Abstract
At deep-sea hydrothermal vents, primary production is carried out by chemolithoautotrophic microorganisms, with the oxidation of reduced sulfur compounds being a major driver for microbial carbon fixation. Dense and highly diverse assemblies of sulfur-oxidizing bacteria (SOB) are observed, yet the principles of niche differentiation between the different SOB across geochemical gradients remain poorly understood. In this study niche differentiation of the key SOB was addressed by extensive sampling of active sulfidic vents at six different hydrothermal venting sites in the Manus Basin, off Papua New Guinea. We subjected 33 diffuse fluid and water column samples and 23 samples from surfaces of chimneys, rocks and fauna to a combined analysis of 16S rRNA gene sequences, metagenomes and real-time in situ measured geochemical parameters. We found Sulfurovum Epsilonproteobacteria mainly attached to surfaces exposed to diffuse venting, while the SUP05-clade dominated the bacterioplankton in highly diluted mixtures of vent fluids and seawater. We propose that the high diversity within Sulfurimonas- and Sulfurovum-related Epsilonproteobacteria observed in this study derives from the high variation of environmental parameters such as oxygen and sulfide concentrations across small spatial and temporal scales.
Introduction
Reduced sulfur compounds are widely distributed in the environment, and sulfur oxidation is one of the most ancient microbial metabolisms (reviewed in Canfield and Raiswell, 1999). The long evolutionary history of sulfur oxidation is reflected in the high diversity of sulfur-oxidizing bacteria (SOB), which inhabit many different environments (reviewed in Canfield and Raiswell, 1999; Friedrich et al., 2005). In aphotic ecosystems, chemolithotrophic SOB are often the main primary producers (for example, Jannasch and Wirsen, 1979; Engel et al., 2003; Nakagawa et al., 2005; Grote et al., 2008). To successfully coexist, SOB have adapted to different ecological niches. The mechanisms of niche partitioning between SOB in, for example, the anoxic water column, sulfidic cave systems and sulfur-oxidizing microbial mats have been well studied (Jørgensen and Revsbech, 1983; Jørgensen and Des Marais, 1986; Macalady et al., 2008; Grünke et al., 2011; Headd and Engel, 2013). A systematic study investigating niche partitioning of co-occurring SOB at hydrothermal vents, where environmental conditions including reduced sulfur compounds concentrations change markedly on very small spatial scales (Tivey, 2004), is still missing.
SOB are ubiquitous in hydrothermal environments. They can be found as free-living microorganisms, but also as ecto- and endosymbionts of vent fauna (reviewed in Nakagawa and Takai, 2008). The key chemolithotrophic SOB at hydrothermal vents belong to the Epsilonproteobacteria and the Gammaproteobacteria (reviewed in Sievert et al., 2008a), whereas sulfur-oxidizing Aquificae only occupy a narrow thermophilic niche (Reysenbach, 2001; Alain et al., 2003; Hugler et al., 2007), and sulfur-oxidizing Archaea (order Sulfolobales) are generally rare in the marine environment (reviewed in Friedrich et al., 2005). Both cultivation-dependent and -independent studies showed that co-occurring Sulfurovum- and Sulfurimonas-related epsilonproteobacterial species are the dominant and most widespread SOB in hydrothermal environments (Inagaki et al., 2003; Lopez-Garcia et al., 2003; Inagaki et al., 2004; Nakagawa et al., 2005; Meyer et al., 2013). While these two groups are phylogenetically distinct, the ecotype differentiation between them remains unresolved (Campbell et al., 2006). The most prominent gammaproteobacterial sulfur oxidizers are Thiomicrospira species, giant mat-forming SOB such as the filamentous Beggiatoa (Jannasch et al., 1985, 1989; Takai et al., 2004; Brazelton and Baross, 2010), and members of the SUP05-clade. The latter ones have been detected in hydrothermal plumes, oxygen minimum zones and symbioses with vent fauna (Sunamura et al., 2004; Duperron et al., 2005; Lesniewski et al., 2012; Anderson et al., 2013; Glaubitz et al., 2013; Marshall and Morris, 2013). Culture-independent studies at hydrothermal vents and other marine sulfidic environments found the SUP05-clade often co-occurring with Epsilonproteobacteria (Sunamura et al., 2004; Labrenz et al., 2007; Bourbonnais et al., 2012; Sheik et al., 2015). Previous studies of marine oxyclines and hydrothermal plume suggested a niche separation between these two groups based on sulfur/oxygen ratio (Schmidtova et al., 2009; Grote et al., 2012; Anderson et al., 2013) analogous to niche differentiation of filamentous gamma- and epsilonproteobacterial SOB in cave systems and sulfidic springs (Anderson, 2001; Macalady et al., 2008; Headd and Engel, 2013). To the best of our knowledge, no study has as yet systematically investigated the shift from SUP05-dominated to Sulfurovum- and Sulfurimonas-dominated microbial communities in the dynamic environments of hydrothermal vents. Thus, our aim was to characterize conditions accompanying this transition, as well as potentially elucidating the ecological difference between Sulfurovum and Sulfurimonas.
At hydrothermal vents, niche partitioning of SOB may have to occur within extremely steep physicochemical gradients (Baross and Hoffman, 1985). At sites of focused discharge, hot hydrothermal fluids (up to 400 °C) enriched in reduced compounds are advected into the surrounding cold oxygenated seawater (Bach et al., 2006), typically forming a turbulent mixing zone with sharp gradients (Tivey, 2004). Studies have shown that microbes flourish on hydrothermal chimneys formed by precipitation of metal sulfides (Harmsen et al., 1997; McCollom and Shock, 1997; Flores et al., 2011; Reeves et al., 2014) and in areas of more diffuse venting (McCollom and Shock, 1997; Amend et al., 2011; Bemis et al., 2012; Meyer et al., 2013). The emerging hydrothermal plumes, however, are rather populated by microoorganisms associated with the background water column (Lesniewski et al., 2012; Anderson et al., 2013; Sheik et al., 2015; Anantharaman et al., 2016).
Here we investigated to what extent niche partitioning of SOB occurs, and which factors may be driving it in hydrothermal environment of the Manus Basin, a back-arc fast-spreading center located between New Britain and New Ireland in the Bismarck Sea. Many Manus Basin hydrothermal fields emit sulfide-rich fluids that are depleted in other energy sources (for example, methane or hydrogen) and offer a variety of different niches for SOB (Scott and Binns, 1995; Reeves et al., 2011b; Yeats et al., 2014; McDermott et al., 2015). We attempted to cover a large span of these niches by collecting venting fluids with different grades of dilution, as well as chimney structures and epibiota detected on vent fauna. Correlating distribution patterns to real-time geochemical data, we were able to assign tentative niches to the key hydrothermal SOB clades. By looking further into the diversity within these clades and into the differentiation of their sulfur oxidation genes, we developed a hypothesis on diversification drivers among closely related epsilonproteobacterial SOB species.
Materials and methods
Site description and sample collection
Samples consisting of fluids, rocks, hydrothermal chimneys and vent fauna were collected during the R/V Sonne expedition SO216 to the Manus Basin in June/July 2011 (Supplementary Figure S1,Supplementary Table S1). Its basaltic to intermediate and felsic lavas generate vigorous venting of sulfidic fluids with varying properties (Binns and Scott, 1993; Reeves et al., 2011a). Venting sites sampled in this study are located at PACManus and North Su hydrothermal fields, at a depth of 1150–1775 m (Supplementary Figure S1, Supplementary Table S1). The basement in both areas is felsic in composition, that is, it consists of silica-rich and oxidized magma that has been excessively degassed (Beier et al., 2015). The vent fluids in the North Su area are variably affected by direct magma degassing (Seewald et al., 2015). The PACManus vent field is also geochemically diverse and fluid compositions are affected by magma degassing and subseafloor mixing with entrained seawater (Reeves et al., 2011b). Fluid samples were collected with the remotely controlled flow-through system KIPS (Kiel Pumping System; Schmidt et al., 2007) mounted on the remotely operated vehicle (ROV) Quest (Marum, Bremen). Samples for metagenome sequencing were collected by pumping fluids directly onto 142 mm diameter cellulose–acetate or polyethersulfone membrane filters (0.22 μm pore size, Millipore, Darmstadt, Germany). Collection time ranged between 13 and 33 min (~3–8 × 108 cells). In addition, fluid samples were collected into 675 ml flasks (Savillex, Eden Prairie, MN, USA). Temperature and pH of all sampled fluids (Supplementary Table S1) were recorded with in-line sensors attached to the KIPS sampling nozzle. In order to record dissolved gas concentrations in real-time (Supplementary Table S1), the inlet of an in situ mass spectrometer (Wankel et al., 2010) was attached alongside the KIPS nozzle.
Rock, hydrothermal chimney and macrofauna samples were collected with the ROV’s hydraulic arm and kept in closed bio-boxes during the ROV’s ascent. Water column samples were collected in Niskin bottles attached to a conductivity-temperature-depth probe.
