Abstract
Despite an increased understanding of functions in sponge microbiomes, the interactions among the symbionts and between symbionts and host are not well characterized. Here we reconstructed the metabolic interactions within the sponge Cymbastela concentrica microbiome in the context of functional features of symbiotic diatoms and the host. Three genome bins (CcPhy, CcNi and CcThau) were recovered from metagenomic data of C. concentrica, belonging to the proteobacterial family Phyllobacteriaceae, the Nitrospira genus and the thaumarchaeal order Nitrosopumilales. Gene expression was estimated by mapping C. concentrica metatranscriptomic reads. Our analyses indicated that CcPhy is heterotrophic, while CcNi and CcThau are chemolithoautotrophs. CcPhy expressed many transporters for the acquisition of dissolved organic compounds, likely available through the sponge’s filtration activity and symbiotic carbon fixation. Coupled nitrification by CcThau and CcNi was reconstructed, supported by the observed close proximity of the cells in fluorescence in situ hybridization. CcPhy facultative anaerobic respiration and assimilation by diatoms may consume the resulting nitrate. Transcriptional analysis of diatom and sponge functions indicated that these organisms are likely sources of organic compounds, for example, creatine/creatinine and dissolved organic carbon, for other members of the symbiosis. Our results suggest that organic nitrogen compounds, for example, creatine, creatinine, urea and cyanate, fuel the nitrogen cycle within the sponge. This study provides an unprecedented view of the metabolic interactions within sponge–microbe symbiosis, bridging the gap between cell- and community-level knowledge.
Introduction
A wide range of animals are found in symbiosis with microorganisms, from basal metazoans, such as marine sponges (phylum Porifera), to humans (phylum Chordata) (Taylor et al., 2007; Cho and Blaser, 2012). Animal–microbe symbioses encompass a variety of acquisition modes (that is, vertical, horizontal and a combination of both; for example, Cary and Giovannoni (1993); Kikuchi et al. (2007); Schmitt et al. (2008)), and levels of diversity (ranging from monoclonal populations to highly complex assemblages; Martens et al. (2003); Schmitt et al. (2012)). Additionally, a gradient of interactions may occur, ranging from competition to mutualism (Dethlefsen et al., 2007; Coyte et al., 2015; Thomas et al., 2016). To account for this spectrum of patterns, Douglas and Werren (2016) have recently argued that host–microbe symbioses are productively approached as ecological communities. Therefore, to further our understanding of the biology of animal–microbe symbioses, research should contemplate the members of the association, that is, the microbial populations and the host, and how they relate to each other.
Marine sponges are filter-feeders found in a wide range of benthic environments, across gradients of depth and latitude (Wulff, 2012). Sponges perform a broad range of functional roles in benthic habitats (Bell, 2008), including the retention of nutrients and energy within the coral reef through the consumption of dissolved organic matter (DOM) and high cell turnover rates (Yahel et al., 2003; de Goeij et al., 2013). In addition to DOM, sponges feed on detritus and microorganisms of diverse sizes (from 0.5 to 70 μm) (Marta et al., 1999; Hadas et al., 2009). Owing to their basal position in metazoans phylogeny and their association with microbial communities, sponges are emerging as models for the evolutionary study of animal–microbe symbiosis (for example, Fan et al., 2012; Ryu et al., 2016).
Microbial communities can account for up to 40% of the sponge tissue volume and often contain novel and uncharacterized diversity (Taylor et al., 2007; Schmitt et al., 2012). Microorganisms are generally found in the animal’s extracellular matrix (mesohyl), although they are not restricted to this location (Taylor et al., 2007). Owing to the lack of culturable representatives of the microbiome, most of the understanding of the functional roles of microbial symbiont have been acquired by approaches either centered on the community, for example, using meta-omics, 16S rRNA gene analysis and compound-uptake experiments, or on specific genomes, pathways and genes (reviewed by Taylor et al., 2007 and Webster and Thomas 2016). Together, these studies have revealed important metabolic attributes of sponge-associated microorganisms, such as different energy acquisition strategies (for example, heterotrophism and autotrophism), complex element cycling, functional equivalence, production of bioactive compounds and nutrient transfer to the host (Hoffmann et al., 2009; Fan et al., 2012; Freeman et al., 2013; Kamke et al., 2013; Wilson et al., 2014). Nevertheless, the metabolic interactions among the microbiome members and between them and the host cells are rarely explored and thus remain largely unknown (Webster and Thomas, 2016).
The sponge Cymbastela concentrica contains consistently dominant and uncharacterized alphaproteobacterial symbionts, particularly within the family Phyllobacteriaceae (Taylor et al., 2004; Fan et al., 2012; Esteves et al., 2016) as well as community members belonging to the Gammaproteobacteria, the Nitrospira and the archaeal phylum Thaumarchaeota among others (Fan et al., 2012; Esteves et al., 2016). In addition, C. concentrica hosts an abundant population of diatoms (Taylor, 2005). The overall community composition and function of the C. concentrica microbiome have been previously characterized on the community level using metagenomics, metaproteomics and metatranscriptomics (Díez-Vives et al., 2016; Thomas et al., 2010; Liu et al., 2011; Fan et al., 2012; Liu et al., 2012). Here we build on these data sets to unveil nutritional interactions in C. concentrica. Towards this goal, we reconstructed three new prokaryotic genomes and estimated their activity by analyzing metatranscriptomic sequences, while metabolic features of the sponge and diatoms were inferred from an eukaryotic transcriptomic data set produced from C. concentrica.
Materials and methods
Genome assembly, binning and annotation
Metagenomic shotgun sequencing reads for three replicates (BBAY40, 41 and 42) of the microbiome of C. concentrica from a previous study (Fan et al., 2012) were co-assembled using the Newbler assembler (Roche, Penzberg, Germany). Sequence reads for each replicate were then mapped back against assembled contigs >500 bp using the Bowtie aligner (Langmead et al., 2009). Coverage file and contigs were then used to bin and refine genomes using the GroopM tool (Imelfort et al., 2014). Genome bins were checked for completeness and contamination using CheckM (Parks et al., 2015) and then annotated using the IMG system (Markowitz et al., 2014). Genome assembly and annotation are avaiable at IMG (http://img.jgi.doe.gov) under the genome IDs 2608642157, 2608642160 and 2626541593. Phylogenetic analysis of 16S rRNA genes and deduced amino-acid sequences for those bins are described in Supplementary Information.
Transcriptomic analysis
We used the metatranscriptomic reads from the microbiome of three C. concentrica individuals (Díez-Vives et al., 2016) to estimate the gene expression of binned genomes. Briefly, mRNA was previously enriched from three individuals (ind1, ind2, and ind3; biological replicates), which were processed with Dynabeads Oligo (dT)25 (Thermo Fisher Scientific, Waltham, MA, USA) and the Ribo-Zero Bacteria Kit (Epicentre, Madison, WI, USA) in technical replicates (aR and bR). Enriched prokaryotic mRNA was sequenced with HiSeq2000 platform (Illumina, San Diego, CA, USA) with 100 bp paired-end reads. Transcript expression for the coding DNA sequences (CDS) of each genome bin was estimated with RSEM v. 1.2.21 (Li and Dewey, 2011) with default parameters, which implements the Bowtie aligner (Langmead et al., 2009). Transcripts per million (TPM) measures were used as estimations of expression (Li et al., 2010; Wagner et al., 2012). Gene expression estimates in each sponge individual were obtained by averaging TPM values of the technical replicates. Only genes with mapped reads (replicate-average TPM>0) coming from at least two of the three sponge individuals were considered expressed. The global expression estimate for each gene was obtained by averaging TPM values (TPMav) across the three sponge individuals.
The analysis of eukaryotic transcripts is described in Supplementary Information. Briefly, transcripts were taxonomically sorted using the Lowest Common Ancestor algorithm implemented in MEGAN v. 5.11.3 (Huson et al., 2011). Peptides deduced from predicted coding sequences were annotated based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology (KO). Expression estimates of KEGG functions were calculated by summing TPM values (TPMsum) across coding sequences.
Localization analysis of symbionts
Three individuals of C. concentrica were collected by SCUBA diving from Botany Bay near Bare Island, Sydney, Australia (33.99222° S, 151.23111° E) during October 2015. Specimens were processed, fixed and dehydrated. Probes specific for the obtained genome bins (that is, CcPhy, CcNi and CcThau; see below) were designed and cells were detected using fluorescence in situ hybridization. Sample processing, probe design and fluorescence in situ hybridization conditions are fully described in Supplementary Information.
