Abstract
In the transition zone of the shifting permafrost border, thaw ponds emerge as hotspots of microbial activity, processing the ancient carbon freed from the permafrost. We analyzed the microbial succession across a gradient of recently emerged to older ponds using three molecular markers: one universal, one bacterial and one fungal. Age was a major modulator of the microbial community of the thaw ponds. Surprisingly, typical freshwater taxa comprised only a small fraction of the community. Instead, thaw ponds of all age classes were dominated by enigmatic bacterial and fungal phyla. Our results on permafrost thaw ponds lead to a revised perception of the thaw pond ecosystem and their microbes, with potential implications for carbon and nutrient cycling in this increasingly important class of freshwaters.
Introduction
As the warming climate pushes the southern border of permafrost northwards, carbon that has been trapped in the permafrost for thousands of years becomes available (Tranvik et al., 2009; Vonk et al., 2012). Thaw ponds emerge as a characteristic feature of thawing permafrost, creating microbial hotspots of carbon cycling (Roiha et al., 2015) and comprising significant sources of greenhouse gas emissions (Abnizova et al., 2012; Negandhi et al., 2013).
The environmental importance of these habitats together with the impact of microbial community composition on carbon degradation (Logue et al., 2016) make it essential to understand the microbial processes and communities in these ponds. Several studies have assessed the underlying microbial communities and their assembly processes (Crevecoeur et al., 2015; Comte et al., 2016; Przytulska et al., 2016). These studies found a bacterial community that resembled the bacterioplankton communities of temperate and boreal lakes but enriched with methanotrophic lineages (Crevecoeur et al., 2015). Still, while the taxonomic composition of thaw pond communities is characterized by these functional groups, it varies significantly across individual thaw ponds, potentially driven by stochastic processes and environmental filtering (Comte et al., 2016). As thaw ponds increase in size with age, often growing from a puddle to a pond within a decade, the age of the pond is likely to be an additional deterministic factor associated with the microbial community composition.
We hypothesize that emerging thaw ponds are initially colonized by a pioneer community, which then undergoes a progressive succession as the pond ages. In this study, we investigated thaw pond communities across a gradient of age classes. To extend previous thaw pond characterization focused on bacterial communities, we used three ribosomal markers (universal SSU, bacterial 16S rRNA gene, and fungal ITS2) to characterize a greater spectrum of the microbial community. We were specifically interested in fungi as they are known to be superior degraders of high molecular weight carbon (Harms et al., 2011), which is the main carbon pool in thaw ponds (Roiha et al., 2015).
Study implementation
The 12 thaw ponds targeted represent three different age categories (emerging, middle-aged, and old) and are located in the Canadian sporadic permafrost area (Figure 1a, Supplementary Methods). The ponds were also sampled vertically according to their thermal stratification (oxic epilimnion, transition zone (metalimnion), and suboxic/anoxic hypolimnion). We extracted DNA from water samples concentrated on 0.22 μm filters and PCR amplified and sequenced three ribosomal markers (the universal V6-8 region, the bacterial V4 region, and the fungal ITS region; Supplementary Methods). We also measured a set of indicators for carbon quantity and quality; and methane and carbon dioxide concentration (Supplementary Figure S1, Supplementary Methods).
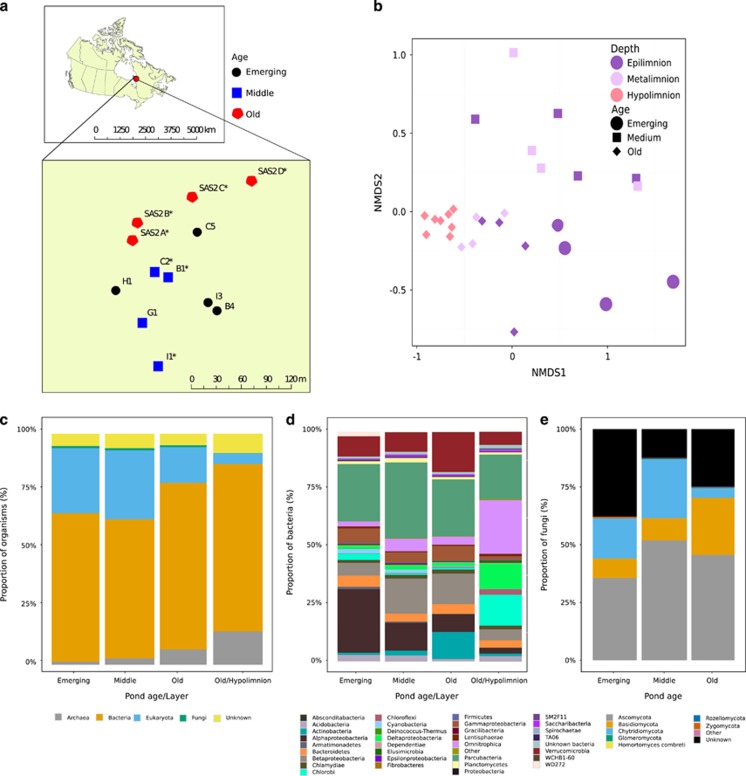
Figure 1.
