Abstract
Soil organic matter (SOM) stocks contain nearly three times as much carbon (C) as the atmosphere and changes in soil C stocks may have a major impact on future atmospheric carbon dioxide concentrations and climate. Over the past two decades, much research has been devoted to examining the influence of warming on SOM decomposition in topsoil. Most SOM, however, is old and stored in subsoil. The fate of subsoil SOM under future warming remains highly uncertain. Here, by combining a long-term field warming experiment and a meta-analysis study, we showed that warming significantly increased SOM decomposition in subsoil. We also showed that a decade of warming promoted decomposition of subsoil SOM with turnover times of decades to millennia in a tall grass prairie and this effect was largely associated with shifts in the functional gene structure of microbial communities. By coupling stable isotope probing with metagenomics, we found that microbial communities in warmed soils possessed a higher relative abundance of key functional genes involved in the degradation of organic materials with varying recalcitrance than those in control soils. These findings suggest warming may considerably alter the stability of the vast pool of old SOM in subsoil, contributing to the long-term positive feedback between the C cycle and climate.
Introduction
A major uncertainty in projecting future atmospheric carbon dioxide (CO2) concentrations and climate is how warming influences decomposition of SOM (Davidson and Janssens, 2006; Stocker et al., 2013). Considerable efforts have been made to assess the effects of warming on SOM decomposition in the surface soil layer (that is, topsoil, 0–20 cm deep) (Davidson and Janssens, 2006; von Lutzow and Kogel-Knabner, 2009; Conant et al., 2011; Crowther et al., 2016). Soil organic C (SOC) in topsoil accounts for one quarter of the global SOC pool and is relatively young, with typical turnover times of decades or less (Hopkins et al., 2012). Topsoil SOM has been deemed to be vulnerable to microbial decomposition under warming (Yergeau et al., 2012; Karhu et al., 2014; Crowther et al., 2016), and hence may have a critical impact on climate (Davidson and Janssens, 2006; Conant et al., 2011; Hopkins et al., 2012). Most SOC is stored in deeper soils (0.2–3 m) (Jobbagy and Jackson, 2000), of which the majority of C is considerably older, with turnover times of centuries to millennia (Trumbore et al., 1996; Rumpel et al., 2002; Fontaine et al., 2007). A significant and long-term feedback to climate would also occur if ongoing and projected warming could considerably alter decomposition of SOM in the subsoil layers (those below the surface soil) of global soils. Yet, the influence of warming on the stability of SOM in subsoil has rarely been examined directly.
It has been traditionally thought that the long residence time of SOM in subsoil is due mainly to the chemical recalcitrance of organic compounds themselves (Rumpel et al., 2002; Davidson and Janssens, 2006; Fontaine et al., 2007; von Lutzow and Kogel-Knabner, 2009), although a recent view has ascribed it to biotic and abiotic protection (Schmidt et al., 2011). Kinetic theory predicts that old SOM, if more chemically recalcitrant, would have higher sensitivities to temperature changes in comparison with young SOM (Knorr et al., 2005; Davidson and Janssens, 2006). But the temperature sensitivity of recalcitrant SOM decomposition has been documented to be either higher than (Knorr et al., 2005; Hakkenberg et al., 2008; Hartley and Ineson, 2008; Craine et al., 2010; Hopkins et al., 2012), similar to or lower than labile SOM (Liski et al., 1999; Giardina and Ryan, 2000; Melillo et al., 2002; Fang et al., 2005; Reichstein et al., 2005), suggesting that other mechanisms through which warming influences the stability of SOM in subsoil have not been fully characterized. The majority of previous studies were short-term soil incubations conducted in the laboratory, which may not accurately represent field conditions. Moreover, most previous work has focused on the relationships between temperature and CO2 emissions (Luo et al., 2001; Zhou et al., 2007; Mahecha et al., 2010; Carey et al., 2016), but did not investigate the microbial mechanisms of SOM decomposition under warming.
Here, we carried out a long-term field experiment and a comprehensive meta-analysis to ascertain the direction and magnitude of the warming effects on subsoil SOM decomposition under field conditions. We then conducted a stable isotope probing (SIP) microcosm experiment, in combination with metagenomic analyses, to elucidate the possible biological underpinnings of the warming effect on SOM decomposition. We hypothesized that experimental warming would (i) increase SOM decomposition in subsoil, and (ii) alter the structure and/or functions of microbial communities in subsoil.
Materials and methods
Field warming experiment
We initiated a long-term field experiment to investigate the responses of a tall grass prairie ecosystem to warming in central Oklahoma, USA (34° 58' 54'' N, 97° 31' 14'' W). This long-term experimental facility was established in November 1999, with a paired, nested factorial design with warming as the main factor (n=6) and clipping as the nested sub-factor. A single infrared heater (Supplementary Figure S1a) was suspended 1.5 m above the ground to elevate soil temperature (Luo et al., 2001; Zhou et al., 2007; Xu et al., 2015). A deep soil collar (a PVC tube with 10 cm in diameter × 70 cm in depth) was installed into an unclipped subplot of each main treatment plot in October, 2001 (Supplementary Figure S1c). Soil samples were taken from the soil profile of 0–25 cm in the deep collar by the end of 11th year growing season using a 5-cm diameter soil corer. Soil C concentrations were determined with a CHN elemental analyzer (Carla Erba and model 2400, Perkin Elmer Co., Norwalk, CT, USA). The 14C contents of SOM were analyzed at the University of Arizona NSF-AMS facility (Donahue, 1995). Detailed descriptions of the experimental site, deep collars, soil sampling and measurements (for example, soil temperature, soil CO2 efflux, soil organic C and radiocarbon analyses) are provided in Supplementary Appendix S1 section 1.
Carbon pool partitioning
We used the mass balance, in combination with the Arrhenius equation and Michaelis–Menten kinetics (Davidson and Janssens, 2006), to develop a model to estimate the relative contribution of different C age pools in decomposed SOM under warming. Detailed descriptions of the theory, model parameterization and mathematical calculations are included in Supplementary Appendix S1 section 2.
Meta-analysis study
A meta-analysis study of the warming effect on SOC changes in the soil profile was conducted using previously published data. We used Web of Science for a thorough search of relevant peer-reviewed articles published before November 2015. We only selected published studies that included the changes of SOC in different soil layers under warming in field. The effect size of warming for each individual observation was estimated by transforming the response ratio (R) with the natural log: lnR=ln 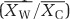 , where
, where 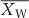 is the mean for warming treatment,
is the mean for warming treatment, 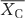 is the mean for ambient treatment. Detailed information for each of selected studies is shown in the Supplementary Appendix S2. Descriptions of data extraction and meta-analysis are provided in Supplementary Appendix S1 section 3.
is the mean for ambient treatment. Detailed information for each of selected studies is shown in the Supplementary Appendix S2. Descriptions of data extraction and meta-analysis are provided in Supplementary Appendix S1 section 3.
