Abstract
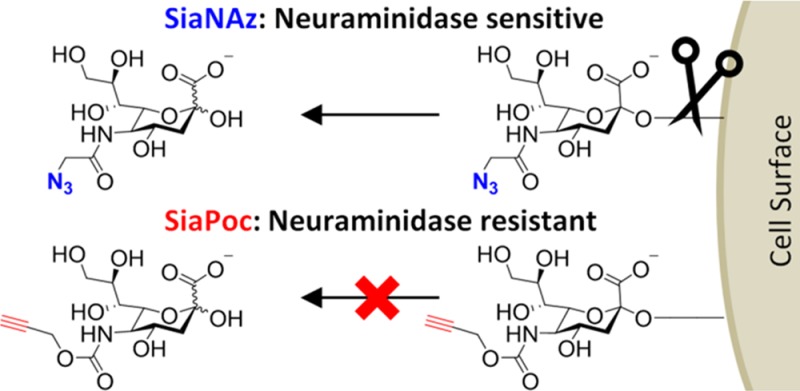
Metabolic incorporation of azide- or alkyne-modified sialic acids into the cellular glycosylation pathway enables the study of sialoglycan expression, localization, and trafficking via bioorthogonal chemistry. Herein, we report that such modifications of the sialic acid sugar can have a profound influence on their hydrolysis by neuraminidases (sialidase). Azidoacetyl (Az)-modified sialic acids were prone to neuraminidase cleavage, whereas propargyloxycarbonyl (Poc)-modified sialic acids were largely resistant to cleavage. Because the influenza virus infection cycle depends on the hydrolysis of host-cell-surface sialic acids, influenza cell-to-cell transmission was strongly reduced in Poc sialic acid glycoengineered host cells. The use of Poc sialic acids may disturb biological processes involving neuraminidase cleavage but also provides perspective for use in applications in which sialic acid hydrolysis is not desired, such as antibody modification, viral infection, etc.
Sialic acids (also known as neuraminic acids) are nine-carbon sugars abundantly expressed at the termini of cell surface glycoconjugates.1 Sialylated glycans are recognized by endogenous and exogenous receptors and play an important role in physiology and pathology, respectively.2 An important tool for the study of the function and localization of glycans in living cells and organisms is metabolic oligosaccharide engineering (MOE).3 In this process, monosaccharides carrying an unnatural modification are supplied to cells and incorporated into glycans at the cell surface. In a second step, the unnatural modification, also called a chemical reporter, can be visualized using bioorthogonal reactions with, for instance, fluorescent molecules.4 This powerful technique can be used to study and to manipulate glycans at the cell surface in various cells and living organisms. Studying biological processes without disturbing them is a major challenge, and hence, the chemical reporter carried by the monosaccharides must preferably be small and biologically inert. This approach is based on earlier work showing that small modifications on the N-acyl chain of N-acetylmannosamines were well-tolerated by the glycosylation machinery and ultimately expressed at the cell surface in the form of modified sialic acids.5 However, these modifications were intended to actively perturb biological processes involving sialic acids, such as pathogenic infection, immune-cell dampening, and neurite outgrowth.6,7 The acyl chain of sialic acid is also a site of natural modifications, which has probably arisen to avoid pathogen recognition.8 Chemical modifications at this site may therefore affect the biological function of sialic acids. Here, we report that azidoacetyl (Az) sialic acids are sensitive to bacterial and viral neuraminidases (also called sialidases), whereas propargyloxycarbonyl (Poc) sialic acids are resistant. Subsequently, incorporation of a propargyloxycarbonyl (Ac5SiaNPoc) into host cells, but not an azidoacetyl (Ac5SiaNAz), markedly reduced neuraminidase-dependent influenza cell-to-cell transmission.