Study location and characteristics of the microbial communities. (a) Map of the thaw ponds. The emerging and middle-aged ponds are located around still existing palsa formations while the old ponds have replaced their respective palsas completely. Palsas are frost heaves containing permanently frozen ice lenses, consisting of an ice core with overlying soil. At the study site, the height of palsas is up to 3 m. Colors depict different age groups, and asterisks indicate ponds that we sampled also vertically. (b) Two-dimensional NMDS (stress =0.11) showing the beta-diversity within the ponds with shapes illustrating pond age and colors sampling depth. Both age and depth were found to influence the universal community assembly (Adonis, R2=0.25, P<0.001 (for age, excluding hypolimnion samples) and R2=0.20, P<0.001 (for depth)). (c) Universal, (d) bacterial and (e) fungal community composition in the epilimnia of ponds representing different stages of succession. The composition is shown at the phylum level for bacteria and fungi and at the domain level for the universal community, except that fungi have been separated from Eukaryota due to their central role in the study. For bacteria, the group ‘Other’ is the sum of phyla containing less than 0.1% of the total community. For the bacterial and universal communities also the taxonomic composition in the hypolimnion of the old ponds is shown. The hypolimnion is the anoxic bottom layer present only in the old ponds. A more detailed taxonomic overview can be found in Supplementary Figure S3.
Results and Discussions
The emerging thaw ponds were significantly different from the older ponds in their environmental parameters. In particular, they differed in their lower pH and higher concentration of dissolved C02, dissolved organic carbon (DOC), refractory carbon, and total nitrogen. Although the methane concentrations were high in hypolimnia of the old ponds, epilimnetic methane was not significantly different between emerging and old ponds. (Supplementary Table S1, Supplementary Figure S1). The organism beta-diversity was significantly correlated with pond age (Figure 1b; Adonis: R2=0.25, P<0.001). The successional progression of thaw ponds was also visible in the relative abundance of the taxonomic groups (Figures 1c–e, Supplementary Figure S2; for a detailed inspection of taxonomic groups, see Supplementary Figure S3). Overall, bacteria constituted the most abundant microbial group followed by Eukarya and Archaea (Figure 1d), both of which can be expected to account for important functional aspects at different depths in the ponds (for example, archaeal methane production at the bottom (Supplementary Figure S4); or algal primary production at the surface). The fungal community ranged between 1 and 3% of the total community and harbored typical freshwater lineages such as Chytridiomycota (Figure 1e; Monchy et al., 2011), but especially in the emerging ponds, which possessed the highest carbon loads, a large part of the community was composed of fungal lineages whose taxonomic resolution could not be resolved with certainty (Figure 1e, Supplementary Table S1). In fact, around 25% of all fungi could only be assigned to kingdom level, and 9% of the fungal reads matched the enigmatic, undescribed fungal lineages identified by Nilsson et al. (2016). In all the pond stages, both the universal and the bacterial markers identified members of candidate phylum Omnitrophica (candidate division OP3) and taxa from the ‘candidate phyla radiation’ (CPR, Brown et al., 2015), in particular Parcubacteria (candidate division OD1) as dominant taxa. These phyla also exhibited the greatest alpha-diversity (Table 1). Typically, these candidate phyla are found in anoxic environments (Peura et al., 2012; Wrighton et al., 2012; Probst et al., 2016), and we can only speculate on their ecological roles in aerobic layers of thaw ponds. The previous thaw pond studies did not recover these poorly known lineages, but instead reported typical freshwater lineages as the dominant bacterioplankton (Crevecoeur et al., 2015, Supplementary Figure S2). Some of the ponds studied here are the same as those examined in the earlier studies (here assigned SAS2 A-B, also sampled in the same season; for example, Crevecoeur et al., 2015). This excludes geographical and seasonal variation as the primary cause for this discrepancy—leaving annual variation or methodological biases as explanations. For example, Rossi et al. (2013) have speculated that previously used primers may have had mismatches within the candidate phyla. We analyzed primer matches in silico and could confirm mismatches of previous primers for at least Omnitrophica and Parcubacteria (Supplementary Table S2).
