Abstract
Extracts of calf thymus have been fractionated to reveal a nuclease activity that specifically cleaves model Holliday junctions in vitro. The products of cleavage are unbranched linear duplex DNA molecules. Using synthetic four-way junctions, we show that the major sites of cutting are diametrically opposed, at sites one nucleotide from the base of the junction. Other types of four-way junctions, including pseudo-cruciform structures and cruciforms extruded from supercoiled plasmids, are also cleaved by the nuclease. The Mr of the partially purified activity, determined by gel filtration, is approximately 75,000. The calf thymus enzyme provides the first example of an endonuclease from a higher eukaryote that acts specifically on branch points in DNA, and indicates that junction-resolving proteins are normal constituents of somatic cells.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barth K. A., Powell D., Trupin M., Mosig G. Regulation of two nested proteins from gene 49 (recombination endonuclease VII) and of a lambda RexA-like protein of bacteriophage T4. Genetics. 1988 Oct;120(2):329–343. doi: 10.1093/genetics/120.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi G. Mechanism of action and structure of acid deoxyribonuclease. Adv Enzymol Relat Areas Mol Biol. 1968;31:1–49. doi: 10.1002/9780470122761.ch1. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E., Beltrame M., Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989 Feb 24;243(4894 Pt 1):1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E. Interaction of a protein from rat liver nuclei with cruciform DNA. EMBO J. 1988 Mar;7(3):843–849. doi: 10.1002/j.1460-2075.1988.tb02883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassuto E., Lightfoot L. A., Howard-Flanders P. Partial purification of an activity from human cells that promotes homologous pairing and the formation of heteroduplex DNA in the presence of ATP. Mol Gen Genet. 1987 Jun;208(1-2):10–14. doi: 10.1007/BF00330415. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Tullius T. D., Kallenbach N. R., Seeman N. C. A Holliday recombination intermediate is twofold symmetric. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4653–4656. doi: 10.1073/pnas.85.13.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. P., Hagerman P. J. Geometry of a branched DNA structure in solution. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7336–7340. doi: 10.1073/pnas.86.19.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Diekmann S., von Kitzing E., Kemper B., Lilley D. M. The structure of the Holliday junction, and its resolution. Cell. 1988 Oct 7;55(1):79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Elborough K. M., West S. C. Specific binding of cruciform DNA structures by a protein from human extracts. Nucleic Acids Res. 1988 May 11;16(9):3603–3616. doi: 10.1093/nar/16.9.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R. A., Detmer K., Rich A. Identification of homologous pairing and strand-exchange activity from a human tumor cell line based on Z-DNA affinity chromatography. Proc Natl Acad Sci U S A. 1988 Jan;85(1):36–40. doi: 10.1073/pnas.85.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganea D., Moore P., Chekuri L., Kucherlapati R. Characterization of an ATP-dependent DNA strand transferase from human cells. Mol Cell Biol. 1987 Sep;7(9):3124–3130. doi: 10.1128/mcb.7.9.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough G. W., Lilley D. M. DNA bending induced by cruciform formation. Nature. 1985 Jan 10;313(5998):154–156. doi: 10.1038/313154a0. [DOI] [PubMed] [Google Scholar]
