Abstract
Microorganisms perform most decomposition on Earth, mediating carbon (C) loss from ecosystems, and thereby influencing climate. Yet, how variation in the identity and composition of microbial communities influences ecosystem C balance is far from clear. Using quantitative stable isotope probing of DNA, we show how individual bacterial taxa influence soil C cycling following the addition of labile C (glucose). Specifically, we show that increased decomposition of soil C in response to added glucose (positive priming) occurs as a phylogenetically diverse group of taxa, accounting for a large proportion of the bacterial community, shift toward additional soil C use for growth. Our findings suggest that many microbial taxa exhibit C use plasticity, as most taxa altered their use of glucose and soil organic matter depending upon environmental conditions. In contrast, bacteria that exhibit other responses to glucose (reduced growth or reliance on glucose for additional growth) clustered strongly by phylogeny. These results suggest that positive priming is likely the prototypical response of bacteria to sustained labile C addition, consistent with the widespread occurrence of the positive priming effect in nature.
Introduction
Soils are a huge reservoir of carbon (C), exceeding plants and the atmosphere combined (Scharlemann et al., 2014). Anthropogenic increases in CO2 are expected to augment primary productivity (Ainsworth and Long, 2005) and thus enhance C transfer from the atmosphere to the soil, which could potentially enhance soil C storage. However, microbial activity responds dynamically to C inputs, where the rate of decomposition of native soil organic C can be reduced or enhanced, a phenomenon known as the ‘priming effect’ (Kuzyakov, 2010). Enhanced mineralization of native soil C, positive priming, has been widely observed across soil types (for example, loam, clay and sand) and in a variety of ecosystems including forests, croplands and grasslands (summarized in Liu et al., 2017). Further, positive priming could offset the ability of increased primary production to sequester C in soils (van Groenigen et al., 2014). The direction (positive or negative) and magnitude of priming in soil is often variable and can be hard to predict (see Luo et al., 2016 and references therein). Consequently, an understanding of the microbial mechanisms that underlie this phenomenon is needed. A current conceptual model posits that priming arises as fast growing r-strategists consume fresh C inputs and indirectly stimulate slower growing k-strategist that then consume additional native soil C (Fontaine et al., 2003; Blagodatskaya and Kuzyakov, 2008; Kuzyakov, 2010; Pascault et al., 2013). These ecological strategies have been further posited to align with phylogenetic groups. For instance, the relative abundance of Acidobacteria declines following nutrient addition (Cleveland et al., 2007; Jenkins et al., 2010) and is negatively correlated with soil organic C (SOC) content (Philippot et al., 2009; Will et al., 2010) suggesting an oligotrophic or k-strategist lifestyle. However, these hypotheses regarding priming have not been rigorously tested.
As with many microbially mediated processes, we lack a deeper understanding of priming because of the difficulty in connecting microbial responses with ecosystem processes. Through modern sequencing techniques, scientists have been able to address the question, ‘Who is there?’, but the next pressing inquiry, ‘What are they doing?’, remains unresolved. Combining phylogenetic markers with taxon-specific functional measurements can disentangle this problem (Poff et al., 2006; Cadotte et al., 2009), yet this has rarely been done for microbial ecology (Thomson et al., 2014; Morrissey et al., 2016).
Here, we used quantitative stable isotope probing (Hungate et al., 2015) to measure taxon-specific and community-level activity patterns in order to connect microbial biodiversity with C mineralization in soil. We hypothesized that following labile C additions microorganisms would exhibit consistent and phylogenetically clustered patterns of C use reflective of their ecological strategy. To address this hypothesis, soil from a ponderosa pine forest was incubated for 6 weeks with or without weekly additions of glucose. Glucose additions were intended to simulate pulses of labile C from root exudates. To assess microbial activity, we used 13C to measure glucose uptake and 18O to determine growth. As water is a universal substrate for DNA synthesis (Schwartz, 2007), a taxon’s genomic 18O content reflects its population growth during the incubation period. Similarly, genomic 13C content provides a quantitative measure of each taxon’s growth on 13C-labeled substrates.
Materials and methods
Experimental design
A portion of the data presented here was previously published in a methods development-oriented article (Hungate et al., 2015). Specifically, data on the 18O and 13C incorporation of bacterial taxa during week 1 was previously presented in Hungate et al. (2015). All other data, including 18O and 13C incorporation by bacterial taxa in week 6, bacterial community composition and soil C cycling measurements are unique to this article. Soil (0–15 cm) was collected from a ponderosa pine meadow (35.41541N, –111.6695W, 2344 m elevation), vegetated with patches of grass on the C. Hart Merriam elevation gradient in November 2012. Soil from this site was previously determined to be a Sponseller (Hendricks, 1985) clay loam with 36.1% clay, 43.5% silt and 20.4% sand (Morrissey et al., 2015). This soil was also characterized as having 2.6±0.1% C content, 0.12±0.01% N content (Dijkstra et al., 2006) and a relatively neutral pH of 6.8 (Blankinship et al., 2010). Soil was sieved through 2 mm mesh, and air-dried at room temperature for ~24 h before initiation of the experiment. Soils were dried to enable the addition of isotopically labeled solutions without saturating the soil.