Results and Discussion
We have previously shown that sialic acids modified at the C-5 position with Ac5SiaNAz or Ac5SiaNPoc are utilized by the cellular sialylation pathway with high selectivity resulting in their incorporation into cell surface sialoglycans (Figure 1).9,10 Both sialic acid analogues are well-tolerated by the cellular sialylation pathway, and these small modifications to the C-5 position are generally thought to be nonintrusive. There is evidence, however, that the modification of sialic acids at the C-5 position affects their recognition and cleavage by bacterial neuraminidases.11 Therefore, we assessed whether cell surface Az and Poc sialic acids can be cleaved by bacterial neuraminidase. Human monocytic THP-1 cells were cultured for 3 days with Ac5SiaNAz or Ac5SiaNPoc and reacted to biotin–alkyne or biotin–azide, respectively, using copper-catalyzed alkyne azide cycloaddition (CuAAc).12−14 The biotin-tagged surface sialoglycans were visualized with fluorescent streptavidin. Confocal images show the dense expression of Az and Poc sialic acids at the cell surface of THP-1 cells (Figure 2a). Quantification of the fluorescence with flow cytometry showed that both sialic acid analogues were incorporated into surface glycans with high efficiency. Ac5SiaNAz (MFI 7979) showed slightly lower incorporation compared to Ac5SiaNPoc (MFI 9345) (Figure 2b).
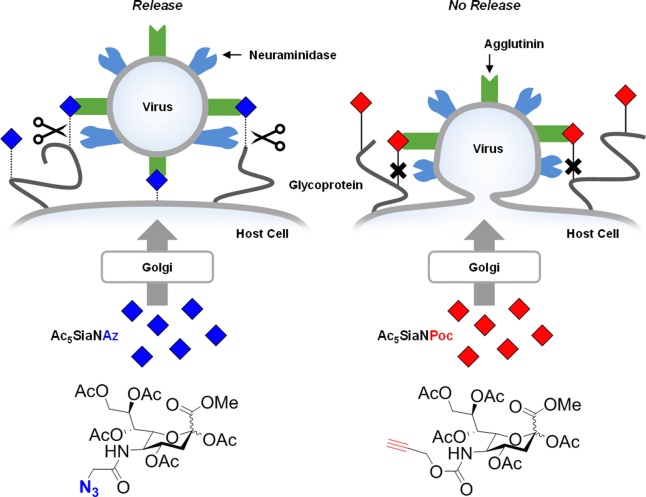
Figure 1.
Schematic presentation of metabolic labeling with Ac5SiaNAz or Ac5SiaNPoc and the resistance of Ac5SiaNPoc against influenza neuraminidase.
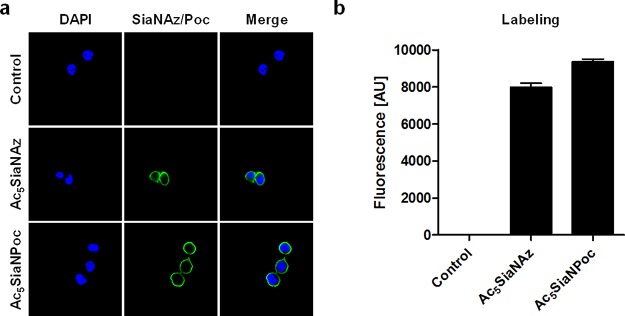
Figure 2.
Metabolic labeling of THP-1 cells. (a) The cells were incubated for 3 days with PBS, 100 μM Ac5SiaNAz, or 100 μM Ac5SiaNPoc and reacted to biotin–alkyne or biotin–azide, respectively, using CuAAC. Biotin-containing sialoglycans were stained with PE–streptavidin and visualized by confocal microscopy. (b) Fluorescence was quantified by flow cytometry, and the mean fluorescence intensity ± SEM is shown (n = 3).
The difference in labeling intensity between Ac5SiaNAz and Ac5SiaNPoc is linked to their different incorporation efficiency and not the labeling step because the CuAAc reaction is faster for Ac5SiaNAz compared to Ac5SiaNPoc.16a Next, it was determined whether surface SiaNAz and SiaNPoc containing glycans can be cleaved by bacterial neuramindase (Figure 3). THP-1 cells cultured for 3 days with Ac5SiaNAz and Ac5SiaNPoc were treated with Clostridium perfringens neuraminidase and then reacted with biotin–alkyne and biotin–azide, respectively, followed by staining with streptavidin–phycoerythrin (PE).17 Flow cytometry analysis revealed that SiaNAz was cleaved from the cells (>64% removal), whereas SiaNPoc could not be removed from the cell surface (<8% removal) (Figure 3a). To evaluate the cleavage selectivity with respect to the linkage type (α-2,3 or α-2,6), the cells were analyzed with the lectins Maackia amurensis lectin (MALII, binding to α-2,3-sialoglycans), Sambucus nigra lectin (SNA-I, binding to α-2,6-sialoglycans), and peanut agglutinin lectin (PNA, binding to terminal β-galactose).