Directly after shipboard retrieval, in situ collected membrane filters were transferred to −80 °C. Fluids collected in flasks were passed through polyethersulfone membrane filters (0.22 μm pore size) and the filters were stored at −20 °C. Retrieved rocks and hydrothermal chimney structures were subsampled and directly frozen at −20 °C until DNA extraction.
Thermodynamic calculations
Gibb’s free energies available from 1mol of substrate were calculated as described in Meier et al. (2016) using concentrations measured with the in situ mass spectrometer instead of activities. To determine the energy available per kg of fluid–water mix, calculated Gibbs’ free energies were multiplied by the concentration of the limiting compound of the reaction.
Modeling of the mixing gradient was performed with the REACT module of the Geochemist’s Workbench software (Aqueous Solutions LLC, Champaign, IL, USA), using the thermodynamic database of Amend et al. (2011) and endmember values for the Fenway vent in the PACManus area from Reeves et al. (2011b).
Metagenome sequencing and assembly
High-molecular-weight genomic DNA for metagenomic analysis was extracted from a quarter of a 142 mm diameter membrane filters as described previously (Meier et al., 2016), with an additional 1 h Proteinase K digestion step (80 μg ml−1 final concentration) at 37 °C and a 2 h incubation at 65 °C after addition of SDS-containing buffer S1 (MO BIO Laboratories, Carlsbad, CA, USA) prior to applying the kit protocol. The genomic DNA was shotgun-sequenced on an Illumina HiSeq2500 sequencer at the Max Planck Genome Center (MPGC, Cologne, Germany) after library construction using the Ovation Ultralow Library system kit (NuGen, San Carlos CA, USA; 15 cycles of amplification). Between 162 and 174 million read pairs were obtained per metagenome.
Raw sequence reads were quality-trimmed, error-corrected and normalized to a k-mer depth of 40 using BBtools (BBmap package v. 33.57 http://sourceforge.net/projects/bbmap/) with default parameters. Bulk assembly of the metagenomes was performed with IDBA_UD v. 1.1.1 with k-mer sizes from 21 to 99 in steps of 10. Full-length 16S rRNA genes were reconstructed from the raw reads using PhyloFlash 2.0 (http://github.com/HRGV/phyloFlash).
16S rRNA gene sequencing and analysis
The DNA was extracted from a 1.5 × 1.5 cm filter piece following the same protocol as used for metagenomic sequencing. The V3–V4 region of the 16S rRNA gene was amplified as described previously (Meier et al., 2016). The amplicons were sequenced on an Illumina MiSeq sequencer at the MPGC. After trimming of 3′-ends with quality below q10, paired end reads were merged using BBmerge (BBmap package v. 33.57 http://sourceforge.net/projects/bbmap/) with a minimum overlap of 50 bp.
Reads were de-multiplexed and randomly subsampled to 5000 reads per sample using Mothur v. 1.34 (Schloss et al., 2009). Reads of the whole data set were decomposed into ‘nodes’ by MED v. 2.0 (Eren et al., 2015) with four discriminant locations and minimum substantive abundance (count of the most abundant sequence in a node) of three. Finally, percentage similarity-independent operational taxonomic units (OTUs) were generated based on representative sequences of MED using SWARM v. 2.1.6 (Mahe et al., 2015). SWARM was run with the ‘fastidious’ option and 20 as the number of sequences in a node for it to be considered ‘big’. The other parameters were kept to their default values. Representative sequences were classified by last common ancestor according to the alignment to the SILVA SSU123 database performed with SINA (Pruesse et al., 2012).
Full-length 16S rRNA genes were amplified using the GM3F and GM4R primer set (Muyzer et al., 1995) and sequenced on a Pacific Biosciences RSII sequencer in a circular consensus mode at the MPGC. Further long 16S rRNA gene sequences were reconstructed from metagenomic reads using PhyloFlash (v. 2.0) with default parameters. Full-length 16S rRNA gene sequences obtained by PacBio sequencing and PhyloFlash reconstruction were quality-trimmed with Mothur v. 1.34 as follows: in a sliding window of 10 bp the average quality should remain above q21 and individual base call quality never fall below q10. Otherwise, the sequence was trimmed at this point. After trimming only sequences over 1000 bp were kept. Subsequently sequences were clustered with vsearch v. 1.9.10 (github.com/torognes/vsearch).
Statistical analysis
All statistical analyses were performed in R using the ‘vegan’ package (Oksanen et al., 2013). Non-parametric permutational multivariate analysis of variance (perMANOVA; Anderson, 2001) was performed with the ‘adonis’ function. Distance-based redundancy analysis was performed with the ‘capsacale’ function. The ‘Simper’ function was used for the similarity percentages breakdown analysis.
Targeted re-assembly of metagenomic bins
Binning of the metagenomes based on differential coverage, tetranucleotide frequencies, taxonomic classification, paired end read mapping and conserved single-copy gene profiles was performed using the Metawatt binning software (v. 3.5.2; Strous et al., 2012). Targeted de novo assemblies of bins of interest were done with the SPAdes assembler v. 3.1.1 (Bankevich et al., 2012) as described in Meier et al. (2016) with three re-assembly rounds per bin. The generated assemblies were automatically annotated with the standard RAST annotation pipeline (Aziz et al., 2008) and further analyzed with the GenDB annotation system (Meyer et al., 2003) using the JCoast frontend (Richter et al., 2008). Completeness and quality of final assemblies were assessed by CheckM (Parks et al., 2015) based on the translated protein sequences exported from RAST and a Proteobacteria-specific set of single-copy marker genes. Average nucleotide identities (ANIs) between the assemblies and to the next sequenced relative were calculated with JSpeciesWS web service (Richter et al., 2015).
The annotation of selected genes referred to in this study was manually inspected: results of RAST annotations were compared to hidden Markov model-based HMMER3 (Eddy, 2011) searches against the Pfam-A database (Finn et al., 2014) and BLASTP searches (Camacho et al., 2009) against the NCBI-nr database.
Orthologous proteins among the SOB genomes were identified by BLAST and OrthoMCL (Li et al., 2003) based FastOrtho tool (http://enews.patricbrc.org/fastortho/) with minimum percent identity set to 10%, minimum of matching amino acids to 20 and otherwise default settings.
Phylogenetic tree construction
Translated soxY genes (encoding sulfur anion carrier protein) identified on contigs of the bins and in the bulk metagenomes were used to construct phylogenetic trees together with 245 SoxY sequences from the UniprotKB database (Magrane and Consortium, 2011) including sequences from isolates of confirmed sulfur oxidizers. Protein sequences were aligned with MAFFT (Katoh and Standley, 2013), using the L-INS-I method and the Blosum62 scoring matrix.
A concatenated alignment of 138 conserved single-copy genes was generated with HMMER3 (Eddy, 2011) implemented in CheckM (Parks et al., 2015).
16S rRNA gene sequences were aligned by SINA (v. 1.3.0, Pruesse et al., 2012) to a curated SILVA SSU123 NR99 database, where all sequences with a pintail value below 50 and alignment quality below 70 were excluded from further analyses. PacBio and PhyloFlash sequences longer than 1200 bp together with high alignment quality (>95) clade represenatative sequences from the SILVA database were used for tree calculations. Shorter metagenomic 16S rRNA gene sequences and OTU-representative Illumina amplicon sequences were added to the calculated trees based on maximum parsimony in ARB (Ludwig et al., 2004). Trees were calculated with various algorithms: neighbor-joining (Ludwig et al., 2004), PhyML (v. 3.1, Guindon et al., 2010), RaxML (v. 8.0.26, Stamatakis, 2014) and FastTree (v. 2.1.9, Price et al., 2009, 2010) to check the stability of the basic topology. Position conservation filters of 20, 25 and 30% were tested for proteins, and 30, 40, 50%, and the bacterial position variability filter of the SILVA SSU123 NR99 database were tested for 16S rRNA sequences.
Nucleotide sequence accession numbers
Raw reads as well as assembled sequences were submitted to the European Nucleotide Archive under the project number PRJEB15554.
Results
Diversity and distribution of SOB in the Manus Basin
The bacterial diversity was assessed by high-throughput 16S rRNA gene amplicon analysis applying minimum entropy decomposition (MED, Eren et al., 2015) and SWARM (Mahe et al., 2015). MED generated a total of 9281 ‘nodes’ for the whole data set, which were further clustered into 1307 OTUs with SWARM.