Stable isotope probing microcosm experiment
We designed a SIP microcosm experiment to ascertain the effects of warming on microbial communities and decomposition. Soils taken from the 20–25 cm soil layer of the deep collar of each plot were split into two 20-g dry mass equivalent aliquots, adjusted to the moisture level of 60% water holding capacity, and then placed in 165-ml jars for laboratory incubation. One jar was treated as the sub-control, the other was added with the uniformly (U) 13C-labeled shoot biomass of Triticum aestivum (ground, 13C abundance > 97%, IsoLife BV, Wageningen, The Netherlands) with a mass ratio of 150:1 (soil: 13C plant material). We used the gradient fractionation method to separate 13C-DNA from 12C-DNA based on a standard protocol (Neufeld et al., 2007). Detailed descriptions of headspace gas sampling, soil DNA extraction, 13C-DNA separation, measurements and calculations are shown in Supplementary Appendix S1 Section 4.
16S rRNA gene sequencing and GeoChip analyses
Pyrosequencing of PCR amplicons that targeted the V4–V8 hypervariable regions (positions 515–1391) of 16S ribosomal RNA gene (rRNA) was performed using a 454 Life Sciences Genome Sequencer FLX titanium instrument (454 Life Sciences, Brandford, CT, USA). Detailed descriptions of amplicon preparations, sequencing and data preprocessing are provided in Supplementary Appendix S1 Section 4.
GeoChip (version 4.0) is a functional gene-array-based high-throughput technology designed for profiling the functional structure, diversity, metabolic potential/activity and dynamics of microbial communities (Tu et al., 2014). GeoChip 4.0 contained a total of 82 047 probes targeting numerous functional genes important to microbial processes. Among all probes, 3% of them targeted 188 archaeal strains, 89% for 4332 bacterial strains, 6% for 420 eukaryotic (mainly fungal) strains, and the remaining for other organisms. GeoChip 4.0 included 8749 probes targeting 33 genes involved in organic compound degradation. Sequence retrieval, probe design, microarray construction and imaging were described previously (Tu et al., 2014). Details for DNA labeling, hybridization, image processing and GeoChip data pre-processing are provided in Supplementary Appendix S1 Section 4.
Data analyses
We employed three different but complementary non-parametric multivariate statistical analysis methods to test statistical differences of microbial communities between warming and control treatments (Legendre and Legendre, 2012; Zhou et al., 2012). The three methods were permutational multivariate analysis of variance using distance matrices (Adonis), analysis of similarities (ANOSIM) and multi-response permutation procedure (MRPP). We also used detrended correspondence analysis (DCA) to assess the similarity of two microbial communities based on 454 pyrosequencing and GeoChip data sets (Legendre and Legendre, 2012). A phylogeny-based metric, called UniFrac (Lozupone and Knight, 2005), was employed to examine changes in the taxonomic structure of microbial communities based on 16S rRNA pyrosequencing data sets. Results from the UniFrac-based principal coordinate analysis (PCA) (data were not shown) were very similar to those generated from DCA, we thus adopted DCA throughout the text. All pyrosequencing data were analyzed based on biological replicates. Detailed analysis of the pyrosequencing data with three technical replicates is beyond the scope of the present work and will be presented in another paper. Data on gas concentrations, SOC content and soil C age in field were analyzed using ANOVA mixed model (Littell et al., 1996). Data on CO2 respiration with repeated measurements either in laboratory or field were analyzed using repeated measures ANOVA. To reduce autocorrelation between repeated measurements, we also included three covariance structures compound symmetric model (CS), the first-order autoregressive mode (AR(1)) and autoregressive with heterogeneous variance in repeated measures ANOVA. Statistical results from repeated measures ANOVA were reported based on the covariance structure that minimized Akaike’s information criterion (AIC) and Bayesian information criterion (BIC) (Littell et al., 1996; Cheng et al., 2011). P-values for the relative abundance of taxa and the signal intensity of functional genes were calculated using paired two-tailed t tests. DCA, Adonis, ANOSIM and MRPP were analyzed using R with the package ‘vegan’ (Oksanen et al., 2007). All statistical analyses were performed using the R software (version 3.1.0, The R Foundation for Statistical Computing, Vienna, Austria). For all tests, P⩽0.05 was considered a statistically significant difference.
Results and discussion
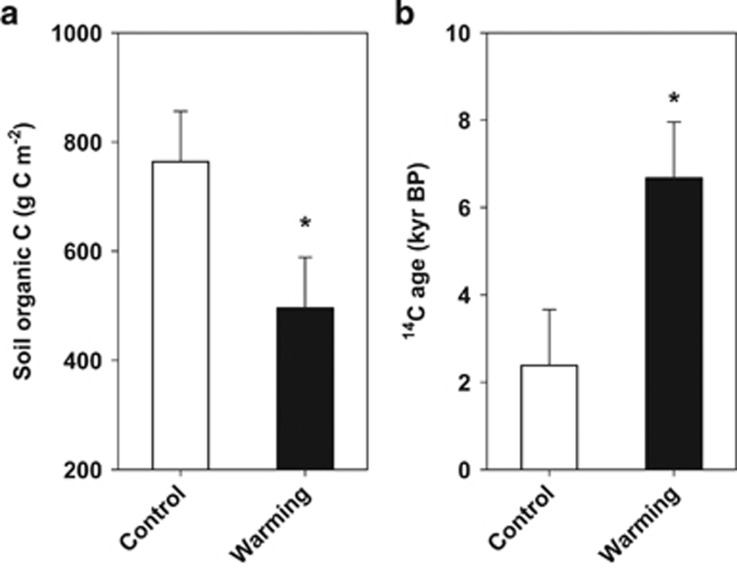
We first investigated the influence of long-term experimental warming on soil C dynamics in the Oklahoma field plots. Warming significantly enhanced soil CO2 efflux from whole plots with an annual mean increase of 126.8 g C m−2 per year during the eleven-year experimental period (Xu et al., 2015). To exclude the impact of new plant-derived C inputs on SOM decomposition, we installed a deep collar into the unclipped subplot of each plot in October 2001 (Supplementary Figures S1b and c). Warming increased soil temperature by ~2 °C at the 5 cm soil depth over a decade of experimental duration (Zhou et al., 2007; Xu et al., 2015), and the magnitude of the heating effects at 25 cm soil depth was comparable to that of 5 cm soil depth adjacent to the deep collars (Supplementary Figure S2). By the end of the eleventh growing season, we determined the changes of SOC in the soil profile within the deep collars. The net loss of soil C contents by warming was 598±69 (s.e.m.) g C m−2 in the 0–20 cm topsoil layer within the deep collars (P<0.05). Warming also significantly reduced soil C contents in the 20–25 cm subsoil layer within the deep collars (P=0.03; Figure 1a). Warming-induced soil C losses may be attributed to alterations in both biotic (for example, microbial respiration) and abiotic factors (for example, leaching, freeze-thaw events) (Davidson and Janssens, 2006). It has been shown that warming could considerably enhance C losses through dissolved organic C leaching (Harrison et al., 2008; Luo et al., 2009) and freeze-thaw (Schuur et al., 2009; Chapin et al., 2011). In combination with our previous measurements (Zhou et al., 2007), we estimated that warming led to an increase of 73±21 (s.e.m.) g C m−2 per year in annual mean soil CO2 efflux from the deep collars (heterotrophic respiration) during the experimental years of 2000–2010. These results indicated that microbial respiration was the dominant pathway of C losses from the soil profile within deep collars.