Results and discussion
Genomes and transcriptomes of the dominant community members in C. concentrica
Three genomes were reconstructed (Table 1) from a previous metagenomic sequence data set of the microbial community of C. concentrica (Fan et al., 2012). Phylogenetic analysis of the 16S rRNA gene sequence recovered from one of the genomes (Ga0068441) placed it with members of the family Phyllobacteriaceae, order Rhizobiales, class Alphaproteobacteria (Supplementary Information; Supplementary Figure S1). Thus the genome will be hereafter referred to as CcPhy (C. concentrica Phyllobacteriaceae). The 16S rRNA gene sequence is only 96% similar to its closest cultured bacterium (Mesorhizobium amorphae CCNWGS0123) and has >99% similarity to sequences that have been previously found in C. concentrica (for example, GenBank: AY942778.1). The cluster represented by these C. concentrica-derived sequences represents a distinct and novel clade within the Phyllobacteriaceae (Supplementary Figure S1). The 16S rRNA gene sequence of the second genome (Ga0068443) was phylogenetically close to other sequences derived from several sponge species (for example, Agelas dilatata, Geodia barretti, Xestospongia testudinaria). These sequences were related yet distinct from sequences of the lineage IV of the Nitrospira genus within the phylum Nitrospirae (Supplementary Information; Supplementary Figure S2). The genome will be hereafter named CcNi. The 16S rRNA gene sequence of the last genome obtained (Ga0078905) clustered within the candidate order Nitrosopumilales (known as group I.1a) (Konneke et al., 2005; Stieglmeier et al., 2014b) within the Thaumarchaeota phylum (Supplementary Information; Supplementary Figure S3). It will be referred as CcThau. The 16S rRNA sequence from CcThau was 99% similar to the sequence from Candidatus Nitrosopumilus sediminis AR2 (GenBank: CP003843.1). According to studies based on OTUs recovered from metagenomic data sets and 16S rRNA gene sequencing, these three organisms are among the most dominant members of the C. concentrica microbiome (Thomas et al., 2010; Fan et al., 2012; Esteves et al., 2016). For example, based on 16S rRNA gene sequencing, the Phyllobacteriaceae OTU may account approximately 17% of all bacteria in the sponge, while the Nitrospira OTU has a relative abundance of ~6% (Thomas et al., 2010). Metagenomic sequencing further indicated that the thaumarchaeal organism has similar relative abundances than the Nitrospira bacterium (Fan et al., 2012).
Table 1. Assembly and annotation statistics of genome bins.
| CcPhy (Ga0068441) | CcNi (Ga0068443) | CcThau (Ga0078905) | |
|---|---|---|---|
| Assembly size (bp) | 2 116 962 | 1 594 656 | 2 161 854 |
| Individual coverage in three replicate metagenomes | 19.5 × /58.4 × /37.5 × | 3.2 × /0.8 × /7.2 × | 0.9 × /15.2 × /2.3 × |
| Estimated genome completeness (%) | 69.17 | 66.56 | 97.12 |
| Estimated contamination (%) | 0.39 | 0 | 4.01 |
| Number of contigs | 918 | 126 | 637 |
| Average GC content (%) | 60.34 | 48.03 | 38.42 |
| Number of CDS | 2411 | 1679 | 2745 |
| Number of rRNA genes | 1 | 3 | 5 |
| Number of tRNA genes | 16 | 25 | 44 |
Abbreviations: CDS, coding DNA sequence; GC, guanine–cytosine; rRNA, ribosomal RNA; tRNA, transfer RNA.
CcThau has the largest genome assembly with a size of 2.16 mega basepairs (Mb), followed by CcPhy with 2.11 Mb and CcNi with 1.59 Mb (Table 1). Genome completeness estimations based on lineage-specific marker genes were higher in CcThau (97.12%) in comparison to CcPhy (69.17%) and CcNi (66.56%). The estimated contamination of genome bins was low, ranging from 0% to 4.01%. GC content varied from 38.42% for CcThau and 48.03% for CcNi to 60.34% for CcPhy. Metatranscriptomic sequencing reads mapped to these genomes at different extent, where most of the reads mapped to the CcThau CDS (88 303±23 580; mean±s.d.), followed by CcPhy (16 540±6128) and CcNi (7054±3542) (Supplementary Table S1). Expressed genes represented 86.7% of the total CDS in CcPhy (2092 of 2411), 80.5% in CcNi (1353 of 1679) and 64.6% in CcThau (1775 of 2745). High gene expression correlation among biological replicates, as estimated by TPM values, indicated low variability between sponge individuals (Supplementary Information; Supplementary Figures S4). Average TPM values (TPMav) of expressed genes across sponge individuals varied from 1.09 to 27 022.43, with a median of 191.54 in CcPhy, 354.2 in CcNi and 49.18 in CcThau (Supplementary Figure S7).
A heterotrophic, facultative anaerobic metabolism of CcPhy
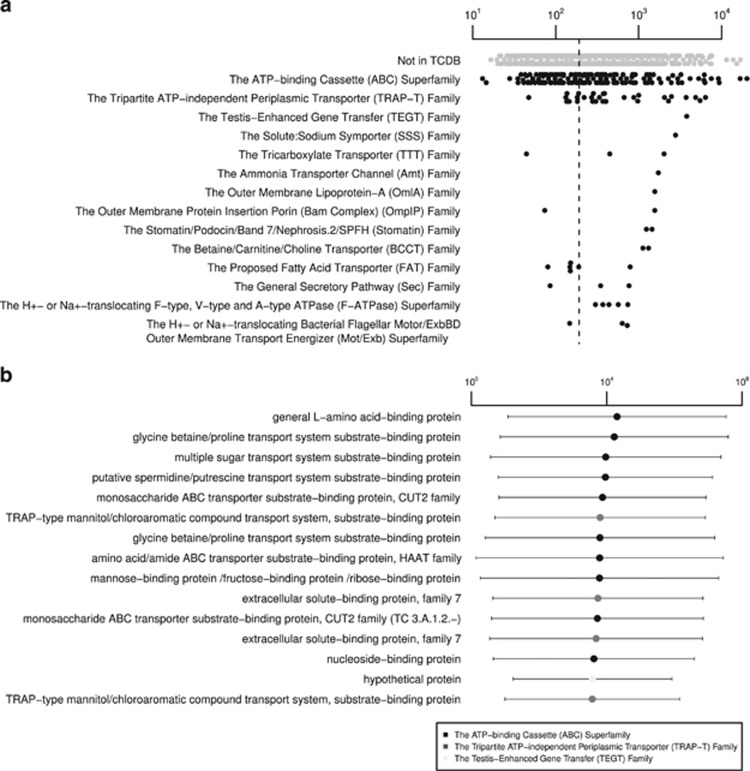
The genomic and transcriptomic data suggest that the population represented by CcPhy has a heterotrophic metabolism, which is broadly consistent with other members of the family Phyllobacteriaceae (Willems, 2014). The CcPhy expressed an almost complete set of genes for glycolysis (Embden–Meyerhof–Parnas pathway) (TPMav 24–343; lower value−higher value), as well as the anabolic enzyme fructose-1,6-bisphosphatase (TPMav 281±39; mean±s.d.) and tricarboxylic acid (TCA) pathways (TPMav 121–1396), which are linked by the pyruvate dehydrogenase complex (TPMav 88–1316) (Figure 1; Supplementary Table S2). Additionally, CcPhy is able to process sugars via the pentose phosphate pathway. In support of a heterotrophic lifestyle, approximately 13.5% of the protein-coding genes (n=328) were involved in membrane transport (according to Transporter Classification Database (TCDB); Supplementary Table S2). From these, 164 CDS were assigned to the adenosine triphosphate (ATP)-binding cassette (ABC) superfamily (TC: 3.A.1), which were among the most expressed genes in the CcPhy transcriptome (Figure 2). The most expressed genes classified by TCDB encoded for components of different transport systems involved in the uptake of general L-amino acids, glycine betaine/proline, multiple sugars, spermidine/putrescine, monosaccharides, mannitol/chloroaromatic compounds, amino acid/amide and manose/fructose/ribose. Genes encoding for Amt-type ammonium transporters (TPMav 1729±351) and the glutamine synthase-glutamine oxoglutarate aminotransferase pathway (TPMav 168–1157) were expressed, allowing for import and assimilation of extracellular ammonium (Javelle et al., 2005).
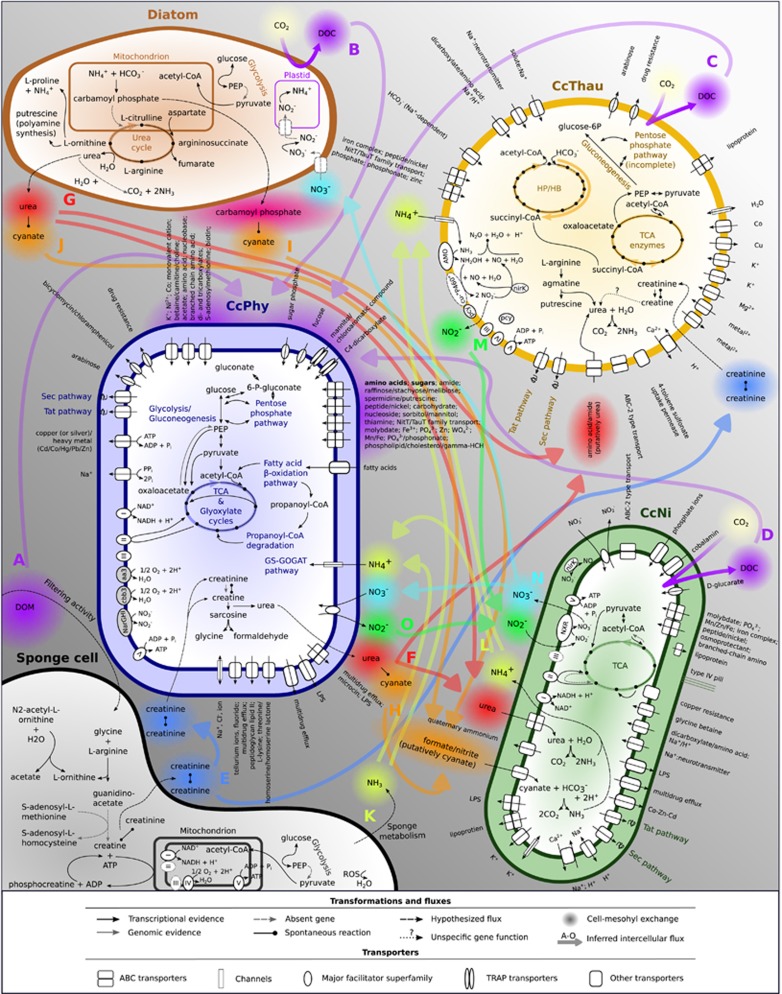
Figure 1.