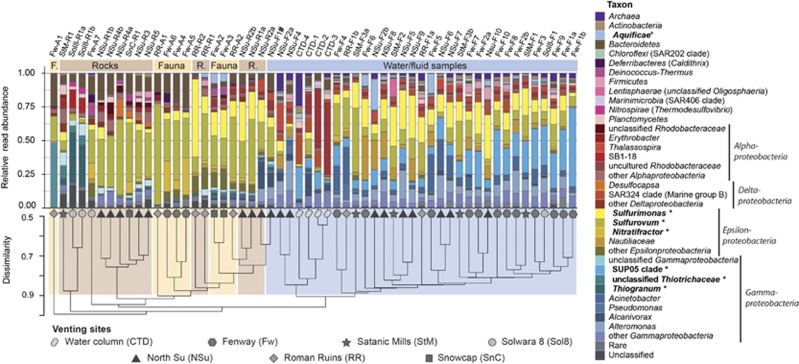
Hierarchical clustering of samples according to their microbial community composition showed distinct patterns based on whether the samples originated from a solid surface or a fluid sample (Figure 1). In contrast, significant clustering based on the venting site was not observed. Sequences affiliating with known SOB were present in all analyzed samples. However, different SOB clades occurred and dominated in different sample types. A perMANOVA revealed that 30% of the community composition variance could be explained by the sample category alone (‘fluid’, ‘water column’, ‘rock surface’, ‘faunal surface’, P=0.0001). Almost all solid surface samples were dominated by sequences assigned to the epsilonproteobacterial genus Sulfurovum (2–74%, on average 30% of all reads; Figure 1). In addition, other sequences classified as sulfur-oxidizing Epsilonproteobacteria (for example, Sulfurimonas: 0–58%, 10% on average, Nitratifractor 0–27%, 4% on average) and uncultured, likely thiotrophic, gammaproteobacterial clades were detected. Sulfurovum-related and Sulfurimonas-related sequences were also present in all collected fluid samples, but in lower relative abundance (on average 24%, compared to 41% on solid surfaces) and with lower proportion of Sulfurovum-related reads (0–47%, on average 11%) in comparison to solid surface samples (2–74%, 30% on average). Few fluid samples also showed increased proportions of 16S rRNA gene sequences affiliated with Aquificeae, a phylum harboring thermophilic SOB (over 1% in 13 of 33 fluid samples, 22% max.). In contrast, sequences related to SUP05-clade Gammaproteobacteria were found almost exclusively in fluid samples, with relative sequence abundances reaching up to 58% (15% on average, 1% min.; Figure 1). In 10 of 23 surface samples SUP05-clade sequences were completely absent, and in the remaining surface samples they accounted for less than 1%. A similarity percentages breakdown (SIMPER) calculated based on relative abundances of OTUs in relation to sample category revealed that most Sulfurovum-related and Sulfurimonas-related OTUs were significantly contributing to the overall community composition difference between fluids and surface samples (P<0.05; Supplementary Figure S2).
Figure 1.
Relative abundances of 16S rRNA gene sequence reads according to their classification. Putative SOB are denoted in bold and are marked with a ‘*’. The cluster dendrogram depicts the average linkage hierarchical clustering based on a Bray-Curtis dissimilarity matrix of community compositions resolved down to MED-node level. #Co-sampling of sediment particles possible.
The diversity of 16S rRNA gene sequences within the three most abundant SOB populations, Sulfurovum-related, Sulfurimonas-related and the SUP05-clade differed significantly (Supplementary Figure S3). Sulfurovum-related sequence reads exhibited the highest level of diversity with 1602 nodes generated by MED, resulting in 149 OTUs generated by SWARM. Sulfurimonas-related SOB were the second most diverse group (1027 MED nodes, 99 OTUs). The SUP05-clade showed comparatively low diversity (515 MED nodes, 24 OTUs), despite high relative abundances in fluid samples (15% on average, 58% max.). Almost full-length 16S rRNA gene sequences retrieved from seven fluid samples (NSu-F2a, CTD-4, RR-F1b, Fw-F1b, Fw-F2a, StM-F2 and StM-F3a) by PacBio amplicon sequencing and targeted 16S rRNA gene reconstruction from metagenomes confirmed the trends emerging from short read analyses. Clustering of long 16S rRNA gene sequences at 94.5% minimum sequence identity level corresponding to a genus-level cutoff according to Yarza et al. (2014) resulted in a number of clusters slightly below the number of OTUs generated by SWARM from short amplicon reads. Again, Sulfurovum-related sequences were more diverse (100 OTUs) than Sulfurimonas-related (85 OTUs), while SUP05-clade sequences (20 OTUs) were the least diverse. A phylogenetic tree reconstruction showed that sequences classified as Sulfurovum and Sulfurimonas form two distinct monophyletic branches with several subclades each (Supplementary Figure S4).
Niche partitioning along an environmental gradient
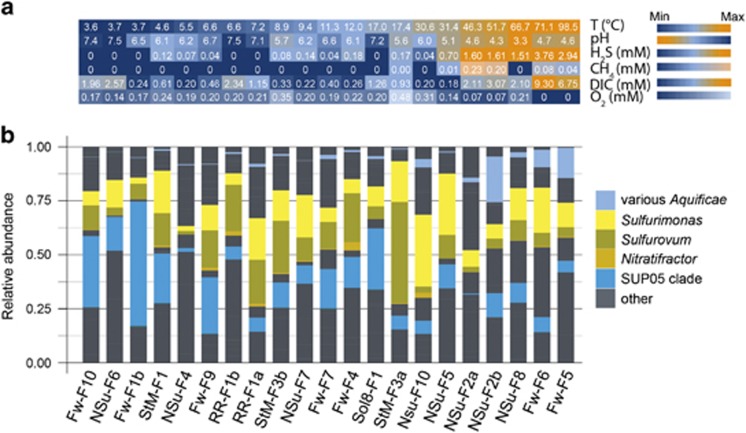
In-line temperature and pH probes as well as an in situ mass spectrometer were used to record the geochemistry of the majority of diffuse fluid samples immediately prior to sample collection (21/29, Figure 2). Most of the recorded parameters exhibit a strong pairwise covariance, while oxygen concentration shows a lesser degree of correlation to other parameters (Figure 2, Supplementary Table S2).
Figure 2.
Distribution of SOB 16S rRNA gene sequences in fluid samples with geochemical data. (a) In situ determined geochemical parameters sorted from cold diluted to more hot and concentrated hydrothermal fluids (left to right) based on temperature. (b) Distribution of putative SOB genera based on 16S rRNA gene amplicon sequences in diffuse fluid samples.
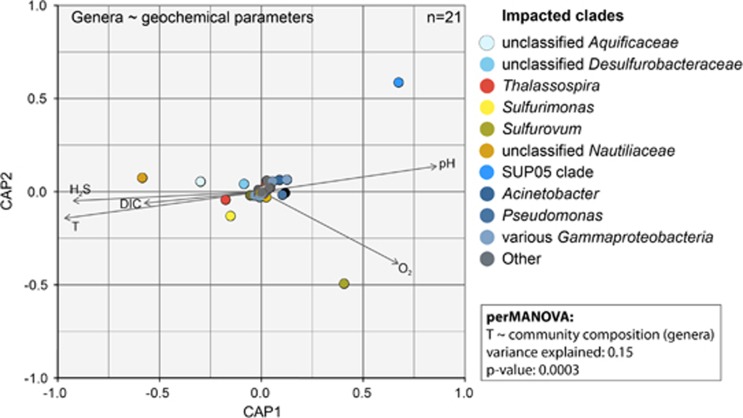
An analysis of SOB clade distribution with respect to geochemical data (Figure 2) indicated that SUP05-clade Gammaproteobacteria preferentially inhabit highly diluted, low sulfide and low-temperature fluids. Aquificae-related sequences were frequent in hot (>50 °C), sulfide-rich fluids. Sulfurimonas-related and Sulfurovum-related Epsilonproteobacteria accounted for a substantial fraction of SOB reads especially in the mixing zone between these two extremes, that is, in areas with temperatures below 40 °C and generally low to moderate sulfide concentrations (0.1–1.0 mmol l−1 sulfide). Distance-based redundancy analysis supported a correlation between abundances of chemolithotrophic microbial clades (for example, Nautiliaceae, Sulfurimonas-related/Sulfurovum-related, Aquificae, and SUP05) and changes in the recorded environmental parameters (Figure 3). A perMANOVA further confirmed that the position of the sample within the geochemical mixing gradient (using temperature as a proxy) could explain 15% of the community composition variance at the genus level (P=0.0003). No significant correlation between community composition on OTU or MED-node level and geochemical parameters was observed.
Figure 3.
Distance-based redundancy analysis (dbRDA) calculated based on a Bray-Curtis dissimilarity matrix and standardized, log-normalized geochemical parameters. The distance matrix was calculated based on relative abundances of microbial genera in 21 fluid samples with geochemical data. Results of a non-parametric permutational multivariate analysis of variance (perMANOVA) are stated in the frame adjacent to the dbRDA panel. perMANOVA was calculated using the ‘adonis’ function of the ‘vegan’ package in R (Oksanen et al., 2013).