To quantify priming, 40 g dry weight soil was weighed into specimen cups, brought up to 60% water holding capacity and placed in 32 oz mason jars (946.3 ml) with septa in the lids to pre-incubate for 7 days. Following this, half of the jars (n=5) received 500 μg C per g soil in 125 μl of a uniformly labeled 13C-glucose solution (992‰), whereas the other half received the same amount of deionized water to serve as a non-amended control. These additions were repeated weekly for 6 weeks with the incubation terminating the end of week 6. As root exudation rates in forests can range from ~200 μg to 3000 μg C per g root per week (for example, Phillips et al., 2011; Yin et al., 2014) our amendment simulated a realistic C flux to root-associated soil. Headspace gas samples for CO2 concentration and δ13C were taken three and four times a week in the control and glucose-amended samples, respectively. Following gas sampling, incubations were left uncovered for ~30 min to allow gas exchange, refreshing O2 and preventing CO2 saturation. Glucose- and soil organic matter-derived CO2-C was partitioned in the glucose-amended samples using mass balance:
where CSOC is CO2-C from native SOC (μg C per g soil), Ctotal is measured CO2-C from glucose-amended samples (μg C per g soil), δtotal is the measured δ13C signature of CO2 from glucose-amended samples, δglucose is the δ13C signature of the glucose solution (992‰) and δSOC is the average δ13C signature from the native SOC measured from the non-amended samples. Priming was calculated as the difference between SOC oxidation of the amended and non-amended samples and reported as CO2-C μg per g soil.
Owing to the expense associated with isotopically labeled substrates, stable isotope probing was conducted in parallel incubations that had less soil and were in smaller containers but were otherwise treated identically to the gas flux incubations described above (Hungate et al., 2015). Although these differences in incubation conditions could affect headspace O2 and CO2 concentrations, we expect these difference to be relatively small because the containers where opened regularly and allowed to exchange with the atmosphere. Briefly, 1 g dry weight soil was weighed into 15 ml Falcon tubes. In week 1, half the samples (n=18) received one of six isotope combinations (n=3): 16O-water only, 18O-water only, 12C-glucose+16O-water, 13C-glucose+16O-water, 12C-glucose+18O-water. The 13C-glucose was at atom fraction 99% and the 18O-water was atom fraction 97%. The other half received unlabeled water or glucose weekly until week 6, when they received one of the above isotope treatments. As with the gas flux incubations, samples were adjusted to 60% water holding capacity and received either 500 μg C per g soil or an equivalent volume of water each week. Samples were incubated with the isotope treatment for 7 days before being frozen at −80 °C. DNA was then extracted using the MP Biomedicals FastDNA spin kit (Solon, OH, USA). Ultracentrifugation, fractionation, quantitative PCR and sequencing were conducted as described in Hungate et al. (2015). Briefly, 5 μg of DNA per sample was separated on a CsCl density gradient via ultracentrifugation at 127 000 g for 72 h at 18 °C using a TLN-100 Rotor in a Optima Max bench top ultracentrifuge (Beckman Coulter, Fullerton, CA, USA). Density fractions (~150 μl) were collected and DNA was purified using isopropanol precipitation. Bacterial 16S ribosomal RNA (rRNA) gene copies in each fraction were quantified using a pan-bacterial broad-coverage quantitative PCR technique (Liu et al., 2012). For fractions containing DNA (9–15 fractions per sample), bacterial 16S rRNA genes were amplified using 338F and 806R primers and sequenced using dual indexing (Fadrosh et al., 2014) and 300-bp paired-end read chemistry on an Illumina MiSeq (Illumina, Inc., San Diego, CA, USA). All sequences and accompanying metadata are available for download from MG-RAST (Meyer et al., 2008) project name hungate_qSIP, project ID 14151.
Data analysis
Processing and quality filtering of sequences were performed as previously described (Hungate et al., 2015). Reads were assigned to their originating samples using 24-bp dual-index barcodes formed by combining the 12-bp barcodes from each paired-end read. Primer sequences were trimmed, and then reads were stitched using FLASH (Fadrosh et al., 2014). The full-length reads were clustered using the UCLUST-based open reference operational taxonomic unit (OTU) picking protocol described in QIIME (Caporaso et al., 2010) against the Greengenes 13_8 97% sequence identity reference database (Caporaso et al., 2010; McDonald et al., 2012). Taxa were analyzed at the ‘species’ level (L7 table in QIIME).
The excess atom fraction (EAF) 18O and 13C were calculated for each taxon as described previously (Hungate et al., 2015). Briefly, a weighted average density was calculated for each taxon’s DNA after incubation in the presence of unlabeled and isotopically enriched substrates (that is, water and glucose) based on its distribution across a CsCl density gradient (data not shown). The shift in weighted average density following incubation with an isotopically enriched substrate can be used to quantify the amount of isotope incorporation based on the theoretically modeled and experimentally verified relationship between isotope incorporation and DNA molecular weight (Hungate et al., 2015). Preliminary data analysis revealed an effect of ultracentrifuge tube on estimates of taxon-weighted average density. This effect is likely a consequence of slight differences in the CsCl density gradients between tubes. To correct this, we assumed that the taxa in the bottom 5% of weighted average density shifts in each treatment did not incorporate any isotope based on the evidence that a significant fraction of soil microorganisms are inactive (Lennon and Jones, 2011; Blagodatskaya and Kuzyakov, 2013). The average difference between the unenriched weighted average density (average of no isotope tubes) and the observed weighted average density for these taxa was then used to quantify and correct for inter-tube variation in weighted average density estimates (as shown in Supplementary Figure S1).
Responses to labile C addition were assessed by comparing the increase in growth upon labile C addition (Δ 18O=18Oglucose – 18Ocontrol) to the 13C uptake. The relationship between these measures provides insight into the taxon-specific and community-level C usage. A decline in growth, indicated by a negative Δ 18O value, would indicate less soil organic matter consumption regardless of 13C uptake. Additional growth using solely the added 13C-glucose would produce a ratio of 13C to Δ 18O that is >2, because 100% of the C atoms in newly synthesized DNA would be derived from glucose, whereas only a fraction of the oxygen used for DNA synthesis comes from water, with the remainder coming from organic substrates (Schwartz, 2007). Escherichia coli grown in pure culture on glucose as the sole C source derives 33% of the oxygen in its DNA from water (Hungate et al., 2015) and consequently would exhibit a 13C/Δ18O≈3. Based on the possibility that some organisms could derive a greater fraction of their oxygen from water, we estimated that growth solely on glucose could produce a 13C/Δ 18O as low as 2 meaning as much as 50% of the oxygen in DNA could be derived from water. This served to ensure our inference of additional soil C consumption (priming) based on a 13C/Δ 18O<2 was conservative. Integrated measures of whole community activity are presented as relative abundance weighted average 13C and Δ18O and the ratio between these values.
