance of arsenic-related genes found in our composite assembly suggests that arsenate is abundant in this environment. However, to show this, further geochemical analysis targeting arsenic redox states at Lō’ihi would be required.
Electron transport chain
Zetaproteobacteria SAGs and isolate genomes encode for a cbb3-type cytochrome c oxidase (Field et al., 2015; Fullerton et al., 2015). M. ferrooxydans PV-1, encodes for subunits I–III (ccoNOP) and appears to be lacking subunit IV (ccoQ) according to Singer et al. (2011). Only the CcoNO subunits were identified in the proteomic profile of M. ferrooxydans PV-1 (Barco et al., 2015). Eleven of the 34 Zetaproteobacteria bins encode all four subunits of the cbb3-type cytochrome c oxidase. Mariprofundus sp. EKF-M39, DIS-1, M. ferrooxydans JV-1 and six of the Zetaproteobacteria SAGs encode all four subunits of the cbb3-type cytochrome c oxidase. Nine of the Zetaproteobacteria genome bins encode for subunits I–III and appear to lack subunit IV. The ccoQ gene product is a membrane-spanning protein of unclear function; ccoN gene encodes for the catalytic subunit and ccoO, a monoheme c-type cytochrome. Only the ccoNO subunits are common to all gene clusters across multiple bacterial phyla (Ducluzeau et al., 2008).
The cbb3-type cytochrome c oxidase has a high affinity for O2 and is predominately used under microaerophilic conditions and may also be used to prevent O2 poisoning (Sievert et al., 2008; Jewell et al., 2016). The aa3-type cytochrome c oxidase is encoded by coxABC where expression is repressed in facultative anaerobes under low oxygen conditions (Pitcher and Watmough, 2004). Ten of the Zetaproteobacteria genome bins contain the coxA gene (aa3-type cytochrome c oxidase), and all but one of these genome bins encodes for the ccoNOP (cbb3-type cytochrome c oxidase) as well. This suggests that like other facultative anaerobes and microaerophiles, Zetaproteobacteria are able to modulate their electron transport chain to account for variable oxygen conditions. Only one of the 24 Zetaproteobacteria SAGs encodes both types of the cytochrome c oxidases (Field et al., 2015).
There is no direct evidence that Zetaproteobacteria can grow anaerobically using nitrate as the terminal electron acceptor; however, a number of other iron-oxidizing Proteobacteria can grow anaerobically this way (Hedrich et al., 2011; Beller et al., 2013). In the Zetaproteobacteria genome bins there was one bin, ZetaBin084, which encoded the respiratory nitrate reductase, NarG. This genome bin also encodes the cbb3 and aa3 type cytochrome c oxidases, that is, both the ccoNO and coxA genes.
Iron oxidation is hypothesized to occur on the outer membrane and is coupled to cytoplasmic and membrane-bound electron transfer proteins (Hedrich et al., 2011; Ilbert and Bonnefoy, 2013). From M. ferrooxydans PV-1 genome analysis, a molybdopterin oxidoreductase (Mob, SPV1_03948) was hypothesized to be important in Fe(II) oxidation (Singer et al., 2011), and showed synteny with two contigs contained in a fosmid library generated from a suction-sample collected from Hiolo South (Singer et al., 2013). This protein was also identified in the top 25 most abundant proteins of M. ferrooxydans PV-1; however, its function in iron oxidation is questionable due to high similarity to proteins found in non-iron oxidizers (Barco et al., 2015). In the Zetaproteobacteria genome bins, similar proteins were detected and annotated by IMG as different molybdopterin-containing oxidoreductases (for example, nitrate reductase NapA; Supplementary Table 4).
