Abstract
The multicellular communities of microorganisms known as biofilms are of high significance in agricultural setting, yet it is largely unknown about the biofilm formed by nitrogen-fixing bacteria. Here we report the biofilm formation by Pseudomonas stutzeri A1501, a free-living rhizospheric bacterium, capable of fixing nitrogen under microaerobic and nitrogen-limiting conditions. P. stutzeri A1501 tended to form biofilm in minimal media, especially under nitrogen depletion condition. Under such growth condition, the biofilms formed at the air–liquid interface (termed as pellicles) and the colony biofilms on agar plates exhibited nitrogenase activity in air. The two kinds of biofilms both contained large ovoid shape ‘cells’ that were multiple living bacteria embedded in a sac of extracellular polymeric substances (EPSs). We proposed to name such large ‘cells’ as A1501 cyst. Our results suggest that the EPS, especially exopolysaccharides enabled the encased bacteria to fix nitrogen while grown under aerobic condition. The formation of A1501 cysts was reversible in response to the changes of carbon or nitrogen source status. A1501 cyst formation depended on nitrogen-limiting signaling and the presence of sufficient carbon sources, yet was independent of an active nitrogenase. The pellicles formed by Azospirillum brasilense, another free-living nitrogen-fixing rhizobacterium, which also exhibited nitrogenase activity and contained the large EPS-encapsuled A1501 cyst-like ‘cells’. Our data imply that free-living nitrogen-fixing bacteria could convert the easy-used carbon sources to exopolysaccharides in order to enable nitrogen fixation in a natural aerobic environment.
Introduction
Biofilms are surface-associated bacterial communities that are predominant in natural environments. Biofilms enhance microbial survival, enabling organisms to adapt to changing conditions collectively instead of as single cells (Costerton et al., 1995). Biofilms are highly structured with bacterial cells embedded in a matrix, which protects bacterial cells from environmental stress or antibacterial factors. The biofilm matrix is usually formed by extracellular polymeric substance (EPS). The composition of EPS for the matrix varies with the biofilms formed by different bacteria, whereas the main components are usually exopolysaccharides, extracellular DNA (eDNA) and proteins (Flemming and Wingender, 2010). Many studies are focusing on the biofilm formation of pathogens, such as Pseudomonas aeruginosa, Candida albicans and Vibrio cholera, etc. (Drenkard and Ausubel, 2002; Hogan and Kolter, 2002; Hall-Stoodley and Stoodley, 2005). In recent years, many reports have focused on the biofilms of beneficial bacteria, such as the plant growth-promoting bacteria Bacillus subtilis (Rosche et al., 2009), which has become a model organism for studying the mechanisms of biofilm formation in soil bacteria (Vlamakis et al., 2013; DeLoughery et al., 2015). However, it remains largely unknown about the biofilm formation by free-living nitrogen-fixing bacteria, an important contributor of natural bioavailable nitrogen on the earth.
Nitrogen-fixing bacteria have been classified into three major groups based upon their relations with plant, including free-living, symbiotic, and root associated (rhizospheric) bacteria (Dixon and Kahn, 2004). The rhizospheric nitrogen-fixing bacteria have drawn considerable attentions because of their wide ecological distribution, good adaptability and their potential to benefit agriculture, especially by increasing productivity of three economical crops, such as corn, wheat and rice (Okon et al., 1977; Revers et al., 2000). Pseudomonas stutzeri, which belongs to the Gamma subclass of Proteobacteria, is widely distributed in diverse ecological niches (Lalucat et al., 2006). The P. stutzeri A1501 strain (formerly identified as Alcalignes faecalis) was one of a few Pseudomonads that can fix nitrogen under microaerobic condition and colonize the rice rhizosphere (You et al., 1991; Vermeiren et al., 1999; Yan et al., 2013). Genome sequencing identified a 49-kb DNA region containing nitrogen fixation related genes on P. stutzeri A1501 chromosome, which was considered as a genomic island acquired from lateral gene transfer (Yan et al., 2008). Many nitrogen-fixing bacteria, such as genera of Rhizobium, Gluconacetobacter and Azospirillum, were found to be able to form biofilms (Rinaudi and Giordano, 2010; Herath et al., 2015). Exopolysaccharides were found to be important for nitrogen-fixing bacteria to colonize the plant root and to form biofilms (Hebbar et al., 1992; Burdman et al., 2000,Meneses et al., 2011). In particular, Azospirillum brasilense is well studied free-living nitrogen-fixing bacterium that can associate with many plants and fix nitrogen under microaerobic condition (Okon et al., 1976; Burdman et al., 2000; Baldani et al., 2014).
In nitrogen-fixing bacteria, the reduction of molecular nitrogen into ammonia is catalyzed by an enzymatic complex termed as nitrogenase, which is irreversibly inactivated by oxygen (Hartmann et al., 1986; Burris and Roberts, 1993). Many oxygen protection mechanisms have been found in the nitrogen-fixing bacteria including enhanced respiration (respiratory protection) as well as the production of extracellular polymers as a barrier to O2 diffusion, and increasing cell size (Inomura et al., 2017). Nitrogen fixation is a high energy-demanding process, the nitrogen fixation (nif) genes are essentially expressed under nitrogen limitation condition in most nitrogen fixers. The nitrogenase synthesis and activity in P. stutzeri A1501 is controlled both by oxygen and ammonia. The nitrogenase activity of A1501 is reversibly inhibited by ammonia at concentrations over 1 mM. Oxygen at >4% repressed the nitrogenase synthesis in A1501 and the enzyme is irreversibly inactivated in air (Desnoues et al., 2003). There have been a large number of studies attempting to improve the nitrogenase activity, including modifying the structures of nitrogenase to increase its tolerance to oxygen, altering the regulatory pathways or reconstructing nitrogen fixation systems, etc. (Wang et al., 2010; Wiig et al., 2012; Yang et al., 2014).
It is now widely accepted that microorganisms tend to live in biofilms in natural environment rather than as single planktonic cells (Flemming and Wingender, 2010). In this work, we have studied the biofilm formation by the rhizospheric bacteria known to fix nitrogen at the free-living state. We have explored the physiological conditions generating biofilm and how biofilm formation benefited the nitrogen-fixing bacteria. Strikingly, we have found that bacterial communities have a long life while grown under the nitrogen-limiting condition. Most importantly, our data imply how free-living nitrogen-fixing bacteria enable nitrogen fixation to occur in a natural aerobic environment, especially in air.
Materials and methods
Bacterial strain and media
P. stutzeri A1501 strain was typically grown in LD medium (Luria–Bertani medium with 2.5 g l−1 of NaCl instead of 10 g l−1) aerobically at 30 °C. For biofilm formation assay and pellicle grown experiments, P. stutzeri A1501 was grown in minimal medium A15 (KH2PO4 0.5 mM, K2HPO4 2.1 mM, NaCl 1.7 mM, MgSO4 1.6 mM, MnSO4 0.07 mM, Fe2(SO4)3 0.02 mM, Na2MoO4 0.04 mM, Lactate 50 mM, (NH4)2SO4 3 mM, pH 6.8) or lactate-containing K medium (KL medium was used in this work to indicate lactate as the carbon source), as previously described (Galimand et al., 1989), with 20 mM of ammonia chloride as sole nitrogen source (KLN+) or without nitrogen source (KLN−). A. brasilense Sp7 was grown in LD-rich medium for culturing and in KL minimal media (including KLN− and KLN+) for biofilm formation. All the following methods used on A. brasilense Sp7 in this study are the same as those on P. stutzeri A1501.