Figure 3.
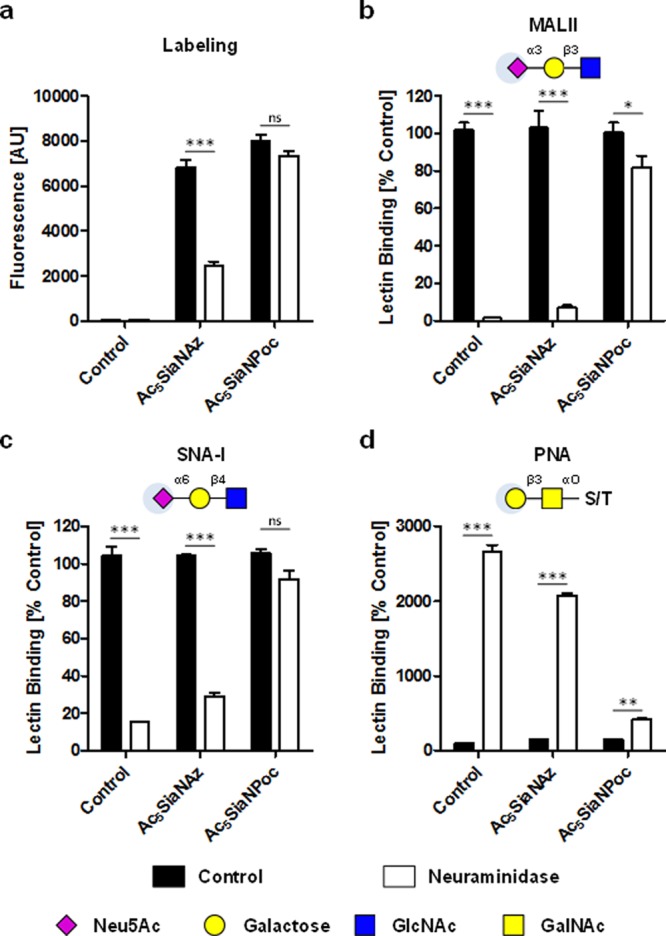
Enzymatic removal of Az and Poc sialic acids from the cell surface of THP-1 cells. Cells incubated for 3 days with PBS, 100 μM Ac5SiaNAz, or 100 μM Ac5SiaNPoc were treated for 1 h with 250mU/mL Clostridium perfringens neuraminidase. Az and Poc sialoglycans were reacted to fluorescent biotin using CuAAC (a), α2,3-sialoglycans were detected with MALII lectin (b), α2,6-sialoglycans were detected with SNA-I lectin (c), and terminal β-galactose was detected with PNA lectin (d). Bar diagrams show mean fluorescence intensity or mean lectin binding normalized to control ± SEM of three independent experiments. MALII: Maackia amurensis lectin; PE: phycoerythrin, PNA: Peanut agglutinin lectin; SEM: standard error of the mean; SNA-I: Sambucus nigra lectin.
Cells incubated with Ac5SiaNAz showed also a strong reduction in sialic acid expression following treatment with neuraminidase (MALII, ↓93%; SNA-I, ↓72%). In contrast, the sialylation of THP-1 cells cultured with Ac5SiaNPoc was barely altered following neuraminidase treatment (MALII, ↓18%; SNA-I, ↓13%). The low sensitivity of Poc sialic acids toward neuraminidase activity was also confirmed by the minor change in PNA binding. Similar findings were obtained when THP-1 cells were treated with neuraminidase derived from Vibrio cholerae or Arthrobacter ureafaciens (Figure S1). Together, these data show that sialic acids modified with an Az group at the C-5 position are substrates for neuraminidases, whereas Poc-modified sialic acids are not. Whether this effect is due to inhibited recognition or hydrolysis of sialic acids by neuraminidases remains to be investigated.