For phylogenetic analysis, a representative sequence for each taxon was aligned with the Greengenes 13_8 97% OTU reference database using BLAST as described in Morrissey and Franklin (2015). The reference sequence with the greatest identity match for each OTU was used for downstream analyses; the median percent identity was 99%. The Greengenes 97% OTU tree was then pruned to contain only the OTUs present in our samples and visualized using the interactive tree of life (Letunic and Bork, 2011). The responses of individual taxa were categorized as described based on their 13C and Δ18O as described below, and the net relatedness index was calculated for each group to see if the members were more or less phylogenetically related than would be expected by chance (Webb et al., 2002) using the ses.mpd function in the picante R package (Kembel et al., 2011). The relative abundance and isotope incorporation (EAF 13C or 18O) of replicates were averaged before the regression and phylogenetic analysis.
All sequence data associated with this project have been deposited at MG-RAST (project accession number 14151, project name ‘hungate_qSIP’), complete metadata are available on Figshare (https://doi.org/10.6084/m9.figshare.4600420.v1).
Results and discussion
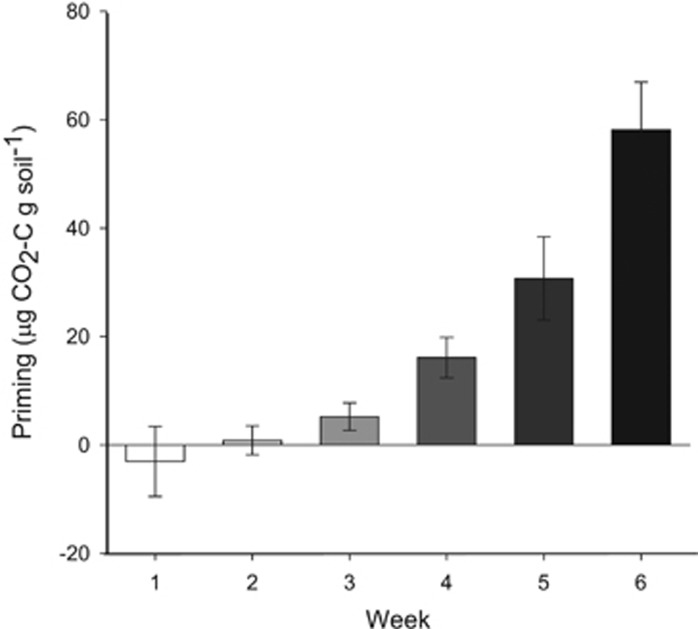
The amount of soil C primed gradually increased over the incubation period (Figure 1), transitioning from slightly negative in week 1, after a single pulse of glucose, to positive in subsequent weeks. Such temporal dynamics are commonly observed in priming experiments and have been hypothesized to arise from microbial community dynamics (Kuzyakov and Bol, 2006; Mau et al., 2015; Wang et al., 2015). Here we show that priming is associated with changes in both the growth and C utilization of individual microbial taxa. Specifically, sustained availability of glucose through repeated pulses alters the utilization of native soil C in the majority of soil bacteria.
Figure 1.
Weekly soil organic matter priming (mean±s.e.), calculated as the difference in soil derived C mineralized in glucose-added and unamended soil. Mineralization rates were evaluated in laboratory microcosms of soil from a ponderosa pine forest receiving weekly additions of glucose or water.
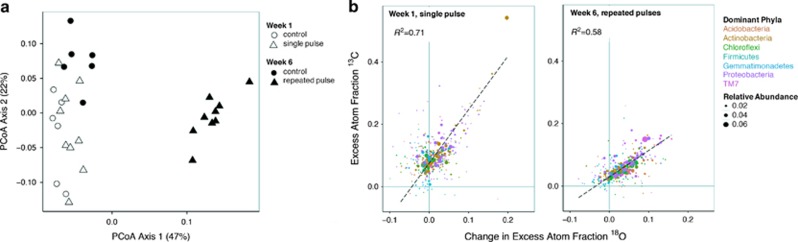
Over the 6-week incubation, bacterial community composition shifted slightly in control soil and substantially in soil with added glucose (Figure 2a; permutational multivariate analyses of variance week × treatment F=9.7, P<0.01). Repeated glucose additions increased the relative abundance of Proteobacteria (specifically α and δ) and TM7, whereas decreasing the proportion of Acidobacteria and Actinobacteria (Supplementary Figure S2).
Figure 2.
Principal coordinates analysis (PCoA) of bacterial community structure (a) and variation in EAF 13C with the change in 18O assimilation because of glucose (b) in response to single and repeated pulses of glucose. For the regression analysis, each taxon’s relative abundance (% 16 S rRNA gene sequences) and taxonomy are indicated using size and color, respectively. The dashed line and coefficient of determination show the results of a regression weighted by relative abundance (% of 16S rRNA gene sequences).
In order to relate taxon-specific activity to priming, we examined the relationship between glucose use (via 13C assimilation) and the change in growth (18O assimilation) because of glucose addition (Δ18O) for all taxa, weighted by their relative abundances (Figure 2b). Increased growth using 13C-glucose as the sole C source would produce a 13C/Δ18O ratio of at least 2. This is because 100% of the C atoms in newly synthesized DNA would be derived from glucose, whereas only a fraction (~50% maximum) of the oxygen used for DNA synthesis comes from water (see Materials and methods, Hungate et al., 2015). Increased growth with lower than expected 13C assimilation from the added glucose (13C/Δ18O<2) is possible by enhanced consumption of SOC. Consequently, the relationship between Δ18O and 13C assimilation in weeks 1 and 6 (Figure 2b) can be used to understand individual and community-level responses to glucose addition and reveal how the aggregated activity of individual taxa mediates the priming of SOC.