Table 1. Diversity measures of the total bacterial community in the ponds, excluding hypolimnion samples, representing different phases of the succession (in italics, ±standard deviation), median of all phyla, and the median diversity of each individual phylum.
| Phylum/age class | Inverse Simpson | Pielou´s evenness |
|---|---|---|
| All ponds | 66.1 | 0.70 |
| Emerging | 56.9±44.9 | 0.75±0.07 |
| Middle-aged | 87.8±55.9 | 0.80±0.05 |
| Old | 87.8±79.0 | 0.79±0.08 |
| Median of all phyla | 11.6 | 0.80 |
| Actinobacteria | 6.9 | 0.67* |
| Alphaproteobacteria | 11.7 | 0.66** |
| Bacteroidetes | 8.7 | 0.68* |
| Betaproteobacteria | 6.0 | 0.58*** |
| Chlamydiae | 34.6*** | 0.87* |
| Chlorobia | 1.7** | 0.42** |
| Cyanobacteria | 3.0 | 0.44*** |
| Gammaproteobacteria | 3.8 | 0.52** |
| Omnitrophica | 32.8** | 0.82 |
| Parcubacteria | 75.9*** | 0.83 |
| Planctomycetes | 25.8*** | 0.89* |
| SM2F11 | 5.0** | 0.73 |
| Unclassified Bacteria | 32.2** | 0.91 |
| Verrucomicrobia | 9.9 | 0.68** |
Only phyla that differ significantly from the median of all phyla are shown (two-sided Wilcoxon signed rank test). Phyla that are considered to be poorly known are highlighted in bold. The results have been corrected for multiple comparisons using Bonferroni correction, and statistical significance is indicated with asterisks; *, <0.05; **, <0.005; ***, <0.0005.
Although our study is somewhat restricted geographically and may not represent the complete circumpolar permafrost border, it shows that permafrost thaw ponds undergo a progressive succession over time with respect to both microbial community composition and the composition of carbon compounds. Given the observation that the carbon pool especially in the emerging and middle stages of pond development is dominated by carbon compounds of terrestrial origin (see HI- and FI-indices and CDOM concentration in Supplementary Figure 1), we hypothesize that the cryptic microorganisms are likely to be involved in the processes associated with thawing permafrost soils.
Conclusions
Our study site, located at the edge of the permafrost line, represents an area central to the global warming discussions due to its sensitivity to temperature change (Abnizova et al., 2012). The microbial diversity in permafrost thaw ponds is to a large extent composed of microbes with enigmatic ecological roles and taxonomic affiliations. Although these ponds share some ecosystem features with lakes, they form a distinct ecosystem type in terms of environmental parameters, organism composition, and slow pond progression. Our findings render thaw pond microbiology unique among surface waters and may establish permafrost thaw ponds as a new ecosystem category. The fact that the ecology of their microbiome cannot be assessed at present only further hints at our incomplete understanding of the processes underlying the carbon and nutrient cycling, both of which lie at the very heart of climate change discussions.
