Abstract
Nitrogen (N) input to the coastal oceans has increased considerably because of anthropogenic activities, however, concurrent increases have not occurred in open oceans. It has been suggested that benthic denitrification in sandy coastal sediments is a sink for this N. Sandy sediments are dynamic permeable environments, where electron acceptor and donor concentrations fluctuate over short temporal and spatial scales. The response of denitrifiers to these fluctuations are largely unknown, although previous observations suggest they may denitrify under aerobic conditions. We examined the response of benthic denitrification to fluctuating oxygen concentrations, finding that denitrification not only occurred at high O2 concentrations but was stimulated by frequent switches between oxic and anoxic conditions. Throughout a tidal cycle, in situtranscription of genes for aerobic respiration and denitrification were positively correlated within diverse bacterial classes, regardless of O2 concentrations, indicating that denitrification gene transcription is not strongly regulated by O2 in sandy sediments. This allows microbes to respond rapidly to changing environmental conditions, but also means that denitrification is utilized as an auxiliary respiration under aerobic conditions when imbalances occur in electron donor and acceptor supply. Aerobic denitrification therefore contributes significantly to N-loss in permeable sediments making the process an important sink for anthropogenic N-inputs.
Introduction
Worldwide, coastal seas receive large amounts of anthropogenic nitrogen (N) inputs through riverine run off and atmospheric deposition (48 and 8 Tg N yr−1, respectively (Galloway et al., 2004)). So far, however, concurrent rises in N have not been observed in the open ocean. This has been attributed to the activity of denitrifying microbes in coastal and continental shelf sediments, which respire much of the fixed inorganic N to N2, whereupon it is lost from the system (Gruber and Galloway, 2008). This denitrification was assumed to primarily occur in cohesive muddy sediments. Recently, however, extensive N-loss has been suggested to occur in the permeable sands that cover 58–70% of the continental shelves (Gihring et al., 2010; Gao et al., 2012; Marchant et al., 2016; Sokoll et al., 2016).
Sandy sediments are characterized by advective porewater flow. Water movement above the rippled sediments forms pressure gradients pumping water rich in oxygen, nitrate and organic matter into the sediment (Huettel et al., 2003). These high fluxes, combined with a microbial biomass of 108–109 cell ml−1 mean that sandy sediments act as biocatalytic filters, in which rates of respiration and organic matter turnover are high (Rusch et al., 2001; de Beer et al., 2005). The extent of advection changes dependent on current flow, sediment movement and exposure of intertidal flats at low tide. This leads to frequent fluctuations in O2 concentrations (Billerbeck et al., 2006; Ahmerkamp et al., 2015). Therefore, although sandy sediments provide high fluxes of electron donors and acceptors (Huettel et al., 2003), facultative anaerobes within them need to be able to rapidly adjust their respiratory strategy. A poorly understood consequence of this seems to be the occurrence of denitrification under oxic conditions, observations of which have been made in both intertidal and subtidal permeable sediments (Rao et al., 2008; Gao et al., 2010; Marchant et al., 2016).
All known marine denitrifiers are facultative anaerobes that can switch from aerobic respiration to anaerobic respiration. The electron transport chain is organized in such a way that the dissimilatory N-oxide reductases (nitrate, nitrite, nitric oxide and nitrous oxide reductase) are ‘plug-in’ branches to the aerobic respiration pathway. These branched electron transport chains, which can also include multiple terminal oxidases, provide respiratory flexibility, as the possession and differential expression of different terminal electron acceptors allows respiration to be adjusted to environmental conditions (Poole and Cook, 2000). Canonically, we assume that dissolved O2 needs to be depleted before facultative anaerobic organisms begin using nitrate (NO3−) as an alternative terminal electron acceptor (Zumft, 1997). This is because physiological, bioenergetic and kinetic considerations all indicate that denitrification should not occur in the presence of oxygen (Chen and Strous, 2013). Nevertheless, the simultaneous respiration of NO3− and O2 has been demonstrated repeatedly in pure cultures (Robertson and Kuenen, 1984; Robertson et al., 1995; Patureau et al., 2000; Kim et al., 2008).
For aerobic denitrification to occur, the N-oxide reductases must be present and functional under oxic conditions. Often this in not the case, as expression of the denitrification pathway is repressed by O2 (Bergaust et al., 2012). However, the inactivation and degradation of denitrification proteins is not instantaneous when O2 is introduced to anaerobic cultures (Qu et al., 2016). Furthermore, basal transcription of denitrification genes in the presence of oxygen is not unprecedented. The level of this basal transcription seems to vary widely across organisms and genes (Robertson and Kuenen, 1990; Zennaro et al., 1993; Baumann et al., 1996; Ka et al., 1997); however, some expression of the denitrification pathway would be favorable for denitrifiers under fluctuating O2 conditions. For example, the rapid transitions from oxic to anoxic conditions that are observed in permeable sediments could result in microbes becoming ‘trapped in anoxia’ if denitrification proteins are not synthesized before O2 (and therefore energy) becomes limiting (Hojberg et al., 1997; Bergaust et al., 2010). The hypothesis that environmental settings with fluctuating O2 concentrations select for aerobic denitrifiers is supported by previous observations of aerobic denitrification in permeable sediments and furthermore by the success that researchers have had in isolating aerobic denitrifiers from other environments where O2 availability fluctuates (Patureau et al., 1994, 2000; Frette et al., 1997). Until now most studies of aerobic denitrification have focused on biochemical and physiological characterization of the process using bacterial isolates (Richardson and Ferguson, 1992; Sears et al., 1997; Ellington et al., 2002). To the best of our knowledge, no study has actually investigated how high the basal transcription and expression of the N-oxide reductases is under oxic conditions in fluctuating marine environments. Nor has any study investigated which members of the microbial community may be responsible for co-respiration of O2 and N-oxides in permeable sediments.
We investigated aerobic denitrification in permeable sediments of the Wadden Sea. These sandy sediments that fringe the North-West European coast are part of the world’s largest tidal flat system and have a vital role in removing anthropogenic N-inputs from land before they reach the North Sea. The aim of our study was to determine the extent to which O2 controls the expression and utilization of the denitrification pathway and to identify the microorganisms carrying out aerobic denitrification.
Materials and methods
Sampling site
Sediment with a permeability of 7.2–9.5 × 10−12 m2, a porosity of 0.35 and a mean grain size of 176 μm was sampled from the Janssand sand flat in the East Frisian Wadden Sea, Germany (see Billerbeck et al., 2006 for a map of the region). The entire flat is inundated with ca 2 m of seawater during high tide, before exposure to the air for 6–8 h during low tide twice per day. While the tide is in, water rich in O2, NO3− and organic matter is pumped into the sediment down to depths of 5 cm, whereas when the tide is out (Huettel and Rusch, 2000; Jansen et al., 2009), O2 within the sediment is rapidly consumed and only penetrates to depths of around 1 cm because of downward drainage of overlying water. NO3− concentrations in the Wadden Sea are rarely limiting, ranging between 2 μM in summer and 100 μM in the winter, with average concentrations around 30 μM (see Gao et al., 2012).
Experimental column set up and sediment acclimatization
Sediment from the upper 5 cm of the sand flat was collected at low tide in April 2012, December 2014 and February 2015, whereupon it was homogenized and returned to the laboratory. Less than 4 h after collection sediment was packed into opaque sediment columns (height, 10 cm, I.D. 10.3 cm) and kept on a simulated tidal cycle for 24–36 h consisting of repeated 6-h ‘inundation’ and ‘exposure’ periods (Supplementary Figure S1.1). During ‘inundation’, seawater (collected from the sampling site) was supplied from the bottom of the core in cycles consisting of a 30-min period where water was pumped in at a rate of 2.5 ml min−1 followed by a 15-min period without pumping. The bottom 2 cm of the core were aerated in this time, whereas no oxygen penetrated into the remaining 8 cm. During the 6-h ‘exposure’ period, no pumping occurred.
Incubation procedure
Rates of O2 consumption and denitrification were determined in the sediment (Supplementary Figure S1.2). Incubations were carried out by exchanging the entire volume of porewater within the sediment core 1.5 times (over 18 min) with aerated seawater amended with 75 μmol l−1 15NO3−. The porewater within the core was subsequently sampled continuously from the core inlet and pumped through a membrane inlet attached to a mass spectrometer (membrane inlet mass spectrometry (MIMS); GAM200, InProcess Instruments, Bremen, Germany), at a rate of 0.4 ml min−1 (leading to a porewater velocity of 0.8 cm h−1). N2 isotopes (28N2, 29N2,30N2), O2 and Ar concentrations were determined every 3–4 s by MIMS over the next 90 min. Therefore, the reaction rates measured were those occurring in the bottom 1.2 cm of the sediment core. Denitrification rates were calculated according to Nielsen (1992), as anammox rates are not significant in these sediments, furthermore previous observations made over multiple seasons indicate that nitrification rates are orders of magnitude lower than denitrification rates.