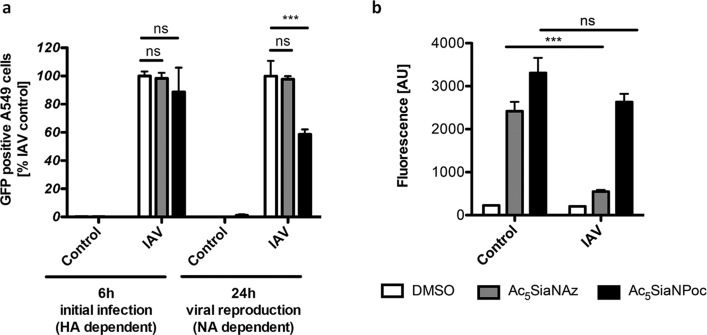
Several pathogens make use of neuraminidases to infect host cells or release host cell sialic acids for their own metabolism.18,19 In particular, influenza virus makes use of the host cell sialic acids to enter and leave the cell. Influenza virus has two major glycoproteins on the outside of the viral particle, hemagglutinin (HA) and neuraminidase (NA).20 Hemagglutinin is a lectin that mediates binding to sialic acids on host cells, whereas neuraminidase is involved in the release of progeny virus from infected cells by cleaving the sialic acid that binds the mature viral particle to the cell surface. Neuraminidase inhibitors including Oseltamivir (Tamiflu) and Zanamivir (Relenza) block neuraminidase enzyme activity and prevent influenza virus reproduction.21−23 Therefore, we assessed whether neuraminidase resistant Poc sialic acid alters influenza reproduction in an A549 cell infection model. In this model, a GFP-reporter influenza virus strain A/PR8-GFP/8/34 was used that expresses GFP in infected cells.24 A549 cells were cultured for 3 days with Ac5SiaNAz or Ac5SiaNPoc, and incorporation of Az and Poc sialic acids as well as cleavage-resistance of Poc sialic acid was confirmed (Figure S2). Next, confluent A549 cells were infected with different dilutions of influenza virus strain A/PR8-GFP/8/34 (IVA) for 1 h and the percentage of GFP-positive A549 cells was determined by flow cytometry 6 and 24 h post-infection, representing the initial infection and the infection of viral progeny, respectively. The initial infection after 6 h did not show a significant difference in the percentage of GFP-positive A549 cells between cells cultured with Ac5SiaNAz, Ac5SiaNPoc, or a solvent control (333× dilution Figure 4a; 1000× dilution, Figure S3).
Figure 4.
Effect of Az and Poc sialic acid on influenza reproduction (333× dilution). A549 cells cultured with either Ac5SiaNAz or Ac5SiaNPoc were treated with influenza virus strain A/PR8-GFP/8/34, and the GFP expression was measured as a readout for successful viral infection after 6 h and reinfection after 24 h (a). Az and Poc sialoglycans were detected 24 h post-infection by reaction with fluorescent biotin using the CuAAC (b). Bar diagrams show mean fluorescence intensity ± SEM (n = 4).
In contrast, after 24 h, the cells incubated with Ac5SiaNPoc showed a significant reduction in GFP-positive A549 cells compared with solvent control or cells incubated with Ac5SiaNAz, indicating that Ac5SiaNPoc, which is resistant to bacterial neuraminidase activity, has a profound effect on virus reproduction (333× dilution, Figure 4a; 1000× dilution, Figure S3). Earlier research has shown that modifications of the N-acyl chain could inhibit influenza infections and led to the hypothesis that longer site chains would sterically hinder binding of hemagglutinin.25 Our experimental data with Ac5SiaNAz or Ac5SiaNPoc after 24 h are indeed in line with earlier observations that sialic acid C-5 modifications can lead to viral resistance. However, the initial infection after 6 h shows no resistance, therefore suggesting a different mechanism. In contrast to the current hypothesis, we found that initial infection (and, therefore, hemagglutinin recognition) is not significantly hindered by longer C-5 modifications such as Az or Poc. Instead, the gain in resistance in the case of Ac5SiaNPoc is probably caused by neuraminidase resistance, thereby inhibiting the release of the viral progeny. This was further confirmed by labeling the Ac5SiaNAz- and Ac5SiaNPoc- treated cells after 24 h of IVA infection (Figure 4b). The Ac5SiaNAz-treated cells showed a significant reduction in labeling signal, indicating the release of SiaNAz after infection. In contrast, Ac5SiaNPoc-treated cells showed very similar labeling levels 24 h post-infection compared to an untreated control. This clearly indicates that SiaNPoc is stable under IVA infection and resists neuraminidase cleavage.