Figure 1.
Warming enhances old soil organic matter (SOM) decomposition in subsoil within deep collars in a tall grass prairie over an 11-year experiment in the Oklahoma site. (a) Total amount of soil organic carbon (SOC). (b) Radiocarbon (14C) age of SOM. The 14C age of SOM is expressed as a unit of one thousand years Before Present (Kyr BP, relative to 1950). Data shown (means±s.e.m.; n=6) are based on the fitted mixed model. * (P⩽0.05) denotes statistically significant effects of warming.
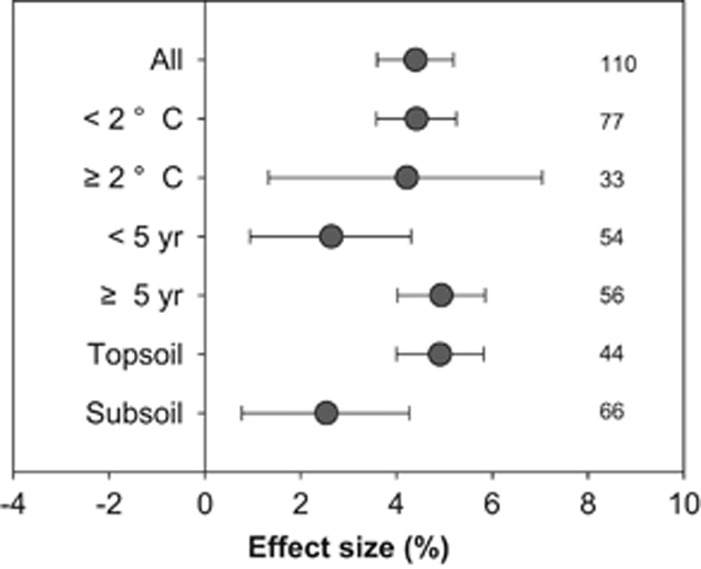
To determine whether the effects of warming on SOM decomposition noted in our Oklahoma prairie grassland were similar to those of previous studies, we conducted a meta-analysis of 14 studies that quantified the changes of SOC across the soil profile in field warming experiments (Supplementary Appendix S1 section 3). Those studies were primarily carried out in the northern hemisphere (Supplementary Figure S3) encompassing grassland, forest, and arctic tundra ecosystems. Warming significantly reduced soil C contents across the soil profile (Figure 2). In most soil layers, temperature elevations by warming were in a range of 1–3 °C (Supplementary Appendix S2). The magnitude of warming effects on soil C reductions (effect size) was comparable between soil layers with temperature elevations < 2 °C and those ⩾ 2 °C (Figure 2). The experimental duration of studies included in our meta-analysis ranged from 1 to 16 years. The effect size for studies with longer experimental time (⩾ 5 yr) was slightly larger than that of short-term experiments (< 5 year; Figure 2). Warming also reduced soil C contents in both top- an subsoil layers, though the effect size for subsoil layers was slightly less than that of topsoil layers (Figure 2). Nevertheless, these results suggested that SOM in the soil profile, even within subsoil layers, was vulnerable to decomposition under experimental warming.
Figure 2.
Meta-analysis of warming effects on SOC changes along the soil depth profile in field studies across the globe. The numbers of observations for all independent observations (‘All’, n=110) and other categories are shown in the figure. Observations were grouped into temperature elevations (< 2 vs ⩾2 °C), experimental durations (< 5 vs ⩾ 5 years) and different soil layers (topsoil vs subsoil). Data obtained on the mean effect sizes 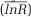 were reported as percentage changes under warming (that is, 100 × (|R-1|)). The warming effect was deemed significant if the 95% confidence intervals did not overlap with 0.
were reported as percentage changes under warming (that is, 100 × (|R-1|)). The warming effect was deemed significant if the 95% confidence intervals did not overlap with 0.
We then examined whether warming influenced old SOM (refer to the average age ⩾ 50 year) decomposition in the subsoil layer of 20–25 cm within the deep collars in the Oklahoma soil plots. We chose to focus solely on this layer for three reasons. First, the sampling design offered a compromise between our current (sampling depth) and future (CO2 efflux monitoring) research objectives. It allowed us a direct, in situ observation on subsoil SOM decomposition with minimized disturbance to the unique experimental unit in this long-term field warming platform. Second, the radiocarbon (14C) contents varied substantially across the soil profile (Rumpel et al., 2002; Fontaine et al., 2007; Jenkinson et al., 2008). Such a short subsoil layer thus helped to minimize the potentially confounding effects of soil profile heterogeneity (Rumpel et al., 2002) and new plant-derived C inputs (Fontaine et al., 2007) on radiocarbon analyses. Third, it was suitable for studying the microbial mechanism of C cycling under climate change since the short subsoil layer had a low spatial and temporal variation in the microbial community structure (Griffiths et al., 2003; Mackelprang et al., 2011). Griffiths et al. studied the influences of depth and sampling time on the bacterial community structure in a grassland and found that microbial communities were relatively stable with less seasonal variation in a 5-cm subsoil layer (for example, 15–20 cm) compared to surface layers (Griffiths et al., 2003). In the present study, we showed that the bulk 14C age of SOM from the subsoil layer of 20–25 cm within the deep collars was several thousand years old (Figure 1b). Most notably, the average 14C age under warming was nearly two times larger than that of control (P=0.02; Figure 1b).