Predicted model of integrated metabolic processes within Cymbastela concentrica. Selected pathways and gene functions are shown. Transporters with at least partial genomic evidence and expression detected were included. Inferred nutrient fluxes (A–O) are discussed in the section ‘Model of integrated metabolism of the microbial community members in C. concentrica’. Expressed genes are listed in Supplementary Tables S2.
Figure 2.
Most expressed CcPhy genes related to membrane transport. (a) CDS assigned to TCDB were grouped by families and super families. Groups were ordered according to maximum TPMav values (x axis), and only the top 15 are shown. For reference, CDS not classified by TCDB are shown (not in TCDB). The median of all expressed genes is represented by the dashed line. (b) Most expressed transport-related genes. Gray scale refer to gene family or superfamily. Genes are ordered according to TPMav values (x axis). Whiskers represent s.d. of biological replicates (n=3).
Genes for the malate synthase and isocitrate lyase (TPMav 211–605) were also expressed. These two enzymes are key for the glyoxylate cycle and their expression is an indication of aerobic growth on fatty acids and acetate (White, 2007). Active uptake of fatty acids could be carried by the product of expressed genes encoding for the proposed fatty acid transporter family (TPMav 80–790). Several genes, whose products are involved in the fatty acid β-oxidation pathway and the downstream propanoyl-CoA degradation pathway were also found expressed (TPMav 88–790) (Supplementary Table S2). Despite existing characterization of the fatty acid contents in sponges and associated bacteria (Gillan et al., 1988; Koopmans et al., 2015), this is to our knowledge the first report of heterotrophic fatty acid-based metabolism of bacteria associated with sponges. Future research focusing on the importance of fatty acids as energy source for sponge microbiome members and the origin of these compounds has the potential to elucidate the ecological relevance of this metabolic feature.
Creatinine degradation via creatine is used by bacteria to assimilate carbon and/or nitrogen (Kim et al., 1986; Shimizu et al., 1986). CcPhy expressed genes related to the degradation of creatine to glycine, that is, creatinase (TPMav 151±25) and the sarcosine oxidase alpha, beta, gamma and delta subunits (TPMav 122–296). A gene encoding for creatinine amidohydrolase was present in the genome but not expressed. Urea is a product of this pathway, which is generated by the hydrolysis of creatine by creatinase (Shimizu et al., 1986). The genome bin CcPhy lacked genes encoding for enzymes involved in the breakdown of urea into ammonium and inorganic carbon (Solomon et al., 2010). Because the genome is not complete and the fact that urease activity has been described in the species of the Phyllobacteriaceae family (for example, Peix et al., 2005), the existence and expression of Phyllobacteriaceae-like urease genes was further investigated for the C. concentrica microbiome. We found transcripts encoding for sequences similar to urease subunits and accessory proteins in the metatranscriptome of C. concentrica (Díez-Vives et al., 2016), but none were related to sequences from Phyllobacteriaceae (Supplementary Information; Supplementary Table S3), further corroborating the absence of urea breakdown in CcPhy.
Members of the Phyllobacteriaceae family are described as having respiratory metabolism with oxygen as the terminal electron acceptor, while anaerobic nitrate respiration and facultative chemolithotrophic metabolism also exist for some members of the family (Willems, 2014). CcPhy has the genetic repertoire to thrive in a variety of oxygenic conditions. The genome encodes for the subunits of two different heme–copper terminal oxidases, the mitochondrial-like aa3-type cytochrome c oxidase and the cbb3-type cytochrome c oxidase. Both these oligomeric cytochrome complexes transfer electrons of the respiratory chain to its final destination, oxygen, but have different O2 affinities and can be differentially regulated according to available oxygen concentrations (Garcia-Horsman et al., 1994; Morris and Schmidt, 2013). For instance, in Bradyrhizobium japonicum (family Bradyrhizobiaceae) the low affinity aa3-type is favored in fully aerobic conditions, while the high affinity cbb3-type is used in microaerobic conditions (Gabel and Maier, 1993; Preisig et al., 1996; Swem et al., 2001).
Genomic data also indicated that, in the absence of O2, CcPhy can perform nitrate (NO3−) respiration, where the membrane-bound nitrate reductase NarGHI complex transfers the electrons to nitrate, reducing it to nitrite (NO2−; Bertero et al., 2003). In this case, it is likely that nitrite is excreted via a nitrate/nitrite transporter NarK as the final product of the CcPhy anaerobic respiration. Nitrite excretion is likely because the genome lacked genes encoding for enzymes capable to further reduce nitrite to nitric oxide (NO), that is, a copper or a cytochrome cd1-containing nitrite reductase (NirK and NirS, respectively), or to ammonium (NH4+), that is, cytochrome c nitrite reductase (NrfA) (Kraft et al., 2011; Zheng et al., 2013). The components and related genes of the cytochrome c oxidases and respiratory nitrate reductase were all expressed and also at similar levels for the aerobic, microaerobic and anaerobic respiratory pathways (Supplementary Figure S8). It is unlikely that any given individual cell within a population performs all three modes of respiration at the same time and therefore our observation indicates that subpopulations within the sponge tissue experience distinct oxygen environments.
Nitrite-dependent chemolithoautotrophy and use of organic nitrogen compounds by CcNi
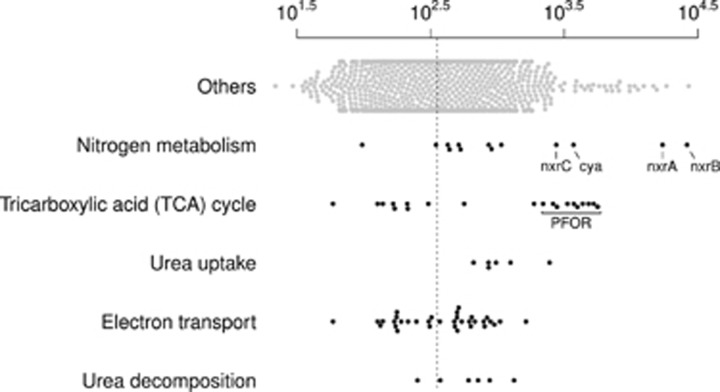
Nitrospira are known to obtain energy by oxidizing nitrite to nitrate using the key enzyme nitrite oxidoreductase (NXR) (Spieck and Bock, 2005). The NXR of Nitrospira was proposed to be a membrane-bound periplasmic complex composed of at least two subunits, the subunits alpha (nxrA) and beta (nxrB) (Spieck et al., 1998; Lucker et al., 2010). Two adjacent genes encoding for nxrA (gene locus Ga0068443_101117) and nxrB (Ga0068443_101118) were found in CcNi (see Supplementary Information for phylogenetic analysis) representing the fourth and second most abundant transcripts in CcNi transcriptome (TPMav 17 252±9822 and TPMav 26 308±13 520, respectively; Figure 3). Electron transport and ATPase encoding genes were also expressed (Supplementary Table S2), supporting a nitrite-dependent chemolithotrophic electron flow (Lucker et al., 2010). Expressed genes related to other nitrogen transformations were also found and are discussed in Supplementary Information. Taken together, these results indicate an active chemolithotrophic conversion of nitrite to nitrate by the CcNi population (Figure 1).
Figure 3.
Expression of selected functional genes of CcNi. TPMav values are shown for genes of selected functions. ‘Others’ stands for genes not grouped in the specific functions shown. The median of all expressed genes is represented by the dashed line. Highlighted genes encode for the nitrite oxidoreductase alpha (nxrA), beta (nxrB), putative C (nxrC) subunits (Supplementary Information), cyanate lyase (cya) and pyruvate ferredoxin oxidoreductase (PFOR).
Few Nitrospira species can decompose urea using the resulting ammonia for nitrification and biomass production, where urea would be imported from the extracellular milieu by the urea ABC transporter and hydrolyzed by urease (Koch et al., 2015; van Kessel et al., 2015). CcNi possess an putative operon with genes related to the transport and decomposition of urea, including those encoding for subunits of urease, urea ABC transporter, as well as auxiliary proteins (Ga0068443_100614-16, Ga0068443_100619-27) (Supplementary Figure S9). Some of these genes were highly expressed, such as gene for the urtA urea transport system substrate-binding protein (TPMav 2458±357), the urease subunit alpha (TPMav 1332±420) and the urtD and urtE urea transport system ATP-binding proteins (TPMav 978–1255; Figure 3). We did not find genomic evidence for the internal production of urea by CcNi, such as genes encoding for agmatinase or arginase. Because nitrite-oxidizing bacteria encode urea-producing enzymes (Palatinszky et al., 2015), the possibility of Nitrospira-like genes encoding for agmatinase or arginase to be expressed in C. concentrica was investigated. Transcripts encoding for sequences similar to agmatinase or arginase were found in the metatranscriptome of C. concentrica (Díez-Vives et al., 2016), but none of them were related to Nitrospira (Supplementary Information; Supplementary Table S4). This result further supports the absence of pathways for internal production of urea in CcNi.
The gene encoding for cyanate lyase (cya) was abundantly expressed in the transcriptome (TPMav 3712±1501) of CcNi (Figure 3). Cyanate lyase decomposes cyanate to CO2 and ammonia in a reaction dependent of bicarbonate (Johnson and Anderson, 1987). Its activity was described in N. moscoviensis and its proposed biological roles include nitrogen assimilation, cyanate detoxification and energy acquisition (Anderson et al., 1990; Palatinszky et al., 2015). The cya gene was localized downstream from two genes encoding for proteins of the formate/nitrite transporter family (TPMav 513–904) (Supplementary Figure S9). Members of the formate/nitrite transporter family (TC 2.A.44) have broad specificity for small monovalent anions (Lu et al., 2013). It is likely that CcNi putative formate/nitrite transporter is also permeable to cyanate, as similarly inferred for the ammonia-oxidizing archaea Candidatus Nitrososphaera gargensis due to the proximity of genes for transporter and enzymatic degradation (Spang et al., 2012).