Biofilm assay and pellicle growth
Crystal violet (CV) assay of biofilm formation was referred to the previous method with modifications (Ma et al., 2006). Overnight culture of A1501 was washed with KLN− medium once and adjusted OD600 to 2.5 with KLN−. Ten microliter of washed culture was inoculated into 150 μl of corresponding media in 96-well PVC plates (Corning Co., New York, NY, USA). Then the plates were incubated under standing conditions in air at 30 °C for requested time (from 4 h to several days). The growth of planktonic bacteria in microtiter wells used for biofilm assay was determined by measuring the OD at A600 nm. For biofilm biomass quantification, the wells were washed with ddH2O for three times. One hundred sixty microliter of 0.1% CV was added into each well and incubated for 10 min. The wells were washed with ddH2O for multiple times until no purple color remained in water. The CV bound to biofilms was solubilized with 30% acetic acid and measured for absorbance at 540 nm using a spectrophotometer (Thermo Scientific, Waltham, MA, USA). For pellicles growth, 100 μl of A1501 overnight culture was inoculated into 1.5 ml of corresponding medium in 24-well plates (NEST Co., Wuxi, China) and incubated at 30 °C for desired time.
Confocal laser scanning microscopy (CLSM) and image acquisition
The air–liquid interface biofilms (pellicles) were grown in glass chambers (Chambered #1.5 German Coverglass System, Nunc, New York, NY, USA) with glass coverslip at the bottom. For CLSM observation, buffer was gently removed from glass chambers to allow the pellicles to drop onto coverslips. The biofilms were either stained by membrane stain FM4-64 (1 μM final concentration, Molecular Probes, Invitrogen, Carlsbad, CA, USA) or live-dead staining kit with SYTO9 and propidium iodide (PI) following the instruction of the manufacturer (Molecular Probes, Invitrogen). Fluorescent images were acquired by a FV1000 CLSM (Olympus, Tokyo, Japan). A Fluoview image browser (Olympus) generated the optical XYZ-sections. CLSM-captured images were subjected to quantitative image analysis using COMSTAT software as previously described (Heydorn et al., 2000).
Scanning electron microscopy (SEM) and transmission electron microscopy (TEM)
Samples for SEM and TEM were fixed with 2.5% glutaraldehyde in phosphate buffer for 1 h at room temperature or overnight at 4 °C. After a series of wash with ddH2O and gradient dehydration with different concentrations of ethanol (50%, 70%, 85%, 95%, 100%), the samples were processed for SEM or TEM examination. For SEM examination, the dehydrated samples were dried in a critical point drier (BAL-TEC CPD030, Bal-Tec, Los Angeles, CA, USA) with liquid carbon dioxide as a transitional fluid and then coated with gold in a sputter coater (BAL-TEC SCD005). The samples were examined with a FEI scanning electron microscope (ESEM FEI QUANTA 200-FEI, Hillsboro, OR, USA). For TEM examination, dehydrated samples were stained with osmic acid for 1 h and then embedded with 50% resin in ethanol at room temperature for 2 h, following with 100% resin embedding at 65 °C overnight. Ultrathin serial sections (70 nm thickness) were cut from resin blocks, followed by uranyl acetate staining, and observed with JEOL JEM-1400 transmission electron microscope (JEOL, Peabody, MA, USA).
EPS extraction and analysis
EPS extracted from P. stutzeri A1501 pellicles was performed as previously described with following modifications (Matthew et al., 2009). Three pieces of pellicles of each sample were collected into 1.5 ml centrifuge tube and washed twice with 0.8% saline. Samples were then resuspended in 100 μl of 0.5 M EDTA, boiled for 10 min at 100 °C. The supernatant from each tube was then sent for analysis of the total amount of protein and exopolysaccharides. The amount of extracted protein was determined using BCA protein assay kit (Pierce, Rockford, IL, USA) following the protocol provided by the manufacturer. Exopolysaccharides was quantitatively determined using phenol-sulfuric method (Dubois et al., 1951). The eDNA preparation from biofilms was performed as described (Rice et al., 2007) with modifications. EDTA extract from biofilms was extracted once with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1). The DNA in aqueous phase was precipitated by 3 vol of ice-cold 100% (vol/vol) ethanol and dissolved in water.
Nitrogenase activity assay
To prepare the culture for inoculation, P. stutzeri A1501 was grown in LD medium overnight and then washed with KLN− medium once and resuspended in KLN− medium. To examine the nitrogenase activity of pellicles or films grown in semisolid media (containing 0.15% agar), 100 μl of A1501 suspension culture was inoculated into 1.5 ml of KLN− liquid media or semisolid KLN− medium. To test the nitrogenase activity of colony biofilm on the KLN− agar (1% agar), 1.5 ml media was placed on the bottom of serum vial. Then 10 μl of 10-fold concentrated A1501 suspension culture was spotted onto the KLN− agar. After 5 days of growth in 30 °C incubator, the nitrogenase activity was assayed in situ using the acetylene reduction method as described previously (Galimand et al., 1989). Ten percentage (vol/vol) of acetylene was injected into serum vial through rubber stopper. The ethylene production was determined at regular intervals by gas chromatography after 4 h of reaction at 30 °C. Nitrogenase activity assay on A. brasilense Sp7 was taken using the same method as mentioned above on P. stutzeri A1501. Nitrogenase activity is expressed as nmol ethylene per hour per OD600 of inoculum. For the assay of nitrogenase inhibition, 20 mM ammonia chloride or glutamate were supplied into the pellicles after 5-day growth. The control samples were added with the same volume of ddH2O.
Results
P. stutzeri A1501 tends to form biofilm under nitrogen-limiting conditions
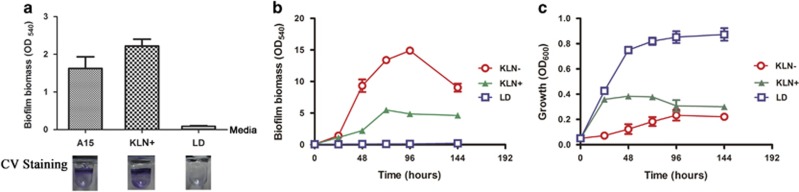
P. stutzeri A1501 biofilm formation was quantitated by microtiter dish biofilm assay using different culture media including LD-rich medium, minimal medium A15 or KL medium. A1501 can form biofilms when grown in minimal media, but not in LD-rich medium (Figures 1a and b). A1501 planktonic cells grew well in LD medium (Figure 1c), thus the absence of biofilm formation in LD medium was not due to bacterial cell growth. We then compared the extent of biofilm formation in KL medium in the absence of nitrogen source (KLN−) or with ammonia as the sole nitrogen source (KLN+). A1501 in KLN− media formed biofilms with highest biomass among the three media (KLN−, KLN+ and LD) during 6 days of growth (Figure 1b). After 2 days of growth, the biofilm biomass of A1501 in KLN− was four times higher than that in KLN+ medium (Figure 1b). We monitored the biofilm biomass and the corresponding OD600 of planktonic culture grown in the same wells over 6 days. A1501 showed the highest biofilm biomass, but the lowest value at OD600 in KLN− medium (Figures 1b and c), suggesting that A1501 tended to form biofilm rather than maintaining planktonic state under the condition lacking nitrogen sources (N− condition). On the contrary, A1501 grown in LD-rich medium stayed mainly in the planktonic state rather than forming biofilms (Figures 1b and c). These results indicated that nitrogen-fixing bacterium A1501 favored biofilm formation under nitrogen depletion condition.
Figure 1.
Biofilm formation of P. stutzeri A1501 in different media. (a) Biofilm biomass of A1501 was determined by CV biofilm assay after 2-day growth with A15, KLN or LD medium, respectively, in 96-well microtiter dish. Shown under each column was a representative well post CV staining. (b) Biofilm formation of P. stutzeri A1501 grown in minimal KLN−, KLN+ and rich LD media in the microtiter dishes during a week of growth. (c) The OD600 of bacteria grown in the corresponding wells for the biofilm assay shown in the graph b.