It has been suggested that changes in the mean 14C ages of bulk SOM result from changes in the 14C contents of SOM pools with different ages (Trumbore, 2009). To examine whether the warming-induced older SOM (Figure 1b) was a consequence of larger losses of millennia-old and/or younger SOM pools, we developed a four-C-age-pool model to calculate changes in decadal, centennial and millennial SOM pools in the subsoil of 20–25 cm layer in the Oklahoma soil plots (see Supplementary Appendix S1 section 2). We estimated that warming led to an increase in the losses of decadal, centennial and millennial soil C contents by a range of 26–30%, 11–33% and 0–36%, respectively, compared to control (Supplementary Figure S4 and Supplementary Table S2). Together, these results revealed that decadal, centennial and even millennial-old SOM pools in the subsoil layer were susceptible to microbial decomposition under a decade of experimental warming in this grassland.
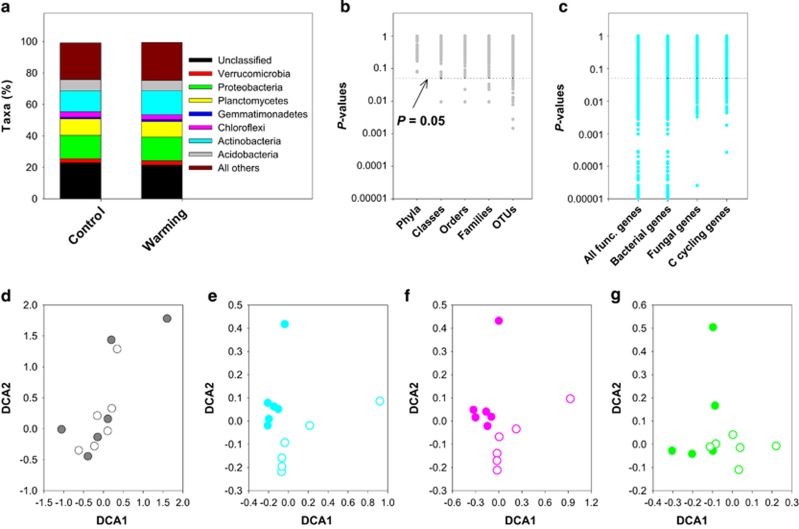
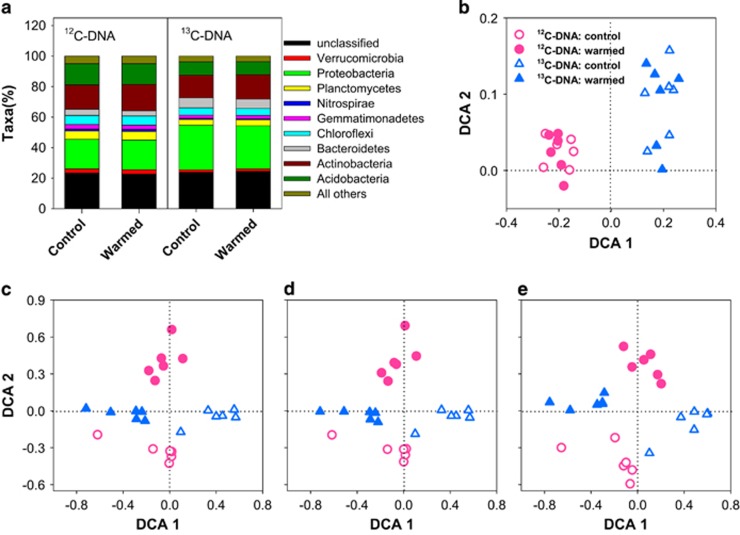
To investigate whether the warming-induced increase in old SOM decomposition observed in-situ (Figure 1b) resulted primarily from alterations in the structure of microbial communities, we determined the taxonomic structure of microbial communities in the 20–25 cm subsoil layer using a multiplexed barcoded pyrosequencing approach (Hamady et al., 2008). The 16S rRNA gene with three barcoded tags was amplified from genomic DNA of each field soil sample. Preprocessed DNA sequences (Supplementary Appendix S1 section 4) were grouped into 97%-identical operational taxonomic units (OTUs). Bacterial OTUs accounted for 99.3% of the total number of detected OTUs. Significance tests from Adonis, ANOSIM and MRPP analyses revealed that warming did not alter the taxonomic structure of bacterial communities (P>0.05; Supplementary Table S1). The relative abundances of bacterial phyla also remained unchanged under warming (Figure 3a), and this pattern maintained at lower taxonomic levels (for example, class, order, family or OTU; Figure 3b). We then used DCA to assess the similarity of bacterial communities among soil plots. DCA ordination revealed that bacterial communities under warming were not separated well from those under control (Figure 3d).
Figure 3.
Effects of warming on taxonomic and functional profiling of microbial communities in subsoil in the Oklahoma site. (a), Relative abundances of dominant phyla (>1% 16S rRNA gene data sets). (b), P-values of warming effects on the relative taxon abundances at different taxonomic levels (16S rRNA gene). (c), P-values of warming effects on all, bacterial, fungal and C cycling functional genes (gene profiles of GeoChip). Data points below the horizontal line in (b) and (c) indicate a statistically significant effect of warming (P⩽0.05). (d–g), DCA ordination of the structure of microbial communities: taxonomic profiles of bacterial communities based on 16S rRNA genes (d), functional profiles of microbial communities based on all functional genes in GeoChip (e), functional profiles of bacterial (f) and fungal (g) communities based on GeoChip. Warming, filled circles. Control, unfilled circles.
Because decomposition is achieved by a diverse repertoire of enzymatic reactions performed by microorganisms (Chapin et al., 2011; Cheng et al., 2012), we reasoned that the warming-induced enhancement in old SOM decomposition may be a consequence of changes in microbial community functions. We employed a microarray-based high-throughput technology, referred to as GeoChip (Zhou et al., 2012; Tu et al., 2014), to assess the functional gene capabilities of microbial communities. The GeoChip (version 4.0) contained 82 074 probes from 410 gene families involved in microbial functional processes such as biogeochemical cycling and environmental stress response, presenting the unique opportunity of detecting functional patterns at the community level. We detected a total of 49 857 gene targets, 85 and 10% of which were targeted, respectively, for bacterial and fungal strains. In contrast to the 16S rRNA gene-based taxonomic composition of bacterial communities, the functional structure of microbial communities were significantly different between warmed and control soils as revealed by Adonis, ANOSIM and MRPP analyses (P<0.05 for all; Supplementary Table S1). In addition, DCA revealed distinct clustering of functional gene profiles of both whole microbial communities (Figure 3e) and bacterial communities (Figure 3f) under warming compared to control.