Proteomic analysis of M. ferrooxydans PV-1 revealed a membrane bound cytochrome that was highly expressed and distantly related to cytochrome c2 of Acidothiobacillus ferrooxydans (Barco et al., 2015; White et al., 2016). It has been proposed that this protein, referred to as Cyc2PV-1, is the site of electron transfer from iron to a cytoplasmic cytochrome (Cyc1PV-1), which was also identified as high-abundant by proteomic analysis. From Cyc1PV-1, electrons are hypothesized to be shuttled into a membrane-bound electron transport chain, terminating with the cbb3-type cytochrome c oxidase. Using the amino acid sequence of Cyc1PV-1 and Cyc2PV-1 to search the composite metagenome, 24 and 41 gene copies, respectively, were identified within the Zetaproteobacteria genome bins (Table 2). The open reading frames most similar to Cyc1PV-1 were annotated as cytochrome c553, whereas the Cyc2PV-1 genes were annotated as hypothetical proteins by IMG (Supplementary Tables 2 and 3). The identification of Cyc1PV-1 and Cyc2PV-1 in our Zetaproteobacteria genome bins, supports the hypothesis that a Cyc2-like protein is the site of iron oxidation, as opposed to the alternative hypothesis using the Mob protein (Hedrich et al., 2011; Singer et al., 2011; Ilbert and Bonnefoy, 2013; Barco et al., 2015). These Cyc2-like proteins were identified in every zOTU detected, indicating their ubiquity across the Zetaproteobacteria, including within the ecologically significant taxa (Table 2).
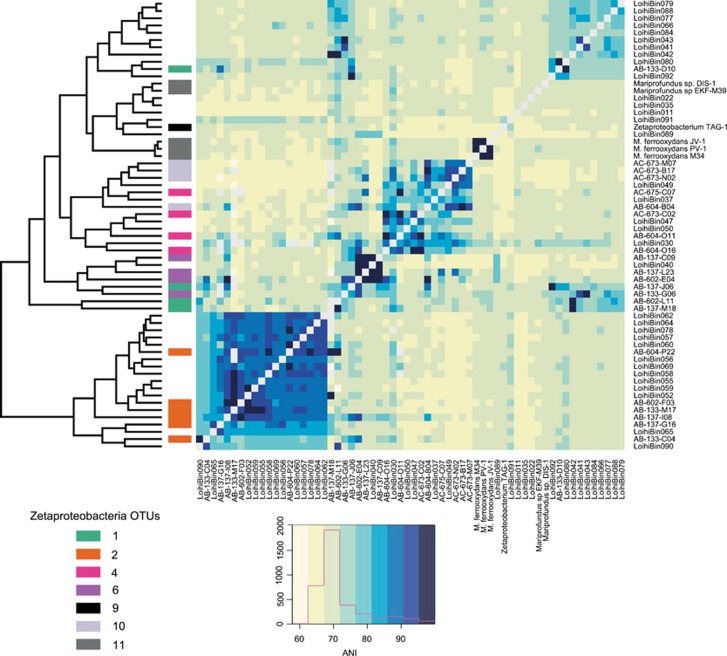
Whole genome comparisons
In this composite metagenome study, there were 249 total SSU rRNA genes recovered. Of these, 41 were contained within Zetaproteobacteria genome bins as determined by CheckM and 37 SSU rRNA genes were identified as Zetaproteobacteria by the RDP classifier (Wang et al., 2007; Parks et al., 2015). Previous studies on Zetaproteobacteria SSU rRNA diversity identified two operational taxonomic units that were ubiquitous across the Pacific Ocean, referred to as zOTUs 1 and 2 (McAllister et al., 2011). Genomes were compared at the nucleotide level to assess genomic diversity across the Zetaproteobacteria genome bins as compared to isolate genomes and SAGs (Figure 6) by ANI. Hierarchical clustering of the genomes based on ANI showed that genome bins most similar to zOTUs 1 and 2 are the most highly represented, with 10 and 13 out of the 34 Zetaproteobacteria genome bins, respectively. Based on Form II RubisCO phylogeny, these zOTUs constitute a single lineage that diverged more recently than any of those that occurred in other lineages (Figure 5). Both these zOTUs were also found to be the most abundant phylotypes detected in microbial mats from Lō’ihi hydrothermal habitats by SAGs and SSU clone library analyses (McAllister et al., 2011; Field et al., 2015). Based on the cluster analysis of ANI comparisons from our Zetaproteobacteria genome bins, this study has shown that zOTU 2 represents a monophyletic cluster and is distinct from all the other zOTU clusters (Figure 6), and based on estimated genome size hints, that genome streamlining may be occurring within this group. This zOTU was also the first to be identified from any hydrothermal system (Moyer et al., 1995). Our whole genome cluster analysis also showed that zOTUs 1, 4, 6 and 10 have much greater genomic dissimilarity (that is, diversity) than what would be expected based on SSU rRNA identity alone. The distribution of Zetaproteobacteria genome bins across the three different groups of mat communities shows that zOTU 2 is the most abundant in both Group I and Group II (that is, both curds and veils) type mats based on gross morphology, representing twisted-stalks and sheaths, respectively. The Group III mats (streamers), which have a low Zetaproteobacterial abundance relative to the other members of the community, included zOTU 11 as the most highly represented within this mat-type (Supplementary Figure 4).
