Abstract
Resistance integrons are bacterial genetic platforms that can capture and express antibiotic resistance genes embedded within gene cassettes. The capture and shuffling of gene cassettes are mediated by the integrase IntI, the expression of which is regulated by the SOS response in Escherichia coli. Gene cassettes are expressed from a common Pc promoter. Despite the clinical and environmental relevance of integrons, the selective forces responsible for their evolution and maintenance are poorly understood. Here, we conducted pairwise competition experiments in order to assess the fitness cost of class 1 integrons in E. coli. We found that integrons are low-cost structures and that their cost is further reduced by their tight regulation. We show that the SOS response prevents the expression of costly integrases whose cost is activity dependent. Thus, when an integron is repressed, its cost depends mostly on the expression of its gene cassettes array and increases with Pc strength and the number of cassettes in the array. Furthermore, different cassettes have different costs. Lastly, we showed that subinhibitory antibiotic concentrations promoted the selection of integron-carrying bacteria, especially those with a strong Pc promoter. These results provide new insights into the evolutionary dynamics of integron-carrying bacterial populations.
Introduction
The development and spread of antibiotic resistance among bacterial pathogens is a major public health issue (Levy and Marshall, 2004). Among the genetic elements involved in this spread, integrons are especially important, given their ability to capture and express resistance genes embedded within gene cassettes (Stokes and Hall, 1989).
Integrons are composed of three key elements: (i) an intI gene encoding an integrase that mediates the insertion, excision and shuffling of gene cassettes; (ii) an attI recombination site where cassettes are inserted; and (iii) a Pc promoter responsible for gene cassette expression (Cambray et al., 2010). Several integron classes have been defined according to their integrase amino acid sequence (Mazel, 2006). Among them, class 1 integrons are highly prevalent in antibiotic-resistant Gram-negative bacteria (Escudero et al., 2015).
In class 1 integrons, intI1 gene expression is regulated by the SOS response (Guerin et al., 2009), a bacterial stress response that can be triggered by various antibiotics (Baharoglu and Mazel, 2011). This regulation involves a specific repressor, LexA protein, which prevents intI1 gene transcription. Upon activation of the SOS response by a stress generating single-strand DNA, the LexA repressor is cleaved and integrase expression is thereby enabled.
Gene cassettes are expressed from the common promoter Pc; at least 13 different Pc variants have been described in class 1 integrons. Four variants predominate, namely PcW, PcH1, PcWTGN-10 and PcS, from the weakest to the strongest (Jové et al., 2010). Moreover, the closer a gene cassette is to the Pc, the stronger its expression (Collis and Hall, 1995). As Pc is located within the intI1 gene, its polymorphism impacts the IntI1 integrase amino acid sequence. Three IntI1 variants are thus associated with the four Pc variants: IntI1R32_H39 (PcW or PcH1), IntI1P32_H39 (PcWTGN-10) and IntI1R32_N39 (PcS). A negative correlation between the strength of the Pc and the activity of its associated integrase has been shown: the weaker the Pc, the more active the integrase (Jové et al., 2010).
Relatively few studies have focused on the evolution dynamics of class 1 integrons, with the exception of studies assessing the effect of antibiotic pressure on gene cassette reshuffling (Barraud and Ploy, 2015; Pérez-Valdespino et al., 2016). Rearrangement of a gene cassette to a position closer to the Pc relies mainly on the excision of upstream cassettes rather than excision and reintegration of the cassette just ahead of the Pc (Barraud and Ploy, 2015). This suggests that gene cassette rearrangements via excision are probably more cost-effective than excision and integration of a distal gene cassette closer to the Pc. Indeed, the ‘cut and paste’ mechanism leading to excision and reintegration of a gene cassette in the first position would require two recombination events, whereas excision of upstream cassettes requires only one recombination event. This raises questions as to the fitness cost associated with integrase activity and gene cassette expression.
Few studies have assessed the fitness cost of integrons. In particular, Starikova et al. (2012) assessed the impact of class 1 integrons in Acinetobacter baylyi. They showed that integron acquisition strongly reduced the fitness of A. baylyi, due to an active integrase and that inactivated integrases rapidly emerged to compensate for this biological cost. These data were used to create a mathematical model simulating the evolution of an integron-carrying bacterial population in response to fluctuating antibiotic pressure (Engelstadter et al., 2016). The main conclusion was that gene cassette reshuffling, necessary to tolerate the fluctuating antibiotic pressure, could compensate for the integrase cost, enabling the maintenance of functional integrases within bacterial populations. However, both studies relied on data retrieved from Acinetobacter which lacks LexA protein (Hare et al., 2012). Yet, evidence of SOS regulation was found for most integrons (Cambray et al., 2011).