After single and repeated pulses of glucose, changes in growth were strongly and positively correlated with glucose assimilation (Figure 2b, both P<0.001) suggesting a direct stimulation of activity within individual taxa. Similarly, our results suggest that the large majority of microorganisms consumed the added glucose, after single and repeated pulses, as their 13C atom percent excess was above zero (Figure 2b). This is consistent with a recent study using RNA-stable isotope probing, which found no differences in fresh organic matter utilization across taxonomic groups (Thomson et al., 2014).
Following a single pulse of glucose, many bacterial taxa (42% of taxa accounting for 39% of 16S rRNA gene sequences) consumed the added glucose without exhibiting increased growth rates (average 13C=0.078 EAF for organisms with Δ18O<0). This suggests that glucose was being consumed in lieu of SOC, a phenomenon often called preferential substrate utilization (Blagodatskaya and Kuzyakov, 2008; Wang et al., 2015). Preferential substrate utilization provides a mechanism by which the introduction of labile C can decrease SOC utilization (negative priming). When considering the community as a whole, the substantial assimilation of labile C (average 13C=0.101 EAF) was accompanied by only a marginal increase in growth rate (average Δ18O=0.016 EAF). This produced a community average 13C/Δ18O of 6.31, much higher than is expected for normal soil organic matter consumption and additional growth using glucose as the sole C source. These data suggest the majority of taxa used the added glucose in lieu of their normal SOC consumption, and very few bacteria increased their use of SOC in response to a single pulse of substrate addition. Consequently, the use of SOC was decreased relative to the control soil, resulting in negative priming, a finding in line with the slightly negative change in SOC mineralization observed during week 1 (Figure 1).
By week 6, after repeated glucose addition, the bacterial community response was consistent with enhanced SOC utilization (positive priming). At this time, fewer organisms consumed glucose without a corresponding increase in growth (22% of taxa accounting for 7% of 16S rRNA gene sequences), and glucose consumption by these organisms was much lower than in week 1 (average 13C=0.032 EAF for organisms with Δ18O<0), indicating a reduction in preferential substrate utilization. When examining the community as a whole, the glucose uptake (average 13C=0.061 EAF) could not account for the increase in growth (average Δ18O =0.039 EAF), indicating that bacteria must have increased their consumption of SOC (average 13C/Δ18O=1.56). Sustained glucose addition enhanced bacterial growth and SOC utilization, thereby priming the decomposition of SOC in week 6 (Figure 1).
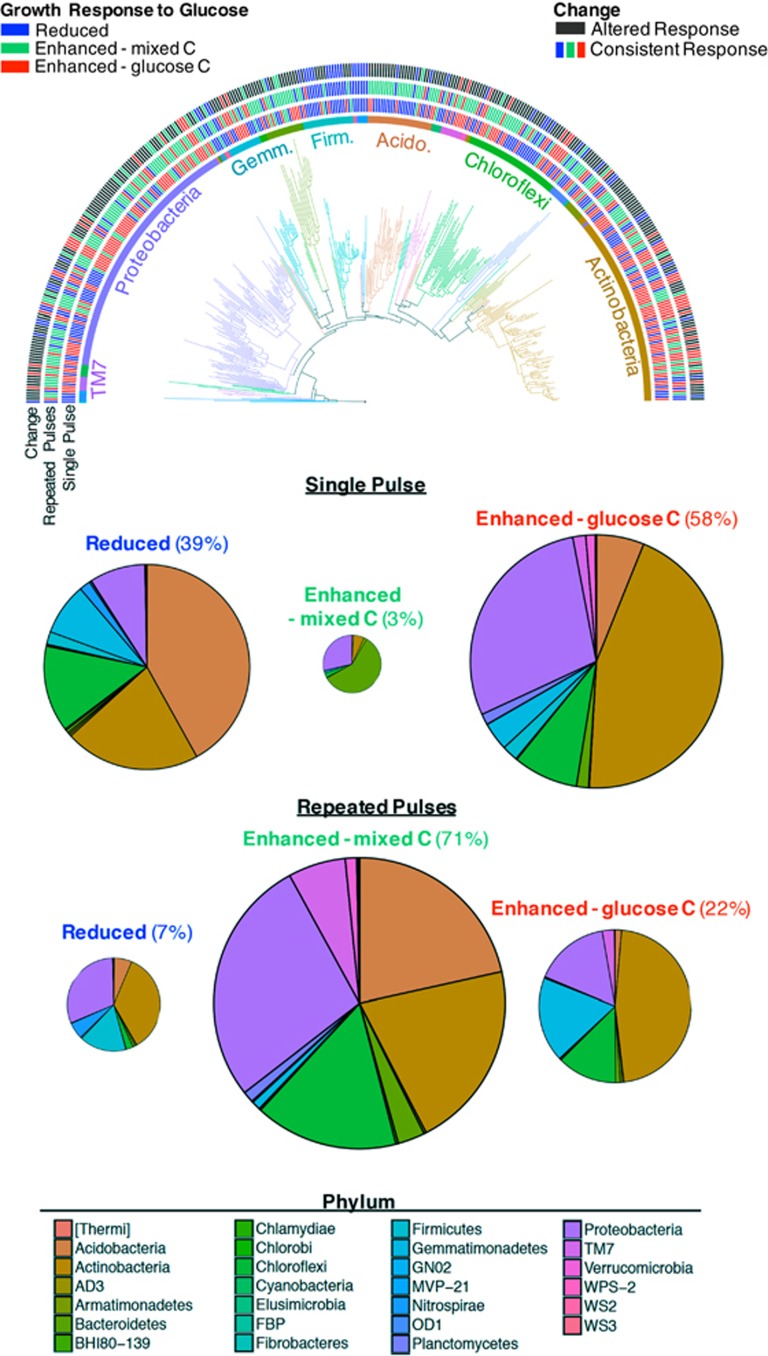
To better understand how individual taxa and phylogenetic groups gave rise to these community-level dynamics, we placed each taxon into one of three categories based on the change in growth rate in response to glucose addition (Δ18O) and the reliance on glucose as a C source for this extra growth (13C): (1) reduced: taxa that exhibited a reduction in growth rate (Δ18O<0), (2) enhanced—glucose C: taxa whose growth rates were enhanced with sufficient 13C consumption, such that there is no evidence of additional soil organic matter usage (Δ18O⩾0 and 13C/Δ18O ⩾2) and (3) enhanced—mixed C: taxa with enhanced growth rates that was at least partly because of use of SOC as a C source (Δ18O⩾0 and 13C/Δ18O<2). Reduced growth is associated with reduced SOC consumption (negative priming). Enhanced growth via the consumption of a mixture of the added labile C and SOC (enhanced—mixed C) indicates positive priming. For organisms that consumed glucose in excess of that necessary to explain their increase in growth (enhanced—glucose C), there are two possibilities: they could have caused no priming, if their glucose consumption was in addition to their normal SOC consumption; or they could have caused negative priming, if their glucose consumption substituted for some of their normal SOC consumption.