- Hsieh P., Meyn M. S., Camerini-Otero R. D. Partial purification and characterization of a recombinase from human cells. Cell. 1986 Mar 28;44(6):885–894. doi: 10.1016/0092-8674(86)90011-5. [DOI] [PubMed] [Google Scholar]
- Ishida R., Akiyoshi H., Takahashi T. Isolation and purification of calcium and magnesium dependent endonuclease from rat liver nuclei. Biochem Biophys Res Commun. 1974 Feb 4;56(3):703–710. doi: 10.1016/0006-291x(74)90662-7. [DOI] [PubMed] [Google Scholar]
- Ishimi Y., Claude A., Bullock P., Hurwitz J. Complete enzymatic synthesis of DNA containing the SV40 origin of replication. J Biol Chem. 1988 Dec 25;263(36):19723–19733. [PubMed] [Google Scholar]
- Jensch F., Kosak H., Seeman N. C., Kemper B. Cruciform cutting endonucleases from Saccharomyces cerevisiae and phage T4 show conserved reactions with branched DNAs. EMBO J. 1989 Dec 20;8(13):4325–4334. doi: 10.1002/j.1460-2075.1989.tb08619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaseelan R., Shanmugam G. Human placental endonuclease cleaves Holliday junctions. Biochem Biophys Res Commun. 1988 Oct 31;156(2):1054–1060. doi: 10.1016/s0006-291x(88)80951-3. [DOI] [PubMed] [Google Scholar]
- KUNITZ M. Crystalline desoxyribonuclease; isolation and general properties; spectrophotometric method for the measurement of desoxyribonuclease activity. J Gen Physiol. 1950 Mar;33(4):349–362. doi: 10.1085/jgp.33.4.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper B., Garabett M. Studies on T4-head maturation. 1. Purification and characterization of gene-49-controlled endonuclease. Eur J Biochem. 1981 Mar 16;115(1):123–131. [PubMed] [Google Scholar]
- Kemper B., Janz E. Function of gene 49 of bacteriophage T4. I. Isolation and biochemical characterization of very fast-sedimenting DNA. J Virol. 1976 Jun;18(3):992–999. doi: 10.1128/jvi.18.3.992-999.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper B., Jensch F., von Depka-Prondzynski M., Fritz H. J., Borgmeyer U., Mizuuchi K. Resolution of Holliday structures by endonuclease VII as observed in interactions with cruciform DNA. Cold Spring Harb Symp Quant Biol. 1984;49:815–825. doi: 10.1101/sqb.1984.049.01.092. [DOI] [PubMed] [Google Scholar]
- Kerr C., Sadowski P. D. The involvement of genes 3,4,5 and 6 in genetic recombination in bacteriophage T7. Virology. 1975 May;65(1):281–285. doi: 10.1016/0042-6822(75)90031-8. [DOI] [PubMed] [Google Scholar]
- Kleff S., Kemper B. Initiation of heteroduplex-loop repair by T4-encoded endonuclease VII in vitro. EMBO J. 1988 May;7(5):1527–1535. doi: 10.1002/j.1460-2075.1988.tb02972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Kemper B. Cruciform-resolvase interactions in supercoiled DNA. Cell. 1984 Feb;36(2):413–422. doi: 10.1016/0092-8674(84)90234-4. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Gally J. A., Edelman G. M. Deoxyribonuclease IV: a new exonuclease from mammalian tissues. Proc Natl Acad Sci U S A. 1969 Feb;62(2):597–603. doi: 10.1073/pnas.62.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Gally J. A., Edelman G. M. Properties of deoxyribonuclease 3 from mammalian tissues. J Biol Chem. 1969 Sep 25;244(18):5014–5019. [PubMed] [Google Scholar]