The pellicles of P. stutzeri A1501 formed at N− condition consist of enlarged ‘cells’ that were multiple bacteria embedded in a sac of EPS
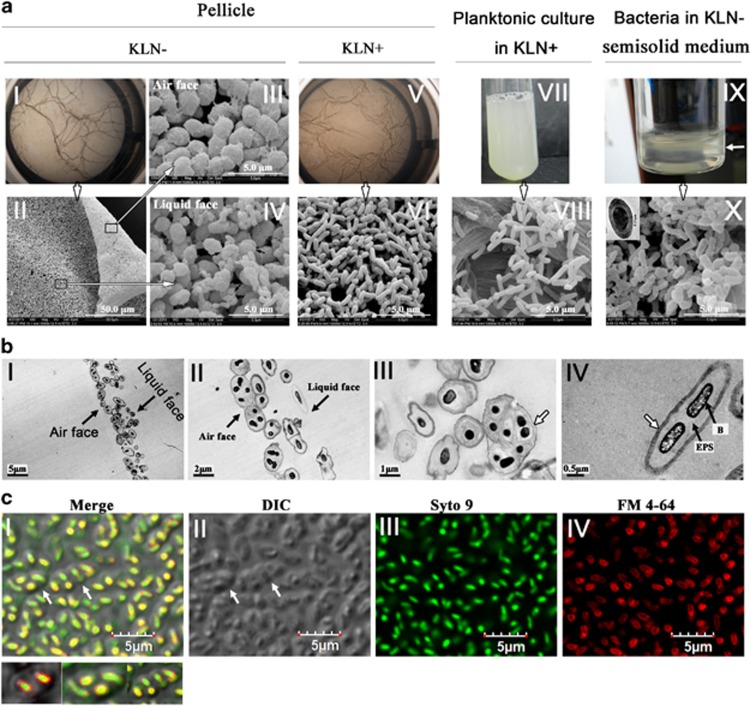
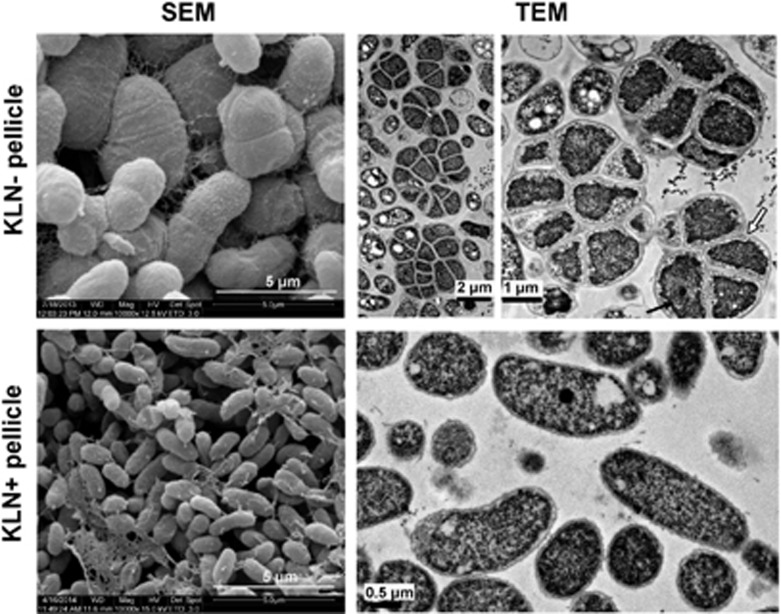
The biofilms formed at the air–liquid interface of standing cultures of soil bacteria are often named as pellicles (Branda et al., 2005, Vlamakis et al., 2013). We then examined the pellicle formation of A1501. A1501 could form pellicles in both KLN+ and KLN− media (Figure 2a). Although grown in KLN+, A1501 could form pellicles, yet the pellicles disassembled and returned to a planktonic existence after 7 days of growth. In contrast, the pellicles formed in KLN− were very stable and could remain as a whole piece of film for 4 weeks. We applied SEM to further analyze the pellicles of A1501. Bacterial morphology appeared dramatically different between pellicles formed at the N+ and N− conditions. Bacteria in pellicles formed in KLN+ medium had regular morphology and similar size as that of planktonic bacteria in a shaking culture in the same medium (Figure 2a, images V–VIII, Table 1). In contrast, most bacterial ‘cells’ in pellicles formed in KLN− medium (N- pellicles) were approximately 2 times longer and 3–4 times wider than A1501 planktonic bacteria (Figure 2a, images I–IV and VII–VIII, Table 1). In addition, the air faces of N− pellicles appeared flat and smooth under SEM and contained mostly large ‘cells’ (Figure 2a, images II–III, Table 1). The liquid faces of N− pellicles appeared rough with a mixture of large ‘cells’ and small cells (Figure 2a, images II and IV). The large ‘cells’ were similar to that found in the air face and the small cells had the similar size in diameter as planktonic bacteria of A1501, yet shorter in length (Figure 2a, images IV and VIII, Table 1), indicating that these small cells are A1501 cells. Some large ‘cells’ on the liquid face of pellicles appeared containing a few small cells, which seemed to be twisted together to form a large ‘cell’ (Figure 2a, indicated by the small black arrows in image IV). To get more detailed information on N− pellicles, we applied TEM and CLSM. TEM results showed that the large ‘cells’ in N− pellicles contained multiple small bacterial cells (two to eight bacteria), which were encased in a sac-like of EPS with light color under TEM (Figure 2b, images I–IV, the edge of EPS sac was indicated by the white arrows). The large EPS-encased ‘cells’ on the air face of pellicles were packed tightly together and most of which consisted of several bacteria (Figure 2b, images I–II). However, on the liquid face, the large ‘cells’ contained mostly a single bacterium (Figure 2b, images I–II). The data analyzed by CLSM further confirmed the results of TEM. To observe pellicles under CLSM, bacterial cell membrane was stained with red fluorescent FM4-64, whereas chromosome DNA was stained in green by SYTO9. The EPS sac containing bacterial cells can be morphologically distinguished by DIC (differential interference contrast microscopy) (Figure 2c image II, the white arrows indicated the edge of EPS sac). The merged images (merged from DIC, green and red fluorescence) showed that about 2–8 small cells could be enclosed in one EPS sac (Figure 2c, image I). Some EPS sacs appeared fused together (Figure 2b image III, indicated by white arrows). Interestingly, the large EPS-encased ‘cells’ can get stained by CV and can also be distinguished from planktonic bacteria by its large size under an optical microscope (Supplementary Figure S1). The large ‘cell’ of A1501 was reminiscent of the cyst in animal system (a sac-like structure filled with cells and liquid or semisolid substance), thus we propose to term it as A1501 cyst.
Figure 2.
Differences in morphology of pellicles and bacterial cell of P. stutzeri A1501 with respect to nitrogen source and oxygen availability. (a) The 2-day-old pellicles of P. stutzeri A1501 grown in KLN−, KLN+ media, and planktonic cells at late log phase in a shaking culture or bacterial film formed in semisolid KLN− medium post 2-day growth (the bacterial film was indicated by a white arrow), and their corresponding SEM images (I and V, the photographs of pellicles; II–IV, SEM images of a N− pellicle; VI, SEM images of a N+ pellicle, VII–VIII, the photo of planktonic culture and its SEM image; IX–X, the bacterial film in semisolid media and it SEM image). A TEM image of a cell is shown at the upper left corner of SEM image of bacterial film (image X) from the semisolid media. (b) TEM pictures of 2-day-old KLN− pellicles. The air face and liquid face of pellicles were indicated in image I and II. The white arrow in image III indicates a larger ‘cell’ that appears formed by fusing several EPS-encased ‘cells’ together. The white arrow in image IV indicates the edge of EPS sac. The black arrows in image IV indicate the EPS and a bacterial cell inside cyst (B, bacteria), respectively. (c) CLSM images of 2-day-old KLN− pellicle. Green fluorescence indicates the nucleic acid in bacteria cells stained by SYTO9 (image III). The bacterial cell membrane was stained in red by FM4-64 (image IV). The edge of each cyst can be identified from the DIC image (indicated by white arrows in images I and II). Three selected large ‘cells’ are shown under the merged image (I).