Compared to the functional structure, the lack of warming effects on the taxonomic composition of bacterial communities could be due to the differences of metagenomic technologies in resolution, sensitivity and/or susceptibility to random sampling artifacts (Zhou et al., 2015). At the OTU level (the lowest taxonomic level, 16S rRNA gene), the relative abundances of only 0.4% of OTUs were altered under warming (P⩽0.05, Figure 3b). At the gene target level (species-strain level, the highest resolution in GeoChip), however, warming significantly changed the signal intensities of 16.2% of all functional genes and 15.6% of bacterial functional genes (P⩽0.05, Figure 3c). Besides, it may be attributed to the phenomenon that functional variation can be independent of taxonomic variation in microbial communities (Shi et al., 2011; Louca et al., 2016). Using a global ocean microbiome dataset (16S rRNA gene), Louca et al. (2016) showed that environmental variables including temperature shaped functional groups, rather than the taxonomic composition within individual functional groups, of bacterial communities. In the present study, we found that warming resulted in alterations in functional gene potentials rather than specific taxonomic groups within bacterial communities (Figure 3). For instance, we found that warming significantly increased the signal intensities of 20.4% of all C cycling genes (P⩽0.05, Figure 3c) and 19.6% of C cycling genes targeting for bacteria (P⩽0.05), suggesting that warming-induced changes in microbial functions occurred at the community level. Though decoupling of function from taxonomy observed in the current and previous (Shi et al., 2011; Louca et al., 2016) studies warranted further investigations, our data did show that warming shifted the functional structure of bacterial communities in subsoil.
Like bacteria, fungi also play a critical role in decomposition (Chapin et al., 2011; Cheng et al., 2012). To explore whether warming altered the structure of fungal communities, we disentangled functional genes targeted for fungi from those for bacteria using GeoChip. We identified 4969 genes targeting for 72 fungal species, and the majority of them belonged to the phyla of Ascomycota and Basidiomycota. Warming significantly increased the signal intensities of 21% of all fungal functional genes and 24.2% of C cycling genes (P⩽0.05; Figure 3c). These results, in combination with our previous observation that warming increased the ratio of fungi to bacteria in soil (Zhang et al., 2005), highlighted the relative importance of fungi in decomposition under warming. But whether fungi and/or specific taxonomic groups were favored or adapted to utilize old SOM remains to be investigated. Analyses of Adonis, ANOSIM and MRPP also showed that warming altered the functional gene structure of fungal communities (P⩽0.05; Supplementary Table S1b). Furthermore, DCA revealed clustering of functional fungal communities by the warming treatment (Figure 3g). Taken together, these results implied that warming-induced old SOM decomposition in subsoil was closely related to the changes in the functional gene structure of microbial communities.
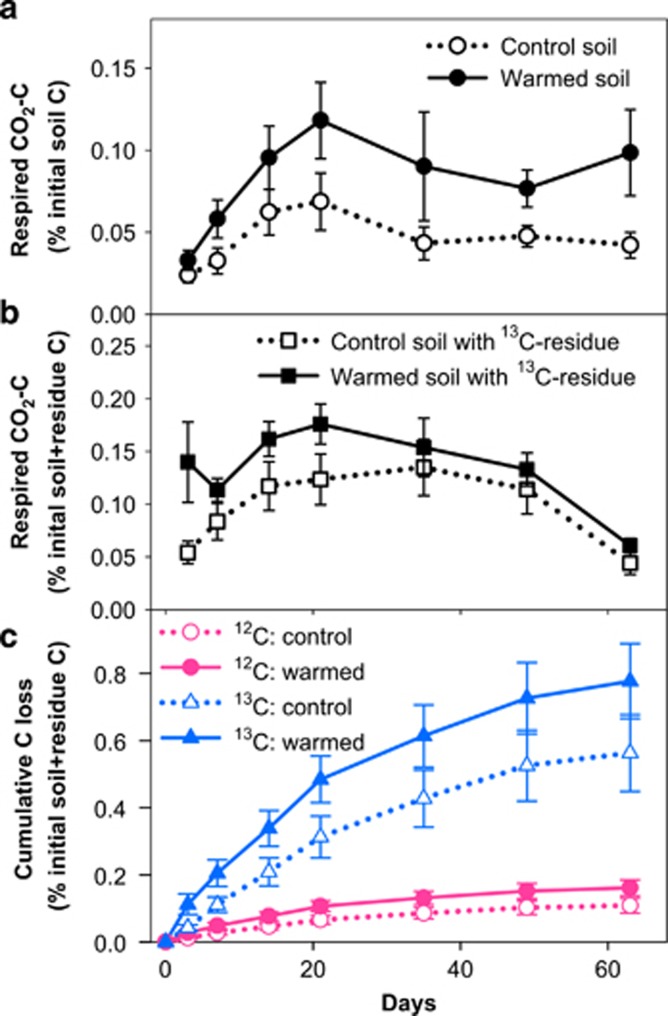
To identify the microorganisms active in old SOM decomposition, we designed a SIP microcosm experiment to track U13C-labeled whole plant materials (T. aestivum) into the active microbial communities (Supplementary Appendix S1 section 4). We treated half of soils of the 20–25 cm subsoil layer from deep collars of warming and control plots with the addition of 13C-labeled plant materials while the remaining half was unamended, and then incubated soils in the laboratory at 25 °C for 9 weeks. For both unamended (Figure 4a) and amended (Figure 4b), the percentages of C loss on the basis of the initial total C in warmed soils were significantly higher than those of control soils during incubation (repeated measures ANOVA, P<0.01 for both). The difference was not statistically significant for the total 13C content of respired CO2 from warmed and control soils with 13C-labeled plant materials (Supplementary Figure S5). A single isotope (that is, 13C), two-source mixing model (Supplementary Appendix S1 section 1) further showed that the percentages of accumulative CO2 respiration from plant residue- and soil-derived C in warmed soils were 38% and 49% higher than those of control soils, respectively (Figure 4c). These results were consistent with the previous view that the activity of degradation enzymes likely depended on microbial communities (Chapin et al., 2011), and suggested that the active microbial communities under warming had a higher potential in degrading organic materials than those under control.
Figure 4.
Decomposition by microbial communities in warmed and control soils: a SIP microcosm experiment. Data are means (n=6)±s.e.m. The y axis denotes the percentage of respired C on the basis of the initial total soil (a) or total soil and residue (b and c) C contents. (a), Respired CO2 from warmed (filled circles) and control (unfilled circles) soils. Warming, P<0.01; time, P<0.01; warming × time, P=0.35; repeated measures ANOVA. (b), Respired CO2 from warmed (filled circles) and control (unfilled circles) soils amended with 13C-labeled residues. Warming, P<0.01; time, P<0.01; warming × time, P=0.63; repeated measures ANOVA. (c), Cumulative C loss by microbial respiration from warmed (solid lines) and control (dotted lines) soils. Red circles: 12C, soil-derived C. Blue triangles: 13C, residue-derived C.