Short-term and long-term exposure experiments
For short-term studies into denitrification and O2 respiration, all cores were exposed to at least four subsequent flushing cycles, which consisted of 18 min when porewater was exchanged and 90 min during which rates were determined (Supplementary Figure S1.2).
For the long-term study, in February 2015 sediment cores were exposed to different O2 regimes for 3 weeks. Six cores were supplied with North Sea seawater containing 20 μmol l−1 NO3−, with two cores assigned to each of three conditions: a ‘tidal cycle’ condition as detailed above, an ‘anoxic’ condition during which the core was supplied with seawater degassed with N2 during the inundation period and an ‘oxic’ condition, in which aerated seawater was constantly pumped through the sediment. After 21 days, the sediment cores were removed from their respective conditions and the rate measurements described above were immediately carried out.
Validation of experimental procedure
The alternation between oxic and anoxic conditions required repeated flushing of sediment cores with aerated, 15NO3− amended water (see Supplementary Figure S6, where it is also shown that the flushing was sufficient to remove 15N-N2 produced from the previous incubation). In order to determine whether the multiple incubations changed the flow paths within the sediment, water saturated alternately with helium or argon was pumped through the core and breakthrough curves determined. Very similar breakthrough curves, obtained from multiple flushings, indicated that the incubation method did not lead to the build-up of preferential flow paths within the sediment core. The breakthrough curves did, however, indicate that the rapid changes in concentration within the sediment, combined with funneling of water between the sediment and the small sampling port in the lid, led to a smearing effect in observed concentrations of O2 and N2. Briefly, when porewater from the edge of the core arrives at the lid, it has to travel toward the central port, therefore it reaches the port later than water from the centre of the core. In effect, this means that water with a shorter residence time mixes with water that has had a longer residence time within the sediment (Supplementary Figure S2). As such, there is a smearing of solute concentrations that become especially visible at O2 concentrations close to zero. This ‘lid-dispersion’ methodological artifact effects the determination of reaction kinetics, as can be observed when MIMS measurements are compared with those made with a microsensor placed directly in the sediment. Such microsensor measurements were conducted in January 2016. Sediment was cored without disturbing its structure and returned to laboratory where it was kept at 4 °C, subsequently the core was percolated with aerated site seawater and O2 consumption rates at 1.5 cm depth within the sediment were determined using a microsensor (as in Polerecky et al., 2005). Using the MIMS method, a low apparent half saturation constant (km) for O2 is observed (24 μM), whereas with the microsensor method the observed km is 2 μmol l−1 (Supplementary Figure S3). Using the breakthrough curves, it was possible to determine the periods during incubations when mixing had the greatest effect on the observed concentration gradients (that is, rates); allowing us to treat such data cautiously (gray bars, Figure 1a).
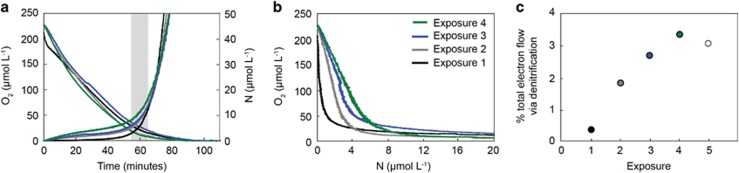
Figure 1.
Dynamics of O2 consumption and production of N by denitrification in sediment exposed to shifts between anoxic and oxic conditions. (a) Consumption of O2 and total N production (Dtot calculated from 29N2 & 30N2 concentrations) in the December incubation, the gray bar indicates where dispersion in the core lid may lead to over or underestimated concentrations. (b) In order to remove errors caused by lid-dispersion, O2 concentrations and N concentrations are compared against each other. (c) Percentage of total electron flow through the denitrification pathway at O2 concentrations above 100 μmol l−1 calculated from linear regressions of the data in b. For the respective regressions, r2 = 0.89, P<0.0001 (exposure 1), r2 = 0.99, P<0.0001 (exposures 2–5). Error bars (±1 s.e.) are smaller than the data points. A fifth exposure is shown that is not plotted in a and b as it overlapped almost entirely with exposure 4.
In situmetagenomics and transcriptomics
Metagenomic and transcriptomic analyses focused specifically on subunit 1 of the marker genes for terminal oxidases (A-class heme-copper oxidase (HCO), B-class HCO, C-class HCO and cyt bd-type oxidases) and the catalytic subunit of the nitrate reductases (napA and narG), and nitrous oxide reductase (nosZ). Metagenomic libraries were constructed with DNA from the upper 2 cm of the intertidal sand flat. Sediment was collected on 24 October 2009 and 23 March 2010. Two of the three March samples were pooled, and the pool was divided into two samples, named March 1 and March 2. Sediment was stored at −20 °C until DNA extraction, which was carried out according to Zhou et al. (1996). For all, 454 sequencing was carried out in two GS FLX Titanium sequencing runs. The four sequence data sets were submitted to the sequence read archive (http://trace.ncbi.nlm.nih.gov/Traces/sra/) under the bioproject PRJNA174601 (accession: SRP015924). The sample accession numbers are SRS365699, SRS365698, SRS365700 and SRS365701. Sediment sampling for transcriptomics was carried out on 21 March 2011 over a tidal cycle, sampling time points were 06:50 hours (low tide), 08:50 hours (late low tide), 10:50 hours (rising tide), 12:50 hours (high tide), 14:50 hours (falling tide) and 16:50 hours (early low tide). Sediment samples were immediately immersed in three volumes of RNA preservation solution (LifeGuardTM Soil Preservation Solution, MoBio, Carlsbad, CA, USA), and stored at 4 °C until RNA extraction (see Supplementary Methods). Complementary DNA libraries generated from the RNA were sequenced on a HiSeq 1500 instrument, in a 1 × 250 bases single end run (Rapid v2 chemistry, Illumina, San Diego, CA, USA). Six metatranscriptome libraries were generated from sediment sampled at six time points over a complete tidal cycle, each of these had between 1.2 and 8 million mRNA sequence reads with a mean length of 180 bp (Supplementary Table S2). The sample accession numbers are SRS417277, SRS417279, SRS417280, SRS417281, SRS417282 and SRS417283
Data analysis—metagenomics
The sequence data sets were pre-processed by removing identical reads with CD-hit (Li and Godzik, 2006) and translated into the correct protein sequences using PRODIGAL (Hyatt et al., 2010). The respective catalytic subunits for each of the four classes of terminal oxidases were identified using profile hidden Markov models and their identity validated following Sousa et al. (2011). Functional marker genes for the denitrification pathway were identified using a pipeline developed in house (see Supplementary Methods for details). The abundance of three housekeeping genes (rpoB, rpoC and recA) was determined using model TIGRFAMs TIGR02027, TIGR02013, TIGR02386. Gene abundances were normalized by gene length and then to average counts of the single copy housekeeping genes, recA, rpoB and rpoC from the same metagenome (Morris and Schmidt, 2013; Dalsgaard et al., 2014). A resulting value of 1 would therefore mean that a gene was of equal abundance to the housekeeping genes. The taxonomic identities of terminal oxidase genes were then inferred after subjecting them to a Blastp search against the NCBI nr database (downloaded on 31 August 2016) using the LCA algorithm in MEGAN 4 (Huson et al., 2011).
Data analysis—tidal cycle transcriptomics
The sequence data sets were pre-processed by Casava v1.8.4 (Illumina, San Diego, CA, USA), and quality trimmed using Trimmomatic (Bolger et al., 2014). SortMeRNA v2.0 (Kopylova et al., 2012) was used to filter out ribosomal RNA reads. From the resulting non-ribosomal RNA reads, identical reads were removed using CD-hit (Li and Godzik, 2006) and translated into protein sequences using PRODIGAL (Hyatt et al., 2010). As before hidden Markov models for napA, narG and nosZ, the catalytic subunits of terminal oxidases and housekeeping genes were used to identify transcripts within the sequence data (see Supplementary Methods). Taxonomic identities of transcripts were inferred using MEGAN 4 (Huson et al., 2011). Counts of terminal oxidase and N-oxide reductase gene transcripts were normalized in the same way as gene abundance.