Table 1. Cell size and nitrogenase comparison of different forms of nitrogen-fixing bacteria grown under different conditions.
| Strain | Culture type and media | Relative nitrogenase activity (%)a | Morphology |
Size |
|
|---|---|---|---|---|---|
| Length (μm) | Diameter (μm) | ||||
| P. stutzeri A1501 | Pellicle in KLN− | 41±2 | LC | 2.53±0.29 | 1.47±0.11 |
| Bacteria inside LC | 1.05±0.07 | 0.43±0.04 | |||
| Free bacteria | 1.71±0.09 | 0.45±0.04 | |||
| biofilm in semisolid KLN− | 100±10 | Bacterial cell | 1.76±0.21 | 0.65±0.08 | |
| Colony on KLN− agar plate | 19±4 | LC | 2.92±0.18 | 1.73±0.12 | |
| Free bacteria | 1.16±0.12 | 0.42±0.07 | |||
| Pellicle in KLN+ | ND | Free bacteria | 1.52±0.19 | 0.44±0.06 | |
| A. brasilense Sp7 | Pellicle in KLN− | 68±9 | LC | 4.59±1.17 | 3.54±0.29 |
| Free bacteria | 2.88±0.49 | 0.99±0.13 | |||
| Bacteria inside LC | 1.57±0.27 | 0.89±0.2 | |||
| Pellicle in KLN+ | ND | Free bacteria | 2.07±0.27 | 0.9±0.04 | |
Abbreviations: ND, the nitrogenase activity was not detectable; LC, large 'cell'.
Relative nitrogenase activity of each sample in the acetylene reduction assay was normalized to that of A1501 semisolid sample (56 nmol h–1 per OD600). The data are reported as average±s.d. of at least three biological replicates.
The EPS sac enables the encased bacteria to fix nitrogen under aerobic growth condition
Under the condition of nitrogen depletion, bacteria have to fix nitrogen to gain nitrogen source for growth. However, A1501 only fixes nitrogen under microaerobic conditions because its nitrogenase is rapidly inactivated after a short-term exposure to oxygen (Desnoues et al., 2003). Thus, A1501 planktonic cells grown in shaking cultures under aerobic conditions did not exhibit nitrogenase activity in KLN− medium because of oxygen inactivation of the nitrogenase (Table 1). However, N− pellicles grown on the air-medium interface showed remarkable nitrogenase activity (41% of the nitrogenase activity of bacteria grown in the semisolid agar) (Table 1), suggesting their ability of fixing nitrogen under aerobic conditions. The A1501 cysts (the EPS-encased bacterial aggregate) were only found in N− pellicles and the fact that N−pellicles expressed nitrogenase activity under aerobic conditions suggested that the EPS sac may provide a suitable microaerobic environment compatible with nitrogenase synthesis and activity similar to the situation encountered in semisolid agar.
The semisolid agar is known to provide the suitable microaerobic environment for nitrogen-fixing rhizobacteria. When A1501 grew in semisolid KLN− media, it formed a thin layer of bacterial film under the surface of medium where had reduced oxygen concentration compatible with nitrogenase functioning (Figure 2a, image IX, the film was indicated by a white arrow). Thus, we examined the film formed by A1501 in the semisolid KLN− medium by SEM. Interestingly, bacteria in the film appeared also slightly wider and longer than planktonic bacteria in a shaking culture although not as large as that in N− pellicles (Figure 2a, image X, Table 1). TEM examination indicated that these bacteria were encased in a 0.1 μm thick layer of EPS (Figure 2a, as indicated in the inset in the corresponding SEM image X). This result showed that A1501 grown at a reduced oxygen condition was also protected by a layer of EPS.
The occurrence of A1501 cysts and the nitrogenase activity detected with the pellicles led us to examine what happened with colony biofilms formed on agar plates. We spotted 5 μl of A1501 culture on a KLN− agar plate and examined the colonies daily for a total of 10 days. A notable increase in the colonies diameter was observed between days 3 and 10 (Figure 3a). SEM examination of a 3-day-old colony showed that there were A1501 cyst-like large ‘cells’ arranged as clusters in the central area of colonies that could be stained with Congo red (Figures 3b and c, the short black arrows indicated a cluster of cysts and the white arrows indicated the bacterial cell), yet few cysts were found in the outer ring of colonies, and almost none on the edges. Most bacteria in the outer ring and edges of colonies had similar size and morphology as planktonic bacteria in a shaking culture (Figure 3b). Most strikingly, the colonies on KLN− agar plate displayed a significant nitrogenase activity (approximately 19% of activity of cultures grown in semisolid agar) (Table 1).
Figure 3.
Colony biofilms and cell morphology of P. stutzeri A1501 grown on KLN− plates. (a) A1501 colony grown on KLN− plate for 3 days and 10 days. (b) SEM of a 3-day colony on KLN− plate and the morphology of bacterial cell in the center, the outer ring and the edge of the colony. (c) The optical microscopy image of large EPS-encased ‘cell’ clusters from A1501 colony grown on a KLN− congo red plate. Two large ‘cell’ clusters are indicated by black arrows. The white arrow indicates the morphology of bacterial cell.
The nitrogenase activity was detected in the pellicles and colonies containing A1501 cysts, suggesting the small bacterial cells inside EPS sac should be alive. To confirm this, we applied live/dead staining to N− pellicles. Green fluorescent dye SYTO9 was utilized to stain bacterial chromosome DNA in living cells, whereas red fluorescent dye, PI was used to indicate dead or dying cells because it only stains eDNA and chromosome DNA of bacteria with compromised membranes. Live/dead staining result showed that 99.8% bacteria in KLN− pellicles were stained in green fluorescence, indicating they were alive (Figure 4a).
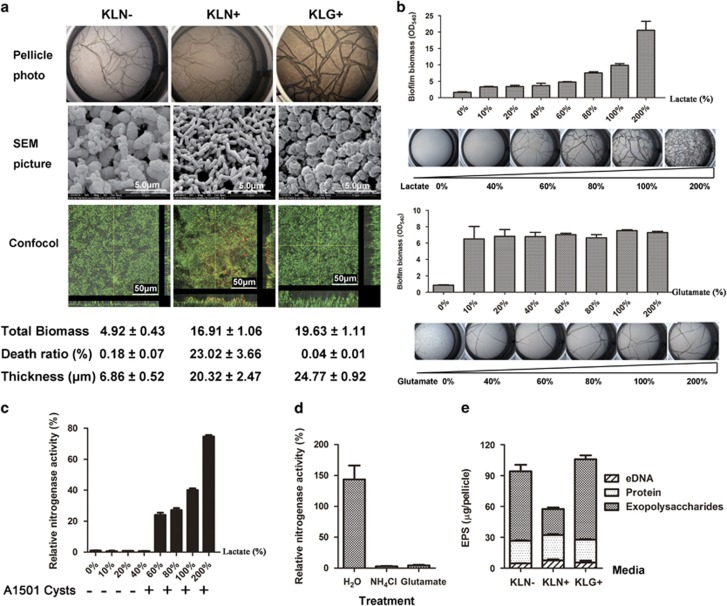
Figure 4.