The difference in correlation to oxygen concentration between Sulfurovum-related and Sulfurimonas-related bacteria is especially noteworthy (Figure 3). While the response of Sulfurimonas-related bacteria to changes in oxygen concentrations was only minute, increased oxygen concentration correlated significantly with higher relative abundance of Sulfurovum-related bacteria. A perMANOVA test confirmed the significant impact of oxygen concentration (P=0.02) and showed relative abundances of Sulfurovum-related species to be most positively affected by increasing oxygen concentrations. Apart from a correlation to the position in the gradient, we also checked for correlation with Gibbs’ free energies available from sulfide oxidation per kg fluid–water mix at a given sampling point (Supplementary Table S1). However, no significant correlation between community composition at any level and the Gibbs’ free energies was observed.
Genomic variability among hydrothermal vent SOB
We sequenced and analyzed the metagenomes of the samples NSu-F2b, NSu-F5, Fw-F1b, Fw-F3 and RR-F1B (Supplementary Figure S5). The NSu-F2b metagenome was obtained from a 52 °C hot acidic fluid (pH=4.3), with high sulfide (1.6 mmol l−1 H2S) and low oxygen (0.07 mmol l−1) concentration. The NSu-F5 metagenome originates from a more diffuse fluid sample (T=31 °C, pH=5.1, H2S: 0.7 mmol l−1, O2: 0.14 mmol l−1). The Fw-F1b, Fw-F3 and RR-F1b metagenomes originate from diffuse venting sites with strong fauna colonization (T=3.7–6.6 °C, pH=6.5–7.5, no detectable H2S and 0.17–0.2 mmol l−1 O2). By multicriteria binning and targeted re-assembly, we obtained 28 bins for the three target groups (11 Sulfurovum-related, 5 Sulfurimonas-related and 12 SUP05-clade; Supplementary Figure S6). Read mapping of the five metagenomes to the bins revealed different distribution patterns and abundances for the epsilonproteobacterial SOB (Supplementary Figure S7). The bins of the SUP05-clade bacteria, in contrast, showed a homogenous distribution pattern. All genomes classified as free-living SUP05-clade bacteria were most abundant in the RR-F1B metagenome, with the exception of SUP05-5, which almost exclusively appeared in the NSu-F5 metagenome. The three SUP05-clade bins classified as sulfur-oxidizing symbionts were all most abundant in the Fw-F3 sample (Supplementary Figure S7) and absent at the two sites devoid of visible macrofauna colonization (Supplementary Figure S5).
The patterns of high 16S rRNA gene diversity among Sulfurovum-related/Sulfurimonas-related bacteria and low diversity within the SUP05-clade were also reflected in the metagenomic data. We compared ANIs between bins and between the bins and available complete genomes according to thresholds suggested by Goris et al. (2007). ANI of Sulfurovum-related bins obtained in this study ranged between 66 and 81% (70% on average, Supplementary Table S3a). SV-10 showed the highest ANI to a cultured representative with 78% to Sulfurovum sp. NBC37-1 (Nakagawa et al., 2007). The five Sulfurimonas bins showed a similar level of diversity, with an average ANI of 72% (68–76% Supplementary Table S3b). SM-5 and SM-6 exhibited the highest ANI (both 75%) to Sulfurimonas autotrophica (Sikorski et al., 2010). Thus, the obtained Sulfurovum-related and Sulfurimonas-related bins span more than just two genera. In contrast, all retrieved SUP05 bins, except SUP05-5, would belong to the genus Candidatus Thioglobus (Marshall and Morris, 2013; Shah and Morris, 2015). Furthermore, SUP05-6 and SUP05-9 as well as SUP05-7 and SUP05-13 would represent different strains of the same species. Recruitment of metagenomic reads to the bins showed that the bins represent sequence-discrete populations (Supplementary Figure S8).
We also compared the metabolic repertoire of all SOB bins with respect to energy-generating pathways, carbon assimilation and adaptations to environmental stress (Supplementary Figure S9). As expected, all Sulfurovum-related and Sulfurimonas-related bins contained genes encoding enzymes of the reverse tricarboxylic acid (rTCA) cycle (ATP-citrate lyase, 2-oxoglutarate synthase and fumarate reductase), whereas the SUP05-clade genomes largely harbored marker genes of the Calvin–Benson–Bassham cycle (for example, RUBISCO encoding cbb genes). The sulfur oxidation multienzyme complex (SOX), terminal cytochrome c oxidases for aerobic respiration (cbb3-type) as well as respiratory nitrate reductase (Nap) genes were found in all three SOB groups. However, all SUP05 bins were lacking the genes encoding SoxCD. Most of the Sulfurovum-related bins (9/11) also contain genes for a complete denitrification pathway, while Sulfurimonas-related and SUP05-clade bins were consistently lacking canonical Nos genes encoding a nitrous-oxide reductase. Only the SUP05-clade bins had the ammonia-forming nitrite reductase (NirD/B), while Sulfurimonas-related and Sulfurovum-related bins contained the NO-producing NirS and some as well the ammonifying NirA nitrite reductase. A unique feature of the Sulfurovum-related and Sulfurimonas-related bins is the presence of a membrane-bound polysulfide reductase (subunit genes psrABC), indicating the potential use of polysulfides as electron acceptor.
Comparing possible adaptations to the environment, we found that the Sulfurovum-related and Sulfurimonas-related bins harbor a much wider range of heavy metal and oxygen stress resistance genes than the SUP05-clade genomes (Supplementary Figure S9). Another feature unique to epsilonproteobacterial SOB bins was the presence of genes for capsular polysaccharide synthesis and export, which were absent in all SUP05 bins. Chemotaxis and flagellar motility were exclusively found in Sulfurimonas-related bins, while both Sulfurimonas-related and Sulfurovum-related bins encoded genes for aerotaxis and type-II and type-IV pili, possibly enabling twitching motility. Flagella and pili can also be used for attachment to surfaces. SUP05-clade bins contained neither pili or flagella, nor chemotaxis genes (Supplementary Figure S9).
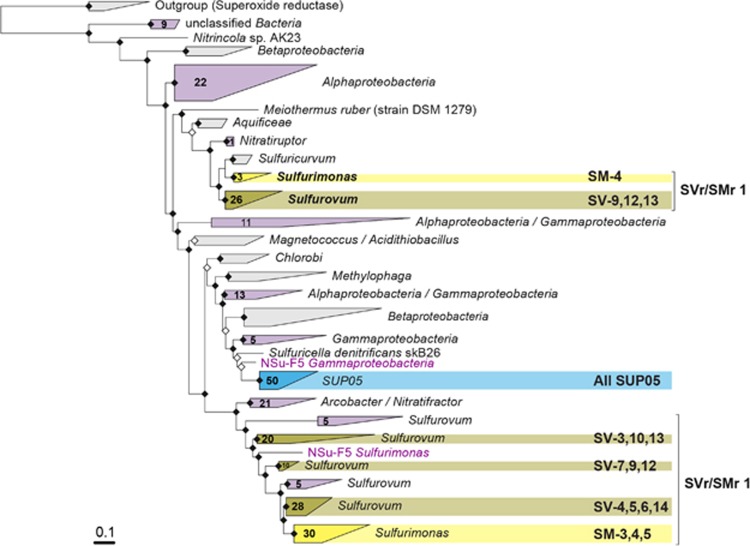
Finally, we looked in detail into the genes encoding the sulfur-oxidizing multienzyme complex (SOX) (Friedrich et al., 2000; Quentmeier and Friedrich, 2001). In Sulfurovum-related and Sulfurimonas-related bins, genes encoding the SOX system were split into two different loci (Supplementary Figure S10), with genes encoding the sulfur anion carrier protein (SoxYZ) present at both. Some of the bins were missing one of the two loci, most probably due to bin incompleteness. A phylogenetic tree of SoxY protein sequences revealed that the two different SoxY proteins encoded by the Sulfurimonas-related and Sulfurovum-related bins as well as Sulfurimonas-related/Sulfurovum-related SoxY sequences from the bulk metagenome assemblies fall into two distinct clusters (Figure 4). One branch contained more conserved SoxY proteins (Sulfurovum-related: 52–100% similarity, Sulfurimonas-related: 68–95% similarity) encoded together with SoxX, Z, A and B. The other branch contained a more diverse group of SoxY proteins (Sulfurovum-related: 21–95% similarity, Sulfurimonas-related: 42–100% similarity) encoded together with SoxZ, C, D and H (Supplementary Figure S10). The single loci encoding the Sox proteins in SUP05-clade bins had a homogeneous structure and all SoxY protein sequences formed a single clade (similarity: 49–100%), closely related to other gammaproteobacterial SoxY proteins (Figure 4).
Figure 4.
Maximum likelihood tree of SoxY amino-acid sequences. In yellow—clusters containing epsilonproteobacterial bins, in blue—SUP05 bins, with respective bins indicated on the right. In purple—clusters containing other sequences encoded in the bulk metagenome assemblies. Numbers on the triangles indicate numbers of SoxY sequences from the bulk metagenome assemblies contained in the cluster. The tree was calculated with PhyML (Guindon et al., 2010) based on positions conserved in at least 25% of the sequences.