Results
Sediment incubations
To gain insights into how and when N2 production occurs in response to O2 fluctuations within permeable sediments, we used an established method combining flow through sediment cores with online MIMS (Rao et al., 2008; Marchant et al., 2014, 2016). This allows simultaneous and high-resolution (<4 s) measurements of O2 and N2 concentrations within the sediment (Figure 1a). When porewater is sampled, different flow path lengths in the lid of the sediment core cause dispersion and thus a slight smearing of O2 and N2 concentrations. When production or consumption rates change quickly, the smearing may bias these changes and, therefore, also the rate; whereas, when rates are constant the smearing has no effect. When the measured O2 and N2 rates are plotted against each other (Figure 1b), the linear parts of the plot indicate when the O2 consumption and N2 production rates are either constant or change at the same pace. All biases from physical mixing (dispersion) are canceled out. The highly non-linear parts of the plot emphasize a strong relative change of O2 consumption over N2 production (gray bar Figure 1a). In Figure 1b, two linear slopes can be distinguished, one at high O2 concentrations and one at low O2 concentrations, reflecting the different denitrification rates under oxic and anoxic conditions. Furthermore, the relative change of O2 over N2 directly reflects the relative partitioning of electron flow between denitrification and aerobic respiration for the consecutive treatments (Figure 1c). Although it is likely that at low O2 concentrations anoxic microsites may have developed within the sediment, at higher O2 concentrations these are very unlikely (see Supplementary Figure S5).
Aerobic and anaerobic denitrification in sediment exposed to rapid fluctuations in O2 availability
Exposure of the freshly collected sediment to rapid fluctuations in O2 availability yielded the same results in each of the three seasons tested (Figure 1 and Supplementary Figure S4). We determined rates in the bottom centimeter of cores previously kept on a tidal cycle, this was the layer of sediment within the core, which was exposed to diel fluctuations in O2 concentrations. A rapid consumption of O2 (maximum rate 6.8 and 5.5 μmol l−1 min−1 in December and February, respectively) and the production of a relatively small amount of N2 gas was observed under oxic conditions (< 0.4 μmol N l−1, or the equivalent of 0.3% of the electron flow). Denitrification rates rose rapidly when O2 concentrations dropped below the apparent half saturation constant (km(O2)) as determined by Michaelis–Menten kinetics (Supplementary Figure S3), stabilizing when sediment became fully anoxic (linear regressions of N production over time during anoxia had r2 >0.99). On average, denitrification rates during anoxia reached 1.08 and 0.8 μmol N l−1 min−1 in December and February, respectively.
After the sediment had been anoxic for 90 min, air-saturated 15NO3− labeled seawater was reintroduced into the sediment. Such oxic–anoxic cycles were repeated multiple times on the same core. The production of N2 at high O2 concentrations increased with each subsequent incubation (Figure 1b), eventually reaching a maximum after four cycles, by which time 3.7 μmol l−1 N was produced, while O2 concentrations were still above 100 μmol l−1, with maximal aerobic denitrification rates of 0.22 and 0.28 μmol N l−1 min−1 at O2 >100 μmol l−1, this was equivalent to 3.4 and 2.8% of the electron flow (February and December, respectively, Figure 1c and Supplementary Figure S4c). After four oxic–anoxic cycles, O2 consumption rates (at O2 concentrations above 100 μmol l−1) in December increased 33% between the first and fourth incubation (Supplementary Table S1). In February, no significant change in O2 consumption rates was observed between incubations. Anoxic denitrification rates also varied as result of repeated incubations, decreasing by 44% between the first and fourth incubation in December, and by 24% between the first and fourth incubation in February (Supplementary Table S1).
Denitrification rates in sediment exposed to long-term oxia, long term anoxia or simulated tidal cycles
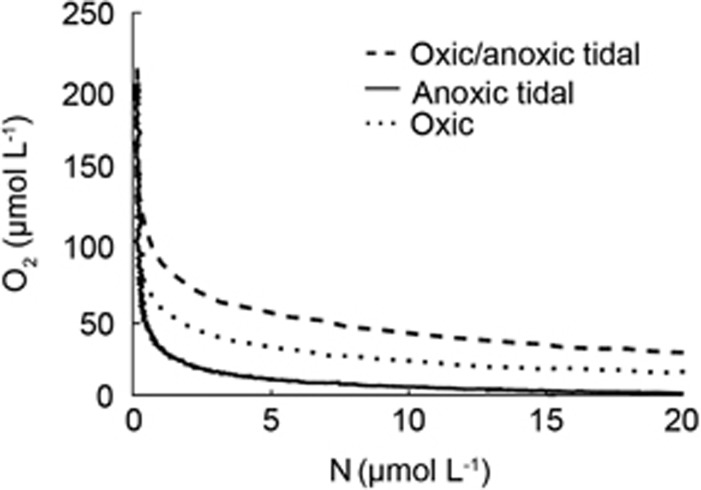
O2 concentrations fluctuate within the sediment at the sand flat over a tidal cycle. In order to determine how these fluctuations have shaped the ability of the microbial community to denitrify, we compared the denitrification rates in sediment exposed to different O2 concentrations over longer time periods. Sediment from the upper layer of the sand flat was kept under alternating oxic/anoxic, constantly oxic or constantly anoxic conditions for 3 weeks. After this time, when aerated, 15NO3− amended water was added to the sediment, O2 consumption was observed in all three conditions (Figure 2). Denitrification also occurred in the sediment from all three conditions. No lag was observed between the onset of anoxia and denitrification in any of the conditions, in fact N2 production began in all the sediments when O2 concentrations were above 50 μmol l−1 (Figure 2).
Figure 2.
Consumption of O2 and production of N by denitrification in sediment cores after exposure to different O2 conditions for three weeks. Sediment was exposed to either constant porewater flushing with oxic water (dotted line), a simulated tidal cycle with anoxic water (solid line) or a simulated tidal cycle with fluctuating oxic and anoxic conditions (dashed line).
Terminal oxidase and denitrification functional gene classification in metagenomes
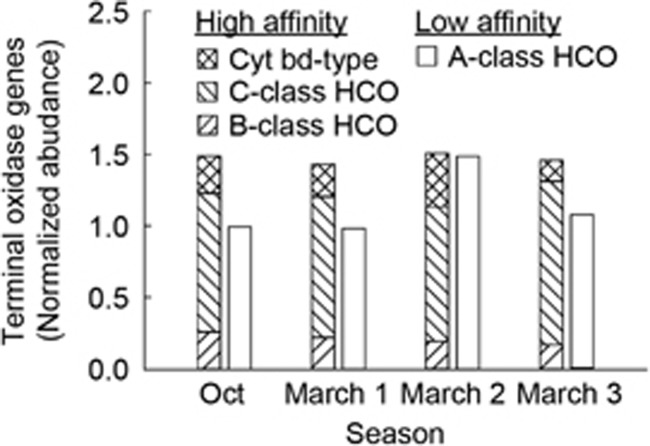
In order to determine how the frequent changes in O2 conditions shape the respiratory strategies of the microbial community within the sediment, in situmetagenomic and metatranscriptomic sampling was carried out. We focused on both denitrification genes and terminal oxidases, which we classified as either high-affinity (B-class HCO’s, C-class HCO’s and cytochrome bd-type oxidases) or low-affinity (A-class HCO’s). Both high- and low-affinity terminal oxidases were detected, with significantly more high-affinity terminal oxidases detected per metagenome (1.47±0.03 s.d.; normalized abundance, where a value of 1 means that genes were of equal abundance to singly copy housekeeping genes) than low-affinity terminal oxidases (1.14±0.24 s.d.) (Figure 3). Taxonomic assignment of the low-affinity HCO’s revealed that between 7% and 20% of reads were Eukaryota, mainly related to the Opistokonta and Stramenopiles.
Figure 3.
Relative abundance of reads matching terminal oxidase genes in four separate metagenomes. Gene abundance was normalized by gene length and against the average abundance of three housekeeping genes within the metagenome (recA, rpoB and rpoC).
Analysis of denitrification marker genes focused on the catalytic subunits of the nitrate and nitrous oxide reductases (napA, narG and nosZ). Reads for all three were detected within the metagenomes in all seasons. nosZ was the most abundant per metagenome (0.31±0.12 normalized abundance) followed by napA (0.19±0.05) and narG (0.08±0.02). There was no overall difference in the normalized abundance of nitrate reductases (napA+narG) and nitrous oxide reductases.