Factors affecting biofilm and pellicle formation of P. stutzeri A1501. (a) Morphology and live/dead staining of A1501 2-day-old pellicles formed in KLN−, KLN+ and KLG+ media. Live bacterial cells were stained by SYTO9 in green and dead cells were stained in red by PI. The corresponding biofilm biomass, thickness and death cell ratio are shown under each image. The corresponding photos of pellicles and SEM images are shown above. (b) Effect of lactate and glutamate concentrations on the biofilm biomass. The concentration of lactate (45 mM) and glutamate (20 mM) in regular KLG+ was normalized as 100%. The biofilm biomass was examined by CV assay after 2 days of growth. (c) The impact of lactate level in KLN− media on the nitrogenase activity and the ability of forming A1501 cysts (indicated as ‘+’ or ‘−’) of A1501 pellicles. Relative nitrogenase activity of each sample was normalized to that of A1501 semisolid sample. (d) Nitrogenase activity of A1501 KLN− pellicles was repressed after treatment with 20 mM of glutamate or ammonia. Control is the sample that treated with ddH2O instead of glutamate or ammonia. The nitrogenase activity was normalized to the level of control sample. (e) EPS composition of A1501 pellicles grown in the KLN−, KLN+ and KLG+ media.
Taken together, our results showed that A1501 could fix nitrogen under three aerobic growth conditions, within pellicles on air–liquid interface of standing culture, in the film of semisolid media and in the colonies of a regular agar plate while the bacteria were encased by EPS. These results suggested that the EPS sac covering bacteria was critical for nitrogen fixation occurring under aerobic conditions.
Factors affecting the biofilm formation and differentiation of A1501 cysts
We first examined the effects of nitrogen sources and carbon sources on the formation of a biofilm and A1501 cyst. Glutamate is known as a poor nitrogen source comparable to a nitrogen-limiting signal although it supports bacterial growth (Huergo and Dixon, 2015). Thus, we tested the effects of glutamate on the A1501 biofilms by replacing ammonia by 20 mM of glutamate in the KL media (KLG+). A1501 formed pellicles in KLG+ that had the highest thickness and biomass (4-fold more than N− pellicles and 1.15-fold of N+ pellicles) and the least dead bacteria compared with pellicles in KLN− and KLN+ (Figure 4a). The morphology of bacteria in KLG+ pellicles was also similar to KLN− pellicles (Figure 4a and Supplementary Figure S2A). The air faces of KLG+ pellicles appeared smooth and contained mainly the A1501 cysts, whereas the liquid face of KLG+ pellicles appeared rough and contained mainly free bacteria and a large amount of EPS (Supplementary Figure S2A, EPS was indicated by a black arrow). The A1501 cysts were also found in the colonies grown on KLG+ agar plate as that on KLN− (Supplementary Figure S2B). Besides glutamate, histidine also promoted biofilm formation and the differentiation of the A1501 cysts, but glutamine or NaNO3 cannot (Supplementary Figures S2C and D, and data not shown). Increasing the concentration of lactate in KLG+ promoted biofilm formation, yet changes of glutamate concentration in KLG+ did not affect the biofilm formation of A1501 (Figure 4b). Glutamate at 2 mM could promote a biofilm with similar biomass as that at 20 mM (Figure 4b). However, the pellicles on KLN− neither showed nitrogenase activity, nor contained the A1501 cysts when the lactate concentration was lower than 40% of the original level (45 mM as 100%) (Figure 4c). A similar phenomenon was also observed when lactate was replaced by glucose (data not shown). These results suggested that sufficient carbon sources and limiting nitrogen sources were critical for the formation of A1501 biofilm and cyst.
The glutamate as a nitrogen source was reported not to repress the nitrogenase activity in some nitrogen-fixing bacteria (Wang et al., 2010). However, we found that the glutamate supplement was able to inhibit the nitrogenase activity of A1501 similarly as ammonia (Figure 4d). In consistence, the pellicles formed on KLG+ did not exhibit nitrogenase activity (data not shown). As shown above, the glutamate as a nitrogen source did promote the formation of biofilms and A1501 cysts. Thus, these results suggest that an active nitrogenase is not required for the formation of biofilms and A1501 cysts.
Finally, we tested whether the oxygen concentration can induce the A1501 cyst formation. We examined the A1501 KLN− pellicles grown at different level of oxygen concentration, 0.5%, 1% and 5%. The A1501 cysts were found in the pellicles when the oxygen level reached 1% or higher, whereas none were found at 0.5% oxygen despite the fact that the nitrogenase activity of pellicles were similar under these three conditions (Supplementary Figure S1B). These results suggested that the oxygen stress may also be an essential factor to induce the A1501 cyst formation.
Exopolysaccharides were the main component of EPS that encased bacteria in A1501 cysts
To identify composition of EPS involved in pellicle formation, we respectively isolated EPS from the pellicles of A1501 grown on KLN−, KLN+ and KLG+ media and compared the amount of total polysaccharides, proteins and eDNA among the EPS samples. The EPS of KLN− pellicles contained 72% of exopolysaccharides and 23% of proteins. In contrast, the EPS of KLN+ pellicle contained only 44% of exopolysaccharides and 43% of proteins (Figure 4e). There were more eDNA in KLN+ pellicles compared with KLN− pellicles, which is consistent with the live-dead staining results from CLSM (Figure 4a). The ratio of dead bacteria was about 120-fold higher in KLN+ pellicles than that in KLN− pellicles (Figure 4a), although the total biomass of the KLN+ pellicle was fourfold higher than that of the KLN− pellicle (Figure 4a). Similar to KLN− pellicles, EPS from KLG+ pellicles contained 74% of exopolysaccharides and similar proportions of extracellular proteins, and eDNA (Figure 4e). The formation of A1501 cyst was promoted by either KLN− or KLG+ medium as shown above (Figures 2–4 and supplementary Figure S2) and the pellicles grown on these two media both contain over 70% of exopolysaccharides, suggesting that exopolysaccharides were the main component of EPS that encased bacteria in the A1501 cyst.
A1501 pellicles grown on KLN− and KLG+ maintained to be a whole piece and had mostly living bacteria (Figure 4a), suggesting that exopolysaccharides may promote the adherence on solid surfaces and the connection between bacteria in addition to block oxygen in air. In addition, the growth conditions that enabled the A1501 cyst formation also enhanced biofilm formation by A1501 in the microtiter dish assay (Figures 1–4,Supplementary Figure S2). Taken together, our data indicated that exopolysaccharide was the main component of the EPS in N− pellicles, which may have major role in connecting bacteria together and providing suitable oxygen conditions for the nitrogen-fixing bacteria.