Nitrospira species are proposed to fix carbon via the reductive TCA (rTCA) cycle (Starkenburg et al., 2006; Lucker et al., 2010, 2013; Koch et al., 2015). The rTCA cycle shares most of its enzymes with the oxidative TCA (oTCA) cycle, with the exception of three key enzymes, which are characteristic of the reductive cycle: ATP citrate lyase, 2-oxoglutarate:ferredoxin oxidoreductase (also named as α-ketogluterate synthase), and fumarate reductase (Berg, 2011). Alternatively, ATP-dependent cleavage of citrate can be carried by citryl-CoA synthetase and citryl-CoA lyase enzymes (Aoshima et al., 2004a,2004b). CcNi encoded enzymes that are shared between the two forms of TCA cycles. In addition, CcNi encoded a pyruvate ferredoxin oxidoreductase, which can act as pyruvate synthase (EC 1.2.7.1), carboxylating acetyl-CoA to pyruvate (Evans et al., 1966; Furdui and Ragsdale, 2000) (Figure 3). However, the characteristic rTCA key enzymes were not annotated in the genome (Figure 1). The annotation of genes encoding for A, B and C subunits of succinate dehydrogenase (complex II) or fumarate reductase (Ga0068443_100221-23) could indicate fumarate reductase activity and be considered an evidence for the rTCA cycle. These two enzymes are structurally similar and carry opposite reactions, that is, succinate oxidation and fumarate reduction (Hagerhall, 1997). Succinate dehydrogenase/fumarate reductase subunit genes are localized immediately adjacent to citrate synthase gene (Ga0068443_100220) in the CcNi genome. Although the mapping of metatranscriptomic reads to CDS does not allow us to verify whether they are co-expressed as an operon, their adjacent localization and similar expression levels (TPMav 58–169) are suggestive of the participation of succinate dehydrogenase/fumarate reductase in the oTCA cycle as succinate dehydrogenase and not fumarate reductase. In the absence of evidence of other carbon fixation pathways (Supplementary Information), we hypothesize that CcNi fix carbon via rTCA cycle as conducted by other Nitrospira members, despite the lack of the key genes of this pathway in the genome bin.
Ammonia-dependent chemolithoautotrophy and use of organic nitrogen compounds by CcThau
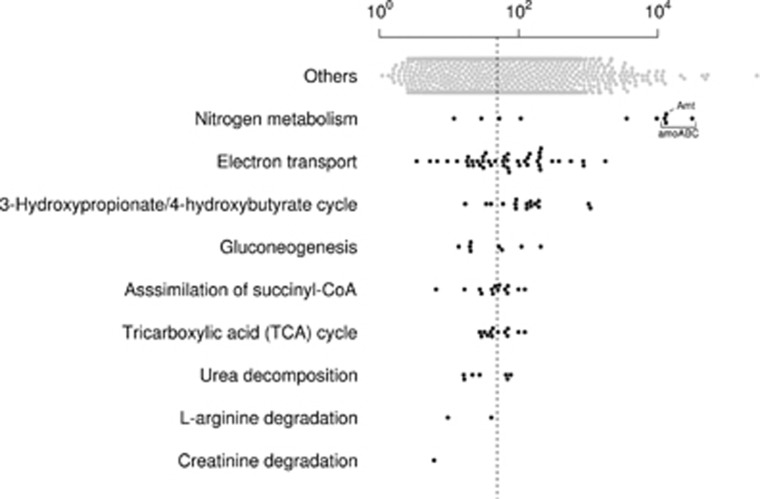
Cultured members of the Thaumarchaeota are able to aerobically oxidase ammonia to nitrite (Stieglmeier et al., 2014a). Genes related to energy acquisition through ammonia oxidation were among the most abundantly transcribed in CcThau (TPMav 3566–31 115, Figures 1 and 4). Ammonia oxidation is carried by ammonia monooxygenase, which in bacteria is composed of three subunits encoded by amoA, amoB and amoC genes (Arp et al., 2007). Additionally, a gene named ‘amoX’ was proposed to be associated with the archaeal ammonia monooxygenase, due to the conserved co-localization with the subunit genes across ammonia-oxidizing archaea (Bartossek et al., 2012). Single copies of genes putatively related to archaeal ammonia monooxygenase were present in pairs, amoA–amoX (Ga0078905_11462, Ga0078905_11463) and amoB–amoC (Ga0078905_103916-17), in the edge of two contigs, possibly forming a single gene array (amoAXCB) as observed in the genomes of Nitrosoarchaeum limnia SFB1 and Nitrosopumilus maritimus SCM1 (Walker et al., 2010; Blainey et al., 2011; Bartossek et al., 2012). The gene encoding for an ammonium transporter (Ga0078905_100724, TPMav 13 184±1736) was estimated to be expressed at an approximate ratio of 1:1 to amoA. In addition, several genes putatively involved in ammonia-dependent chemolithotrophy in Thaumarchaeota were expressed at different levels by CcThau (Kozlowski et al., 2016), including genes encoding for nitrite reductase (NO-forming) (TPMav 107–9653), plastocyanins (TPMav 37–1761), copper-binding proteins of the plastocyanin/azurin family (TPMav 19–200), ubiquinol–cytochrome c reductase cytochrome b subunit (TPMav 304±26), multicopper oxidase (TPMav 18±4) and ATPase subunits (TPMav 26–847). These findings are consistent with the observation of a large transcriptional effort invested in genes related to ammonia-dependent chemolithotrophy by archaeal populations in marine sponges (Radax et al., 2012; Moitinho-Silva et al., 2014).
Figure 4.
Expression of selected functional genes of CcThau. TPMav values are shown for genes of selected functions. ‘Others’ stands for genes not grouped in the specific functions shown. The median of all expressed genes is represented by the dashed line. Highlighted genes encode for the ammonia monooxygenase subunits alpha (amoA), beta (amoB) and C (amoC) subunits as well as ammonium transporter (Amt).
Genomic analysis of several Thaumarchaea species revealed the presence of genes related to transport, production and degradation of urea (Palatinszky et al., 2015). Urea can be used by Thaumarchaeota members as source of ammonia, as shown by biochemical assays of cultivated species (Lehtovirta-Morley et al., 2016), and as carbon source, as inferred from 14C-labeled urea incorporation of prokaryotic communities of Artic waters (Alonso-Saez et al., 2012). The genes encoding for urease subunits, as well as auxiliary proteins, were present in two separate contigs of CcThau but lowly expressed (TPMav 16–77). No encoded urea transporter was found. A set of genes encoding for amino acid/amide ABC transporter of the hydrophobic amino-acid uptake transporter (HAAT) family (TCDB:3.A.1.4), which includes urea transporters (Hosie et al., 2002), were found in the genome. Thus it is conceivable that the encoded products may participate in urea transport. Urea can also be internally produced from the degradation of the amino-acid L-arginine (Shaibe et al., 1985), and the genes encoding for arginine decarboxylase and agmatinase were expressed at low levels (TPMav 10–40).
The expression of a gene encoding for creatinine amidohydrolase, albeit at low levels (TPMav 6±7), further indicates that CcThau degrades creatinine. Creatinine amidohydrolase genes are also present in other members of the candidate order Nitrosopumilales, for example, Nitrosopumilus maritimus SCM1 (Nmar_0222) (Walker et al., 2010). The gene encoding for a creatinase, which would carry the downstream hydrolysis of creatine to sarcosine and urea (Shimizu et al., 1986), was however not found in the CcThau genome bin. The most similar sequence to characterized creatinases from bacteria species was a gene encoding for a Xaa-Pro aminopeptidase (Ga0078905_10021), which is structurally related to creatinase (Supplementary Information; Supplementary Table S5; Bazan et al., 1994). This similarity, however, does not provide enough evidence that CcThau converts creatinine to creatine in order to produce urea and, consequently, ammonia. Therefore, it is unclear in which biochemical context creatinine degradation is carried out by CcThau.
An almost complete set of genes for the 3-hydroxypropionate/4-hydroxybutyrate (HP/HB) pathway was expressed (Figure 1; Supplementary Table S2). The HP/HB pathway is an autotrophic CO2 fixation cycle recently discovered in archaea and present in all Thaumarchaeota species characterized so far (Berg et al., 2007; Bayer et al., 2016). CcThau uses the same set of enzymes as N. maritimus, including 3-hydroxypropionyl-CoA synthetase (ADP-forming) (TPMav 173±53) and 4-hydroxybutyryl-CoA synthetase (ADP-forming) (TPMav 90±31) (Konneke et al., 2014). In one cycle of the HP/HB pathway, two bicarbonate molecules are converted to one acetyl-coA. It is possible that the CcThau HP/HB pathway mainly connects to the central carbon metabolism via succinyl-CoA, which is produced from the generated acetyl-CoA and another pair of bicarbonate molecules by an additional half turn of the cycle (Estelmann et al., 2011). With the exception of the gene for fumarase, for which no orthologous was identified, CcThau showed the expression of the enzymatic repertoire capable to further transform succinyl-coA into precursor metabolites, such as oxaloacetate, phosphoenolpyruvate (PEP), pyruvate and 2-oxoglutarate (TPMav 6–122). This set includes enzymes that are components of the oxidative TCA cycle. In addition, genes encoding for enzymes of the rTCA cycle were also found expressed, such as citrate lyase subunit beta/citryl-CoA lyase and 2-oxoglutarate ferredoxin oxidoreductase (TPMav 32–39). In Thaumarchaeota, oTCA cycle was assumed to be used for anaplerotic reactions and the capacity of Thaumarchaeota to fix carbon via rTCA cycle was not demonstrated (Spang et al., 2012; Zhalnina et al., 2014; Stieglmeier et al., 2014a). Mixotrophic growth has been reported in the Thaumarchaeota phylum (for example, Qin et al., 2014). For CcThau, the possible uptake of urea and amino acids by the amino acid/amide ABC transporter of the HAAT family and the degradation of creatinine by creatinine amidohydrolase were the only evidence to suggest the use of external organic carbon sources (Supplementary Information).