Acknowledgments
We thank the CEN Whapmagoostui-Kuujjuarapik Station and Claude Tremblay for the use of laboratory space and transportation to the SAS2 site. Karólína Einarsdottir, Maxime Wauthy, David-Alexandre Gauthier, Felix Faucher, and Julien Lebrun are acknowledged for their help with the sampling, Caroline Ponsonby for the map of the sampling site, Pilar Lopez-Hernandez for her help with the molecular laboratory work, Alexander Eiler for processing the bacterial amplicon data, and Jerome Comte for comments on an early draft of the manuscript. The funding was provided by the Academy of Finland (grant 265902 to SP), the International Network for Terrestrial Research and Monitoring in the Arctic (INTERACT; grant to SP), and the Marie Skłodowska-Curie actions (grant CRYPTRANS to CW). Financial support was further provided by the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Canada Research Chair program.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Abnizova A, Siemens J, Langer M, Boike J. (2012). Small ponds with major impact: the relevance of ponds and lakes in permafrost landscapes to carbon dioxide emissions. Global Biogeochem Cy 26: GB2041. [Google Scholar]
- Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A et al. (2015). Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523: 208–211. [DOI] [PubMed] [Google Scholar]
- Comte J, Monier A, Crevecoeur S, Lovejoy C, Vincent WF. (2016). Microbial biogeography of permafrost thaw ponds across the changing northern landscape. Ecography 39: 609–618. [Google Scholar]
- Crevecoeur S, Vincent WF, Comte J, Lovejoy C. (2015). Bacterial community structure across environmental gradients in permafrost thaw ponds: methanotroph-rich ecosystems. Front Microbiol 6: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms H, Schlosser D, Wick LY. (2011). Untapped potential: exploiting fungi in bioremediation of hazardous chemicals. Nat Rev Microbiol 9: 177–192. [DOI] [PubMed] [Google Scholar]
- Logue JB, Stedmon CA, Kellerman AM, Nielsen NJ, Andersson AF, Laudon H et al. (2016). Experimental insights into the importance of aquatic bacterial community composition to the degradation of dissolved organic matter. ISME J 10: 533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monchy S, Sanciu G, Jobard M, Rasconi S, Gerphagnon M, Chabé M et al. (2011). Exploring and quantifying fungal diversity in freshwater lake ecosystems using rDNA cloning/sequencing and SSU tag pyrosequencing. Environ Microbiol 13: 1433–1453. [DOI] [PubMed] [Google Scholar]
- Negandhi K, Laurion I, Whiticar MJ, Galand PE, Xu X, Lovejoy C. (2013). Small thaw ponds: an unaccounted source of methane in the Canadian High Arctic. PLoS ONE 8: e78204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson RH, Wurzbacher C, Bahram M, Coimbra VRM, Larsson E, Tedersoo L et al. (2016). Top 50 most wanted fungi. MycoKeys 12: 29–40. [Google Scholar]
- Peura S, Eiler A, Bertilsson S, Nykänen H, Tiirola M, Jones RI. (2012). Distinct and diverse anaerobic bacterial communities in boreal lakes dominated by candidate division OD1. ISME J 6: 1640–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AJ, Castelle CJ, Singh A, Brown CT, Anantharaman K, Sharon I et al. (2016). Genomic resolution of a cold subsurface aquifer community provides metabolic insights for novel microbes adapted to high CO2 concentrations. Environ Microbiol 19: 459–474. [DOI] [PubMed] [Google Scholar]
- Przytulska A, Comte J, Crevecoeur S, Lovejoy C, Laurion I, Vincent WF. (2016). Phototrophic pigment diversity and picophytoplankton in permafrost thaw lakes. Biogeosciences 13: 13–26. [Google Scholar]
- Roiha T, Laurion I, Rautio M. (2015). Carbon dynamics in highly net heterotrophic subarctic thaw ponds. Biogeosciences 12: 7223–7237. [Google Scholar]
- Rossi PG, Laurion I, Lovejoy C. (2013). Distribution and identity of Bacteria in subarctic permafrost thaw ponds. Aquat Microb Ecol 69: 231–245. [Google Scholar]
- Tranvik LJ, Downing JA, Cotner JB, Loiselle SA, Striegl RG, Ballatore TJ et al. (2009). Lakes and reservoirs as regulators of carbon cycling and climate. Limnol Oceanogr 54: 2298–2314. [Google Scholar]
- Vonk JE, Sanchez-Garcia L, van Dongen BE, Alling V, Kosmach D, Charkin A et al. (2012). Activation of old carbon by erosion of coastal and subsea permafrost in arctic Siberia. Nature 489: 137–140. [DOI] [PubMed] [Google Scholar]
- Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC et al. (2012). Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 28: 1661–1665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.