Conclusions
In conclusion, we demonstrate that metabolic engineering with Poc-modified sialic acids confers resistance toward neuraminidases. Consequently, the incorporation of Poc sialic acids into host cells reduced neuraminidase-dependent influenza cell-to-cell transmission. Furthermore, these findings indicate that, depending on the chemical reporter used, metabolic oligosaccharide engineering may not be biologically inert. The neuraminidase resistance conferred by Poc sialic acids may disturb biological processes involving neuraminidase cleavage but could be applied in situations in which sialic acid hydrolysis is not desired, such as therapeutic protein modification, viral infection, etc.
Acknowledgments
This work was supported by a VENI grant (no. 722.013.004) from The Netherlands Organization for Scientific Research (NWO) awarded to T.J.B., The Netherlands Research School for Chemical Biology (NRSCB), and a KWF grant (no. KUN2015-7604) awarded to G.J.A, T.J.B, and C.B.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconjchem.7b00224.
Additional details on materials and methods. Figures showing the removal of Az and Poc sialic acids, Poc sialic acid sialidase resistance, and the effect of Az and Poc sialic acid on influenza reproduction. (PDF)
Author Contributions
⊥ T.H. and C.B. contributed equally.
The authors declare no competing financial interest.
Footnotes
The difference in incorporation may be higher than the minor difference in fluorescence seen as the upper detection limit was reached for Ac5SiaNPoc. In contrast to our previously published comparison of Ac5SiaNAz and Ac5SiaNPoc (see ref (9)), the click reaction was now done at 37 °C instead of at room temperature, which greatly increased the fluorescence signal and gave usual click efficiencies of 80% or higher (see ref (10)).
Supplementary Material
References
- Altheide T. K.; Hayakawa T.; Mikkelsen T. S.; Diaz S.; Varki N.; Varki A. (2006) System-wide Genomic and Biochemical Comparisons of Sialic Acid Biology Among Primates and Rodents: evidence for two modes of rapid evolution. J. Biol. Chem. 281, 25689–25702. 10.1074/jbc.M604221200. [DOI] [PubMed] [Google Scholar]
- Varki A. (2008) Sialic acids in human health and disease. Trends Mol. Med. 14, 351–360. 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube D. H.; Bertozzi C. R. (2003) Metabolic oligosaccharide engineering as a tool for glycobiology. Curr. Opin. Chem. Biol. 7, 616–625. 10.1016/j.cbpa.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Cheng B.; Xie R.; Dong L.; Chen X. (2016) Metabolic Remodeling of Cell-Surface Sialic Acids: Principles, Applications, and Recent Advances. ChemBioChem 17, 11–27. 10.1002/cbic.201500344. [DOI] [PubMed] [Google Scholar]
- Wratil P. R.; Horstkorte R.; Reutter W. (2016) Metabolic Glycoengineering with N-Acyl Side Chain Modified Mannosamines. Angew. Chem., Int. Ed. 55, 9482–9512. 10.1002/anie.201601123. [DOI] [PubMed] [Google Scholar]
- Kelm S.; Schauer R.; Manuguerra J. C.; Gross H. J.; Crocker P. R. (1994) Modifications of cell surface sialic acids modulate cell adhesion mediated by sialoadhesin and CD22. Glycoconjugate J. 11, 576–585. 10.1007/BF00731309. [DOI] [PubMed] [Google Scholar]
- Buttner B.; Kannicht C.; Schmidt C.; Loster K.; Reutter W.; Lee H. Y.; Nohring S.; Horstkorte R. (2002) Biochemical engineering of cell surface sialic acids stimulates axonal growth. J. Neurosci. 22, 8869–8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angata T.; Varki A. (2002) Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 102, 439–469. 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- Büll C.; Heise T.; Beurskens D. M.; Riemersma M.; Ashikov A.; Rutjes F. P.; van Kuppevelt T. H.; Lefeber D. J.; den Brok M. H.; Adema G. J.; Boltje T. J. (2015) Sialic Acid Glycoengineering Using an Unnatural Sialic Acid for the Detection of Sialoglycan Biosynthesis Defects and On-Cell Synthesis of Siglec Ligands. ACS Chem. Biol. 10, 2353–2363. 10.1021/acschembio.5b00501. [DOI] [PubMed] [Google Scholar]