An analysis of the sulfur anion-binding domains of the two epsilonproteobacterial SoxY clusters (Supplementary Figure S11) showed that sulfur-binding cysteine was conserved in all sequences, whereas the other amino acids of the sulfur carrying ‘swinging arm’ (Sauve et al., 2007) were only completely conserved in the first, less diverse Sulfurimonas-related/Sulfurovum-related SoxY cluster. Compared to the canonical GGCGG sequence in the SoxY ‘swinging arm’ of Paracoccus panthotrophus, SoxY from the Sulfurimonas-related/Sulfurovum-related cluster was missing one C-terminal glycine resulting in the amino-acid sequence GGCG. In the second, more-diverse SoxY cluster of Sulfurimonas-related/Sulfurovum-related sequences the sulfur-binding cysteine was followed by a variable position most frequently occupied by glutamic acid and a rather conserved glycine (GGCEG). The five AAs preceding the ‘swinging arm’ were completely conserved in the first SoxY cluster and variable in the second (Supplementary Figure S11).
Discussion
In the Manus Basin three clades of SOB with different degrees of intragroup diversity dominate the free-living microbial populations at the venting sites: the low-diversity SUP05-clade Gammaproteobacteria, the more diverse Sulfurimonas-related Epsilonproteobacteria and highly diverse Sulfurovum-related Epsilonproteobacteria. While SUP05-clade bacteria were preferentially found under cold, low-sulfide conditions, Sulfurovum-related and Sulfurimonas-related SOB dominated in moderate temperature fluids with elevated sulfide concentrations. A succession of different SOB clades mainly occurring along geochemical gradients was already reported earlier for sediments, as well as terrestrial and limnic environments (Jørgensen and Revsbech, 1983; Jørgensen and Des Marais, 1986; Macalady et al., 2008; Grünke et al., 2011; Headd and Engel, 2013). Thereby, in particular, sulfide and oxygen concentrations were identified as determining factors for niche partitioning between gamma- and epsilonproteobacterial SOB (Macalady et al., 2008; Headd and Engel, 2013). In marine environments, SUP05-clade Gammaproteobacteria have often been found co-occurring with sulfur-oxidizing Epsilonproteobacteria in diffuse hydrothermal vent fluids and oxyclines (Sunamura et al., 2004; Labrenz et al., 2007; Grote et al., 2008; Bourbonnais et al., 2012; Glaubitz et al., 2013; Sheik et al., 2015). For anoxic water column environments, it was hypothesized that a niche separation based on sulfide and oxygen concentrations may also apply to Epsilonproteobacteria and the SUP05-clade (Schmidtova et al., 2009; Grote et al., 2012). Here, we report a statistically supported correlation between SOB clade abundances and the degree of mixing of hydrothermal fluids with admixed seawater, resolving the niches of SUP05-clade and Epsilonproteobacteria SOB in dynamic hydrothermal environments.
Other studies of SOB at hydrothermal vents also report the presence and co-occurrence of multiple Epsilonproteobacteria genera (Nakagawa et al., 2005; Opatkiewicz et al., 2009; Huber et al., 2010; Flores et al., 2012; Akerman et al., 2013). A novel finding of this study is the positive correlation between oxygen concentrations and Sulfurovum-related sequence abundances. Despite the use of the oxygen-sensitive rTCA cycle for carbon fixation (Hugler et al., 2005; Campbell et al., 2006; Nakagawa and Takai, 2008), Sulfurovum-related species in the Manus Basin seem to thrive best at relatively high oxygen levels (~200 μmol l−1, Figure 2). The detection of genes encoding capsular polysaccharide production and export, genes for aerobic terminal oxidases, various genes involved in reducing oxidative stress and aerotaxis-related genes in almost all of the reconstructed epsilonproteobacterial bins (Supplementary Figure S9) underline the adaptation of Sulfurovum-related bacteria to oxygenated environments. Aerobic growth enabled via an adapted version of the 2-oxoglutarate synthase was shown for thermophilic Hydrogenibacter (phylum Aquificae; Yamamoto et al., 2006). In addition, nitrite oxidizers of the genus Nitrospira use the rTCA cycle for CO2 fixation under aerobic conditions (Lücker et al., 2010). Apart from the possibility of having adapted rTCA cycle enzymes, Sulfurovum might get access to a more oxic niche by growing in thick sheath-protected filaments (Stokke et al., 2015) creating anoxic microenvironments for optimal carbon fixation. In contrast, although exhibiting a similar genetic repertoire, no clear correlation with oxygen concentration could be observed for Sulfurimonas-related species. We therefore hypothesize that difference in adaptation to increased oxygen concentrations may be an important mechanism for niche partitioning between Sulfurovum-related and Sulfurimonas-related SOB at hydrothermal vent sites. Sulfurovum-related SOB were also found to dominate the sampled solid surfaces, whereas Sulfurimonas-related SOB seemed to be more abundant in the fluids. Whether the preference for growth on solid surfaces is a niche-partitioning factor remains open, considering that diffuse venting fluids hardly represent a stable planktonic environment for microorganisms. Therefore, ‘fluid-specific’ epsilonproteobacterial OTUs may also thrive attached to solid surfaces in areas close to diffuse fluid outlets or in subsurface areas (Akerman et al., 2013; Meyer et al., 2013). Both Sulfurovum-related and Sulfurimonas-related bins have the genomic potential for surface attachment, like pili and flagella genes.
In this study we also observed a high level of diversity within the Sulfurovum-related and Sulfurimonas-related clades confirming observations of previous studies (Huber et al., 2007, 2010; Meyer et al., 2013). The convergent results of independent short and long 16S rRNA gene analyses combined with metagenome analysis as well as the low diversity observed for SUP05 strongly support that the observed high diversity of Sulfurovum and Sulfurimonas is not a method-related artifact (Quince et al., 2009; Kunin et al., 2010) and provide insights on its potential drivers. The environmental 16S rRNA gene sequence diversity currently attributed to these genera in databases like SILVA (Quast et al., 2013), as well as Sulfurovum-related and Sulfurimonas-related sequences recovered in this study, rather constitute family-level clades according to identity-based thresholds suggested by Yarza et al. (2014). The average nucleotide identities of the recovered Sulfurovum-related bins support the existence of multiple, closely related genera (Konstantinidis and Tiedje, 2005; Goris et al., 2007). The SUP05-clade sequences recovered in this study, in contrast, look rather uniform. The number of 16S rRNA OTUs is low and their phylogenetic distances small. In addition, the ANI of the retrieved bins is rather high. We hypothesize that the difference in microdiversification between the SUP05-clade and Sulfurovum-related/Sulfurimonas-related SOB might be explained by their position in the ‘flat’ versus the ‘steep’ part of the geochemical gradients occurring at hydrothermal vents (Figure 5). Microdiversity of highly abundant organisms occupying a broad niche had previously been attributed either to slightly different adaptations to varying environmental conditions, such as light intensity in the case of the phototrophic Prochlorococcus (Moore et al., 1998; Urbach et al., 1998), or to differentiation with respect to physiological roles, as in the case of hydrothermal Methanosarcinales biofilms (Brazelton et al., 2011). In our case, the varying concentrations of electron donors and acceptors, for example, reduced sulfur compounds and oxygen or nitrate could be the main driver for diversification.
Figure 5.
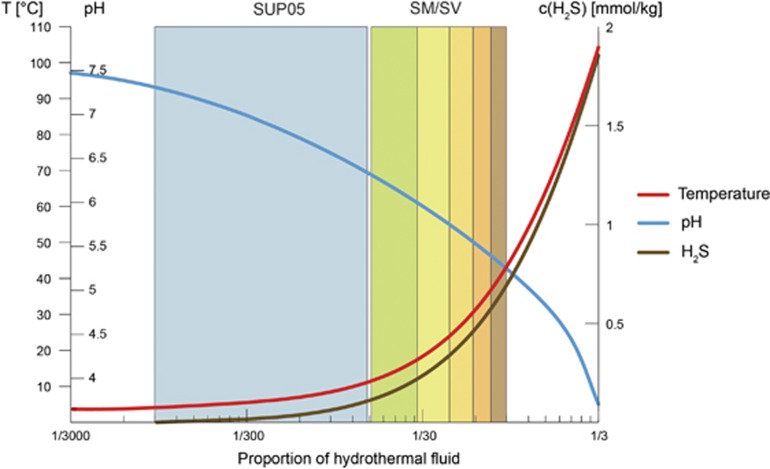
Schematic placement of niches of SUP05 (blue) and Sulfurimonas/Sulfurovum-related SOB (SM/SV; green to brown) in the mixing gradient. Being placed in the steep part of the gradient, Sulfurimonas and Sulfurovum-related bacteria are exposed to higher amplitudes of variation of environmental parameters, for example, hydrogen sulfide concentrations, which leads to diversification into different species, each one with its own microniche.