In situtranscription of terminal oxidases and functional genes for denitrification across a tidal cycle
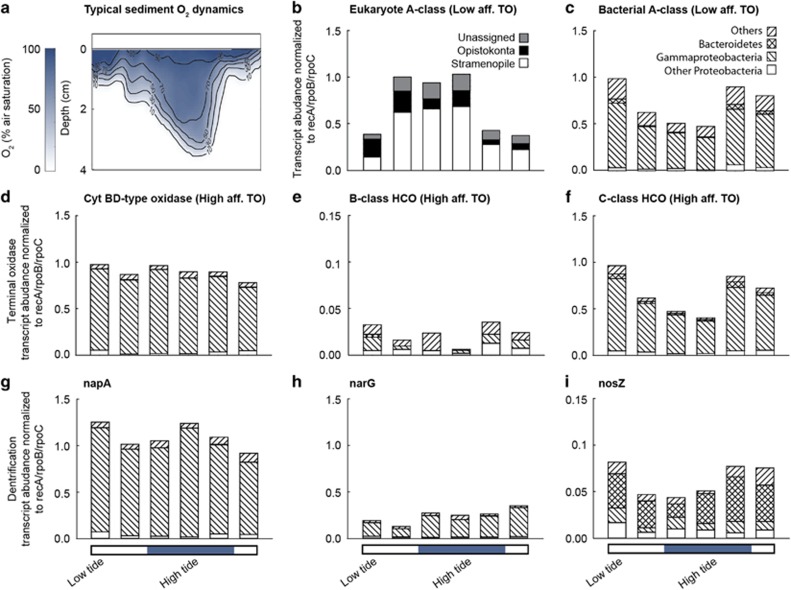
Transcriptomic analyses focused on the marker genes for terminal oxidases and functional genes for denitrification over a 12-h tidal cycle, during which the sediment was sampled once every 2 h. Sampling took place on a cloudy day, and as such solar irradiance measured at the nearby Spiekeroog Time Series Station (Reuter et al., 2009) peaked at 255 μmol m−2 s−1 at 1400 hours. As the flat was inundated with water during the day, almost no light would have reached the sediment as the water was turbid. At the three sampling points when the tide was out (early morning and late afternoon), solar irradiance was between 83 and 110 μmol m−2 s−1. In the metagemore low-affinity terminal oxidase reads were dominated by reads taxonomically affiliated to bacteria whereas, in the transcriptomes, half of the total low-affinity terminal oxidase transcripts were most similar to eukaryotic sequences from the Stramenopile and the Opistokonta. Eukaryote-associated low-affinity terminal oxidase gene transcription varied over a tidal cycle, with higher relative transcription at late low tide, rising tide and high tide (Figure 4b). This did not match the period when the tide was in (rising tide, high tide and falling tide), or the period when the tide was out (early low tide, late low tide and rising tide).
Figure 4.
Relative abundance of transcripts matching terminal oxidases and denitrification genes in the top 2 cm of the sediment over a tidal cycle. Transcript abundance was normalized by gene length and against the average transcript abundance of three housekeeping genes within the metatranscriptomes (recA, rpoB and rpoC). (a) Typical O2 profile within the sediment over a tidal cycle, (b, c) low-affinity terminal oxidases. (d, e, f) High-affinity terminal oxidases, (g) periplasmic nitrate reductase, (h) membrane-bound nitrate reductase and (i) nitrous oxide reductase.
Of the bacteria-associated low-affinity terminal oxidase transcripts, the most common taxonomic assignment was to Proteobacteria; specifically the class Gammaproteobacteria. As Gammproteobacteria reads were so abundant (up to 1300 per metatranscriptome), further taxonomic assignment was carried out to the order level, where the majority of sequences fell into the orders Alteromonodales, Enterobacteriales, Oceanospirillales and Vibrionales (Supplementary Table S3a). Over the tidal cycle, bacteria-associated low-affinity terminal oxidase gene transcription was inverse to that of the Eukaryotes and was relatively higher at falling tide, early low tide and low tide (Figure 4c). The same trend was observed at class level, particularly in the Bacteroidetes and Gammaproteobacteria. At the gammproteobacterial order level, higher transcription of A-class HCO was also visible at falling tide, early low tide and low tide in Alteromonodales and Vibrionales (Supplementary Table S3a), however, there was a different pattern in the Enterobacteriales; transcription of low-affinity terminal oxidase in this order was highest from rising tide to falling tide.
Of the high-affinity terminal oxidase transcripts, Cyt bd-type oxidases were the most expressed, followed by C-class HCO’s (Figures 4d–f), only minor transcription of B-class HCO’s was observed. Transcription of the high-affinity terminal oxidases followed similar patterns to those of the low-affinity terminal oxidases. Most high-affinity transcripts were taxonomically assigned as Gammaproteobacteria (Figure 4f), specifically to the classes Alteromonodales and Vibrionales (Supplementary Table S3b–d). Furthermore, transcription was higher at falling tide, early low tide and low tide. As such, when high-affinity terminal oxidase transcription was compared with low-affinity terminal oxidase transcription over the tidal cycle, there was a positive correlation, this correlation was especially strong for the low-affinity terminal oxidases and the C-Class high-affinity HCO’s of the Bacteroidetes, Alteromondales and Vibrionales (Supplementary Table S5b), and between the low-affinity terminal oxidases and Cyt bd-type high-affinity terminal oxidases of Actinobacteria, Enterobacteriales and Oceanospiralles. Negative correlations between the transcription of high- and low-affinity terminal oxidases were never observed.
In situtranscription of functional genes for denitrification across a tidal cycle
Transcriptomic analyses of denitrification marker genes focused on the transcription of the nitrate and nitrous oxide reductases (napA, narG and nosZ). As the periplasmic nitrate reductase (for which napA is the catalytic subunit) is hypothesized to be used during aerobic denitrification, differences in napA and narG transcription may indicate whether specific taxonomic groups are responsible for aerobic denitrification, whereas nitrous oxide reductase is of particular interest as it mediates the last step in denitrification. Denitrification genes were transcribed within the sediment at all sampling points over the tidal cycle. The most common taxonomic assignment of transcripts for napA and narG was Proteobacteria; specifically the class Gammaproteobacteria (Figures 4g and h and Supplementary Table S4a and b). The majority of sequences fell into the orders Altermonodales, Enterobacteriales, Oceanospiralles, Pseudomonodales and Vibrionales. Other transcripts were commonly associated to the phyla Actinobacteria and Planctomycetes. nosZ transcripts were mostly assigned to the phyla Bacteriodetes (Figure 4i and Supplementary Table S4c). When the sum of all napA transcripts were taken together, there was no clear trend over the tidal cycle (Figure 4g). Neither was there a clear trend when the napA transcripts from individual phyla, class or order were analyzed. Similar results were obtained for narG transcripts. Overall, napA transcripts were four times more abundant than narG transcripts (Figures 4g–h). This was driven by high abundances of Alteromonodales and Vibrionales napA transcripts, however, in the case of the Actinobacteria, Alphaproteobacteria and Enterobacteriales, narG transcripts were significantly more abundant (Supplementary Table S4a and b). napA transcript abundance correlated positively with narG transcript abundance within the phyla Bacteroidetes and the order Enterobacteriales (Pearson correlation, P<0.001 for both), no other correlations were observed (Supplementary Table S5a). When the sum of all nosZ transcripts were taken together, there was a significant difference in transcript abundance over the tidal cycle, nosZ transcripts were less abundant at late low tide, rising tide and high tide compared with those from falling tide, early low tide and low tide (Student's t-test P<0.001). This was mainly driven by changes in the abundance of Bacteroidetes-associated nosZ transcripts (Figure 4i).
Denitrification functional gene transcription in comparison with terminal oxidase transcription
In order to compare the transcription patterns of napA, narG and nosZ with the transcription patterns of the terminal oxidases, the data for each gene pair was compared on individual XY plots at phyla, class and order level and Pearson correlation values determined (Supplementary Table S5b–e). There was only one case, the Betaproteobacteria, where transcription of denitrification functional genes (in this case napA) negatively correlated with the transcription of a Betaproteobacteria terminal oxidase (high-affinity C-class HCO) (Supplementary Table S5d). In contrast, many positive correlations were observed between terminal oxidases and denitrification enzymes at phyla, class and order level. Within the Bacteroidetes-associated transcripts (which dominated the nosZ counts), nosZ normalized transcript abundance always positively correlated with the abundance of Bacteroidetes low-affinity terminal oxidases, as well as with the high-affinity C-class HCO’s and cyt bd-type terminal oxidases (Figures 4c, f and i). Similarly, Bacteroidetes napA and narG normalized transcript abundances positively correlated with both Bacteroidetes low-affinity terminal oxidase and high-affinity C-class HCO transcription (Supplementary Table S5). Positive correlations between napA, narG, and terminal oxidases were also common within the Gammaproteobacterial classes (Supplementary Table S5).
Discussion
High fluxes of electron acceptors and donors within permeable sandy sediments mean that they have the potential to act as biocatalytic filters, mediating high rates of respiration and remineralization. As such permeable sediments have been suggested to contribute extensively to marine N-loss (Gao et al., 2012; Huettel et al., 2014; Marchant et al., 2016; Sokoll et al., 2016). The extent of this N-loss may be further increased by denitrification occurring in the presence of oxygen. Here, we examined the transcription and utilization of the denitrification pathway in natural sediment communities from an intertidal sand flat where we hypothesize that fluctuating O2 conditions leads to co-respiration of O2 and N-oxides.