Few organisms enhanced their consumption of SOC (enhanced—mixed C) in response to a single pulse of glucose. Instead, most either reduced their growth or exhibited enhanced growth using the added glucose (Figures 2 and 3). However, after repeated glucose additions, the number of microbial taxa with enhanced growth based on a mix of glucose and SOC (enhanced—mixed C) increased substantially: 64% of bacterial taxa changed categories in response to multiple C pulses, with the majority of those (163 of 237, 69%) transitioning from reduced or enhanced—labile C to enhanced—mixed C (Figure 3), in other words, transitioning to an isotopic signature indicative of priming.
Figure 3.
Phylogeny of bacterial taxa and each taxon’s growth response to single and repeated pulses of glucose. Phyla are indicated by color and dominant groups are annotated. Growth responses during weeks 1 and 6, as well as the change in response between these weeks, are designated using color strips. Pie charts display the proportion (%) and phylum-level composition of 16S rRNA gene sequences in each response category.
Taxa involved in the positive priming of SOC (that is, enhanced—mixed C) were not phylogenetically clustered at any time (Table 1, Figure 3). Similarly, the widespread transition in response to repeated glucose addition was not phylogenetically constrained. This suggests that enhanced SOC utilization in response to sustained labile C addition (priming) is a prototypical bacterial activity in soil that does not require any specialized ecophysiological characteristics, but instead results from fundamental requirements for balanced microbial growth. It is perhaps not surprising that a diverse majority of microorganisms use both C sources given that glucose is considered a relatively universal substrate and soil organic matter consists of a variety of compounds. Our findings support a scenario where the initial pulse of glucose addition alleviates C limitation and is used directly for growth. During this period, other nutrients are likely to be quickly immobilized (Buchkowski et al., 2015; Sawada et al., 2015), which, in turn, triggers the production of extracellular enzymes (Blagodatskaya and Kuzyakov, 2008). These enzymes then gradually facilitate the priming of native soil C by liberating monomeric C as microbes ‘mine’ for other macro- and micronutrients, producing the positive priming response we observed after multiple pulses of glucose. Thus, our results substantiate microbe-based models developed to explain soil C cycling dynamics (Schimel and Weintraub, 2003; Kuzyakov, 2010) and are congruent with the nitrogen mining hypothesis (for example, Fontaine and Barot, 2005; Chen et al., 2014; Zhang et al., 2016).
Table 1. Phylogenetic clustering, assessed using the NRI, in the growth responses of bacteria to single and repeated pulses of glucose, as well as the consistency in response.
| NRI | P-value | |
|---|---|---|
| Single pulse | ||
| Reduced | 0.00 | 0.52 |
| Enhanced—mixed C | −1.21 | 0.89 |
| Enhanced—glucose C | 2.04 | 0.04* |
| Repeated pulses | ||
| Reduced | 1.88 | 0.03* |
| Enhanced—mixed C | −2.03 | 0.99 |
| Enhanced—glucose C | 2.21 | 0.01* |
| Change | ||
| Altered response | −2.43 | 0.99 |
| Consistent response | ||
| Reduced | 1.95 | 0.03* |
| Enhanced—mixed C | 0.17 | 0.58 |
| Enhanced—glucose C | 3.98 | <0.01* |
Abbreviations: C, carbon; NRI, net relatedness index.
*Significant, α=0.05.
However, these results stand in opposition to our hypothesis that microorganisms exhibit consistent and phylogenetically clustered patterns of C use reflective of their ecological strategy. Instead we found that many organisms switched from using glucose C to using a mixture of glucose and SOC for additional growth, indicating plasticity in their C use (Germida et al., 1998; Haei et al., 2011). Bacteria are known to regulate their growth and substrate use in culture (for example, Reizer et al., 1984; Kotte et al., 2010; Saier, 2014), but this has been difficult to observe directly in situ. However, nucleic acid labeling techniques have shown that many microbial taxa can use a variety of C substrates in their natural assemblages (Goldfarb et al., 2011; Mayali et al., 2014; Thomson et al., 2014). The metabolic flexibility observed in this study—where many microbial taxa can effectively compete for labile C while also having the capacity to utilize presumably more recalcitrant soil organic matter—provides insight into the ecology of these organisms. Specifically, it suggests that the majority of organisms in our soil cannot be neatly defined as copiotrophs specializing on labile C or as oligotrophs specializing on the more recalcitrant organic matter (sensu Fontaine et al., 2003). Instead these organisms are metabolically flexible enough to adjust their C use to suit the changing resource environment. Consequently, our results highlight the need to characterize the physiological flexibility of microorganisms in their natural environments in order to understand the functional relevance of microbial community composition.