Metabolic features of diatoms and the sponge
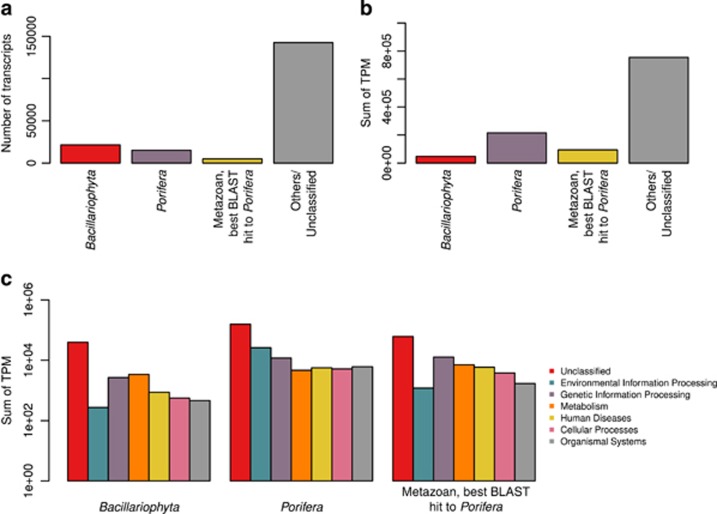
From the 184 053 eukaryotic transcripts recovered from C. concentrica (Díez-Vives et al., 2016), the number of transcripts assigned to diatoms (Bacillariophyta) was similar to the combined number of transcripts classified as Porifera or as metazoans with best BLAST hit against sequences from sponges (Figure 5a). Higher expression, estimated from the sum of TPM values, was found for the host compared with the diatom symbiont (Figure 5b). Most transcripts could not be functionally classified for both organisms. Environmental information processing, genetic information processing and metabolism were the pathways with highest overall sums of TPM (Figure 5c). The comprehensive description of the diatom and sponge transcriptomes is beyond the scope of this study and therefore we focused here on key metabolic pathways.
Figure 5.
Expression of diatom and sponge transcripts in Cymbastela concentrica. (a) Transcripts taxonomically classified as diatoms (Bacillariophyta) or sponge (Porifera or Metazoan with best BLAST hit to sponge sequences) were sorted from other eukaryotic transcripts recovered from C. concentrica. (b) Overall expression of diatom and sponge is indicated by the sum of transcripts’ TPM. (c) Deduced peptide sequences from transcripts’ coding sequences were used for KEGG functional classification. Expression of KEGG functions was inferred from the sum of TPM values across coding sequences.
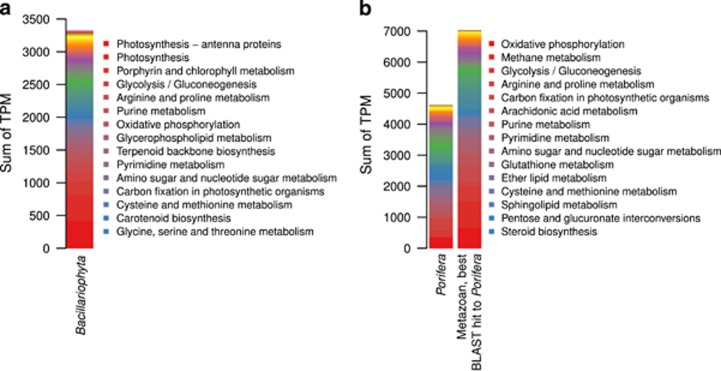
The diatom symbionts invested a large transcriptional effort in functions for energy harvesting via photosynthesis and consumption via glycolysis (Figure 6a; Supplementary Table S6). In addition, functions related to arginine and proline metabolism were found to be abundantly expressed by the diatoms, in particular prolyl 4-hydroxylase (TPMsum 46, Supplementary Table S6), the enzyme that carries out posttranslational modifications of proline (Gorres and Raines, 2010). The expression of carbamoyl-phosphate synthase, argininosuccinate synthase, argininosuccinate lyase and arginase (TPMsum 3–10) are indicative of an active urea cycle in the diatom symbionts. Additional support for this metabolic cycle is provided by the expression of ornithine cyclodeaminase (TPMsum 4) and ornithine decarboxylase (TPMsum 5), which produce proline and putrescine from ornithine, respectively. The urea cycle in diatoms is proposed to redistribute and turnover inorganic carbon and nitrogen coming from catabolism and/or photorespiration (Allen et al., 2011). With respect to sources of nitrogen, the diatoms expressed functions related to nitrate assimilation (nitrate reductase (NADPH), ferredoxin-nitrite reductase, TPMsum 2–5) and production of ammonia via nitrite generated from the oxygenation of alkyl nitronates (nitronate monooxygenase TPMsum 1). The co-expression of these pathways at different intensities has also been recently observed in metatranscriptomes of Skeletonema spp. recovered from surface seawater (Alexander et al., 2015) and indicates the simultaneous use of multiple sources of nitrogen by the diatom symbiont.
Figure 6.
Expressed metabolic functions in diatoms and the sponge. Sums of TPM are shown for KEGG metabolic pathways found in (a) diatoms (Bacillariophyta) and (b) sponge (Porifera and Metazoan or Metazoan with best BLAST hit to sponge sequences). Legends list the top expressed pathways.
Sponge cells abundantly expressed functions of the oxidative phosphorylation (for example, cytochrome c oxidase subunit 6a, TPMsum 418, Supplementary Table S7) and glycolysis (glyceraldehyde 3-phosphate dehydrogenase, TPMsum 418) (Figure 6b). The expression of functions assigned to methane metabolism was largely due to antioxidant enzymes, catalase (TPMsum 468) and peroxiredoxin 6, 1-Cys peroxiredoxin (TPMsum 353), which may protect the sponge cells from reactive oxygen species (Tate et al., 1995; Chen et al., 2000). The most abundant functions related to arginine and proline metabolism were acetylornithine deacetylase (TPMsum 237) and glycine amidinotransferase (TPMsum 94). Both enzymes carry out reactions that produces L-ornithine, the former from N2-acetyl-L-ornithine and H2O and the latter from L-arginine and glycine. The reaction of glycine amidinotransferase is the first of the two-step creatine biosynthesis (Wyss and Kaddurah-Daouk, 2000). Along with creatine, creatinine is expected to be found in sponge cells, as it is a product of spontaneous, non-enzymatic conversion of the former. In addition, the sponge expressed a creatine kinase function (TPMsum 34), which reversibly converts creatine and ATP to phosphocreatine and ADP. This result is in support of previous detection of creatine kinase activity in the tissue of the sponge Tethya aurantia (Ellington, 2000; Sona et al., 2004). In sponges, the creatine kinase/phosphocreatine system was proposed as an intracellular spatial buffer of energy, connecting the ATP produced in the mitochondria with distant cytosolic sites of ATP consumption (Sona et al., 2004; Ellington and Suzuki, 2007).
Model of integrated metabolism of the microbial community members in C. concentrica
Based on the genomic and transcriptomic information of these three prokaryotic symbionts and the expression of eukaryotic transcripts assigned to diatoms and sponges, we developed a metabolic model for the sponge–microbe symbiosis in which nutrient fluxes between cells and the extracellular milieu, that is the sponge mesohyl, were inferred from our data set (see Figure 1).
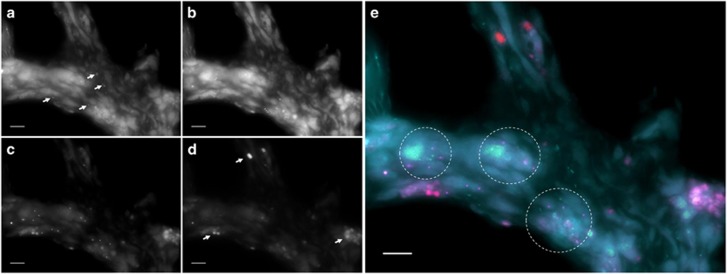
As shown above, CcPhy is a heterotrophic bacterium with the capacity to uptake a range of organic and inorganic compounds. Such nutrients are likely found across the sponge tissue, which is consistent with a scattered distribution of CcPhy cells throughout the mesohyl (see Figure 7; Supplementary Figure S10). Considering the high relative abundance of Phyllobacteriaceae within the symbiont community (Fan et al., 2012; Esteves et al., 2016), CcPhy may thus be a major contributor to uptake and utilization of DOM by the microbiome. The DOM used by CcPhy may come from the sponge’s filtering activity (Figure 1a; de Goeij et al., 2008, 2013) or from diatoms (Figure 1b) that were found throughout the mesohyl (Figure 7), especially considering that axenic cultures of diatoms have been shown to release between 5% and 21% of their total fixed carbon as dissolved organic carbon (Wetz and Wheeler, 2007). In support of this, photosynthesis functions were abundantly expressed in the diatom transcriptome (Figure 6a). Another source of carbon for the members of the sponge–microbe symbiosis may be from the autotrophic metabolism of CcThau and CcNi (Figures 1c and d, see above).
Figure 7.