Simultaneous O2 consumption and denitrification in permeable sediments
To study how the fluctuating O2 concentrations in permeable sediments relate to the simultaneous respiration of oxygen and N-oxides, we collected sediment from an intertidal sand flat (Janssand) in the Wadden Sea. Over a tidal cycle, the upper 3–5 cm of sediment at the sand flat is generally oxic while the tide is in because of advective porewater transport and anoxic while the tide is out (Billerbeck et al., 2006). To imitate this, we simulated 3–4 tidal cycles. This ensured that microbial community in the bottom 2 cm of the core was exposed to O2 for part of the day, whereas the rest of the core was anoxic the whole time because of the high O2 consumption rates in the sediment (Supplementary Figure S1). Rate measurements were performed at the end of a simulated ‘tide in’ cycle, when the core had been supplied with oxic water for 6 h. At this point, N2 production was observed in the sediment at O2 concentrations above 100 μmol l−1, indicating that denitrification was never fully inhibited by the presence of O2 (Figure 1).
Although the initial incubation imitated the large-scale changes in O2 concentrations over a tidal cycle (that is, oxic or anoxic), it did not take into account the more complex O2 dynamics that occur within the sediment at high tide. When the sand flat is inundated, advection of O2-rich water depends on bottom water current velocities, which change over the tidal cycle (for Janssand the highest velocities occur during early rising tide and the lowest during high tide; Gao et al., 2012). Furthermore, when velocities are high enough, rippled permeable sediments start to move, burying previously oxic sediment and exposing previously anoxic sediment to O2 (Ahmerkamp et al., 2015). When repeated incubations simulating these rapid changes in O2 concentrations were carried out, the amount of N2 produced at O2 concentrations above 100 μmol l−1 increased by more than an order of magnitude (Figure 1). Such a substantial production of N2 (>3.5 μmol l−1) is a clear indication that aerobic denitrification was occurring (see Supplementary Figure S5 for discussion concerning the low likelihood that denitrification was occurring solely in anoxic microniches). The continued occurrence of denitrification after a switch from anaerobic to aerobic conditions has been observed in pure culture, where it is postulated that denitrification enzymes are not immediately repressed and there is still a marginal flow of electrons to the N-oxides (Qu et al., 2016). Due to the rapid transitions from anoxia to oxia that occurred here, it is highly likely that there was electron flow to denitrification proteins synthesised during anoxia. Intriguingly, such a production of μmol l−1 concentrations of N2 in the presence of high O2 concentrations has not been observed before in marine environments. In fact, in O2 minimum zones, the other main site of N-loss in the marine environment, denitrification can be reversibly inhibited at O2 concentrations above 2 μmol l−1 (Dalsgaard et al., 2014). Furthermore, the stimulation of aerobic denitrification by repeated switches between oxia and anoxia has not been observed before in culture or in environmental samples.
The stimulation of aerobic denitrification in response to the switches in O2 concentrations does not seem to be because of changes in gene transcription or protein levels. An increase in denitrification enzymes could have been induced by the production of nitrite or nitric oxide during the incubation, however, we saw no evidence for such an induction, as the anaerobic denitrification rates did not increase across the incubations. Therefore, the stimulation seems to be a metabolic response to the changing environmental conditions, for example, substrate availability. We attribute the increase in aerobic denitrification upon repeated shifts between oxia and anoxia to increased electron donor supply, which would result from aerobic breakdown of complex organic matter that was anoxic during the previous 36-h period (that is, everything above the lower 2 cm). Electron donors rather than electron acceptors (O2) are more likely to be limiting respiration within the sediment, however, as a consequence of the rapid switches between oxia and anoxia, electron donor supply to the lower 1.5 cm of the sediment would have increased (because of the re-oxygenation of the anoxic sediment immediately adjacent). An increase in electron donor supply is supported by the increase in oxygen consumption rates observed over the repeated incubations in the December experiment. It is likely that the products of remineralization that are formed in response to re-oxygenation after anoxia, as well as the less degraded organic matter from the previously anoxic sediment percolate into the lower 1.5 cm during the measurement period. The repeated incubations would push the system progressively into a state where electron donor supply is greater than electron acceptor supply.
Imbalances in the supply of electron donor and acceptors are known to stimulate aerobic denitrification (Sears et al., 1997; Ellington et al., 2002; Richardson, 2008). Here we found that electron flow through the entire denitrification pathway at O2 concentrations above 100 μmol l−1 rose from <1% of the total electron flow (denitrification and aerobic respiration combined) during the first incubation to 4% in the last incubation. When reduced carbon sources were supplied to a pure culture and aerobic nitrate reduction was monitored, electron flow to nitrite was around 1% under oxic conditions (Sears et al., 1997; Ellington et al., 2002). Although these results are not directly comparable to those obtained here as we were measuring full denitrification (which represents the transfer of 2.5 more electrons per mole NO3− than reduction of NO3− to NO2−), the increase in electron flow we observed is in the same range.
We propose that in the environment, imbalances in electron donor and electron acceptor supply that stimulate aerobic denitrification are likely to occur because of both the dynamic supply of O2 into the sediment and the trapping of organic matter. Although fluxes of oxygen, nitrate and organic carbon into the sediment are high, the depth to which organic carbon (in the form of particulate organic matter) penetrates is determined not just by the advective flow of water, but also by the trapping of particulate organic matter in the sediment matrix (Ehrenhauss et al., 2004a, b). Particulate organic matter is rapidly filtered out in the upper centimeter of the sediment and remobilized when sediment starts to move. Such resuspension events expose previously buried particulate organic matter to the sediment surface (Pilditch and Miller, 2006).
Adaptations in the microbial community leading to aerobic denitrification
It is evident that for aerobic denitrification to occur, the entire denitrification pathway must be functional in the presence of O2. After a 3-week exposure to constantly oxic conditions, denitrification began within the sediment before the sediment became anoxic for the first time, with maximal denitrification rates reached <10 min after the sediment became anoxic (Figure 2). It is unlikely that this was a result of reactivation of denitrification enzymes formed in response to anoxia 3 weeks before. Although there is evidence that denitrification enzymes are not immediately repressed and degraded in cultures upon reintroduction of oxygen (Qu et al., 2016), we expect the bacterial community would be turned over in 3 weeks. For example, Böer et al. (2009) estimated that the bacterial biomass in subtidal sands was turned over in 2–8 days. Furthermore, it is unlikely that denitrification resulted from de-novo induction and expression of the entire denitrification pathway, as this would have had to occur within 30 min of oxygen levels dropping below air saturation. In pure cultures, when induction of the denitrification pathway is stimulated by a transition from oxic to anoxic conditions, maximal denitrification rates are only observed after periods of 5–24 h (Baumann et al., 1996, 1997; Qu et al., 2016). Therefore, it seems that the denitrification observed resulted from basal expression of denitrification genes. The rate of anaerobic denitrification that occurred in the sediment previously kept oxic was a third of the rates measured in sediment that had been kept constantly anoxic or exposed to tidal cycle conditions. The maintenance costs associated with expressing the denitrification pathway under oxic conditions are significant, generally leading to strong regulation of denitrification gene expression by O2 (Bergaust et al., 2008, 2010; Bakken et al., 2012). We hypothesize, however, that the rapid fluctuations in O2 concentrations that occur within these sediments may have shaped the community over longer time scales, reducing the regulatory role of oxygen by increasing basal expression of the denitrification pathway. After all, in permeable sediments, where O2 availability is variable, there are also significant costs associated with late induction of the denitrification pathway.
To gain further insights into how the frequent oscillations in O2 within these sediments shape microbial respiration strategies, we carried out metabolic profiling to examine the respiration pathways present and the transcription of these pathways over a tidal cycle. We sampled sediment directly from the sand flat over an entire tidal cycle in order to capture the natural responses of the community respiratory gene transcription to the large diurnal changes in in situO2 concentrations. Diverse terminal oxidase (which catalyze the terminal electron transfer in aerobic respiration) and denitrification pathway marker genes were present within DNA extracted from the upper layer of sediment (Figure 3). When normalized to single copy housekeeping genes, the abundance of the terminal oxidases was in the same range as that found in soils, but higher than the abundance observed in environments where O2 concentrations are more stable (that is, the open ocean or animal guts; Morris and Schmidt, 2013). In such stable environments, which are either constantly oxic, or constantly anoxic, either low- or high-affinity terminal oxidases tend to dominate (Morris and Schmidt, 2013). In contrast, within the permeable sediments studied here there was a similar relative abundance of low-affinity A-class HCO’s and high-affinity C-class HCO’s. The high abundance and even distribution of high- and low-affinity terminal oxidases indicate that the aerobes within the intertidal permeable sediment are more likely to possess multiple terminal oxidases than in stable environments. This would confer greater metabolic versatility and enable the microbial community to better exploit the fluctuating O2 concentrations (Poole and Cook, 2000).