Figure 6.
Hierarchical clustering heatmap and dendrogram of ANI of Zetaproteobacteria genome bins, and isolate Zetaproteobacteria genomes. Genome self-comparisons and where ANI could not be determined are presented in light gray. Genome bins were confirmed by average amino acid identity.
Using this hierarchical cluster analysis approach, patterns of metabolic potential across zOTUs can also be realized. The only two bins with a nifH gene (ZetaBin035 and ZetaBin089) were also most closely related to isolates that are able to fix nitrogen. All cultured isolates remain within the same tight cluster, including the type strain M. ferrooxydans PV-1, possibly indicating a narrow range of selection pressure resulting from our present culturing techniques. Furthermore, there were few Zetaproteobacteria genome bins with similarity to any cultured isolates, suggesting environmental parameters are poorly mimicked in the lab. In general, the RubisCO protein relationships and genome relationships identified by ANI were conserved (that is, similar). None of the genome bins within zOTU 2 contained genes for the aa3-type cytochrome c oxidase, further supporting adaptation to the low O2 levels found at Lō’ihi hydrothermal habitats.
Conclusions
Coverage analysis of our composite metagenome indicates that carbon is fixed primarily by Zetaproteobacteria containing Form II RubisCO. Through an assessment of the diversity of Form II RubisCO genes and the abundance of cbb3-type cytochrome c oxidase genes, many Zetaproteobacteria show an adaptation to life at very low oxygen levels in conjunction with high-ferrous iron and dissolved CO2 levels. Denitrification is less common, and our data indicates that bioavailable nitrogen is primarily metabolized. Nearly all of the Zetaproteobacteria genome bins contain genes for the detoxification of arsenate as well as representatives from each of the other classes that were detected in these microbial mat communities. This shows that metagenomics analyses can also provide insights into geochemical conditions. The lineage represented by zOTU 2 is monophyletic suggesting an ancestral bottleneck during its more recent evolutionary history. This zOTU is also the most prevalent of our Zetaproteobacteria genome bins, indicating it is the most ecologically successful manifestation of both sheath and stalk morphology. Through the use of genome-resolved metagenomics, we have better constrained patterns observed in metabolic potential and divergence across many of the Zetaproteobacteria growing within microbial mats at Lō’ihi Seamount.
Acknowledgments
We thank the captain and crew of the R/V Kilo Moana (KM0923) along with the entire ROV Jason II operations team for their assistance with sample collection during our October 2009 cruise to Lō’ihi. We also thank Carl Kaiser and the AUV Sentry operations team for mapping the summit of Lō’ihi Seamount, during our March 2013 cruise, which was not an easy task. We are extremely grateful to David Clague and Jenny Paduan of the Monterey Bay Aquarium Research Institute (MBARI) for exceptionally adroit multibeam data processing, making our efforts in mapping possible. This work was funded in part by Western Washington University’s Office of Research and Sponsored Programs, by the Biology Alumni Student Research Fellowship, by the Fouts Foundation for student research enhancement and by the National Science Foundation award OCE 1155756 (to CLM).