We considered communities that had assimilated 13C-labeled substrates into their nucleic acids to be active in decomposition (Dumont and Murrell, 2005). Active microbial communities were identified by sequencing 13C-DNA (buoyant density of 1.71–1.75 g ml−1, ‘heavy DNA’) that was separated from 12C-DNA (buoyant density of 1.67–1.70 g ml−1, ‘light DNA’) using ultracentrifugation. We used the gradient fractionation method to retrieve DNA from 12 fractions. Gel image showed that 13C-DNA was mainly distributed in fractions 9–11 (panels I and II in Supplementary Figure S6a) and well separated from that of 12C-DNA (fractions 5–8, panel II in Supplementary Figure S6a). Measurements of the buoyant density of each fraction confirmed a successful DNA gradient formation which was critical for the subsequent DNA retrieval and downstream analysis (Supplementary Figure S6b). DCA revealed a strong clustering by the 16S rRNA gene profiles of active microbial communities (13C-DNA) (Figure 5b). Even though, we did not observe any difference in the relative abundances of bacterial phyla between warmed and control soils (Figure 5a). This trend maintained at lower taxonomic levels as well (for example, class, order, family or OTU; data not shown). Significance tests by the three multivariate statistical analyses also showed that the taxonomic structure of both active (13C-DNA) and total (12C-DNA) communities based on 16S sequences were not significantly different between warmed and control soils (Supplementary Table S1a). In contrast, the functional gene-based GeoChip analysis showed that communities associated with warming were evidently clustered together (Figure 5c) and differed significantly from controls within both active and total soil communities (Supplementary Table S1b). These patterns persisted in the functional gene structure of both bacterial (Figure 5d, Supplementary Table S1b) and fungal (Figure 5e, Supplementary Table S1b) communities.
Figure 5.
Taxonomic and functional structures of microbial communities in warmed and control soils in the SIP microcosm experiment. (a), Relative abundances of dominant phyla (>1% 16S rRNA gene). 12C-DNA, total communities, light fractions isolated during SIP. 13C-DNA, active communities, heavy fractions isolated during SIP. (b–e), Taxonomic (b, 16S rRNA gene) and functional (c–e; GeoChip) structures of total (12C-DNA; red circles) and active (13C-DNA; blue circles) microbial communities. (b), Bacterial communities of 16S gene. (c), All functional genes in GeoChip. (d), Functional genes targeted for bacteria. (e), Functional genes targeted for fungi. Warmed soils, filled symbols. Control soils, unfilled symbols.
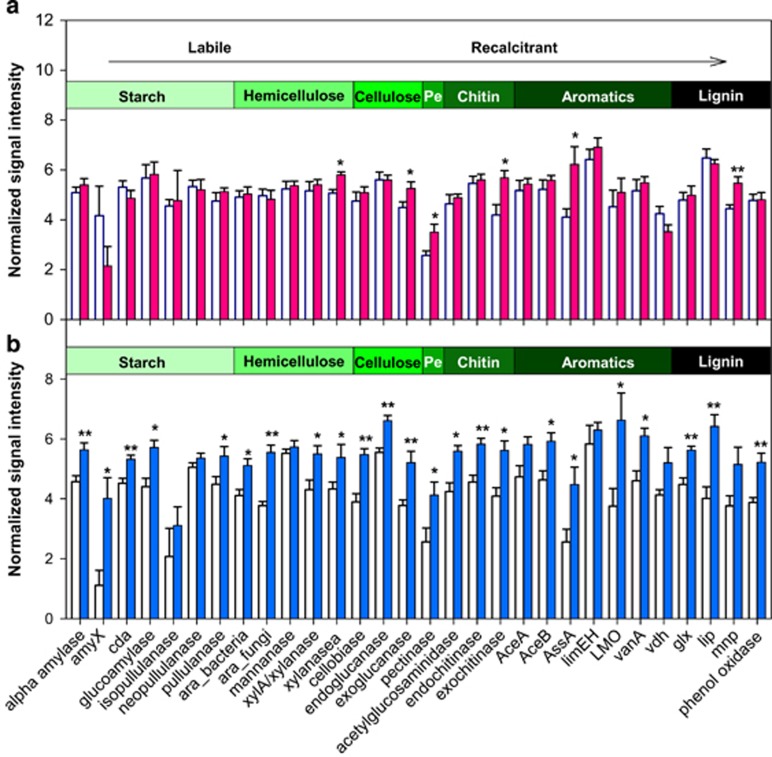
To link the changes in the functional profile of microbial communities to decomposition, we probed specific genes coding for enzymes involved in organic compound degradation. The 13C-labeled T. aestivum residue consisted of organic compounds that spanned a wide range of recalcitrance from the labile starch to the recalcitrant lignin, allowing us to examine how active microbial communities utilized substrates with different recalcitrance. We detected, on average, nearly 3200 probes within 29 gene families responsible for organic compound degradation in both warmed and control samples. Warming slightly increased the abundances of all genes on average by 7% in total soil communities (12C-DNA), with a larger effect occurred in gene categories involved in recalcitrant organic compound (pectin, chitin, aromatics and lignin) degradation (Figure 6a). Microbial communities from warmed soils active in assimilating 13C from plant residues possessed a higher relative abundance of genes associated with decomposition than control soils, with an average increase of 41% across all C degradation genes (Figure 6b). Taken together, these results suggested that warming significantly increased the functional genes coding for the degradation of various organic compounds with distinct chemical recalcitrance within active microbial communities.
Figure 6.
Key C degradation genes in warmed and control soils in the SIP microcosm experiment. (a), 12C-DNA, total communities, light fractions isolated during SIP. (b), 13C-DNA, active communities, heavy fractions isolated during SIP. Warmed soils, filled bars. Control soils, blank bars. Signal intensities were the average abundances of detected genes. Organic compounds shown in the horizontal bars are typical in soil and/or plant residues. Pe, an abbreviation of ‘pectin’. The arrow denotes the increasing chemical recalcitrance of organic compounds. *P⩽0.05) and **P⩽0.01) are statistically significant effects of warming.