Microbes encoding multiple terminal oxidases are able to fine tune the expression of their high- and low-affinity terminal oxidases in response to O2 concentrations (Tseng et al., 1996; Poole and Cook, 2000; Otten et al., 2001; Arai, 2011). However, we could find no evidence for differential transcription between high- and low-affinity bacterial terminal oxidases across a tidal cycle as O2 concentrations changed (Figures 4c–f). This contrasts with other environmental studies, where switches in expression of low- and high-affinity terminal oxidases have been observed over oxic–anoxic gradients. However, in other studies, the changes in O2 concentration occurred at time scales of weeks, or over spatial scales of meters (Schunck et al., 2013; Hewson et al., 2014; Eggleston et al., 2015; Kalvelage et al., 2015). In permeable sediments, changes in O2 concentration happen over minutes and hours and at spatial scales of millimeters to centimeters.
In fact, our evidence indicates that bacterial terminal oxidase transcription is not correlated with changes in O2 availability over a tidal cycle in Wadden Sea permeable sediments (Figure 4). Instead, bacterial low-affinity terminal oxidase transcription was strongly and positively correlated to high-affinity terminal oxidase transcription at the class and order level, with transcription of both dropping at late low tide, rising tide and high tide. These sampling points represent very different O2 concentrations within the sediment, at late low tide O2 availability is low within the sediment, having been consumed while the tide is out, whereas at rising tide and high tide O2 availability increases rapidly as the sand flat is inundated with water and O2 is supplied by advection.
Interestingly, the transcription of eukaryote low-affinity terminal oxidases also did not correlate well with O2 concentrations, however, it was the inverse of the bacterial terminal oxidase transcription. This could reflect differences in the energy demands of the bacteria and eukaryotes over the tidal cycle, and suggests that the respiratory apparatus of the eukaryotes in the sediment is regulated by other factors. These could be related to circadian rhythms or light, as most of the identified eukaryotes were Stramenopiles, and most likely diatoms, which are abundant in the surface layer of the sediment (Stief et al., 2013; Marchant et al., 2014). The time period when the eukaryote terminal oxidases were most expressed was during the day, when light levels at the sediment surface would have been minimal because of high cloud coverage and water turbidity. During this time, it is likely that the eukaryotes were mainly carrying out respiration, whereas when the tide is out and oxygen concentrations are low, the eukaryotes may instead mainly perform photosynthesis. Repeated sampling during a period when high tide falls overnight could help to unravel this observation.
Genes for the denitrification pathway were also expressed at all time points across the tidal cycle regardless of the ambient O2 concentration (Supplementary Table S4). The transcription of the marker gene for the periplasmic nitrate reductase (napA) was almost four times greater than the transcription of that for the membrane bound nitrate reductase (narG). Furthermore, napA was twice as common in the metagenomes than narG. Predominance of Nap in certain marine systems has been attributed to its high affinity for nitrate (Smith et al., 2007; Dong et al., 2009; Vetriani et al., 2014). However, nitrate is rarely limiting in the permeable sediments of the Wadden Sea (Gao et al., 2012). This suggests that the dominance of Nap is likely because of the physiological role that it is predicted to play during aerobic denitrification, where it is typically used rather than Nar (Bell et al., 1990; Richardson and Ferguson, 1992).
Strikingly, there was no correlation between the transcription pattern of napA, narG and nosZ and the diurnal changes in O2 levels within the sediment. Instead, at the phylum, class and order level, transcription of denitrification genes correlated with that of the high- and low-affinity terminal oxidases (Figure 4). A positive correlation between high-affinity terminal oxidases and denitrification gene transcription may not be unexpected, as both are known to be expressed under low O2 conditions. However, the concurrent upregulation of low-affinity terminal oxidase and denitrification gene transcription strongly indicates that the broad changes in O2 availability over a tidal cycle do not control transcription of the respiratory pathways.
The distribution of napA and narG transcription and the co-transcription patterns observed between denitrification genes and terminal oxidases persisted even at the class and order level. This evidence, combined with the observations that a large proportion of the community appear to constitutively express the denitrification pathway, strongly suggests that no-one particular group of organisms is responsible for aerobic denitrification within these sediments, but rather the trait is selected for throughout the community. This supports evidence from phylogeny analysis of cultured aerobic denitrifiers, which have been shown to belong to diverse proteobacterial classes, as well as the Actinobacteria and Firmicute phyla (Ji et al., 2015).
Environmental implications and conclusions
Within the permeable sediments studied here, aerobic denitrification at high O2 concentrations occurred because of a relaxed regulation by O2 on transcription of respiratory genes, as well as metabolic regulation of electron flow to the denitrification pathway in the presence of oxygen. Such a relaxation has likely occurred in order to allow microbes to exploit the large but periodic supply of electron acceptors that are supplied by advective porewater flow (Figure 5). Here, we have focused on intertidal sediments that are exposed to large fluctuations in O2 availability over a tidal cycle. However, even permanently inundated subtidal sandy sediments experience large fluctuations in O2 depending on flow velocity and sediment redistribution (Ahmerkamp et al., 2015). We have observed denitrification occurring immediately when subtidal sediments kept oxic for a month were allowed to become anoxic (unpublished data). Therefore, it is likely the same decrease in regulation by O2 occurs in subtidal sediments, driving the aerobic denitrification that has been reported previously (Rao et al., 2008; Marchant et al., 2016). Although aerobic denitrification contributes little to the overall energy conservation by respiration in permeable sediments, in terms of N2 production it can be substantial. Here we observed aerobic denitrification rates that at times were up to 60% of the maximal anaerobic denitrification rate—rates that are among the highest measured in the marine environment (Gihring et al., 2010; Gao et al., 2012). Sandy sediments cover between 50% and 70% of the continental shelves (Emery, 1968; Hall, 2002), and the growing consensus is that they are extremely important sites of N-loss (Huettel et al., 2014; Devol, 2015; Sokoll et al., 2016). It therefore appears that aerobic denitrification may not just be a fascinating process in regards to the kinetics and bioenergetics of branched respiratory pathways, but an important process in the marine N cycle.
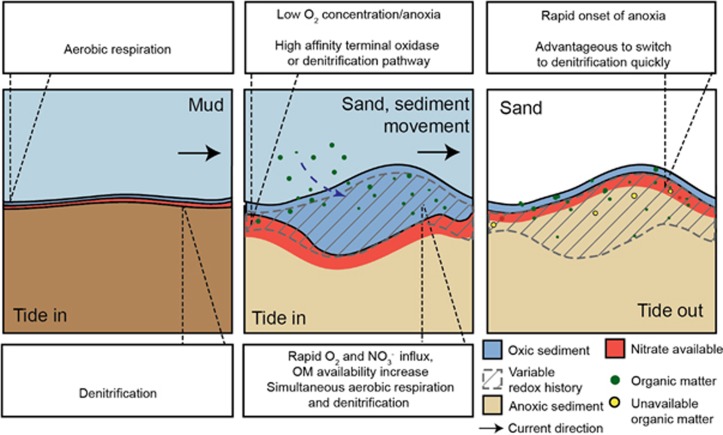
Figure 5.
Schematic overview of the dynamic conditions that lead to constitutive expression of multiple respiration pathways in permeable sands and aerobic denitrification. In muddy sediments, supply of electron acceptors and donors relies on diffusion from the overlying water, O2 and NO3− are consumed within millimeters. The supply of organic matter and NO3− varies on seasonal scales—for example, organic matter reaching the sediment may be higher during phytoplankton blooms. In permeable sands, O2, NO3− and organic matter are advected into the sediment at velocities orders of magnitude higher than diffusion, allowing penetration depths to increase to centimeters even while respiration rates remain high. In contrast to muds, sands are more dynamic. When current velocities increase sediment starts to move, changing O2 and NO3− penetration depths and re-exposing organic matter buried in anoxic sediment to oxic conditions. Furthermore, intertidal sediments are exposed to the air for hours each day when the tide is out, at which point they are limited by diffusion and drainage of water through sand flats. These dynamics results in the upper layer of the sediment being exposed to variable redox conditions. Under such rapid fluctuations, it is favorable to constantly express a suite of respiratory pathways in order to take advantage of the abundant energy sources. Furthermore, when previously anoxic sediment becomes oxic, it is likely that denitrification proteins expressed under oxic conditions are still functional, which coincides with organic matter that was previously refractory under anoxic conditions becoming available to microbial degradation again. Such rapid imbalances between electron donor and electron acceptor supply are likely responsible for the observed use of the denitrification pathway under oxic conditions.