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Andrews S. (2010), FastQC: A quality control tool for high throughput sequence data. Available at: http://wwwbioinformaticsbabrahamacuk/projects/fastqc.
- Beller HR, Zhou P, Legler TC, Chakicherla A, Kane S, Letain TE et al. (2013). Genome-enabled studies of anaerobic, nitrate-dependent iron oxidation in the chemolithoautotrophic bacterium Thiobacillus denitrificans. Front Microbiol 4: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco RA, Emerson D, Sylvan JB, Orcutt BN, Meyers MEJ, Ramírez GA et al. (2015). New insight into microbial iron oxidation as revealed by the proteomic profile of an obligate iron-oxidizing chemolithoautotroph. Appl Environ Microbiol 81: 5927–5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Emerson D, Luther GW III. (2016. a). The role of microaerophilic Fe-oxidizing micro-organisms in producing banded iron formations. Geobiology 14: 509–528. [DOI] [PubMed] [Google Scholar]
- Chan CS, Fakra SC, Emerson D, Fleming EJ, Edwards KJ. (2011). Lithotrophic iron-oxidizing bacteria produce organic stalks to control mineral growth: implications for biosignature formation. ISME J 5: 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, McAllister SM, Leavitt AH, Glazer BT, Krepski ST, Emerson D. (2016. b). The architecture of iron microbial mats reflects the adaptation of chemolithotrophic iron oxidation in freshwater and marine environments. Front Microbiol 7: 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE, Moyer CL. (2008). Extreme spatial and temporal variability of hydrothermal microbial mat communities along the Mariana Island Arc and southern Mariana back-arc system. J Geophys Res 113: B08S15. [Google Scholar]
- Druschel GK, Emerson D, Sutka R, Suchecki P, Luther GW III. (2008). Low-oxygen and chemical kinetic constraints on the geochemical niche of neutrophilic iron (II) oxidizing microorganisms. Geochim Cosmochim Acta 72: 3358–3370. [Google Scholar]
- Ducluzeau A-L, Ouchane S, Nitschke W. (2008). The cbb3 oxidases are an ancient innovation of the domain bacteria. Mol Biol Evol 25: 1158–1166. [DOI] [PubMed] [Google Scholar]
- Emerson D, Rentz JA, Lilburn TG, Davis RE, Aldrich H, Chan CS et al. (2007). A novel lineage of Proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PLoS One 2: e667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson D, Moyer CL. (2010). Microbiology of Seamounts: common patterns observed in community structure. Oceanography 23: 148–163. [Google Scholar]
- Emerson D, Field EK, Chertkov O, Davenport KW, Goodwin L, Munk C et al. (2013). Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front Microbiol 4: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field EK, Kato S, Findlay AJ, MacDonald DJ, Chiu BK, Luther GW III et al. (2016). Planktonic marine iron oxidizers drive iron mineralization under low-oxygen conditions. Geobiology 14: 499–508. [DOI] [PubMed] [Google Scholar]
- Field EK, Sczyrba A, Lyman AE, Harris CC, Woyke T, Stepanauskas R et al. (2015). Genomic insights into the uncultivated marine Zetaproteobacteria at Loihi Seamount. ISME J 9: 857–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming EJ, Davis RE, McAllister SM, Chan CS, Moyer CL, Tebo BM et al. (2013). Hidden in plain sight: discovery of sheath-forming, iron-oxidizing Zetaproteobacteria at Loihi Seamount, Hawaii, USA. FEMS Microbiol Ecol 85: 116–127. [DOI] [PubMed] [Google Scholar]
- Fullerton H, Hager KW, Moyer CL. (2015). Draft genome sequence of Mariprofundus ferrooxydans strain JV-1, isolated from Loihi Seamount, Hawaii. Genome Announc 3: e01118–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaby JC, Buckley DH. (2012). A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS One 7: e42149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer BT, Rouxel OJ. (2009). Redox speciation and distribution within diverse iron-dominated microbial habitats at Loihi Seamount. Geomicrobiol J 26: 606–622. [Google Scholar]