Looking at genes encoding the sulfur anion-binding SoxY protein (Quentmeier and Friedrich, 2001; Sauve et al., 2007), which had been reported to be the most highly expressed gene of the SOX complex in vent Epsilonproteobacteria (Dahle et al., 2013), we found two different homologs in Sulfurovum-related and Sulfurimonas-related bins, as well as in the bulk metagenome contigs classified as Sulfurovum or Sulfurimonas. The two different SoxY loci correspond to the ones found in Sulfurovum and Sulfurimonas isolates (Sievert et al., 2008a, b). Protein alignments showed that, while the sulfur-binding domain and five preceding amino acids are highly conserved in one version of SoxY, some of the amino acids surrounding the sulfur-binding cysteine were variable in the other SoxY version. These differences in a specific location of the protein are an indication of functional diversification, possibly an adaptation to bind a different sulfur-containing molecule. The variability of the second SoxY could represent adaptations to differing sulfur compound concentrations, which appear as a spatial gradient within pores of a hydrothermal chimney wall (Tivey, 2004; Flores et al., 2011), or as a temporal variation caused by differences in venting intensity (Figure 5). A similar hypothesis was raised with respect to SoxB gene diversity correlating to geochemical gradients in a sulfidic spring (Headd and Engel, 2013). However, SoxB diversity reported by Headd and Engel (2013) correlates with the distribution of major sulfur-oxidizing bacterial clades. In contrast, we found a high variability of the SoxY gene within the two epsilonproteobacterial clades of Sulfurimonas and Sulfurovum. Spatial and temporal variation of reduced compound concentrations makes the habitat of Sulfurovum-related/Sulfurimonas-related SOB one of ‘intermediate disturbance’ (Conell, 1978; Huston, 1979; Flöder and Sommer, 1999; Roxburgh et al., 2004). This might be the main cause for the high diversity within the Sulfurovum and Sulfurimonas populations (Figure 5).
Compared to the diverse SoxY proteins of Sulfurovum and Sulfurimonas, the SoxY of SUP05-clade SOB is conserved and uniform. In accordance, we found the SUP05-clade to be abundant in the ‘flat’, highly diluted end of the mixing gradient (Figure 5). We hypothesize that these stable conditions characterized, for example, by constantly low reduced sulfur concentrations provide only a limited niche space. As a consequence, the diversity of the SUP05-clade and its SoxY protein are low.
By using a combination of 16S rRNA gene and metagenomics analyses, in situ physicochemical measurements, as well as a substantial number of samples, we have advanced the understanding of the distribution and niche partitioning of SOB. Another important outcome of this study is a significant refinement of testable hypotheses on niche adaptation of gammaproteobacterial and epsilonproteobacterial SOB. We propose that the high diversity of co-occurring Sulfurovum and Sulfurimonas-related species is caused by high spatiotemporal variation of environmental parameters, for example, sulfide concentrations. Steep mixing gradients and temporal alterations of the fluid flow divide the general niche of Sulfurimonas and Sulfurovum into multiple microniches driving diversification and preventing selection of one subtype over the other. Systematic investigation of microbial populations in other environments characterized by steep spatiotemporal gradients of seemingly simple energy sources could help elucidate whether high diversity is a common theme of such settings.
Acknowledgments
We would like to thank officers, crew, shipboard scientific party and the technical team of the ROV Quest 4000 m (MARUM) on R/V Sonne cruise SO216, for their invaluable assistance. The cruise SO216 with R/V Sonne was an integral part of the Cluster of Excellence of the MARUM ‘The Ocean in the Earth System, Research Area GB: Geosphere-Biosphere Interactions’ funded by the German Research Foundation (DFG). We thank Richard Reinhard, Bruno Huettel and the team of the Max Planck Genome Centre in Cologne for sequencing, and Harald Gruber-Vodicka and Hanno Teeling for help with computational analyses. Further, we thank Nicole Krombholz and Kathrin Büttner for excellent technical assistance in the Molecular Ecology department. This work was supported by the Max Planck Society. This work was further supported by U.S. National Science Foundation under Grant Nos. IOS-1257755 and OCE-1536653 to Girguis.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Akerman NH, Butterfield DA, Huber JA. (2013). Phylogenetic diversity and functional gene patterns of sulfur-oxidizing subseafloor Epsilonproteobacteria in diffuse hydrothermal vent fluids. Front Microbiol 4: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alain K, Rolland S, Crassous P, Lesongeur F, Zbinden M, le Gall C et al. (2003). Desulfurobacterium crinifex sp. nov., a novel thermophilic, pinkish-streamer forming, chemolithoautotrophic bacterium isolated from a Juan de Fuca Ridge hydrothermal vent and amendment of the genus Desulfurobacterium. Extremophiles 7: 361–370. [DOI] [PubMed] [Google Scholar]
- Amend JP, McCollom TM, Hentscher M, Bach W. (2011). Catabolic and anabolic energy for chemolithoautotrophs in deep-sea hydrothermal systems hosted in different rock types. Geochim Cosmochim Acta 75: 5736–5748. [Google Scholar]
- Anantharaman K, Breier JA, Dick GJ. (2016). Metagenomic resolution of microbial functions in deep-sea hydrothermal plumes across the Eastern Lau Spreading Center. ISME J 10: 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46. [Google Scholar]
- Anderson RE, Beltran MT, Hallam SJ, Baross JA. (2013). Microbial community structure across fluid gradients in the Juan de Fuca Ridge hydrothermal system. FEMS Microbiol Ecol 83: 324–339. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach W, Edwards KJ, Hayes JM, Huber JA, Sievert SM, Sogin ML. (2006). Energy in the dark: fuel for life in the deep ocean and beyond. Eos Trans Am Geophys Union 87: 73–78. [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baross JA, Hoffman SE. (1985). Submarine hydrothermal vents and associated gradient environments as sites for the origin and evolution of life. Origins Life Evol Biosph 15: 327–345. [Google Scholar]
- Beier C, Bach W, Turner S, Niedermeier D, Woodhead J, Erzinger J et al. (2015). Origin of silicic magmas at spreading centres - an example from the South East Rift, Manus Basin. J Petrol 56: 255–271. [Google Scholar]
- Bemis K, Lowell RP, Farough A. (2012). Diffuse flow on and around hydrothermal vents at mid-ocean ridges. Oceanography 25: 182–191. [Google Scholar]
- Binns RA, Scott SD. (1993). Actively forming polymetallic sulfide deposits associated with felsic volcanic-rocks in the Eastern Manus back-arc basin, Papua-New-Guinea. Econ Geol 88: 2226–2236. [Google Scholar]
- Bourbonnais A, Juniper SK, Butterfield DA, Devol AH, Kuypers MMM, Lavik G et al. (2012). Activity and abundance of denitrifying bacteria in the subsurface biosphere of diffuse hydrothermal vents of the Juan de Fuca Ridge. Biogeosciences 9: 4661–4678. [Google Scholar]
- Brazelton WJ, Baross JA. (2010). Metagenomic comparison of two Thiomicrospira lineages inhabiting contrasting deep-sea hydrothermal environments. PLoS One 5: e13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazelton WJ, Mehta MP, Kelley DS, Baross JA. (2011). Physiological differentiation within a single-species biofilm fueled by serpentinization. MBio 2: e00127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Engel AS, Porter ML, Takai K. (2006). The versatile epsilon-Proteobacteria: key players in sulphidic habitats. Nat Rev Microbiol 4: 458–468. [DOI] [PubMed] [Google Scholar]
- Canfield DE, Raiswell R. (1999). The evolution of the sulfur cycle. Am J Sci 299: 697–723. [Google Scholar]
- Conell JH. (1978). Diversity in tropical rain forests and coral reefs. Science 199: 1302–1310. [DOI] [PubMed] [Google Scholar]
- Dahle H, Roalkvam I, Thorseth IH, Pedersen RB, Steen IH. (2013). The versatile in situ gene expression of an Epsilonproteobacteria-dominated biofilm from a hydrothermal chimney. Environ Microbiol Rep 5: 282–290. [DOI] [PubMed] [Google Scholar]
- Duperron S, Nadalig T, Caprais JC, Sibuet M, Fiala-Medioni A, Amann R et al. (2005). Dual symbiosis in a Bathymodiolus sp. mussel from a methane seep on the Gabon continental margin (Southeast Atlantic): 16S rRNA phylogeny and distribution of the symbionts in gills. Appl Environ Microbiol 71: 1694–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. (2011). Accelerated profile HMM searches. PLoS Comput Biol 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel AS, Lee N, Porter ML, Stern LA, Bennett PC, Wagner M. (2003). Filamentous "Epsilonproteobacteria" dominate microbial mats from sulfidic cave springs. Appl Environ Microbiol 69: 5503–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML. (2015). Minimum entropy decomposition: Unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J 9: 968–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR et al. (2014). Pfam: the protein families database. Nucleic Acids Res 42: D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flöder S, Sommer U. (1999). Diversity in planktonic communities: an experimental test of the intermediate disturbance hypothesis. Limnol Oceanogr 44: 1114–1119. [Google Scholar]