Currently, estimates of N-loss from permeable sediments are poorly constrained (Huettel et al., 2014), largely because of the difficulties of modeling biogeochemical transformations in such complex environments. Estimates of O2 and N fluxes from permeable sediments are, however, improving constantly because of the application of two-dimensional reactive transport models (Cardenas et al., 2008; Ahmerkamp et al., 2015; Sokoll et al., 2016). The occurrence of aerobic denitrification is currently not considered in these biogeochemical models, which rely on the kinetics of reaction rates for aerobic respiration and denitrification and so far do not simulate the transport of organic matter into the sediment. We have shown that denitrification rates are governed not only by kinetics but also by metabolic responses to changing electron donor and acceptor availability. If, as seems likely, the potential for aerobic denitrification is inherently selected for by the dynamic conditions that characterize permeable sands, then aerobic denitrification should be considered in permeable sediment models going forward.
Acknowledgments
We thank the Marine Sensor Systems group at the ICBM, Oldenburg for providing data from the Time Series Station at Spiekeroog. G Klockgether for technical assistance, D de beer for assistance with the microsensor profiles and F Widdel for discussions concerning the data. This work was financially supported by the Max Planck Society and the DFG-Research Center/Cluster of Excellence 'The Ocean in the Earth System' at the University of Bremen.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Ahmerkamp S, Winter C, Janssen F, Kuypers MMM, Holtappels M. (2015). The impact of bedform migration on benthic oxygen fluxes. J Geophys Res Biogeosci 120: 2229–2242. [Google Scholar]
- Arai H. (2011). Regulation and function of versatile aerobic and anaerobic respiratory metabolism inPseudomonasaeruginosa. Front Microbiol 2: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken LR, Bergaust L, Liu B, Frostegård Å. (2012). Regulation of denitrification at the cellular level: a clue to the understanding of N2O emissions from soils. Philos Trans R Soc B Biol Sci 367: 1226–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann B, Snozzi M, Zehnder AJB, vanderMeer JR. (1996). Dynamics of denitrification activity of Paracoccus denitrificans in continuous culture during aerobic-anaerobic changes. J Bacteriol 178: 4367–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann B, van der Meer JR, Snozzi M, Zehnder AJ. (1997). Inhibition of denitrification activity but not of mRNA induction in Paracoccus denitrificans by nitrite at a suboptimal pH. Antonie van Leeuwenhoek 72: 183–189. [DOI] [PubMed] [Google Scholar]
- Bell LC, Richardson DJ, Ferguson SJ. (1990). Periplasmic and membrane-bound respiratory nitrate reductases in Thiosphaera pantotropha. FEBS Lett 265: 85–87. [DOI] [PubMed] [Google Scholar]
- Bergaust L, Shapleigh J, Frostegard A, Bakken L. (2008). Transcription and activities of NOx reductases in Agrobacterium tumefaciens: the influence of nitrate, nitrite and oxygen availability. Environ Microbiol 10: 3070–3081. [DOI] [PubMed] [Google Scholar]
- Bergaust L, Mao Y, Bakken LR, Frostegård Å. (2010). Denitrification response patterns during the transition to anoxic respiration and posttranscriptional effects of suboptimal pH on nitrous oxide reductase in Paracoccus denitrificans. Appl Environ Microbiol 76: 6387–6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergaust L, van Spanning RJM, Frostegård Å, Bakken LR. (2012). Expression of nitrous oxide reductase in Paracoccus denitrificans is regulated by oxygen and nitric oxide through FnrP and NNR. Microbiology 158: 826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billerbeck M, Werner U, Polerecky L, Walpersdorf E, de Beer D, Huettel M. (2006). Surficial and deep pore water circulation governs spatial and temporal scales of nutrient recycling in intertidal sand flat sediment. Mar Ecol-Prog Ser 326: 61–76. [Google Scholar]
- Böer SI, Arnosti C, van Beusekom JEE, Boetius A. (2009). Temporal variations in microbial activities and carbon turnover in subtidal sandy sediments. Biogeosciences 6: 1149–1165. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics: btu170 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas MB, Cook PL, Jiang H, Traykovski P. (2008). Constraining denitrification in permeable wave-influenced marine sediment using linked hydrodynamic and biogeochemical modeling. Earth Planet Sci Lett 275: 127–137. [Google Scholar]
- Chen JW, Strous M. (2013). Denitrification and aerobic respiration, hybrid electron transport chains and co-evolution. Biochim Biophys Acta-Bioenerg 1827: 136–144. [DOI] [PubMed] [Google Scholar]
- Dalsgaard T, Stewart FJ, Thamdrup B, De Brabandere L, Revsbech NP, Ulloa O et al. (2014). Oxygen at nanomolar levels reversibly suppresses process rates and gene expression in anammox and denitrification in the oxygen minimum zone off Northern Chile. mBio 5: e01966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer D, Wenzhofer F, Ferdelman TG, Boehme SE, Huettel M, van Beusekom JEE et al. (2005). Transport and mineralization rates in North Sea sandy intertidal sediments, Sylt-Romo Basin, Wadden Sea. Limnol Oceanogr 50: 113–127. [Google Scholar]
- Devol AH. (2015). Denitrification, anammox, and N(2) production in marine sediments. Ann Rev Mar Sci 7: 403–423. [DOI] [PubMed] [Google Scholar]
- Dong LF, Smith CJ, Papaspyrou S, Stott A, Osborn AM, Nedwell DB. (2009). Changes in benthic denitrification, nitrate ammonification, and anammox process rates and nitrate and nitrite reductase gene abundances along an estuarine nutrient gradient (the Colne Estuary, United Kingdom). Appl Environ Microbiol 75: 3171–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggleston EM, Lee DY, Owens MS, Cornwell JC, Crump BC, Hewson I. (2015). Key respiratory genes elucidate bacterial community respiration in a seasonally anoxic estuary. Environ Microbiol 17: 2306–2318. [DOI] [PubMed] [Google Scholar]
- Ehrenhauss S, Witte U, Buhring SI, Huettel M. (2004. a). Effect of advective pore water transport on distribution and degradation of diatoms in permeable North Sea sediments. Mar Ecol Prog Ser 271: 99–111. [Google Scholar]
- Ehrenhauss S, Witte U, Janssen F, Huettel M. (2004. b). Decomposition of diatoms and nutrient dynamics in permeable North Sea sediments. Cont Shelf Res 24: 721–737. [Google Scholar]
- Ellington MJ, Bhakoo KK, Sawers G, Richardson DJ, Ferguson SJ. (2002). Hierarchy of carbon source selection in Paracoccus pantotrophus: strict correlation between reduction state of the carbon substrate and aerobic expression of the nap operon. J Bacteriol 184: 4767–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery KO. (1968). Relict sediments on continental shelves of world. AAPG Bull 52: 445–464. [Google Scholar]
- Frette L, Gejlsbjerg B, Westermann P. (1997). Aerobic denitrifiers isolated from an alternating activated sludge system. FEMS Microbiol Ecol 24: 363–370. [Google Scholar]
- Galloway JN, Dentener FJ, Capone DG, Boyer EW, Howarth RW, Seitzinger SP et al. (2004). Nitrogen cycles: past, present, and future. Biogeochemistry 70: 153–226. [Google Scholar]
- Gao H, Schreiber F, Collins G, Jensen MM, Kostka JE, Lavik G et al. (2010). Aerobic denitrification in permeable Wadden Sea sediments. ISME J 4: 417–426. [DOI] [PubMed] [Google Scholar]
- Gao H, Matyka M, Liu B, Khalili A, Kostka JE, Collins G et al. (2012). Intensive and extensive nitrogen loss from intertidal permeable sediments of the Wadden Sea. Limnol Oceanogr 57: 185–198. [Google Scholar]
- Gihring TM, Canion A, Riggs A, Huettel M, Kostka JE. (2010). Denitrification in shallow, sublittoral Gulf of Mexico permeable sediments. Limnol Oceanogr 55: 43–54. [Google Scholar]
- Gruber N, Galloway JN. (2008). An Earth-system perspective of the global nitrogen cycle. Nature 451: 293–296. [DOI] [PubMed] [Google Scholar]
- Hall SJ. (2002). The continental shelf benthic ecosystem: current status, agents for change and future prospects. Environ Conserv 29: 350–374. [Google Scholar]