- Büll C.; Heise T.; van Hilten N.; Pijnenborg J. F.; Bloemendal V.; Gerrits L.; Kers-Rebel E. D.; Ritschel T.; den Brok M. H.; Adema G. J.; Boltje T. J. (2017) Steering Siglec-Sialic Acid Interactions on Living Cells using Bioorthogonal Chemistry. Angew. Chem., Int. Ed. 56, 3309–3313. 10.1002/anie.201612193. [DOI] [PubMed] [Google Scholar]
- Cao H. Z.; Li Y. H.; Lau K.; Muthana S.; Yu H.; Cheng J. S.; Chokhawala H. A.; Sugiarto G.; Zhang L.; Chen X. (2009) Sialidase substrate specificity studies using chemoenzymatically synthesized sialosides containing C5-modified sialic acids. Org. Biomol. Chem. 7, 5137–5145. 10.1039/b916305k. [DOI] [PubMed] [Google Scholar]
- Tornøe C. W.; Christensen C.; Meldal M. (2002) Peptidotriazoles on Solid Phase: [1,2,3]-Triazoles by Regiospecific Copper(I)-Catalyzed 1,3-Dipolar Cycloadditions of Terminal Alkynes to Azides. J. Org. Chem. 67, 3057–3064. 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- Rostovtsev V. V.; Green L. G.; Fokin V. V.; Sharpless K. B. (2002) A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective “Ligation” of Azides and Terminal Alkynes. Angew. Chem., Int. Ed. 41, 2596–2599. . [DOI] [PubMed] [Google Scholar]
- Kennedy D. C.; McKay C. S.; Legault M. C. B.; Danielson D. C.; Blake J. A.; Pegoraro A. F.; Stolow A.; Mester Z.; Pezacki J. P. (2011) Cellular Consequences of Copper Complexes Used To Catalyze Bioorthogonal Click Reactions. J. Am. Chem. Soc. 133, 17993–18001. 10.1021/ja2083027. [DOI] [PubMed] [Google Scholar]
- Speers A. E.; Cravatt B. F. (2004) Profiling Enzyme Activities In Vivo Using Click Chemistry Methods. Chem. Biol. 11, 535–546. 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Möller H.; Böhrsch V.; Bentrop J.; Bender J.; Hinderlich S.; Hackenberger C. P. R. (2012) Glycan-Specific Metabolic Oligosaccharide Engineering of C7-Substituted Sialic Acids. Angew. Chem., Int. Ed. 51, 5986–5990. 10.1002/anie.201108809. [DOI] [PubMed] [Google Scholar]
- Lewis A. L.; Lewis W. G. (2012) Host sialoglycans and bacterial sialidases: a mucosal perspective. Cell. Microbiol. 14, 1174–1182. 10.1111/j.1462-5822.2012.01807.x. [DOI] [PubMed] [Google Scholar]
- Stencel-Baerenwald J. E.; Reiss K.; Reiter D. M.; Stehle T.; Dermody T. S. (2014) The sweet spot: defining virus-sialic acid interactions. Nat. Rev. Microbiol. 12, 739–749. 10.1038/nrmicro3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K.; Aramini J. M.; Ma L. C.; Krug R. M.; Arnold E. (2010) Structures of influenza A proteins and insights into antiviral drug targets. Nat. Struct. Mol. Biol. 17, 530–538. 10.1038/nsmb.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubareva L. V.; Kaiser L.; Hayden F. G. (2000) Influenza virus neuraminidase inhibitors. Lancet 355, 827–835. 10.1016/S0140-6736(99)11433-8. [DOI] [PubMed] [Google Scholar]
- von Itzstein M. (2007) The war against influenza: discovery and development of sialidase inhibitors. Nat. Rev. Drug Discovery 6, 967–974. 10.1038/nrd2400. [DOI] [PubMed] [Google Scholar]
- Palese P.; Compans R. W. (1976) Inhibition of Influenza Virus Replication in Tissue Culture by 2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA): Mechanism of Action. J. Gen. Virol. 33, 159–163. 10.1099/0022-1317-33-1-159. [DOI] [PubMed] [Google Scholar]
- Manicassamy B.; Manicassamy S.; Belicha-Villanueva A.; Pisanelli G.; Pulendran B.; Garcia-Sastre A. (2010) Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl. Acad. Sci. U. S. A. 107, 11531–11536. 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler O. T.; Herrmann M.; von der Lieth C. W.; Stehling P.; Reutter W.; Pawlita M. (1998) Elongation of the N-acyl side chain of sialic acids in MDCK II cells inhibits influenza A virus infection. Biochem. Biophys. Res. Commun. 253, 437–442. 10.1006/bbrc.1998.9650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.