As suggested above, our results contradict the hypothesis that one group of organisms consume the labile C (copiotrophic or r-strategist), and this activity indirectly stimulates a taxonomically distinct group of microorganisms (oligotrophic or K-strategist), that consume additional SOC (Fontaine et al., 2003; Blagodatskaya et al., 2007; Kuzyakov, 2010; Pascault et al., 2013). Rather, we found that most taxa whose growth was increased by glucose addition ended up consuming a mix of glucose and SOC (Figures 2b and 3), suggesting a direct stimulation of activity within individual taxa.
Although priming exhibited no phylogenetic clustering among the taxa involved, other responses to glucose addition were phylogenetically clustered (Table 1, Figure 3). After a single pulse of glucose, only the enhanced—glucose C group was clustered, with the majority of taxa in several broad taxonomic assemblages, including Actinobacteria (65% of member taxa), Verrucomicrobia (78%) and β-Proteobacteria (79%) exhibiting this response. Similarly, after repeated pulses of glucose, both the reduced and enhanced—glucose C groups were phylogenetically clustered. Many of the taxa that were reduced or enhanced using the glucose after multiple pulses also exhibited those responses initially (after a single pulse), suggesting a consistent response to glucose over the course of the experiment (Figure 3). A persistent reduction in growth upon glucose addition was seen in most Firmicutes (52%) and in all members of Nitrospirae. Enhanced growth using glucose was consistently observed within multiple Actinobacteria families, including Micromonosporaceae (57%), Nocardioidaceae (67%) and Streptomycetaceae (75%).
Phylogenetic clustering of organisms in the reduced and enhanced—glucose C groups suggests that these responses to labile C addition are brought about by physiological or ecological traits that are evolutionarily conserved. Reduced growth after labile C addition could reflect an oligotrophic ecological strategy, as low-resource-adapted organisms are expected to be outcompeted by copiotrophs in nutrient-rich environments (Fierer et al., 2007). This could underlie the reduced growth of Firmicutes observed in this experiment; these taxa were primarily within the class Bacilli, a diverse group containing both relatively copiotrophic and oligotrophic ‘species’ (Moreno-Letelier et al., 2011; Valvidia-Anistro et al., 2015). Within Bacilli, these growth strategies have been found to sort by phylogeny in cultured representatives (Mitsui et al., 2007). This phylogenetic clustering in growth response could arise from conserved physiological attributes, and indeed characteristics consistent with adaptation to a low-resource environment have been described in Bacilli spp., including small genome size (Moreno-Letelier et al., 2011), and the ability to maintain near-zero-specific growth rates (Overkamp et al., 2015). In the case of Nitrospirae, reduced growth may have resulted from glucose-induced reductions in nitrogen availability, which take place rapidly after labile C addition to soil, creating a nitrogen-scarce environment (Buchkowski et al., 2015; Sawada et al., 2015). Nitrospirae may be particularly sensitive to reductions in N availability as these organisms use nitrogen for both energy and biomass production.
The taxa that consistently enhanced their growth using only the added glucose (enhanced—glucose C) were also phylogenetically clustered (Table 1). These included Actinobacteria within the order Actinomycetales, previously shown to exhibit enhanced growth with labile C (Goldfarb et al., 2011), and favor rhizosphere environments (Smalla et al., 2001; DeAngelis et al., 2009). Phylogenetic clustering of organisms that prefer labile C could reflect a conserved genomic basis for opportunism within soil bacteria (Evans and Wallenstein, 2014) or sensitive catabolite repression of alternate C degrading metabolic pathways (Kremling et al., 2015).
Although we found agreement between the activity of bacterial taxa and the priming of soil C, this picture is likely incomplete as other soil heterotrophs, most notably fungi, were not examined. Similarly, because the current work only considered a single ecosystem and utilized experimental microcosms, additional research is necessary to determine whether our findings can be widely generalized to natural systems. However, using this technique, we were able to quantify taxon-specific activities in situ, and gain a clearer picture of how individual bacterial ‘species’ respond to C addition. In the soil, we assessed, positive priming was caused by the majority of soil bacteria, organisms that were not phylogenetically constrained, suggesting that the priming effect may not depend upon specialized phylogenetic groups. Our findings also suggest that many bacterial taxa can exhibit plasticity with regard to C use, changes in C use that could underlie the emergence of positive priming following the addition of labile C. Examining these dynamics in a broad range of soils could test whether this ubiquity of priming across bacteria biodiversity contributes to the widespread occurrence of positive priming in soil.