Visualization of microorganisms associated with Cymbastela concentrica by fluorescence in situ hybridization. (a) Rod-shaped cells (white arrows) were observed from hybridization with a Pacific Blue-labeled CcPhy-specific probe. Signals from (b) fluorescein-isothiocyanate-labeled CcNi-specific and (c) Cy3-labeled CcThau-specific probes are shown. (d) Red autofluorescence signals from diatoms were captured (arrows). (e) Clusters of CcNi-specific (green) and CcThau-specific (magenta) probes in the sponge mesohyl are highlighted in the merged color image by circles. CcPhy cells (cyan) did not specifically co-localize with CcNi or CcThau cells. Autofluorescence of the sponge tissue (light blue/cyan) served as a reference for the structure of the sponge mesohyl. Scale bars, 10 μm.
Creatine and creatinine are products of the sponge metabolism, as indicated by the expression transcripts encoding for glycine amidinotransferase and creatine kinase. Although it is unknown whether sponge cells exchange creatine between different cells and tissues as described in higher animals (Wyss and Kaddurah-Daouk, 2000), it is conceivable that these compounds are accessible to the microbial community (Figure 1e). The possibility that creatine and creatinine are actively used by the microbiome is supported by the expression of genes able to degrade creatine to glycine by CcPhy and the encoded creatinine amidohydrolase in CcPhy and CcThau genomes. The degradation of these host metabolites by the symbionts is likely widespread, as inferred by the previous observation of related genes in the metagenome of six sponges, including C. concentrica (Fan et al., 2012). Here we could assign this function to be actively used by specific members of the microbiome.
The metabolism of CcPhy, diatom, CcNi and CcThau are linked by the production and consumption of urea. CcPhy internally generates urea from creatine degradation, which can potentially accumulate and diffuse through the bacterial membrane (Figure 1f; Lodish et al., 2000). This assumption is further supported by the fact that heterotrophic marine bacteria have been observed to be net producer of urea even in the presence of hydrolytic activity (Cho et al., 1996; Berg and Jorgensen, 2006). The diatom symbiont is another potential source of urea as suggested by the expression of the urea cycle (Figure 1g). The Nitrospira population represented by CcNi may benefit directly from the urea in the extracellular milieu, as it abundantly expresses the genes related to the transport and degradation of urea. Despite the internal production of urea via L-arginine degradation, CcThau may also uptake urea via the amino acid/amide ABC transporter of the HAAT family. Additionally, urea can spontaneously dissociate to cyanate (Marier and Rose, 1964; Guilloton and Karst, 1985). Similarly, cyanate is formed from carbamoyl phosphate (Allen and Jones, 1964), which is an intermediate of the urea cycle in diatoms. Therefore, urea indirectly links CcPhy and the diatom symbiont to CcNi via cyanate (Figures 1h–j), which could be imported and decomposed to CO2 and ammonia as indicated by the expression of cyanate lyase genes in CcNi.
Urease genes were also found in other sponge-associated microbial genomes and microbiomes (Hallam et al., 2006; Siegl et al., 2011; Bayer et al., 2014; Liu et al., 2016). Together with the phylogenetic diversity found for ureC genes and transcripts found in the sponge Xestospongia testudinaria (Su et al., 2013), these data indicate that urea degradation is a common trait of sponge-associated microorganisms. Although urea can be found in benthic environments (Crandall and Teece, 2012), our data points to the possibility of urea being derived from host metabolites, that is, via bacterial metabolism of creatine and creatinine, and from intracellular metabolism of sponge symbionts, such as diatoms.
Pumping activity and host metabolism have been regarded as sources of ammonia in the sponge tissue (Ribes et al., 2012; Figure 1k). Additionally, urea and cyanate are degraded by CcNi, which is likely a source of ammonia (Figure 1l; Koch et al., 2015). Our analysis indicates that CcPhy takes up ammonium for nitrogen assimilation, while CcThau oxidizes ammonia to generate energy, releasing nitrite (Figure 1m). Nitrite in turn serves as the energy source for CcNi, which expressed the genes necessary for nitrite oxidation. Fluorescence in situ hybridization analysis showed that CcThau and CcNi are often localized in close proximity (Figure 7), supporting such a metabolic interaction. Nitrification, a process in which the oxidation of ammonia is coupled with the oxidation of nitrite, is a common theme in sponge microbiology, being the subject of many studies (Taylor et al., 2007). Based on the use of urea and cyanate as indirect sources of ammonia in nitrification carried out by co-cultures of Nitrospira and ammonia-oxidizing bacteria (Koch et al., 2015; Palatinszky et al., 2015) and the expression profile of CcNi and CcThau, we propose that these compounds should be considered when modeling nitrification in marine sponges.
The end product of the combined aerobic nitrification by CcThau and CcNi is nitrate, which would support anaerobic nitrate respiration by CcPhy (Figure 1n, see above). Such a coupling is possible if the two processes and their associated population are temporally and/or spatially separated by variations in oxygen concentration or redox potential, which have been described to occur in sponges (Schläppy et al., 2010; Lavy et al., 2016). The facultative nature of the CcPhy metabolism supports its temporal separation from the aerobic metabolisms CcThau and CcNi, while the observation that CcPhy rarely co-occured with the CcThau and CcNi cells would indicate also a spatial separation of their metabolisms (see Figure 7). The incomplete denitrification pathway of CcPhy would also yield nitrite (Figure 1o), which again could serve as an energy source for CcNi, thus creating a ‘mini’ N cycle at the interface of aerobic and anaerobic micro-habitats. An alternative path for the N cycle within C. concentrica is the nitrate assimilation by diatoms (Figure 1n).
Conclusion
We described the lifestyle of prominent members of C. concentrica sponge–microbe symbiosis, that is, associated prokaryotes, diatoms and the host, by analyzing their transcriptional activities. Although connected metabolic pathways are expected to occur based on the complexity of nutrient cycles within sponges (Hoffmann et al., 2009), our study is unprecedented in modeling how the physiology of microbiome members are integrated. We thus predicted how and which members can generate, retain, recycle and compete for biochemical resources. In addition, we considered the gene expression of the sponge and diatoms, unveiling the nutritional basis for biological interactions within the sponge–microbe symbiosis. Finally, we demonstrated the power of a holistic approach to further our understanding of the complexity of animal–microbe symbioses.
Acknowledgments
We acknowledge the financial support of the Betty and Gordon Moore Foundation and the Australian Research Council. We thank Dr Tamsin Peters and Jadranka Nappi for sample collection. We also thank Dr Ute Hentschel for hosting GB at the GEOMAR Helmholtz Centre for Ocean Research. MTJ was supported by grants of the German Excellence Initiative to the Graduate School of Life Sciences, University of Wuerzburg.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Alexander H, Jenkins BD, Rynearson TA, Dyhrman ST (2015). Metatranscriptome analyses indicate resource partitioning between diatoms in the field. Proc Natl Acad Sci USA 112: E2182–E2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen AE, Dupont CL, Obornik M, Horak A, Nunes-Nesi A, McCrow JP et al (2011). Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473: 203–207. [DOI] [PubMed] [Google Scholar]
- Allen CM, Jones ME (1964). Decomposition of carbamylphosphate in aqueous solutions. Biochemistry 3: 1238–1247. [DOI] [PubMed] [Google Scholar]
- Alonso-Saez L, Waller AS, Mende DR, Bakker K, Farnelid H, Yager PL et al (2012). Role for urea in nitrification by polar marine Archaea. Proc Natl Acad Sci USA 109: 17989–17994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson PM, Sung YC, Fuchs JA (1990). The cyanase operon and cyanate metabolism. FEMS Microbiol Rev 7: 247–252. [DOI] [PubMed] [Google Scholar]
- Aoshima M, Ishii M, Igarashi Y (2004. a). A novel enzyme, citryl-CoA synthetase, catalysing the first step of the citrate cleavage reaction in Hydrogenobacter thermophilus TK-6. Mol Microbiol 52: 751–761. [DOI] [PubMed] [Google Scholar]
- Aoshima M, Ishii M, Igarashi Y (2004. b). A novel enzyme, citryl-CoA lyase, catalysing the second step of the citrate cleavage reaction in Hydrogenobacter thermophilus TK-6. Mol Microbiol 52: 763–770. [DOI] [PubMed] [Google Scholar]
- Arp DJ, Chain PS, Klotz MG (2007). The impact of genome analyses on our understanding of ammonia-oxidizing bacteria. Annu Rev Microbiol 61: 503–528. [DOI] [PubMed] [Google Scholar]
- Bartossek R, Spang A, Weidler G, Lanzen A, Schleper C (2012). Metagenomic analysis of ammonia-oxidizing archaea affiliated with the soil group. Front Microbiol 3: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer B, Vojvoda J, Offre P, Alves RJ, Elisabeth NH, Garcia JA et al (2016). Physiological and genomic characterization of two novel marine thaumarchaeal strains indicates niche differentiation. ISME J 10: 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer K, Moitinho-Silva L, Brummer F, Cannistraci CV, Ravasi T, Hentschel U (2014). GeoChip-based insights into the microbial functional gene repertoire of marine sponges (high microbial abundance, low microbial abundance) and seawater. FEMS Microbiol Ecol 90: 832–843. [DOI] [PubMed] [Google Scholar]
- Bazan JF, Weaver LH, Roderick SL, Huber R, Matthews BW (1994). Sequence and structure comparison suggest that methionine aminopeptidase, prolidase, aminopeptidase P, and creatinase share a common fold. Proc Natl Acad Sci USA 91: 2473–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JJ (2008). The functional roles of marine sponges. Estuar Coastal Shelf Sci 79: 341–353. [Google Scholar]
- Berg GM, Jorgensen NOG (2006). Purine and pyrimidine metabolism by estuarine bacteria. Aquat Microb Ecol 42: 215–226. [Google Scholar]