- Guilbaud R, Poulton SW, Butterfield NJ, Zhu M, Shields-Zhou GA. (2015). A global transition to ferruginous conditions in the early Neoproterozoic oceans. Nat Geosci 8: 466–470. [Google Scholar]
- Hedrich S, Schlömann M, Johnson DB. (2011). The iron-oxidizing proteobacteria. Microbiol 157: 1551–1564. [DOI] [PubMed] [Google Scholar]
- Hernandez JM, Baker SH, Lorbach SC, Shively JM, Tabita FR. (1996). Deduced amino acid sequence, functional expression, and unique enzymatic properties of the form I and form II ribulose bisphosphate carboxylase/oxygenase from the chemoautotrophic bacterium Thiobacillus denitrificans. J Bacteriol 178: 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland HD. (2006). The oxygenation of the atmosphere and oceans. Phil Trans R Soc Lond B Biol Sci 361: 903–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hügler M, Sievert SM. (2011). Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Ann Rev Mar Sci 3: 261–289. [DOI] [PubMed] [Google Scholar]
- Ilbert M, Bonnefoy V. (2013). Insight into the evolution of the iron oxidation pathways. Biochim Biophys Acta 1827: 161–175. [DOI] [PubMed] [Google Scholar]
- Inoue A, Anraku M, Nakagawa S, Ojima T. (2016). Discovery of a novel alginate lyase from Nitratiruptor sp. SB155-2 thriving at deep-sea hydrothermal vents and identification of the residues responsible for its heat stability. J Biol Chem 291: 15551–15563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesser KJ, Fullerton H, Hager KW, Moyer CL. (2015). Quantitative PCR analysis of functional genes in iron-rich microbial mats at an active hydrothermal vent system (Lō’ihi Seamount, Hawai’i). Appl Environ Microbiol 81: 2976–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell TN, Karaoz U, Brodie EL, Williams KH, Beller HR. (2016). Metatranscriptomic evidence of pervasive and diverse chemolithoautotrophy relevant to C, S, N and Fe cycling in a shallow alluvial aquifer. ISME J 10: 2106–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S et al. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konhauser KO, Newman DK, Kappler A. (2005). The potential significance of microbial Fe (III) reduction during deposition of Precambrian banded iron formations. Geobiology 3: 167–177. [Google Scholar]
- Langmead B, Salzberg SL. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12. [Google Scholar]
- McAllister SM, Davis RE, McBeth JM, Tebo BM, Emerson D, Moyer CL. (2011). Biodiversity and emerging biogeography of the neutrophilic iron-oxidizing Zetaproteobacteria. Appl Environ Microbiol 77: 5445–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta MP, Butterfield DA, Baross JA. (2003). Phylogenetic diversity of nitrogenase (nifH genes in deep-sea and hydrothermal vent environments of the Juan de Fuca Ridge. Appl Environ Microbiol 69: 960–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Dombard DAR, Amend JP, Osburn MR. (2013). Microbial diversity and potential for arsenic and iron biogeochemical cycling at an arsenic rich, shallow-sea hydrothermal vent (Tutum Bay, Papua New Guinea). Chem Geol 348: 37–47. [Google Scholar]
- Moyer CL, Dobbs FC, Karl DM. (1995). Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol 61: 1555–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer CL, Tiedje JM, Dobbs FC, Karl DM. (1998). Diversity of deep-sea hydrothermal vent Archaea. Deep Sea Res II 45: 303–317. [Google Scholar]