We demonstrate, for the first time, that warming-stimulated decomposition of old SOM in subsoil is associated with a shift in the functional gene structure of microbial communities. Though to generalize whether this finding is applicable to far deeper soil layers or other ecosystems requires further studies, this study is valuable because it provides experimental evidence of the microbial mechanisms at play in warming-induced increases in SOM decomposition in subsoil. When young and old SOM coexist, warming enhances decomposition by stimulating genes coding for enzymes that process degradation of both young and old organic compounds, no matter whether they are labile or recalcitrant. When old SOM dominates, as observed in subsoil of deep collars in our Oklahoma grass prairie, warming still promotes the decomposition of old SOM by stimulating genes coding for old, recalcitrant organic compound degradation. Our findings, thus, are not consistent with the previous view that the warming effects on SOM decomposition largely depend on the chemical properties of SOM itself (Davidson and Janssens, 2006; Craine et al., 2010; Hopkins et al., 2012). Rather, our data imply that warming could enhance the decomposition of a wide range of organic compounds, irrespective of their recalcitrance.
Our findings that warming increases decades- to millennia-old SOM decomposition are consistent with and extend previous studies (Schuur et al., 2009; Hopkins et al., 2012). Warming-induced permafrost thaw enhanced old C emissions by 40–78% from the soil profile compared to minimally thawed areas in the arctic tundra ecosystem in Alaska (Schuur et al., 2009). In a short-term laboratory incubation study, elevated temperature increased decades-old SOM decomposition in surface soils from two temperate forest sites (Hopkins et al., 2012). Findings from both of these studies are consistent with our results, indicating that ongoing and projected warming may substantially alter the stability of old SOM in terrestrial soils. One caveat to this conclusion is that the warming effects on old SOM decomposition might operate in tandem with other controlling factors. For instance, new C inputs from roots (Fontaine et al., 2007) and mycorrhizal fungi (Cheng et al., 2012) likely reinforce the magnitude of the warming effect on old SOM decomposition in subsoil through priming mechanisms. Additionally, concurrent reductions in soil moisture under warming (Luo et al., 2001; Zhou et al., 2007) might lessen the warming effects on old SOM decomposition in dry areas such as grasslands in the west of North America, but could aggravate it in deeper soil layers through increasing oxygenation in wet regions such as arctic tundra (Schuur et al., 2009), temperate wetlands and tropical forests. Nevertheless, our results raise an important question about predictions of future atmospheric CO2 and climate by considering changes in the functional gene structure of microbial communities and their consequences for old SOM decomposition in soils.
Acknowledgments
We thank three anonymous reviewers for their constructive inputs, S Hu for thoughtful comments and B Sherry for field assistance. LC is supported by the NSFC Outstanding Young Scientist Award and the Thousand Talents Plan. This work was supported by the US Department of Energy (DE-SC0004601, DE-SC0010715), the National Natural Science Foundation of China (NSFC, 31370487, 31422010 and 31670501) and the Zhejiang Provincial NSFC (LR14C030001). 454 pyrosequencing GeoChip data are available at http://ieg.ou.edu/4download.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Carey JC, Tang J, Templer PH, Kroeger KD, Crowther TW, Burton AJ et al. (2016). Temperature response of soil respiration largely unaltered with experimental warming. Proc Natl Acad Sci USA 113: 13797–13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin FS, Matson PA, Mooney HA. (2011) Principles of Terrestrial Ecosystem Ecology, 2nd edn. Springer-Verlag: New York, USA. [Google Scholar]
- Cheng L, Booker FL, Burkey KO, Tu C, Shew HD, Rufty TW et al. (2011). Soil microbial responses to elevated CO2 and O3 in a nitrogen-aggrading agroecosystem. PLoS One 6: e21377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Booker FL, Tu C, Burkey KO, Zhou L, Shew HD et al. (2012). Arbuscular mycorrhizal fungi increase organic carbon decomposition under elevated CO2. Science 337: 1084–1087. [DOI] [PubMed] [Google Scholar]
- Conant RT, Ryan MG, Ågren GI, Birge HE, Davidson EA, Eliasson PE et al. (2011). Temperature and soil organic matter decomposition rates – synthesis of current knowledge and a way forward. Global Change Biol 17: 3392–3404. [Google Scholar]
- Craine JM, Fierer N, McLauchlan KK. (2010). Widespread coupling between the rate and temperature sensitivity of organic matter decay. Nature Geosci 3: 854–857. [Google Scholar]
- Crowther TW, Todd-Brown KEO, Rowe CW, Wieder WR, Carey JC, Machmuller MB et al. (2016). Quantifying global soil carbon losses in response to warming. Nature 540: 104–108. [DOI] [PubMed] [Google Scholar]
- Davidson EA, Janssens IA. (2006). Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 440: 165–173. [DOI] [PubMed] [Google Scholar]
- Donahue DJ. (1995). Radiocarbon analysis by accelerator mass spectrometry. Int J Mass Spectrom Ion Processes 143: 235–245. [Google Scholar]
- Dumont MG, Murrell JC. (2005). Stable isotope probing-linking microbial identity to function. Nat Rev Microbiol 3: 499–504. [DOI] [PubMed] [Google Scholar]
- Fang CM, Smith P, Moncrieff JB, Smith JU. (2005). Similar response of labile and resistant soil organic matter pools to changes in temperature. Nature 433: 57–59. [DOI] [PubMed] [Google Scholar]
- Fontaine S, Barot S, Barre P, Bdioui N, Mary B, Rumpel C. (2007). Stability of organic carbon in deep soil layers controlled by fresh carbon supply. Nature 450: 277–280. [DOI] [PubMed] [Google Scholar]
- Giardina CP, Ryan MG. (2000). Evidence that decomposition rates of organic carbon in mineral soil do not vary with temperature. Nature 404: 858–861. [DOI] [PubMed] [Google Scholar]
- Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. (2003). Influence of depth and sampling time on bacterial community structure in an upland grassland soil. FEMS Microbiol Ecol 43: 35–43. [DOI] [PubMed] [Google Scholar]
- Hakkenberg R, Churkina G, Rodeghiero M, Börner A, Steinhof A, Cescatti A. (2008). Temperature sensitivity of the turnover times of soil organic matter in forests. Ecol Appl 18: 119–131. [DOI] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. (2008). Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5: 235–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison AF, Taylor K, Scott A, Poskitt J, Benham D, Grace J et al. (2008). Potential effects of climate change on DOC release from three different soil types on the Northern Pennines UK: examination using field manipulation experiments. Global Change Biol 14: 687–702. [Google Scholar]