The formation of A1501 cyst is reversible in response to the changes in the carbon or nitrogen source status
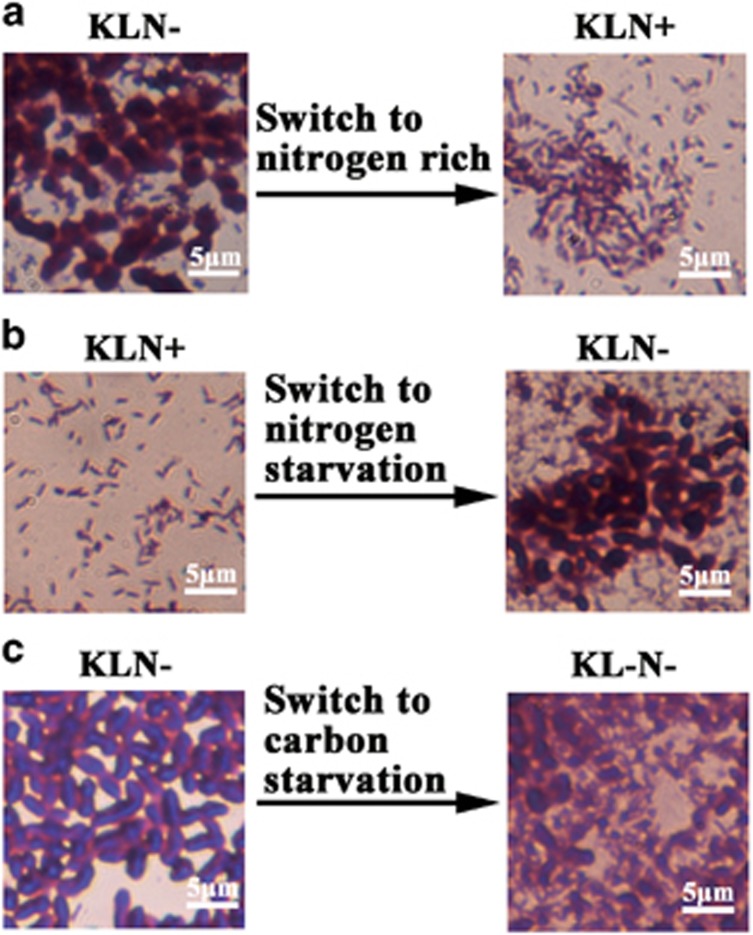
As shown above, nitrogen starvation and a sufficient amount of carbon source induced the formation of A1501 cyst. To investigate whether the supplied nitrogen source or carbon starvation can change or disassemble the A1501 cysts, the pellicles of A1501 were stained with CV and observed under optical microscope. When adding 20 mM of ammonia chloride to a pellicle formed on KLN− media, free bacterial cells were released from the A1501 cysts and the pellicle was disassembled to planktonic bacteria in 2 days (Figure 5a), indicating that the ammonia supplement would cause the release of bacteria from the A1501 cysts. On the other hand, the A1501 cysts could form while a KLN+ pellicle was washed off the ammonia nutrient and grown in KLN− medium for 7 days, suggesting that the A1501 cyst formation was reversible (Figure 5b). The carbon starvation also caused the disassembling of N− pellicles and the bacteria released from EPS sac, suggesting the exopolysaccharide in pellicles could serve as a carbon source for bacteria (Figure 5c). These results indicated that the formation of A1501 cysts was a dynamic process in response to the nutrient status. These data also suggested another important role of exopolysaccharide in A1501 cyst as a way of storing excess carbon when bacteria were growing under a condition with sufficient carbon sources, implying that forming the A1501 cysts may also contribute to carbon sequestration by converting the easy-used carbon sources to the slow-degraded exopolysaccharides, which may open a new avenue for biological nitrogen-fertilizer development on slowing the global warming.
Figure 5.
Changes in A1501 cyst morphology in P. stutzeri A1501 pellicles in response to the variation of carbon and nitrogen status in the media. The pellicles were stained with CV and imaged by optical microscopy. (a) Adding 20 mM of ammonia to the 2-day-old KLN− pellicle medium caused the disassembling of the A1501 cysts. (b) Removing nitrogen from KLN+ medium could induce the A1501 cyst formation. (c) Removing the carbon source from the KLN− medium to provide a carbon starvation signal (KL−N−) caused the disassembling of the A1501 cyst in a pellicle.
The large ‘cells’(EPS-encased bacterial aggregate) were also found in pellicles formed by A. brasilense, a typical nitrogen-fixing rhizobacterium
In order to know whether other rhizosphere nitrogen-fixing bacteria, such as A. brasilense can form biofilm and the A1501 cyst-like structure in nitrogen-limiting media, we grew A. brasilense Sp7 in standing KLN− or KLN+ medium. Sp7 could form pellicles on these media. Similar to A1501, Sp7 pellicles grown on the KLN− media had mostly the large-size ‘cell’ on its air face (Figure 6, upper left panel, Table 1). TEM examination confirmed that the large-size ‘cells’ were a sac of EPS-encased bacterial cells, the number of bacteria in the EPS Sac varied from 1 to 12 (Figure 6, upper middle and right panels, a large ‘cell’ was indicated by a white arrow). The large-size ‘cells’ of Sp7 found in N− pellicles were morphologically similar to the cyst previously described in Azospirillum species (Sadasivan and Neyra, 1985; Sadasivan and Neyra, 1987; Bleakley et al., 1988). Sp7 pellicles grown on KLN+ media had regular size bacteria similar to that in a shaking culture (Figure 6, lower panel, Table 1). Most importantly, N− pellicles of Sp7 formed on air-media interface also displayed nitrogenase activity (Table 1), an observation not previously reported. In summary, our data suggest that bacterial capsulation or forming A1501 cyst may be a common strategy in these two nitrogen-fixing bacteria to enable nitrogen fixation under the air.
Figure 6.
SEM and TEM images of Azospirillum brasilense Sp7 pellicles formed in KLN− and KLN+ media. The A1501 cyst-like large ‘cells’ (EPS-encased bacterial aggregate) were found in KLN− pellicles, but not from KLN+ pellicles. Black arrow in TEM picture of KLN− pellicle indicates the bacteria cell inside the EPS sac, and white arrow indicates the edge of EPS sac. Two TEM images of A1501 cyst-like structures from Sp7 KLN− pellicle were shown in the upper panel.
Discussion
Biofilm formation is established as a general strategy that bacteria have developed to colonize specific niches, including the plant rhizosphere (Ramey et al., 2004). In this work, we investigated the physiological conditions for the formation of biofilms by the free-living nitrogen-fixing rhizobacteria, P. stutzeri A1501 and A. brasilense. Analysis of physiological conditions enabling P. stutzeri A1501 to form and disassemble biofilms revealed that the most stable and long surviving biofilms were formed when sufficient carbon sources were available and the nitrogen source was limiting (N−) or poorly assimilated (such as glutamate or histidine), showing that environment stresses often induce bacteria biofilm formation as reported previously in other bacteria (Berleman and Bauer, 2004; Huerta et al., 2016). Most interestingly, we found that multiple bacterial cells were embedded in a sac of EPS, which appeared like a large ovoid ‘cell’ under SEM and optical microscope. Such ovoid large ‘cells’ of A1501 were reminiscent of what was designed as cysts in animal systems (Zadik et al., 2011), thus we propose to term it as A1501 cyst. A1501 forms cysts on pellicles at the air–liquid interface and colony biofilms on agar plate under the aerobic condition with sufficient carbon source and limiting nitrogen source. The pellicles and colony biofilms both show nitrogenase activity, suggesting that the formation of cysts likely enables bacterial nitrogen fixation to occur while grown under the aerobic condition. A large amount of exopolysaccharides was found in the EPS of A1501 cysts, which may function as a barrier to block excess oxygen in air and provide a suitable microaerobic condition for bacteria inside the EPS to fix nitrogen. The pellicles formed by A. brasilense Sp7 also contain cyst-like EPS-encased bacterial aggregate and exhibit remarkable nitrogenase activity. These imply that the aerobic nitrogen-fixing bacteria can fix nitrogen at the free-living state in the natural aerobic environment under certain physiological conditions (for example, within a biofilm).
Capsulation and aggregation observed during flocculation were thought to be a mechanism aiding bacteria against unfavorable oxygen conditions (Berg et al., 1980; Bible et al., 2015). Nitrogen fixation is generally considered an energy costing process. However, a recent report of Inomura et al. (2017) on nitrogen-fixing bacteria Azotobacter vinelandii has revealed that the direct energetic cost of nitrogen fixation is small relative to the cost of managing intracellular oxygen. By quantifying the costs and benefits of several potential oxygen protection mechanisms present in nature, they have shown that the production of EPS as a barrier to O2 diffusion leads to higher growth efficiencies than respiratory protection. This is consistent with our finding that exopolysaccharide was the main component of EPS sac that encased bacteria in the A1501 cysts. Our data are also in favor that N-limiting signals very likely induce the production of exopolysaccharide. This exopolysaccharide may also enhance bacterial adherence to surfaces and connection between bacteria because A1501 formed the best biofilms on microtiter dishes at N-limiting growth conditions and the cysts were tightly connected together in N− pellicles. Cellulolytic bacteria were also reported to produce large amounts of exopolysaccharide in response to N limitation (Young et al., 2012). Therefore, the production of exopolysaccharide could be a general response of bacteria to N-limiting growth condition.