- Berg IA, Kockelkorn D, Buckel W, Fuchs G (2007). A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318: 1782–1786. [DOI] [PubMed] [Google Scholar]
- Berg IA (2011). Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol 77: 1925–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertero MG, Rothery RA, Palak M, Hou C, Lim D, Blasco F et al (2003). Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat Struct Biol 10: 681–687. [DOI] [PubMed] [Google Scholar]
- Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR (2011). Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS ONE 6: e16626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary SC, Giovannoni SJ (1993). Transovarial inheritance of endosymbiotic bacteria in clams inhabiting deep-sea hydrothermal vents and cold seeps. Proc Natl Acad Sci USA 90: 5695–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-W, Dodia C, Feinstein SI, Jain MK, Fisher AB (2000). 1-Cys peroxiredoxin, a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. J Biol Chem 275: 28421–28427. [DOI] [PubMed] [Google Scholar]
- Cho B, Park M, Shim J, Azam F (1996). Significance of bacteria in urea dynamics in coastal surface waters. Mar Ecol Prog Ser 142: 19–26. [Google Scholar]
- Cho I, Blaser MJ (2012). The human microbiome: at the interface of health and disease. Nat Rev Genet 13: 260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyte KZ, Schluter J, Foster KR (2015). The ecology of the microbiome: networks, competition, and stability. Science 350: 663–666. [DOI] [PubMed] [Google Scholar]
- Crandall JB, Teece MA (2012). Urea is a dynamic pool of bioavailable nitrogen in coral reefs. Coral Reefs 31: 207–214. [Google Scholar]
- de Goeij JM, van den Berg H, van Oostveen MM, Epping EHG, van Duyl FC (2008). Major bulk dissolved organic carbon (DOC) removal by encrusting coral reef cavity sponges. Mar Ecol Prog Ser 357: 139–151. [Google Scholar]
- de Goeij JM, van Oevelen D, Vermeij MJ, Osinga R, Middelburg JJ, de Goeij AF et al (2013). Surviving in a marine desert: the sponge loop retains resources within coral reefs. Science 342: 108–110. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA (2007). An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449: 811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez-Vives C, Moitinho-Silva L, Nielsen S, Reynolds D, Thomas T. (2016).Expression of eukaryotic-like protein in the microbiome of sponges. Mol Ecol; e-pub ahead of print 30 December 2016; doi:10.1111/mec.14003. [DOI] [PubMed]
- Douglas AE, Werren JH (2016). Holes in the hologenome: why host-microbe symbioses are not holobionts. MBio 7: e02099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington WR (2000). A dimeric creatine kinase from a sponge: implications in terms of phosphagen kinase evolution. Comp Biochem Physiol B Biochem Mol Biol 126: 1–7. [DOI] [PubMed] [Google Scholar]
- Ellington WR, Suzuki T (2007). Early evolution of the creatine kinase gene family and the capacity for creatine biosynthesis and membrane transport. Subcell Biochem 46: 17–26. [DOI] [PubMed] [Google Scholar]
- Estelmann S, Hugler M, Eisenreich W, Werner K, Berg IA, Ramos-Vera WH et al (2011). Labeling and enzyme studies of the central carbon metabolism in Metallosphaera sedula. J Bacteriol 193: 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves AI, Amer N, Nguyen M, Thomas T (2016). Sample processing impacts the viability and cultivability of the sponge microbiome. Front Microbiol 7: 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MC, Buchanan BB, Arnon DI (1966). A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proc Natl Acad Sci USA 55: 928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Reynolds D, Liu M, Stark M, Kjelleberg S, Webster NS et al (2012). Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc Natl Acad Sci USA 109: E1878–E1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman CJ, Thacker RW, Baker DM, Fogel ML (2013). Quality or quantity: is nutrient transfer driven more by symbiont identity and productivity than by symbiont abundance? ISME J 7: 1116–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdui C, Ragsdale SW (2000). The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood-Ljungdahl pathway. J Biol Chem 275: 28494–28499. [DOI] [PubMed] [Google Scholar]
- Gabel C, Maier RJ (1993). Oxygen-dependent transcriptional regulation of cytochrome aa3 in Bradyrhizobium japonicum. J Bacteriol 175: 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Horsman JA, Barquera B, Rumbley J, Ma J, Gennis RB (1994). The superfamily of heme-copper respiratory oxidases. J Bacteriol 176: 5587–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillan FT, Stoilov IL, Thompson JE, Hogg RW, Wilkinson CR, Djerassi C (1988). Fatty acids as biological markers for bacterial symbionts in sponges. Lipids 23: 1139–1145. [DOI] [PubMed] [Google Scholar]
- Gorres KL, Raines RT (2010). Prolyl 4-hydroxylase. Crit Rev Biochem Mol Biol 45: 106–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilloton M, Karst F (1985). A spectrophotometric determination of cyanate using reaction with 2-aminobenzoic acid. Anal Biochem 149: 291–295. [DOI] [PubMed] [Google Scholar]
- Hadas E, Shpigel M, Ilan M (2009). Particulate organic matter as a food source for a coral reef sponge. J Exp Biol 212: 3643–3650. [DOI] [PubMed] [Google Scholar]
- Hagerhall C (1997). Succinate: quinone oxidoreductases. Variations on a conserved theme. Biochim Biophys Acta 1320: 107–141. [DOI] [PubMed] [Google Scholar]
- Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM et al (2006). Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol 4: e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann F, Radax R, Woebken D, Holtappels M, Lavik G, Rapp HT et al (2009). Complex nitrogen cycling in the sponge Geodia barretti. Environ Microbiol 11: 2228–2243. [DOI] [PubMed] [Google Scholar]
- Hosie AH, Allaway D, Galloway CS, Dunsby HA, Poole PS (2002). Rhizobium leguminosarum has a second general amino acid permease with unusually broad substrate specificity and high similarity to branched-chain amino acid transporters (Bra/LIV) of the ABC family. J Bacteriol 184: 4071–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC (2011). Integrative analysis of environmental sequences using MEGAN4. Genome Res 21: 1552–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imelfort M, Parks D, Woodcroft BJ, Dennis P, Hugenholtz P, Tyson GW (2014). GroopM: an automated tool for the recovery of population genomes from related metagenomes. PeerJ 2: e603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelle A, Thomas G, Marini AM, Kramer R, Merrick M (2005). In vivo functional characterization of the Escherichia coli ammonium channel AmtB: evidence for metabolic coupling of AmtB to glutamine synthetase. Biochem J 390: 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WV, Anderson PM (1987). Bicarbonate is a recycling substrate for cyanase. J Biol Chem 262: 9021–9025. [PubMed] [Google Scholar]
- Kamke J, Sczyrba A, Ivanova N, Schwientek P, Rinke C, Mavromatis K et al (2013). Single-cell genomics reveals complex carbohydrate degradation patterns in poribacterial symbionts of marine sponges. ISME J 7: 2287–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Hosokawa T, Fukatsu T (2007). Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl Environ Microbiol 73: 4308–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Shimizu S, Yamada H (1986). Sarcosine oxidase involved in creatinine degradation in Alcaligenes denitrificans subsp dentrificans J9 and Arthrobacter spp J5 and J11. Agric Biol Chem 50: 2811–2816. [Google Scholar]
- Koch H, Lucker S, Albertsen M, Kitzinger K, Herbold C, Spieck E et al (2015). Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc Natl Acad Sci USA 112: 11371–11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546. [DOI] [PubMed] [Google Scholar]
- Konneke M, Schubert DM, Brown PC, Hugler M, Standfest S, Schwander T et al (2014). Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc Natl Acad Sci USA 111: 8239–8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M, van Rijswijk P, Boschker HT, Marco H, Martens D, Wijffels RH (2015). Seasonal variation of Fatty acids and stable carbon isotopes in sponges as indicators for nutrition: biomarkers in sponges identified. Mar Biotechnol (NY) 17: 43–54. [DOI] [PubMed] [Google Scholar]
- Kozlowski JA, Stieglmeier M, Schleper C, Klotz MG, Stein LY (2016). Pathways and key intermediates required for obligate aerobic ammonia-dependent chemolithotrophy in bacteria and Thaumarchaeota. ISME J 10: 1836–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft B, Strous M, Tegetmeyer HE (2011). Microbial nitrate respiration—genes, enzymes and environmental distribution. J Biotechnol 155: 104–117. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy A, Keren R, Yahel G, Ilan M (2016). Intermittent hypoxia and prolonged suboxia measured in situ in a marine sponge. Front Mar Sci 3: 263. [Google Scholar]
- Lehtovirta-Morley LE, Ross J, Hink L, Weber EB, Gubry-Rangin C, Thion C et al (2016). Isolation of ‘Candidatus Nitrosocosmicus franklandus’, a novel ureolytic soil archaeal ammonia oxidiser with tolerance to high ammonia concentration. FEMS Microbiol Ecol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN (2010). RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26: 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Li J, Feng G, Li Z (2016). New genomic insights into ‘Entotheonella’ symbionts in Theonella swinhoei: mixotrophy, anaerobic adaptation, resilience, and interaction. Front Microbiol 7: 1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Fan L, Zhong L, Kjelleberg S, Thomas T (2012). Metaproteogenomic analysis of a community of sponge symbionts. ISME J 6: 1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MY, Kjelleberg S, Thomas T (2011). Functional genomic analysis of an uncultured delta-proteobacterium in the sponge Cymbastela concentrica. ISME J 5: 427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J (2000) Section 15.1. Diffusion of Small Molecules Across Phospholipid Bilayers. Molecular Cell Biology 4th ednFreeman & Co.: New York, NY, USA, p1084. [Google Scholar]