- Flores GE, Shakya M, Meneghin J, Yang ZK, Seewald JS, Geoff Wheat C et al. (2012). Inter-field variability in the microbial communities of hydrothermal vent deposits from a back-arc basin. Geobiology 10: 333–346. [DOI] [PubMed] [Google Scholar]
- Flores GE, Campbell JH, Kirshtein JD, Meneghin J, Podar M, Steinberg JI et al. (2011). Microbial community structure of hydrothermal deposits from geochemically different vent fields along the Mid-Atlantic Ridge. Environ Microbiol 13: 2158–2171. [DOI] [PubMed] [Google Scholar]
- Friedrich CG, Bardischewsky F, Rother D, Quentmeier A, Fischer J. (2005). Prokaryotic sulfur oxidation. Curr Opin Microbiol 8: 253–259. [DOI] [PubMed] [Google Scholar]
- Friedrich CG, Quentmeier A, Bardischewsky F, Rother D, Kraft R, Kostka S et al. (2000). Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J Bacteriol 182: 4677–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaubitz S, Kiesslich K, Meeske C, Labrenz M, Jurgens K. (2013). SUP05 dominates the gammaproteobacterial sulfur oxidizer assemblages in pelagic redoxclines of the central Baltic and Black Seas. Appl Environ Microbiol 79: 2767–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57: 81–91. [DOI] [PubMed] [Google Scholar]
- Grote J, Jost G, Labrenz M, Herndl GJ, Jurgens K. (2008). Epsilonproteobacteria represent the major portion of chemoautotrophic bacteria in sulfidic waters of pelagic redoxclines of the Baltic and Black Seas. Appl Environ Microbiol 74: 7546–7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote J, Schott T, Bruckner CG, Glockner FO, Jost G, Teeling H et al. (2012). Genome and physiology of a model epsilonproteobacterium responsible for sulfide detoxification in marine oxygen depletion zones. Proc Natl Acad Sci USA 109: 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünke S, Felden J, Lichtschlag A, Girnth AC, De Beer D, Wenzhofer F et al. (2011). Niche differentiation among mat-forming, sulfide-oxidizing bacteria at cold seeps of the Nile Deep Sea Fan (Eastern Mediterranean Sea). Geobiology 9: 330–348. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Harmsen HJM, Prieur D, Jeanthon C. (1997). Distribution of microorganisms in deep-sea hydrothermal vent chimneys investigated by whole-cell hybridization and enrichment culture of thermophilic subpopulations. Appl Environ Microbiol 63: 2876–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Headd B, Engel AS. (2013). Evidence for niche partitioning revealed by the distribution of sulfur oxidation genes collected from areas of a terrestrial sulfidic spring with differing geochemical conditions. Appl Environ Microbiol 79: 1171–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber JA, Cantin HV, Huse SM, Welch DB, Sogin ML, Butterfield DA. (2010). Isolated communities of Epsilonproteobacteria in hydrothermal vent fluids of the Mariana Arc seamounts. FEMS Microbiol Ecol 73: 538–549. [DOI] [PubMed] [Google Scholar]
- Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA et al. (2007). Microbial population structures in the deep marine biosphere. Science 318: 97–100. [DOI] [PubMed] [Google Scholar]
- Hugler M, Wirsen CO, Fuchs G, Taylor CD, Sievert SM. (2005). Evidence for autotrophic CO2 fixation via the reductive tricarboxylic acid cycle by members of the epsilon subdivision of Proteobacteria. J Bacteriol 187: 3020–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugler M, Huber H, Molyneaux SJ, Vetriani C, Sievert SM. (2007). Autotrophic CO2 fixation via the reductive tricarboxylic acid cycle in different lineages within the phylum Aquificae: evidence for two ways of citrate cleavage. Environ Microbiol 9: 81–92. [DOI] [PubMed] [Google Scholar]
- Huston M. (1979). A general hypothesis of species diversity. Am Natural 113: 81–101. [Google Scholar]
- Inagaki F, Takai K, Nealson KH, Horikoshi K. (2004). Sulfurovum lithotrophicum gen. nov.,sp. nov., a novel sulfur-oxidizing chemolithoautotroph within the epsilon-Proteobacteria isolated from Okinawa Trough hydrothermal sediments. Int J Syst Evol Microbiol 54: 1477–1482. [DOI] [PubMed] [Google Scholar]
- Inagaki F, Takai K, Kobayashi H, Nealson KH, Horikoshi K. (2003). Sulfurimonas autotrophica gen. nov., sp. nov., a novel sulfur-oxidizing epsilon-proteobacterium isolated from hydrothermal sediments in the Mid-Okinawa Trough. Int J Syst Evol Microbiol 53: 1801–1805. [DOI] [PubMed] [Google Scholar]
- Jannasch HW, Wirsen CO. (1979). Chemosynthetic primary production at East Pacific sea floor spreading centers. Bioscience 29: 592–598. [Google Scholar]
- Jannasch HW, Nelson DC, Wirsen CO. (1989). Massive natural occurrence of unusually large bacteria (Beggiatoa sp.) at a hydrothermal deep-sea vent site. Nature 342: 834–836. [Google Scholar]
- Jannasch HW, Wirsen CO, Nelson DC, Robertson LA. (1985). Thiomicrospira crunogena sp. nov., a colorless, sulfur-oxidizing bacterium from a deep-sea hydrothermal vent. Int J Syst Bacteriol 35: 422–424. [Google Scholar]
- Jørgensen BB, Revsbech NP. (1983). Colorless sulfur bacteria, Beggiatoa spp. and Thiovulum spp., in O2 and H2S microgradients. Appl Environ Microbiol 45: 1261–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen BB, Des Marais DJ. (1986). Competition for sulfide among colorless and purple sulfur bacteria in cyanobacterial mats. FEMS Microbiol Ecol 38: 179–186. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. (2005). Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA 102: 2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. (2010). Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12: 118–123. [DOI] [PubMed] [Google Scholar]
- Labrenz M, Jost G, Jurgens K. (2007). Distribution of abundant prokaryotic organisms in the water column of the central Baltic Sea with an oxic-anoxic interface. Aquat Microb Ecol 46: 177–190. [Google Scholar]
- Lesniewski RA, Jain S, Anantharaman K, Schloss PD, Dick GJ. (2012). The metatranscriptome of a deep-sea hydrothermal plume is dominated by water column methanotrophs and lithotrophs. ISME J 6: 2257–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert SJJ, Roos DS. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia P, Duperron S, Philippot P, Foriel J, Susini J, Moreira D. (2003). Bacterial diversity in hydrothermal sediment and epsilonproteobacterial dominance in experimental microcolonizers at the Mid-Atlantic Ridge. Environ Microbiol 5: 961–976. [DOI] [PubMed] [Google Scholar]
- Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B et al. (2010). A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci USA 107: 13479–13484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macalady JL, Dattagupta S, Schaperdoth I, Jones DS, Druschel GK, Eastman D. (2008). Niche differentiation among sulfur-oxidizing bacterial populations in cave waters. ISME J 2: 590–601. [DOI] [PubMed] [Google Scholar]
- Magrane M, , Consortium, U. (2011). UniProt Knowledgebase: a hub of integrated protein data. Database 2011: bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahe F, Rognes T, Quince C, de Vargas C, Dunthorn M. (2015). Swarm v2: highly-scalable and high-resolution amplicon clustering. PeerJ 3: e1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall KT, Morris RM. (2013). Isolation of an aerobic sulfur oxidizer from the SUP05/Arctic96BD-19 clade. ISME J 7: 452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollom TM, Shock EL. (1997). Geochemical constraints on chemolithoautotrophic metabolism by microorganisms in seafloor hydrothermal systems. Geochim Cosmochim Acta 61: 4375–4391. [DOI] [PubMed] [Google Scholar]
- McDermott JM, Ono S, Tivey MK, Seewald JS, Shanks WC, Solow AR. (2015). Identification of sulfur sources and isotopic equilibria in submarine hot-springs using multiple sulfur isotopes. Geochim Cosmochim Acta 160: 169–187. [Google Scholar]
- Meier DV, Bach W, Girguis PR, Gruber-Vodicka HR, Reeves EP, Richter M et al. (2016). Heterotrophic Proteobacteria in the vicinity of diffuse hydrothermal venting. Environ Microbiol 18: 4348–4368. [DOI] [PubMed] [Google Scholar]
- Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J et al. (2003). GenDB - an open source genome annotation system for prokaryote genomes. Nucleic Acids Res 31: 2187–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JL, Akerman NH, Proskurowski G, Huber JA. (2013). Microbiological characterization of post-eruption 'snowblower' vents at Axial Seamount, Juan de Fuca Ridge. Front Microbiol 4: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LR, Rocap G, Chisholm SW. (1998). Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393: 464–467. [DOI] [PubMed] [Google Scholar]
- Muyzer G, Teske A, Wirsen CO, Jannasch HW. (1995). Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol 164: 165–172. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Takai K. (2008). Deep-sea vent chemoautotrophs: diversity, biochemistry and ecological significance. FEMS Microbiol Ecol 65: 1–14. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Takaki Y, Shimamura S, Reysenbach AL, Takai K, Horikoshi K. (2007). Deep-sea vent epsilon-proteobacterial genomes provide insights into emergence of pathogens. Proc Natl Acad Sci USA 104: 12146–12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Takai K, Inagaki F, Hirayama H, Nunoura T, Horikoshi K et al. (2005). Distribution, phylogenetic diversity and physiological characteristics of epsilon-Proteobacteria in a deep-sea hydrothermal field. Environ Microbiol 7: 1619–1632. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Michin PR, O'Hara RB et al. (2013). vegan: Community Ecology Package. R package version 2.0-10. Available at http://CRAN.R-project.org/package=vegan.