- Hewson I, Eggleston EM, Doherty M, Lee DY, Owens M, Shapleigh JP et al. (2014). Metatranscriptomic analyses of plankton communities inhabiting surface and subpycnocline waters of the Chesapeake Bay during oxic-anoxic-oxic transitions. Appl Environ Microbiol 80: 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojberg O, Binnerup SJ, Sorensen J. (1997). Growth of silicone-immobilized bacteria on polycarbonate membrane filters, a technique to study microcolony formation under anaerobic conditions. Appl Environ Microbiol 63: 2920–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel M, Rusch A. (2000). Transport and degradation of phytoplankton in permeable sediment. Limnol Oceanogr 45: 534–549. [Google Scholar]
- Huettel M, Roy H, Precht E, Ehrenhauss S. (2003). Hydrodynamical impact on biogeochemical processes in aquatic sediments. Hydrobiologia 494: 231–236. [Google Scholar]
- Huettel M, Berg P, Kostka JE. (2014). Benthic exchange and biogeochemical cycling in permeable sediments. Annu Rev Mar Sci 6: 23–51. [DOI] [PubMed] [Google Scholar]
- Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. (2011). Integrative analysis of environmental sequences using MEGAN4. Genome Res 21: 1552–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform 11: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen S, Walpersdorf E, Werner U, Billerbeck M, Bottcher ME, de Beer D. (2009). Functioning of intertidal flats inferred from temporal and spatial dynamics of O2, H2S and pH in their surface sediment. Ocean Dyn 59: 317–332. [Google Scholar]
- Ji B, Yang K, Zhu L, Jiang Y, Wang H, Zhou J et al. (2015). Aerobic denitrification: a review of important advances of the last 30 years. Biotechnol Bioprocess Eng 20: 643–651. [Google Scholar]
- Ka JO, Urbance J, Ye RW, Ahn TY, Tiedje JM. (1997). Diversity of oxygen and N-oxide regulation of nitrite reductases in denitrifying bacteria. FEMS Microbiol Lett 156: 55–60. [DOI] [PubMed] [Google Scholar]
- Kalvelage T, Lavik G, Jensen MM, Revsbech NP, Löscher C, Schunck H et al. (2015). Aerobic microbial respiration in oceanic oxygen minimum zones. PLoS One 10: e0133526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Jeong S-Y, Yoon SJ, Cho SJ, Kim YH, Kim MJ et al. (2008). Aerobic denitrification of Pseudomonas putida AD-21 at different C/N ratios. J Biosci Bioeng 106: 498–502. [DOI] [PubMed] [Google Scholar]
- Kopylova E, Noé L, Touzet H. (2012). SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28: 3211–3217. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. [DOI] [PubMed] [Google Scholar]
- Marchant HK, Lavik G, Holtappels M, Kuypers MMM. (2014). The fate of nitrate in intertidal permeable sediments. PLoS One 9: e104517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant HK, Holtappels M, Lavik G, Ahmerkamp S, Winter C, Kuypers MMM. (2016). Coupled nitrification–denitrification leads to extensive N loss in subtidal permeable sediments. Limnol Oceanogr 61: 1033–1048. [Google Scholar]
- Morris RL, Schmidt TM. (2013). Shallow breathing: bacterial life at low O2. Nat Rev Micro 11: 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen LP. (1992). Denitrification in sediment determined from nitrogen isotope pairing. FEMS Microbiol Ecol 86: 357–362. [Google Scholar]
- Otten MF, Stork DM, Reijnders WN, Westerhoff HV, Van Spanning RJ. (2001). Regulation of expression of terminal oxidases in Paracoccus denitrificans. Eur J Biochem 268: 2486–2497. [DOI] [PubMed] [Google Scholar]
- Patureau D, Davison J, Bernet N, Moletta R. (1994). Denitrification under various aeration conditions in Comamonas sp. strain SGLY2. FEMS Microbiol Ecol 14: 71–78. [Google Scholar]
- Patureau D, Zumstein E, Delgenes JP, Moletta R. (2000). Aerobic denitrifiers isolated from diverse natural and managed ecosystems. Microbial Ecol 39: 145–152. [DOI] [PubMed] [Google Scholar]
- Pilditch CA, Miller DC. (2006). Phytoplankton deposition to permeable sediments under oscillatory flow: effects of ripple geometry and resuspension. Cont Shelf Res 26: 1806–1825. [Google Scholar]
- Polerecky L, Franke U, Werner U, Grunwald B, de Beer D. (2005). High spatial resolution measurement of oxygen consumption rates in permeable sediments. Limnol Oceanogr Methods 3: 75–85. [Google Scholar]
- Poole RK, Cook GM. (2000). Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv Microbial Physiol 43: 165–224. [DOI] [PubMed] [Google Scholar]
- Qu Z, Bakken LR, Molstad L, Frostegård Å, Bergaust LL. (2016). Transcriptional and metabolic regulation of denitrification in Paracoccus denitrificans allows low but significant activity of nitrous oxide reductase under oxic conditions. Environ Microbiol 18: 2951–2963. [DOI] [PubMed] [Google Scholar]
- Rao AMF, McCarthy MJ, Gardner WS, Jahnke RA. (2008). Respiration and denitrification in permeable continental shelf deposits on the South Atlantic Bight: N2: Ar and isotope pairing measurements in sediment column experiments. Cont Shelf Res 28: 602–613. [Google Scholar]
- Reuter R, Badewien TH, Bartholomä A, Braun A, Lübben A, Rullkötter J. (2009). A hydrographic time series station in the Wadden Sea (southern North Sea). Ocean Dyn 59: 195–211. [Google Scholar]
- Richardson D. (2008) Structural and functional flexibility of bacterial respiromes. In:El-Sharoud W (ed), Bacterial Physiology. Springer: Berlin Heidelberg, Germany, pp 97–128. [Google Scholar]
- Richardson DJ, Ferguson SJ. (1992). The influence of carbon substrate on the activity of the periplasmic nitrate reductase in aerobically grown Thiosphaera pantotropha. Arch Microbiol 157: 535–537. [Google Scholar]
- Robertson LA, Kuenen JG. (1984). Aerobic denitrification - old wine in new bottles. Antonie Van Leeuwenhoek J Microbiol 50: 525–544. [DOI] [PubMed] [Google Scholar]
- Robertson LA, Kuenen JG. (1990). Combined heterotrophic nitrification and aerobic denitrification in Thiosphaera pantotropha and other bacteria. Antonie van Leeuwenhoek 57: 139–152. [DOI] [PubMed] [Google Scholar]
- Robertson LA, Dalsgaard T, Revsbech NP, Kuenen JG. (1995). Confirmation of aerobic denitrification in batch cultures, using gas-chromatography and N15 mass spectrometry. FEMS Microbiol Ecol 18: 113–119. [Google Scholar]
- Rusch A, Forster S, Huettel M. (2001). Bacteria, diatoms and detritus in an intertidal sandflat subject to advective transport across the water-sediment interface. Biogeochemistry 55: 1–27. [Google Scholar]
- Schunck H, Lavik G, Desai DK, Großkopf T, Kalvelage T, Löscher CR et al. (2013). Giant hydrogen sulfide plume in the oxygen minimum zone off Peru supports chemolithoautotrophy. PLoS One 8: e68661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears HJ, Spiro S, Richardson DJ. (1997). Effect of carbon substrate and aeration on nitrate reduction and expression of the periplasmic and membrane-bound nitrate reductases in carbon-limited continuous cultures of Paracoccus denitrificans Pd1222. Microbiology 143: 3767–3774. [DOI] [PubMed] [Google Scholar]
- Smith CJ, Nedwell DB, Dong LF, Osborn AM. (2007). Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl Environ Microbiol 73: 3612–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoll S, Lavik G, Sommer S, Goldhammer T, Kuypers MMM, Holtappels M. (2016). Extensive nitrogen loss from permeable sediments off North-West Africa. J Geophys Res Biogeosci 121: 1144–1157. [Google Scholar]
- Sousa FL, Alves RJ, Pereira-Leal JB, Teixeira M, Pereira MM. (2011). A bioinformatics classifier and database for heme-copper oxygen reductases. PLoS One 6: e19117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stief P, Kamp A, de Beer D. (2013). Role of diatoms in the spatial-temporal distribution of intracellular nitrate in intertidal sediment. PLoS One 8: e73257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C-P, Albrecht J, Gunsalus RP. (1996). Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J Bacteriol 178: 1094–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetriani C, Voordeckers JW, Crespo-Medina M, O'Brien CE, Giovannelli D, Lutz RA. (2014). Deep-sea hydrothermal vent Epsilonproteobacteria encode a conserved and widespread nitrate reduction pathway (Nap). ISME J 8: 1510–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zennaro E, Ciabatti I, Cutruzzola F, D'Alessandro R, Silvestrini MC. (1993). The nitrite reductase gene of Pseudomonas aeruginosa: effect of growth conditions on the expression and construction of a mutant by gene disruption. FEMS Microbiol Lett 109: 243–250. [DOI] [PubMed] [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. (1996). DNA recovery from soils of diverse composition. Appl Environ Microbiol 62: 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft WG. (1997). Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61: 533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.