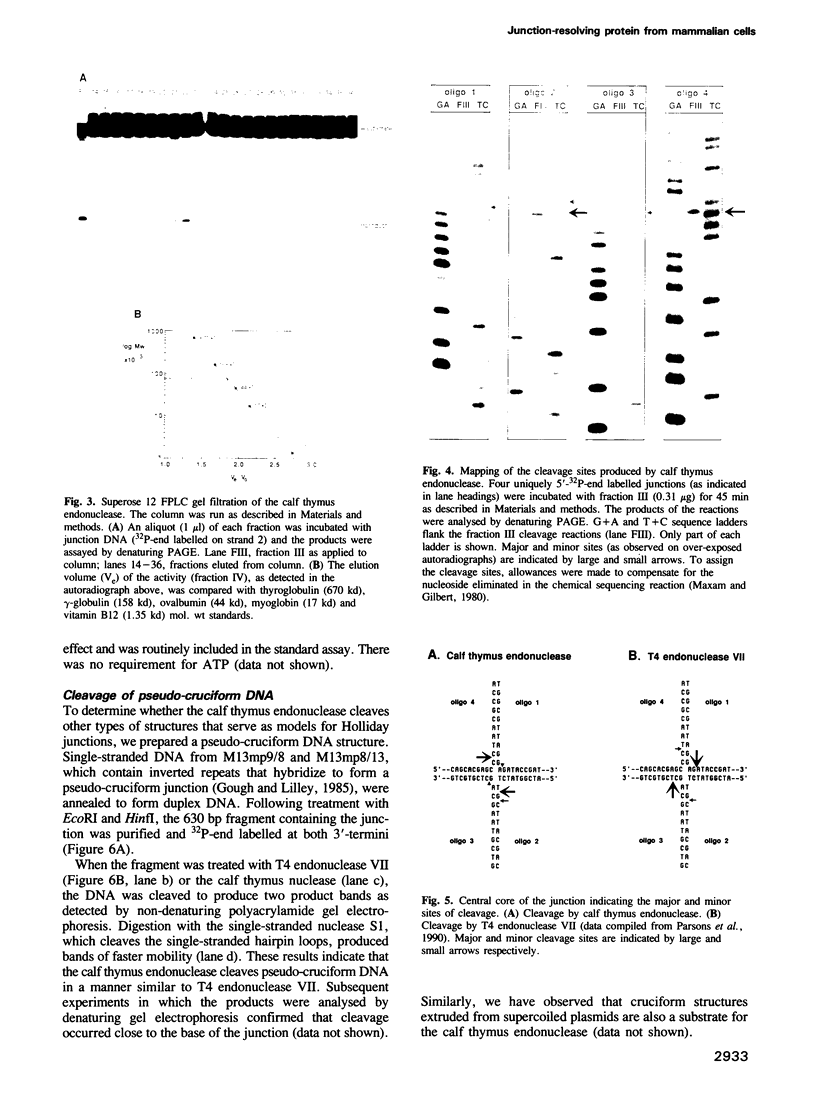
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miyazaki J., Ryo Y., Minagawa T. Involvement of gene 49 in recombination of bacteriophage T4. Genetics. 1983 May;104(1):1–9. doi: 10.1093/genetics/104.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Mueller J. E., Kemper B., Cunningham R. P., Kallenbach N. R., Seeman N. C. T4 endonuclease VII cleaves the crossover strands of Holliday junction analogs. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9441–9445. doi: 10.1073/pnas.85.24.9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B., Jones C., Kemper B., West S. C. Enzymatic formation and resolution of Holliday junctions in vitro. Cell. 1990 Jan 26;60(2):329–336. doi: 10.1016/0092-8674(90)90747-3. [DOI] [PubMed] [Google Scholar]
- Parsons C. A., Kemper B., West S. C. Interaction of a four-way junction in DNA with T4 endonuclease VII. J Biol Chem. 1990 Jun 5;265(16):9285–9289. [PubMed] [Google Scholar]
- Parsons C. A., West S. C. Resolution of model Holliday junctions by yeast endonuclease is dependent upon homologous DNA sequences. Cell. 1988 Feb 26;52(4):621–629. doi: 10.1016/0092-8674(88)90474-6. [DOI] [PubMed] [Google Scholar]
- Pedrini A. M., Ranzani G., Pedrali Noy G. C., Spadari S., Falaschi A. A novel endonuclease of human cells specific for single-stranded DNA. Eur J Biochem. 1976 Nov 1;70(1):275–283. doi: 10.1111/j.1432-1033.1976.tb10979.x. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Kolodner R. Partial purification of an enzyme from Saccharomyces cerevisiae that cleaves Holliday junctions. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7247–7251. doi: 10.1073/pnas.82.21.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto Y., Ogawa H. Intermediates in genetic recombination of bacteriophage T7 DNA. Biological activity and the roles of gene 3 and gene 5. J Mol Biol. 1978 Nov 5;125(3):255–273. doi: 10.1016/0022-2836(78)90402-3. [DOI] [PubMed] [Google Scholar]
- Waldman A. S., Liskay R. M. Resolution of synthetic Holliday structures by an extract of human cells. Nucleic Acids Res. 1988 Nov 11;16(21):10249–10266. doi: 10.1093/nar/16.21.10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. C., Körner A. Cleavage of cruciform DNA structures by an activity from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6445–6449. doi: 10.1073/pnas.82.19.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. C. Processing of recombination intermediates in vitro. Bioessays. 1990 Apr;12(4):151–154. doi: 10.1002/bies.950120402. [DOI] [PubMed] [Google Scholar]
- de Massy B., Studier F. W., Dorgai L., Appelbaum E., Weisberg R. A. Enzymes and sites of genetic recombination: studies with gene-3 endonuclease of phage T7 and with site-affinity mutants of phage lambda. Cold Spring Harb Symp Quant Biol. 1984;49:715–726. doi: 10.1101/sqb.1984.049.01.081. [DOI] [PubMed] [Google Scholar]
- de Massy B., Weisberg R. A., Studier F. W. Gene 3 endonuclease of bacteriophage T7 resolves conformationally branched structures in double-stranded DNA. J Mol Biol. 1987 Jan 20;193(2):359–376. doi: 10.1016/0022-2836(87)90224-5. [DOI] [PubMed] [Google Scholar]