- Hartley IP, Ineson P. (2008). Substrate quality and the temperature sensitivity of soil organic matter decomposition. Soil Biol Biochem 40: 1567–1574. [Google Scholar]
- Hopkins FM, Torn MS, Trumbore SE. (2012). Warming accelerates decomposition of decades-old carbon in forest soils. Proc Natl Acad Sci USA 109: E1753–E1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson DS, Poulton PR, Bryant C. (2008). The turnover of organic carbon in subsoils. Part 1. Natural and bomb radiocarbon in soil profiles from the Rothamsted long-term field experiments. Eur J Soil Sci 59: 391–399. [Google Scholar]
- Jobbagy EG, Jackson RB. (2000). The vertical distribution of soil organic carbon and its relation to climate and vegetation. Ecol Appl 10: 423–436. [Google Scholar]
- Karhu K, Auffret MD, Dungait JAJ, Hopkins DW, Prosser JI, Singh BK et al. (2014). Temperature sensitivity of soil respiration rates enhanced by microbial community response. Nature 513: 81–84. [DOI] [PubMed] [Google Scholar]
- Knorr W, Prentice IC, House JI, Holland EA. (2005). Long-term sensitivity of soil carbon turnover to warming. Nature 433: 298–301. [DOI] [PubMed] [Google Scholar]
- Legendre P, Legendre l. (2012) Numerical Ecology 3rd edn. Elsevier: Oxford, UK. [Google Scholar]
- Liski J, Ilvesniemi H, Makela A, Westman CJ. (1999). CO2 emissions from soil in response to climatic warming are overestimated - The decomposition of old soil organic matter is tolerant of temperature. Ambio 28: 171–174. [Google Scholar]
- Littell RC, Milliken GA, Strooup WW, Wolfinger RD. (1996) SAS System for Mixed Models. SAS Institute Inc.: Cary, NC, USA. [Google Scholar]
- Louca S, Parfrey LW, Doebeli M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353: 1272–1277. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo CY, Xu GP, Wang YF, Wang SP, Lin XW, Hu YG et al. (2009). Effects of grazing and experimental warming on DOC concentrations in the soil solution on the Qinghai-Tibet plateau. Soil Biol Biochem 41: 2493–2500. [Google Scholar]
- Luo YQ, Wan SQ, Hui DF, Wallace LL. (2001). Acclimatization of soil respiration to warming in a tall grass prairie. Nature 413: 622–625. [DOI] [PubMed] [Google Scholar]
- Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria KL, Blazewicz SJ et al. (2011). Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480: 368–371. [DOI] [PubMed] [Google Scholar]
- Mahecha MD, Reichstein M, Carvalhais N, Lasslop G, Lange H, Seneviratne SI et al. (2010). Global Convergence in the Temperature Sensitivity of Respiration at Ecosystem Level. Science 329: 838–840. [DOI] [PubMed] [Google Scholar]
- Melillo JM, Steudler PA, Aber JD, Newkirk K, Lux H, Bowles FP et al. (2002). Soil warming and carbon-cycle feedbacks to the climate system. Science 298: 2173–2176. [DOI] [PubMed] [Google Scholar]
- Neufeld JD, Vohra J, Dumont MG, Lueders T, Manefield M, Friedrich MW et al. (2007). DNA stable-isotope probing. Nat Protocols 2: 860–866. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ et al. (2007). The vegan package. Community Ecology Package.
- Reichstein M, Katterer T, Andren O, Ciais P, Schulze ED, Cramer W et al. (2005). Temperature sensitivity of decomposition in relation to soil organic matter pools: critique and outlook. Biogeosciences 2: 317–321. [Google Scholar]
- Rumpel C, Kogel-Knabner I, Bruhn F. (2002). Vertical distribution, age, and chemical composition of organic, carbon in two forest soils of different pedogenesis. Org Geochem 33: 1131–1142. [Google Scholar]
- Schmidt MWI, Torn MS, Abiven S, Dittmar T, Guggenberger G, Janssens IA et al. (2011). Persistence of soil organic matter as an ecosystem property. Nature 478: 49–56. [DOI] [PubMed] [Google Scholar]
- Schuur EAG, Vogel JG, Crummer KG, Lee H, Sickman JO, Osterkamp TE. (2009). The effect of permafrost thaw on old carbon release and net carbon exchange from tundra. Nature 459: 556–559. [DOI] [PubMed] [Google Scholar]
- Shi Y, Tyson GW, Eppley JM, DeLong EF. (2011). Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J 5: 999–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker TF, Qin D, Plattner G-K, Tignor M, Allen SK, Boschung J et al. (2013) Climate Change 2013: The Physical Science Basis. Cambridge University Press: Cambridge, United Kingdom and New York, NY, USA. [Google Scholar]
- Trumbore S. (2009). Radiocarbon and Soil Carbon Dynamics. Annu Rev Earth Planet Sci 37: 47–66. [Google Scholar]
- Trumbore SE, Chadwick OA, Amundson R. (1996). Rapid exchange between soil carbon and atmospheric carbon dioxide driven by temperature change. Science 272: 393–396. [Google Scholar]
- Tu Q, Yu H, He Z, Deng Y, Wu L, Van Nostrand JD et al. (2014). GeoChip 4: a functional gene-array-based high-throughput environmental technology for microbial community analysis. Mol Ecol Resour 14: 914–928. [DOI] [PubMed] [Google Scholar]
- von Lutzow M, Kogel-Knabner I. (2009). Temperature sensitivity of soil organic matter decomposition-what do we know? Biol Fertility Soils 46: 1–15. [Google Scholar]
- Xu X, Shi Z, Li D, Zhou X, Sherry RA, Luo Y. (2015). Plant community structure regulates responses of prairie soil respiration to decadal experimental warming. Global Change Biol 21: 3846–3853. [DOI] [PubMed] [Google Scholar]
- Yergeau E, Bokhorst S, Kang S, Zhou J, Greer CW, Aerts R et al. (2012). Shifts in soil microorganisms in response to warming are consistent across a range of Antarctic environments. ISME J 6: 692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Parker K, Luo Y, Wan S, Wallace L, Hu S. (2005). Soil microbial responses to experimental warming and clipping in a tallgrass prairie. Global Change Biol 11: 266–277. [Google Scholar]
- Zhou J, Xue K, Xie J, Deng Y, Wu L, Cheng X et al. (2012). Microbial mediation of carbon-cycle feedbacks to climate warming. Nat Clim Change 2: 106–110. [Google Scholar]
- Zhou J, He Z, Yang Y, Deng Y, Tringe SG, Alvarez-Cohen L. (2015). High-throughput metagenomic technologies for complex microbial community analysis: open and closed formats. Mbio 6: e0228814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wan SQ, Luo YQ. (2007). Source components and interannual variability of soil CO2 efflux under experimental warming and clipping in a grassland ecosystem. Global Change Biol 13: 761–775. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.