It is critical for a bacterium to maintain the sustainable growth of its community in the natural environment. The colony biofilms in some degree could mimic the biofilms of soil bacteria formed on the surface of soil or plant roots. We have shown that the colony biofilms of A1501 grown on the KLN− plates can fix nitrogen in air. In such colonies, the A1501 cysts were found in the central region of colonies and bacteria on the edge of colonies appeared to be similar to bacteria in planktonic culture. The cysts in the center of colonies through their nitrogen-fixing activity may provide a nitrogen source for bacteria on the edge of the colonies to grow as we proposed that only cells within cysts were able to fix nitrogen under the air. In A1501 N− pellicles, the air surface was built with big cysts, which were tightly attached to each other, like a shield and may have a major role in blocking oxygen from air. This allows the fast growth of bacteria in liquid face, which is indicated by the free bacteria released from the EPS sac and frequently observed dividing bacteria inside the EPS sac (Supplementary Figure S3). The multiple bacteria embedded in a sac of EPS may be a way for bacteria to efficiently use nutrients and space to get maximum growth, meanwhile, the growth of several bacteria can use some oxygen that may help to make a suitable environment for nitrogenase. Taken together, our data imply that bacteria could share nutrients and help each other to ensure the best growth of their communities in the natural aerobic environment.
Bacterial cyst was described long ago in Azotobacter, a genus phylogenetically close to Pseudomonas spp. (Rediers et al., 2004). The cysts were generally termed as the dormant prokaryotic forms that are usually embedded by EPS, absence of division, with long-term maintenance of viability and heat resistance, and devoid of metabolic activity (Sadoff, 1975). A bacterial cyst usually contains only a bacterial cell. By extension, ovoid shaped cells reported for the other free-living nitrogen-fixing species such Azopsirillum spp. (Sadasivan and Neyra, 1987; Bleakley et al., 1988) and Rhodopsirillum centenum (Berleman and Bauer, 2004) were also considered as cysts like. The N limitation usually induces the cyst formation in A. brasilense. Exopolysaccharide is also the main component of EPS encapsulated the cyst of A. brasilense (Pope and Wyss, 1970; Berg et al., 1980; Sadasivan and Neyra, 1985). The A1501 cysts described in this study also have slower growth rate than the bacteria grown under the conditions with ammonia. However, the cell division was found in the A1501 cysts and 1 to several bacteria could be encased in a cyst. The most important character of the A1501 cysts is their nitrogen-fixing ability. We have shown that A. brasilense Sp7 can form the A1501 cyst-like structure that is capable of fixing nitrogen in air, looks like a large ovoid ‘cell’ under SEM, yet are EPS-encased bacterial aggregate. Such ovoid shape ‘cells’ of A. brasilense is also morphologically similar to the previously described Azospirillum cysts. In addition, several reports have shown that cyst forms of Azospirillum species are not dormant. Berg et al. (1980) reported that encapsulated forms of A. brasilense in sugarcane callus, were not dormant. Colonization of root surfaces by Azospirillum is accompanied by morphological change with the formation of non-motile ovoid cells metabolically active (Assmus et al., 1995; Pereg-Gerk et al., 1998), similar to those observed during the cell aggregation and flocculation process leading to cyst formation (Sadasivan and Neyra, 1985). Thus, in Azospirillum, the differentiation of ovoid sessile cells surrounded by capsular material enabling anchoring to and colonization of the root surface could be compatible with nitrogenase activity, in particular under nitrogen-limiting condition. Although further investigates are certainly required to clarify whether A1501-like structure can be form in other free-living nitrogen-fixing bacteria, such as Azotobacter vinelandii. We would like to propose to revise the definition of a cyst formed by nitrogen-fixing bacteria as EPS-encased a bacterial cell or bacterial aggregate that often appear like enlarged ovoid cell shape, a form that provides protection for bacteria from oxygen, desiccation, and enables bacteria to fix nitrogen under the aerobic natural environment.
In summary, we have shown in this study that the formation of A1501 cysts was a dynamic process in response to the nutrient status, especially the concentration of carbon and nitrogen sources. Our data imply that forming cyst-like cells can be a general strategy for soil bacteria to survive under N-limiting and aerobic conditions, and may be also a common way for free-living nitrogen fixing bacteria to protect their nitrogenase from oxygen in air. Furthermore, forming the EPS-encased cysts may not only ensure the nitrogen fixation to occur in natural environment, but also contribute to carbon sequestration by converting the easy-used carbon sources to exopolysaccharides, which usually were not easily degraded. Our data also open new prospects for biological nitrogen-fertilizer development.
Acknowledgments
We thank Dr Min Lin from the Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, for providing P. stutzeri strain. Mr Chunli Li and Ms Jingnan Liang from the Institute of Microbiology, Chinese Academy of Sciences for technical support on SEM and TEM, respectively. This work was supported by the National Basic Research Program of China (973 Program 2014CB846002 to LM and 2015CB150600), and the National Natural Science Foundation of China (31570126 to LM and 31200044 to DW). This work was conducted with the support of Institut Pasteur, Paris, France.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Assmus B, Hutzler P, Kirchhof G, Amann R, Lawrence JR, Hartmann A. (1995). In situ localization of Azospirillum brasilense in the rhizosphere of wheat with fluorescently labeled, rRNA-targeted oligonucleotide probes and scanning confocal laser microscopy. Appl Environ Microbiol 61: 1013–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldani JI, Reis VM, Videira SS, Boddey LH, Baldani VL. (2014). The art of isolating nitrogen-fixing bacteria from non-leguminous plants using N-free semi-solid media: a practical guide for microbiologists. Plant Soil 384: 19. [Google Scholar]
- Berg RH, Tyler ME, Novick NJ, Vasil V, Vasil IK. (1980). Biology of azospirillum-sugarcane association: enhancement of nitrogenase activity. Appl Environ Microbiol 39: 642–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleman JE, Bauer CE. (2004). Characterization of cyst cell formation in the purple photosynthetic bacterium Rhodospirillum centenum. Microbiology 150: 383–390. [DOI] [PubMed] [Google Scholar]
- Bible AN, Khalsa-Moyers GK, Mukherjee T, Green CS, Mishra P, Purcell A et al. (2015). Metabolic adaptations of Azospirillum brasilense to oxygen stress by cell-to-cell clumping and flocculation. Appl Environ Microbiol 81: 8346–8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleakley BH, Gaskins MH, Hubbell DH, Zam SG. (1988). Floc formation by Azospirillum lipoferum grown on poly-β-hydroxybutyrate. Appl Environ Microbiol 54: 2986–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Vik S, Friedman L, Kolter R. (2005). Biofilms: the matrix revisited. Trends Microbiol 13: 20–26. [DOI] [PubMed] [Google Scholar]
- Burdman S, Okon Y, Jurkevitch E. (2000). Surface characteristics of Azospirillum brasilense in relation to cell aggregation and attachment to plant roots. Critical Rev Microbiol 26: 91–110. [DOI] [PubMed] [Google Scholar]
- Burris RH, Roberts GP. (1993). Biological nitrogen fixation. Annu Rev Nutr 13: 317–335. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. (1995). Microbial biofilms. Annu Rev Microbiol 49: 711–745. [DOI] [PubMed] [Google Scholar]
- DeLoughery A, Dengler V, Chai Y, Losick R. (2015). Biofilm formation by Bacillus subtilis requires an endoribonuclease-containing multisubunit complex that controls mRNA levels for the matrix gene repressor SinR. Mol Microbiol. 99: 425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoues N, Lin M, Guo X, Ma L, Carreno-Lopez R, Elmerich C. (2003). Nitrogen fixation genetics and regulation in a Pseudomonas stutzeri strain associated with rice. Microbiology 149: 2251–2262. [DOI] [PubMed] [Google Scholar]
- Dixon R, Kahn D. (2004). Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol 2: 621–631. [DOI] [PubMed] [Google Scholar]
- Drenkard E, Ausubel FM. (2002). Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416: 740–743. [DOI] [PubMed] [Google Scholar]
- Dubois M, Gilles K, Hamilton JK, Rebers PA, Smith F. (1951). A colorimetric method for the determination of sugars. Nature 168: 167. [DOI] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. (2010). The biofilm matrix. Nat Rev Microbiol 8: 623–633. [DOI] [PubMed] [Google Scholar]
- Galimand M, Perroud B, Delorme F, Paquelin A, Vieille C, Bozouklian H et al. (1989). Identification of DNA regions homologous to nitrogen fixation genes nifE, nifUS and fixABC in Azospirillum brasilense Sp7. J Gen Microbiol 135: 1047–1059. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Stoodley P. (2005). Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol 13: 7–10. [DOI] [PubMed] [Google Scholar]
- Hartmann A, Fu H, Burris RH. (1986). Regulation of nitrogenase activity by ammonium chloride in Azospirillum spp. J Bacteriol 165: 864–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbar KP, Gueniot B, Heyraud A, Colinmorel P, Heulin T, Balandreau J et al. (1992). Characterization of exopolysaccharides produced by rhizobacteria. Appl Microbiol Biot 38: 248–253. [Google Scholar]
- Herath H, Menikdiwela K, Igalavithana A, Seneviratne G. (2015). Developed fungal-bacterial biofilms having nitrogen fixers: universal biofertilizers for legumes and non-legumesde Bruijn FJ(ed). Biol Nitrogen Fixation. Wiley-Blackwell Press: Hoboken, NJ, USA, pp 1041–1046.
- Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK et al. (2000). Quantification of biofilm structures by the novel computer program comstat. Microbiology 146: 2395–2407. [DOI] [PubMed] [Google Scholar]
- Hogan DA, Kolter R. (2002). Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296: 2229–2232. [DOI] [PubMed] [Google Scholar]
- Huergo LF, Dixon R. (2015). The emergence of 2-oxoglutarate as a master regulator metabolite. Microbiol Mol Biol Rev 79: 419–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta JM, Aguilar I, Lopez-Pliego L, Fuentes-Ramirez LE, Castaneda M. (2016). The role of the ncRNA RgsA in the oxidative stress response and biofilm formation in Azotobacter vinelandii. Curr Microbiol 72: 671–679. [DOI] [PubMed] [Google Scholar]
- Inomura K, Bragg J, Follows MJ. (2017). A quantitative analysis of the direct and indirect costs of nitrogen fixation: a model based on Azotobacter vinelandii. ISME J 11: 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalucat J, Bennasar A, Bosch R, Garcia-Valdes E, Palleroni NJ. (2006). Biology of Pseudomonas stutzeri. Microbiol Mol Biol Rev 70: 510–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. (2006). Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol 188: 8213–8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew SB, Irina S, Evgueny V, Haiping L, April BS, Stephen HR et al. (2009). Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol 73: 622–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneses CH, Rouws LF, Simões-Araújo JL, Vidal MS, Baldani JI. (2011). Exopolysaccharide production is required for biofilm formation and plant colonization by the nitrogen-fixing endophyte Gluconacetobacter diazotrophicus. Mol Plant-Microbe Interact 24: 1448–1458. [DOI] [PubMed] [Google Scholar]
- Okon Y, Albrecht SL, Burris RH. (1976). Factors affecting growth and nitrogen fixation of Spirillum lipoferum. J Bacteriol 127: 1248–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okon Y, Houchins JP, Albrecht SL, Burris RH. (1977). Growth of Spirillum lipoferum at constant partial pressures of oxygen, and the properties of its nitrogenase in cell-free extracts. J Gen Microbiol 98: 87–93. [DOI] [PubMed] [Google Scholar]
- Pereg-Gerk L, Paquelin A, Gounon P, Kennedy IR, Elmerich C. (1998). A transcriptional regulator of the LuxR-UhpA family, FlcA, controls flocculation and wheat root surface colonization by Azospirillum brasilense Sp7. Mol Plant-Microbe Interact 11: 177–187. [DOI] [PubMed] [Google Scholar]
- Pope LM, Wyss O. (1970). Outer layers of the Azotobacter vinelandii cyst. J Bacteriol 102: 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey BE, Koutsoudis M, von Bodman SB, Fuqua C. (2004). Biofilm formation in plant–microbe associations. Curr Opin Microbiol 7: 602–609. [DOI] [PubMed] [Google Scholar]
- Rediers H, Vanderleyden J, De Mot R. (2004). Azotobacter vinelandii: a Pseudomonas in disguise? Microbiology 150: 1117–1119. [DOI] [PubMed] [Google Scholar]
- Revers LF, Passaglia LM, Marchal K, Frazzon J, Blaha CG, Vanderleyden J et al. (2000). Characterization of an Azospirillum brasilense Tn5 mutant with enhanced N2 fixation: the effect of ORF280 on nifH expression. FEMS Microbiol Lett 183: 23–29. [DOI] [PubMed] [Google Scholar]
- Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS et al. (2007). The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci USA 104: 8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaudi LV, Giordano W. (2010). An integrated view of biofilm formation in rhizobia. FEMS Microbiol Lett 304: 1–11. [DOI] [PubMed] [Google Scholar]
- Rosche B, Li XZ, Hauer B, Schmid A, Buehler K. (2009). Microbial biofilms: a concept for industrial catalysis? Trends Biotechnol 27: 636–643. [DOI] [PubMed] [Google Scholar]
- Sadasivan L, Neyra CA. (1985). Flocculation in Azospirillum brasilense and Azospirillum lipoferum: exopolysaccharides and cyst formation. J Bacteriol 163: 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadasivan L, Neyra CA. (1987). Cyst production and brown pigment formation in aging cultures of Azospirillum brasilense ATCC 29145. J Bacteriol 169: 1670–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff HL. (1975). Encystment and germination in Azotobacter vinelandii. Bacteriological Rev 39: 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeiren H, Willems A, Schoofs G, de Mot R, Keijers V, Hai W et al. (1999). The rice inoculant strain Alcaligenes faecalis A15 is a nitrogen-fixing Pseudomonas stutzeri. Sys Appl Microbiol 22: 215–224. [DOI] [PubMed] [Google Scholar]
- Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. (2013). Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11: 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang Y, Welch E, Li J, Roberts GP. (2010). Elimination of Rubisco alters the regulation of nitrogenase activity and increases hydrogen production in Rhodospirillum rubrum. Int J Hydrogen Energy 35: 7377–7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiig JA, Hu YL, Lee CC, Ribbe MW. (2012). Radical SAM-dependent carbon insertion into the nitrogenase M-cluster. Science 337: 1672–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Yang J, Dou Y, Chen M, Ping S, Peng J et al. (2008). Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc Natl Acad Sci USA 105: 7564–7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Lu W, Chen M, Wang J, Zhang W, Zhang Y et al. (2013). Genome transcriptome analysis and functional characterization of a nitrogen-fixation island in root-associated Pseudomonas stutzeri. Mol Micro Ecol Rhizosp 1 and 2: 851–863. [Google Scholar]
- Yang J, Xie X, Wang X, Dixon R, Wang YP. (2014). Reconstruction and minimal gene requirements for the alternative iron-only nitrogenase in Escherichia coli. Proc Natl Acad Sci USA 111: E3718–E3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You CB, Song HX, Wang JP, Lin M, Hai WL. (1991). Association of Alcaligenes faecalis with wetland rice. Plant Soil 137: 81–85. [Google Scholar]
- Young JM, Leschine SB, Reguera G. (2012). Reversible control of biofilm formation by Cellulomonas spp. in response to nitrogen availability. Enviro Microbiol 14: 594–604. [DOI] [PubMed] [Google Scholar]
- Zadik Y, Yitschaky O, Neuman T, Nitzan DW. (2011). On the self-resolution nature of the buccal bifurcation cyst. J Oral Maxillofac Surg 69: e282–e284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.