- Lu W, Du J, Schwarzer NJ, Wacker T, Andrade SL, Einsle O (2013). The formate/nitrite transporter family of anion channels. Biol Chem 394: 715–727. [DOI] [PubMed] [Google Scholar]
- Lucker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B et al (2010). A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci USA 107: 13479–13484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucker S, Nowka B, Rattei T, Spieck E, Daims H (2013). The genome of nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front Microbiol 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marier JR, Rose D (1964). Determination of cyanate, and a study of its accumulation in aqueous solutions of urea. Anal Biochem 7: 304–314. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Pillay M et al (2014). IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res 42: D560–D567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marta R, Rafel C, Josep-Maria G (1999). Natural diet and grazing rate of the temperate sponge Dysidea avara (Demospongiae, Dendroceratida) throughout an annual cycle. Mar Ecol Prog Ser 176: 179–190. [Google Scholar]
- Martens EC, Heungens K, Goodrich-Blair H (2003). Early colonization events in the mutualistic association between Steinernema carpocapsae nematodes and Xenorhabdus nematophila bacteria. J Bacteriol 185: 3147–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitinho-Silva L, Seridi L, Ryu T, Voolstra CR, Ravasi T, Hentschel U (2014). Revealing microbial functional activities in the Red Sea sponge Stylissa carteri by metatranscriptomics. Environ Microbiol 16: 3683–3698. [DOI] [PubMed] [Google Scholar]
- Morris RL, Schmidt TM (2013). Shallow breathing: bacterial life at low O(2). Nat Rev Microbiol 11: 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatinszky M, Herbold C, Jehmlich N, Pogoda M, Han P, von Bergen M et al (2015). Cyanate as an energy source for nitrifiers. Nature 524: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peix A, Rivas R, Trujillo ME, Vancanneyt M, Velazquez E, Willems A (2005). Reclassification of Agrobacterium ferrugineum LMG 128 as Hoeflea marina gen. nov., sp. nov. Int J Syst Evol Microbiol 55: 1163–1166. [DOI] [PubMed] [Google Scholar]
- Preisig O, Zufferey R, Thony-Meyer L, Appleby CA, Hennecke H (1996). A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J Bacteriol 178: 1532–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Amin SA, Martens-Habbena W, Walker CB, Urakawa H, Devol AH et al (2014). Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci USA 111: 12504–12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radax R, Rattei T, Lanzen A, Bayer C, Rapp HT, Urich T et al (2012). Metatranscriptomics of the marine sponge Geodia barretti: tackling phylogeny and function of its microbial community. Environ Microbiol 14: 1308–1324. [DOI] [PubMed] [Google Scholar]
- Ribes M, Jimenez E, Yahel G, Lopez-Sendino P, Diez B, Massana R et al (2012). Functional convergence of microbes associated with temperate marine sponges. Environ Microbiol 14: 1224–1239. [DOI] [PubMed] [Google Scholar]
- Ryu T, Seridi L, Moitinho-Silva L, Oates M, Liew YJ, Mavromatis C et al (2016). Hologenome analysis of two marine sponges with different microbiomes. BMC Genomics 17: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schläppy ML, Weber M, Mendola D, Hoffmann F, de Beer D (2010). Heterogeneous oxygenation resulting from active and passive flow in two Mediterranean sponges, Dysida avara and Chondrosia reniformis. Limnol Oceanogr 55: 1289–1300. [Google Scholar]
- Schmitt S, Angermeier H, Schiller R, Lindquist N, Hentschel U (2008). Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl Environ Microbiol 74: 7694–7708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Tsai P, Bell J, Fromont J, Ilan M, Lindquist N et al (2012). Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J 6: 564–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaibe E, Metzer E, Halpern YS (1985). Metabolic pathway for the utilization of L-arginine, L-ornithine, agmatine, and putrescine as nitrogen sources in Escherichia coli K-12. J Bacteriol 163: 933–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Kim J, Shinmen Y, Yamada H (1986). Evaluation of two alternative metabolic pathways for creatinine degradation in microorganisms. Arch Microbiol 145: 322–328. [Google Scholar]
- Siegl A, Kamke J, Hochmuth T, Piel J, Richter M, Liang C et al (2011). Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J 5: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon C, Collier J, Berg G, Glibert P (2010). Role of urea in microbial metabolism in aquatic systems: a biochemical and molecular review. Aquatic Microbial Ecol 59: 67–88. [Google Scholar]
- Sona S, Suzuki T, Ellington WR (2004). Cloning and expression of mitochondrial and protoflagellar creatine kinases from a marine sponge: implications for the origin of intracellular energy transport systems. Biochem Biophys Res Commun 317: 1207–1214. [DOI] [PubMed] [Google Scholar]
- Spang A, Poehlein A, Offre P, Zumbragel S, Haider S, Rychlik N et al (2012). The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol 14: 3122–3145. [DOI] [PubMed] [Google Scholar]
- Spieck E, Ehrich S, Aamand J, Bock E (1998). Isolation and immunocytochemical location of the nitrite-oxidizing system in nitrospira moscoviensis. Arch Microbiol 169: 225–230. [DOI] [PubMed] [Google Scholar]
- Spieck E, Bock E (2005). The lithoautotrophic nitrite-oxidizing bacteria In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, Boone DR, Vos PD et al (eds). Bergey’s Manual of Systematic Bacteriology. Springer Science+Business Media: New York, USA, pp 149–153. [Google Scholar]
- Starkenburg SR, Chain PS, Sayavedra-Soto LA, Hauser L, Land ML, Larimer FW et al (2006). Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl Environ Microbiol 72: 2050–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglmeier M, Alves RJE, Schleper C (2014. a) The Phylum Thaumarchaeota In:Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F(eds). The Prokaryotes: Other Major Lineages of Bacteria and the Archaea 4th edn.Springer-Verlag: Berlin, Heidelberg, Germany. [Google Scholar]
- Stieglmeier M, Klingl A, Alves RJ, Rittmann SK, Melcher M, Leisch N et al (2014. b). Nitrososphaera viennensis gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota. Int J Syst Evol Microbiol 64: 2738–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Jin L, Jiang Q, Sun W, Zhang F, Li Z (2013). Phylogenetically diverse ureC genes and their expression suggest the urea utilization by bacterial symbionts in marine sponge Xestospongia testudinaria. PLoS ONE 8: e64848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem LR, Elsen S, Bird TH, Swem DL, Koch HG, Myllykallio H et al (2001). The RegB/RegA two-component regulatory system controls synthesis of photosynthesis and respiratory electron transfer components in Rhodobacter capsulatus. J Mol Biol 309: 121–138. [DOI] [PubMed] [Google Scholar]
- Tate Jr DJ, Miceli MV, Newsome DA (1995). Phagocytosis and H2O2 induce catalase and metallothionein gene expression in human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci 36: 1271–1279. [PubMed] [Google Scholar]
- Taylor MW, Schupp PJ, Dahllof I, Kjelleberg S, Steinberg PD (2004). Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ Microbiol 6: 121–130. [DOI] [PubMed] [Google Scholar]
- Taylor MW (2005) The ecology of marine sponge-associated bacteria Ph.D. thesisThe University of New South Wales: Sydney, Australia. [Google Scholar]
- Taylor MW, Radax R, Steger D, Wagner M (2007). Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev 71: 295–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Rusch D, DeMaere MZ, Yung PY, Lewis M, Halpern A et al (2010). Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J 4: 1557–1567. [DOI] [PubMed] [Google Scholar]
- Thomas T, Moitinho-Silva L, Lurgi M, Bjork JR, Easson C, Astudillo-Garcia C et al (2016). Diversity, structure and convergent evolution of the global sponge microbiome. Nat Commun 7: 11870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel MA, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJ, Kartal B et al (2015). Complete nitrification by a single microorganism. Nature 528: 555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Lynch VJ (2012). Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci 131: 281–285. [DOI] [PubMed] [Google Scholar]
- Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ et al (2010). Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA 107: 8818–8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NS, Thomas T (2016). The sponge hologenome. MBio 7: e00135-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetz MS, Wheeler PA (2007). Release of dissolved organic matter by coastal diatoms. Limnol Oceanogr 52: 798–807. [Google Scholar]
- White D (2007), The Physiology and Biochemistry of Prokaryotes. Oxford University Press: New York, NY, USA.
- Willems A (2014) The family Phyllobacteriaceae. In:Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F(eds). The Prokaryotes: Alphaproteobacteria and Betaproteobacteria 4th edn.Springer: Berlin, Germany, pp 355–418. [Google Scholar]
- Wilson MC, Mori T, Ruckert C, Uria AR, Helf MJ, Takada K et al (2014). An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506: 58–62. [DOI] [PubMed] [Google Scholar]
- Wulff J (2012). Ecological interactions and the distribution, abundance, and diversity of sponges. Adv Mar Biol 61: 273–344. [DOI] [PubMed] [Google Scholar]
- Wyss M, Kaddurah-Daouk R (2000). Creatine and creatinine metabolism. Physiol Rev 80: 1107–1213. [DOI] [PubMed] [Google Scholar]
- Yahel G, Sharp JH, Marie D, Hase C, Genin A (2003). In situ feeding and element removal in the symbiont-bearing sponge Theonella swinhoei: bulk DOC is the major source for carbon. Limnol Oceanogr 48: 141–149. [Google Scholar]
- Zhalnina KV, Dias R, Leonard MT, Dorr de Quadros P, Camargo FA, Drew JC et al (2014). Genome sequence of Candidatus Nitrososphaera evergladensis from group I.1b enriched from Everglades soil reveals novel genomic features of the ammonia-oxidizing archaea. PLoS One 9: e101648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Wisedchaisri G, Gonen T (2013). Crystal structure of a nitrate/nitrite exchanger. Nature 497: 647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.