Acknowledgments
This research was supported by grants from the National Science Foundation (EAR-1124078 and DEB-1321792) and the Department of Energy's Biological Systems Science Division, Program in Genomic Science.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Ainsworth EA, Long SP. (2005). What have we learned from 15 years of free‐air CO2 enrichment (FACE)? A meta‐analytic review of the responses of photosynthesis, canopy properties and plant production to rising CO2. New Phytol 165: 351–372. [DOI] [PubMed] [Google Scholar]
- Blagodatskaya E, Kuzyakov Y. (2008). Mechanisms of real and apparent priming effects and their dependence on soil microbial biomass and community structure: critical review. Biol Fert Soils 45: 115–131. [Google Scholar]
- Blagodatskaya E, Kuzyakov Y. (2013). Active microorganisms in soil: critical review of estimation criteria and approaches. Soil Biol Biochem 67: 192–211. [Google Scholar]
- Blagodatskaya EV, Blagodatsky SA, Anderson TH, Kuzyakov Y. (2007). Priming effects in chernozem induced by glucose and N in relation to microbial growth strategies. Appl Soil Ecol 37: 95–105. [Google Scholar]
- Blankinship JC, Brown JR, Dijkstra P, Allwright MC, Hungate BA. (2010). Response of terrestrial CH4 uptake to interactive changes in precipitation and temperature along a climatic gradient. Ecosystems 13: 1157–1170. [Google Scholar]
- Buchkowski RW, Schmitz OJ, Bradford MA. (2015). Microbial stoichiometry overrides biomass as a regulator of soil carbon and nitrogen cycling. Ecology 96: 1139–1149. [DOI] [PubMed] [Google Scholar]
- Cadotte MW, Cavender-Bares J, Tilman D, Oakley TH. (2009). Using phylogenetic, functional and trait diversity to understand patterns of plant community productivity. PLoS One 4: e5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Senbayram M, Blagodatsky S, Myachina O, Dittert K, Lin X et al. (2014). Soil C and N availability determine the priming effect: microbial N mining and stoichiometric decomposition theories. Glob Change Biol 20: 2356–2367. [DOI] [PubMed] [Google Scholar]
- Cleveland CC, Nemergut DR, Schmidt SK, Townsend AR. (2007). Increases in soil respiration following labile carbon additions linked to rapid shifts in soil microbial community composition. Biogeochemistry 82: 229–240. [Google Scholar]
- DeAngelis KM, Brodie EL, DeSantis TZ, Anderson GL, Lindow SE, Firestone MK. (2009). Selective progressive response of soil microbial community to wild oat roots. ISME J 3: 168–178. [DOI] [PubMed] [Google Scholar]
- Dijkstra P, Ishizu A, Doucett R, Hart SC, Schwartz E, Menyailo OV et al. (2006). 13C and 15N natural abundance of the soil microbial biomass. Soil Biol Biochem 38: 3257–3266. [Google Scholar]
- Evans SE, Wallenstein MD. (2014). Climate change alters ecological strategies of soil bacteria. Ecol Lett 17: 155–164. [DOI] [PubMed] [Google Scholar]
- Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM et al. (2014). An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Bradford MA, Jackson RB. (2007). Toward an ecological classification of soil bacteria. Ecology 88: 1354–1364. [DOI] [PubMed] [Google Scholar]
- Fontaine S, Barot S. (2005). Size and functional diversity of microbe populations control plant persistence and long‐term soil carbon accumulation. Ecol Lett 8: 1075–1087. [Google Scholar]
- Fontaine S, Mariotti A, Abbadie L. (2003). The priming effect of organic matter: a question of microbial competition? Soil Biol Biochem 35: 837–843. [Google Scholar]
- Germida JJ, Siciliano SD, Seib AM. (1998). Phenotypic plasticity of Pseudomonas aureofaciens (lacZY) introduced into and recovered from field and laboratory microcosm soils. FEMS Microbiol Ecol 27: 133–139. [Google Scholar]
- Goldfarb KC, Karoz U, Hanson CA, Santee CA, Bradford MA, Treseder KK et al. (2011). Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Front Microbiol 2: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haei M, Rousk J, Ilstedt U, Öquist M, Bååth E, Laudon H. (2011). Effects of soil frost on growth, composition and respiration of the soil microbial decomposer community. Soil Biol Biochem 43: 2069–2077. [Google Scholar]
- Hendricks DM. (1985) Arizona Soils. In: Haney RA (eds). College of Agriculture, University of Arizona: Tucson, AZ.
- Hungate BA, Mau RL, Schwartz E, Caporaso JG, Dijkstra P, van Gestel N et al. (2015). Quantitative microbial ecology through stable isotope probing. Appl and Environ Microb 81: 7570–7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins SN, Rushton SP, Lanyon CV, Whiteley AS, Waite IS, Brookes PC et al. (2010). Taxon-specific responses of soil bacteria to the addition of low level C inputs. Soil Biol Biochem 42: 1624–1631. [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD et al. (2011). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26: 1463–1464. [DOI] [PubMed] [Google Scholar]
- Kotte O, Zaugg JB, Heinemann M. (2010). Bacterial adaptation through distributed sensing of metabolic fluxes. Mol Syst Biol 6: 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremling A, Geiselmann J, Ropers D, De Jong H. (2015). Understanding carbon catabolite repression in Escherichia coli using quantitative models. Trends Microbial 23: 99–109. [DOI] [PubMed] [Google Scholar]
- Kuzyakov Y, Bol R. (2006). Sources and mechanisms of priming effect induced in two grassland soils amended with slurry and sugar. Soil Biol Biochem 38: 747–758. [Google Scholar]
- Kuzyakov Y. (2010). Priming effects: interactions between living and dead organic matter. Soil Biol Biochem 42: 1363–1371. [Google Scholar]
- Lennon JT, Jones SE. (2011). Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol 9: 119–130. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. (2011). Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39: W475–W478 gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, Aziz M, Kachur S, Hsueh P-R, Huang Y-T, Keim P et al. (2012). BactQuant: an enhanced broad-coverage bacterial quantitative realtime PCR assay. BMC Microbiol 12: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XJA, Sun J, Mau RL, Finley BK, Compson ZG, van Gestel N et al. (2017). Labile carbon input determines the direction and magnitude of the priming effect. Appl Soil Ecol 109: 7–13. [Google Scholar]