- Opatkiewicz AD, Butterfield DA, Baross JA. (2009). Individual hydrothermal vents at Axial Seamount harbor distinct subseafloor microbial communities. FEMS Microbiol Ecol 70: 413–424. [DOI] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. (2010). FastTree 2 - approximately maximum-likelihood trees for large alignments. PLoS ONE 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glockner FO. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentmeier A, Friedrich CG. (2001). The cysteine residue of the SoxY protein as the active site of protein-bound sulfur oxidation of Paracoccus pantotrophus GB17. FEBS Lett 503: 168–172. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM et al. (2009). Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Method 6: 639–641. [DOI] [PubMed] [Google Scholar]
- Reeves EP, Prieto X, Hentscher M, Rosner M, Seewald JS, Hinrichs KU et al. (2011. a). Phase separation, degassing and anomalous methane at the Menez Gwen hydrothermal field. Mineral Mag 75: 1702. [Google Scholar]
- Reeves EP, Yoshinaga MY, Pjevac P, Goldenstein NI, Peplies J, Meyerdierks A et al. (2014). Microbial lipids reveal carbon assimilation patterns on hydrothermal sulfide chimneys. Environ Microbiol 16: 3515–3532. [DOI] [PubMed] [Google Scholar]
- Reeves EP, Seewald JS, Saccocia P, Bach W, Craddock PR, Shanks WC et al. (2011. b). Geochemistry of hydrothermal fluids from the PACMANUS, Northeast Pual and Vienna Woods hydrothermal fields, Manus Basin, Papua New Guinea. Geochim Cosmochim Acta 75: 1088–1123. [Google Scholar]
- Reysenbach AL. (2001) Phylum BI. Aquificae phy. nov. In: Boone DR, Castenholz RW, Garrity GM (eds). Bergey’s Manual of Systematic Bacteriology. Springer: New York, NY, USA. [Google Scholar]
- Richter M, Rossello-Mora R, Oliver Glockner F, Peplies J. (2015). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32: 929–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Lombardot T, Kostadinov I, Kottmann R, Duhaime MB, Peplies J et al. (2008). JCoast - a biologist-centric software tool for data mining and comparison of prokaryotic (meta)genomes. BMC Bioinformatics 9: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxburgh SH, Shea K, Wilson JB. (2004). The intermediate disturbance hypothesis: patch dynamics and mechanisms of species coexistence. Ecology 85: 359–371. [Google Scholar]
- Sauve V, Bruno S, Berks BC, Hemmings AM. (2007). The SoxYZ complex carries sulfur cycle intermediates on a peptide swinging arm. J Biol Chem 282: 23194–23204. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K, Koschinsky A, Garbe-Schönberg D, de Carvalho LM, Seifert R. (2007). Geochemistry of hydrothermal fluids from the ultramafic-hosted Logatchev hydrothermal field, 15 degrees N on the Mid-Atlantic Ridge: temporal and spatial investigation. Chem Geol 242: 1–21. [Google Scholar]
- Schmidtova J, Hallam SJ, Baldwin SA. (2009). Phylogenetic diversity of transition and anoxic zone bacterial communities within a near-shore anoxic basin: Nitinat Lake. Environ Microbiol 11: 3233–3251. [DOI] [PubMed] [Google Scholar]
- Scott SD, Binns RA. (1995). Hydrothermal processes and contrasting styles of mineralization in the western Woodlark and eastern Manus basins of the western Pacific. Geol Soc Lond Spec Publ 87: 191–205. [Google Scholar]
- Seewald JS, Reeves EP, Bach W, Saccocia PJ, Craddock PR, Shanks WC et al. (2015). Submarine venting of magmatic volatiles in the Eastern Manus Basin, Papua New Guinea. Geochim Cosmochim Acta 163: 178–199. [Google Scholar]
- Shah V, Morris RM. (2015). Genome sequence of “Candidatus Thioglobus autotrophica” strain EF1, a chemoautotroph from the SUP05 clade of marine Gammaproteobacteria. Genome Announce 3: e01156–e01115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheik CS, Anantharaman K, Breier JA, Sylvan JB, Edwards KJ, Dick GJ. (2015). Spatially resolved sampling reveals dynamic microbial communities in rising hydrothermal plumes across a back-arc basin. ISME J 9: 1434–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert SM, Hügler M, Taylor CD, Wirsen CO. (2008. a). Sulfur oxidation at deep-sea hydrothermal vents. In: Dahl C, Friedrich CG (eds), Microbial Sulfur Metabolism. Springer: Berlin, Heidelberg, pp 238–258. [Google Scholar]
- Sievert SM, Scott KM, Klotz MG, Chain PS, Hauser LJ, Hemp J et al. (2008. b). Genome of the epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl Environ Microbiol 74: 1145–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski J, Munk C, Lapidus A, Ngatchou Djao OD, Lucas S, Glavina Del Rio T et al. (2010). Complete genome sequence of Sulfurimonas autotrophica type strain (OK10T). Stand Genomic Sci 3: 194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokke R, Dahle H, Roalkvam I, Wissuwa J, Daae FL, Tooming-Klunderud A et al. (2015). Functional interactions among filamentous Epsilonproteobacteria and Bacteroidetes in a deep-sea hydrothermal vent biofilm. Environ Microbiol 17: 4063–4077. [DOI] [PubMed] [Google Scholar]
- Strous M, Kraft B, Bisdorf R, Tegetmeyer HE. (2012). The binning of metagenomic contigs for microbial physiology of mixed cultures. Front Microbiol 3: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunamura M, Higashi Y, Miyako C, Ishibashi J, Maruyama A. (2004). Two Bacteria phylotypes are predominant in the Suiyo seamount hydrothermal plume. Appl Environ Microbiol 70: 1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K, Hirayama H, Nakagawa T, Suzuki Y, Nealson KH, Horikoshi K. (2004). Thiomicrospira thermophila sp. nov., a novel microaerobic, thermotolerant, sulfur-oxidizing chemolithomixotroph isolated from a deep-sea hydrothermal fumarole in the TOTO caldera, Mariana Arc, Western Pacific. Int J Syst Evol Microbiol 54: 2325–2333. [DOI] [PubMed] [Google Scholar]
- Tivey MK. (2004) Environmental conditions within active seafloor vent structures: sensitivity to vent fluid composition and fluid flow. In: Wilcock WSD, DeLong EF, Kelley DS, Baross JA, Cary SC (eds). The Subseafloor Biosphere at Mid-ocean Ridges. American Geophysical Union: Washington D. C., pp 137–152. [Google Scholar]
- Urbach E, Scanlan DJ, Distel DL, Waterbury JB, Chisholm SW. (1998). Rapid diversification of marine picophytoplankton with dissimilar light-harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (Cyanobacteria). J Mol Evol 46: 188–201. [DOI] [PubMed] [Google Scholar]
- Wankel SD, Joye SB, Samarkin VA, Shah SR, Friederich G, Melas-Kyriazi J et al. (2010). New constraints on methane fluxes and rates of anaerobic methane oxidation in a Gulf of Mexico brine pool via in situ mass spectrometry. Deep Sea Res 57: 2022–2029. [Google Scholar]
- Yamamoto M, Arai H, Ishii M, Igarashi Y. (2006). Role of two 2-oxoglutarate:ferredoxin oxidoreductases in Hydrogenobacter thermophilus under aerobic and anaerobic conditions. FEMS Microbiol Lett 263: 189–193. [DOI] [PubMed] [Google Scholar]
- Yarza P, Yilmaz P, Pruesse E, Glockner FO, Ludwig W, Schleifer KH et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12: 635–645. [DOI] [PubMed] [Google Scholar]
- Yeats CJ, Parr JM, Binns RA, Gemmell JB, Scott SD. (2014). The SuSu Knolls hydrothermal field, eastern Manus Basin, Papua New Guinea: an active submarine high-sulfidation copper-gold system. Econ Geol 109: 2207–2226. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.