- Park S-J, Ghai R, Martín-Cuadrado A-B, Rodríguez-Valera F, Jung M-Y, Kim J-G et al. (2012). Draft genome sequence of the sulfur-oxidizing bacterium ‘Candidatus Sulfurovum sediminum’ AR, which belongs to the Epsilonproteobacteria. J Bacteriol 194: 4128–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Leung HCM, Yiu SM, Chin FYL. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28: 1420–1428. [DOI] [PubMed] [Google Scholar]
- Pitcher RS, Watmough NJ. (2004). The bacterial cytochrome cbb3 oxidases. Biochim Biophys Acta 1655: 388–399. [DOI] [PubMed] [Google Scholar]
- Planavsky N, Rouxel O, Bekker A, Shapiro R, Fralick P, Knudsen A. (2009). Iron-oxidizing microbial ecosystems thrived in late Paleoproterozoic redox-stratified oceans. Earth Planet Sci Lett 286: 230–242. [Google Scholar]
- Rassa AC, McAllister SM, Safran SA, Moyer CL. (2009). Zeta-Proteobacteria dominate the colonization and formation of microbial mats in low-temperature hydrothermal vents at Loihi Seamount, Hawaii. Geomicrobiol J 26: 623–638. [Google Scholar]
- Raven JA, Evans MCW, Korb RE. (1999). The role of trace metals in photosynthetic electron transport in O2-evolving organisms. Photosyn Res 60: 111–149. [Google Scholar]
- Resing JA, Sedwick PN, German CR, Jenkins WJ, Moffett JW, Sohst BM et al. (2015). Basin-scale transport of hydrothermal dissolved metals across the South Pacific Ocean. Nature 523: 200–203. [DOI] [PubMed] [Google Scholar]
- Rodriguez-R LM, Konstantinidis KT. (2016). The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints 4: e1900v1. Available at: https://doi.org/10.7287/peerj.preprints.1900v1. [Google Scholar]
- Richter M, Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106: 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JJ, Breier JA, Luther GW III, Emerson D. (2015). Microbial iron mats at the Mid-Atlantic Ridge and evidence that Zetaproteobacteria may be restricted to iron-oxidizing marine systems. PLoS One 10: e0119284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedwick PN, McMurtry GM, Macdougall JD. (1992). Chemistry of hydrothermal solutions from Pele’s Vents, Loihi Seamount, Hawaii. Geochim Cosmochim Acta 56: 3643–3667. [Google Scholar]
- Sievert SM, Scott KM, Klotz MG, Chain PS, Hauser LJ, Hemp J et al. (2008). Genome of the epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl Environ Microbiol 74: 1145–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert SM, Vetriani C. (2012). Chemoautotrophy at deep-sea vents: past, present, and future. Oceanography 25: 218–233. [Google Scholar]
- Sikorski J, Munk C, Lapidus A, Djao ODN, Lucas S, Del Rio TG et al. (2010). Complete genome sequence of Sulfurimonas autotrophica type strain (OK10T. Stand Genomic Sci 3: 194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer E, Emerson D, Webb EA, Barco RA, Kuenen JG, Nelson WC et al. (2011). Mariprofundus ferrooxydans PV-1 the first genome of a marine Fe(II) oxidizing Zetaproteobacterium. PLoS One 6: e25386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer E, Heidelberg JF, Dhillon A, Edwards KJ. (2013). Metagenomic insights into the dominant Fe (II) oxidizing Zetaproteobacteria from an iron mat at Lō'ihi, Hawai'i. Front Microbiol 4: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS. (2008). Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J Exp Bot 59: 1515–1524. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheat CG, Jannasch HW, Plant JN, Moyer CL, Sansone FJ, McMurtry GM. (2000). Continuous sampling of hydrothermal fluids from Loihi Seamount after the 1996 event. J Geophys Res 105: 19353–19367. [Google Scholar]
- White GF, Edwards MJ, Gomez-Perez L, Richardson DJ, Butt JN, Clarke TA. (2016). Mechanisms of bacterial extracellular electron exchange. Adv Microb Physiol 68: 87–138. [DOI] [PubMed] [Google Scholar]
- Wu Y-W, Simmons BA, Singer SW. (2016). MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32: 605–607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.