- Luo Z, Wang E, Sun OJ. (2016). A meta-analysis of the temporal dynamics of priming soil carbon decomposition by fresh carbon inputs across ecosystems. Soil Biol Biochem 101: 96–103. [Google Scholar]
- Mau RL, Liu CM, Aziz M, Schwartz E, Dijkstra P, Marks JC et al. (2015). Linking soil bacterial biodiversity and soil carbon stability. ISME J 9: 1477–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayali X, Weber PK, Mabery S, Pett-Ridge J. (2014). Phylogenetic patterns in the microbial response to resource availability: amino acid incorporation in San Francisco Bay. PloS One 9: e95842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M et al. (2008). The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform 9: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui H, Gorlach K, Lee HJ, Hattori R, Hattori T. (2007). Incubation time and media requirements of culturable bacteria from different phylogenetic groups. J Microbiol Meth 30: 103–110. [Google Scholar]
- Moreno-Letelier A, Olemdo G, Eguiarte LE, Martinez-Castilla L, Souza V. (2011). Parallel evolution and horizontal gene transfer of the pst operon in Firmicutes from oligotrophic environments. Int J Evol Biol 2011: 781642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey EM, McHugh TA, Preteska L, Hayer M, Dijkstra P, Hungate BA et al. (2015). Dynamics of extracellular DNA decomposition and bacterial community composition in soil. Soil Biol Biochem 86: 42–49. [Google Scholar]
- Morrissey EM, Franklin RB. (2015). Evolutionary history influences the salinity preference of bacterial taxa in wetland soils. Front Microbiol 6: 1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey EM, Mau RL, Schwartz E, Caparosa JG, Dijkstra P, van Gestel N et al. (2016). Phylogenetic organization of bacteria activity. ISME J 10: 2336–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overkamp W, Ercan O, Herber M, Maris AJ, Kleerebezum M, Kuipers OP. (2015). Physiological and cell morphology adaptation of Bacillus subtilis at near‐zero specific growth rates: a transcriptome analysis. Environ Microbiol 17: 346–363. [DOI] [PubMed] [Google Scholar]
- Pascault N, Ranjard L, Kaisermann A, Bachar D, Christen R, Terrat S et al. (2013). Stimulation of different functional groups of bacteria by various plant residues as a driver of soil priming effect. Ecosystems 16: 810–822. [Google Scholar]
- Philippot L, Bru D, Saby N, Čuhel J, Arrouays D, Šimek M et al. (2009). Spatial patterns of bacterial taxa in nature reflect ecological traits of deep branches of the 16S rRNA bacterial tree. Environ Microbiol 11: 3096–3104. [DOI] [PubMed] [Google Scholar]
- Phillips RP, Finzi AC, Bernhardt ES. (2011). Enhanced root exudation induces microbial feedbacks to N cycling in a pine forest under long‐term CO2 fumigation. Ecol Lett 14: 187–194. [DOI] [PubMed] [Google Scholar]
- Poff NL, Olden JD, Vieira NK, Finn DS, Simmons MP, Kondratieff BC. (2006). Functional trait niches of North American lotic insects: traits-based ecological applications in light of phylogenetic relationships. J N Am Benthol Soc 25: 730–755. [Google Scholar]
- Reizer JO, Novotny MJ, Stuiver IN, Saier MH. (1984). Regulation of glycerol uptake by the phosphoenolpyruvate-sugar phosphotransferase system in Bacillus subtilis. J Bacteriol 159: 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH. (2014) Mechanisms and Regulation of Carbohydrate Transport in Bacteria. Academic Press Inc.: Orlando, FL, USA. [Google Scholar]
- Sawada K, Funakawa S, Toyota K, Kosaki T. (2015). Potential nitrogen immobilization as influenced by available carbon in Japanese arable and forest soils. Soil Sci Plant Nutr 61: 917–926. [Google Scholar]
- Scharlemann JP, Tanner EV, Hiederer R, Kapos V. (2014). Global soil carbon: understanding and managing the largest terrestrial carbon pool. Carbon Manage 5: 81–91. [Google Scholar]
- Schimel JP, Weintraub MN. (2003). The implications of exoenzyme activity on microbial carbon and nitrogen limitation in soil: a theoretical model. Soil Biol Biochem 35: 549–563. [Google Scholar]
- Schwartz E. (2007). Characterization of growing microorganisms in soil by stable isotope probing with H218O. Appl Environ Microbiol 73: 2541–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S et al. (2001). Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl Environ Microbiol 67: 4742–4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson BC, Ostle NJ, McNamara NP, Oakley S, Whiteley AS, Bailey MJ et al. (2014). Plant soil interactions alter carbon cycling in an upland grassland soil. Front Microbiol 4: 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvidia-Anistro JA, Eguiarte-Fruns LE, Delgado-Sapién G, Márquez-Zacarías P, Gasca-Pineda J, Learned J et al. (2015). Variability of rRNA operon copy number and growth rate dynamics of Bacillus isolated from an extremely oligotrophic aquatic ecosystem. Front Microbiol 6: 1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Groenigen KJ, Qi X, Osenberg CW, Luo Y, Hungate BA. (2014). Faster decomposition under increased atmospheric CO2 limits soil carbon storage. Science 344: 508–509. [DOI] [PubMed] [Google Scholar]
- Wang H, Boutton TW, Xu W, Hu G, Jiang P, Bai E. (2015). Quality of fresh organic matter affects priming of soil organic matter and substrate utilization patterns of microbes. Sci Rep 5: 10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. (2002). Phylogenies and community ecology. Annu Rev Ecol Syst 33: 475–505. [Google Scholar]
- Will C, Thürmer A, Wollherr A, Nacke H, Herold N, Schrumpf M et al. (2010). Horizon-specific bacterial community composition of German grassland soils, as revealed by pyrosequencing-based analysis of 16S rRNA genes. Appl Environ Microbiol 76: 6751–6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Wheeler E, Phillips RP. (2014). Root-induced changes in nutrient cycling in forests depend on exudation rates. Soil Biol Biochem 78: 213–221. [Google Scholar]
- Zhang W, Chao L, Yang Q, Wang Q, Fang Y, Wang S. (2016). Litter quality mediated nitrogen effect on plant litter decomposition regardless of soil fauna presence. Ecology 97: 2834–2843. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.