The aim of the present study was thus to assess the fitness cost of class 1 integrons in E. coli, a species in which integrase expression is regulated by the SOS response. We show that class 1 integrons are low-cost structures and that their cost is further reduced by the repressive system. LexA protein prevents the expression of costly integrases, whose cost is linked to their catabolic activity. We also show that the fitness of a given integron is dependent both on Pc polymorphism and the gene cassettes content. Finally, we show that subinhibitory antibiotic concentrations can select integron-carrying bacteria. These results provide new insights into the evolutionary dynamics of integron-carrying bacterial populations.
Materials and methods
Bacteria and plasmids
The bacteria and plasmids used in this study are listed in Table 1. Strains were grown in Luria-Bertani broth (LB) (MoBio Laboratories, Carlsbad, CA, USA) or Davis Minimal (DM) broth (Sigma Aldrich, St Louis, MO, USA) supplemented with 25 μg ml−1 glucose (DM25 medium). Kanamycin (Km, 25 mg l−1) and tobramycin (Tob, various concentrations) were added when necessary.
Table 1. Strains and plasmids.
| No. | Strains or plasmids | Genotype or description | Source |
|---|---|---|---|
| Strains | |||
| Escherichia coli MG1656 | MG1655 (K-12F– λ– ilvG– rfb-50 rph-1) ΔlacMluI | Espéli et al. (2001) | |
| Escherichia coli MG1656 λatt::gfp | MG1656 strain containing the gfpmut3 gene at the λatt site | This study | |
| Escherichia coli MG1656 Δcpxl::km-frt | cpxl gene replaced by the km-frt cassette in strain MG1656 | Beloin et al. (2004) | |
| Plasmids | |||
| pKOBEGA | λ red recombination system expression plasmid | Chaveroche et al. (2000) | |
| pCP20 | FLP flippase expression plasmid | Cherepanov and Wackernagel (1995) | |
| pZE1-gfpmut3 | gfpmut3 gene under the control of the constitutive pλR promoter | Gift from C Guet | |
| pZE-km-frt-gfpmut3 | km-frt cassette amplified from MG1656Δcpxl::km-frt using primers 7-8 (Supplementary Table S1) and cloned into pZE1R-gfpmut3 (SacI–XhoI). | This study | |
| p1W | integron intI1-aac(6′)-Ib-dfrA15-aadA1-catB9 with PcW promoter cloned into pSU38ΔtotlacZ | Barraud and Ploy (2015) | |
| p1S | integron intI1-aac(6′)-Ib-dfrA15-aadA1-catB9 with PcS promoter cloned into pSU38ΔtotlacZ | Barraud and Ploy (2015) | |
| p1WTGN-10 | integron intI1-aac(6′)-Ib-dfrA15-aadA1-catB9 with PcWTGN-10 promoter cloned into pSU38ΔtotlacZ | Barraud and Ploy (2015) | |
| p16Sa | integron intI1-aac(6′)-Ib-dfrA15-aadA1-catB9-aac(6′)-Ib-dfrA15-aadA1-catB9 cloned into pSU38ΔtotlacZ | Barraud and Ploy (2015) | |
| p8741 | pBAD::intI1Y312F | Loot et al. (2014) | |
| pZA2 and derivatives | |||
| 1 | pZA2 | pZA2 plasmid, Kmr | Lutz and Bujard (1997) |
| 2 | pW-4GC | integron intI1-aadA1-catB9-aac(6′)-Ib-dfrA15 with PcW promoter cloned into pZA2 | This study |
| 3 | pH1-4GC | integron intI1-aadA1-catB9-aac(6′)-Ib-dfrA15 with PcH1 promoter cloned into pZA2 | This study |
| 4 | pWTGN-10-4GC | integron intI1-aadA1-catB9-aac(6′)-Ib-dfrA15 with PcWTGN-10 promoter cloned into pZA2 | This study |
| 5 | pS-4GC | integron intI1-aadA1-catB9-aac(6′)-Ib-dfrA15 with PcS promoter cloned into pZA2 | This study |
| 6 | pW PintI*-4GC | pW-4GC with inactivated PintI promoter (no intI1 expression) | This study |
| 7 | pH1 PintI*-4GC | pH1-4GC with inactivated PintI promoter (no intI1 expression) | This study |
| 8 | pWTGN-10 PintI*-4GC | pWTGN-10-4GC with inactivated PintI promoter (no intI1 expression) | This study |
| 9 | pS PintI*-4GC | pS-4GC with inactivated PintI promoter (no intI1 expression) | This study |
| 10 | pW*PintI*-4GC | pW-4GC with inactivated Pc promoter and inactivated PintI promoter (no intI1 expression nor cassette expression) | This study |
| 11 | pW*-4GC | pW-4GC with inactivated PcW promoter (silent mutation, IntI1R32-H39 integrase variant, no cassette expression) | This study |
| 12 | pWTGN-10*-4GC | pWTGN-10-4GC with inactivated PcWTGN-10 promoter (silent mutation, IntI1P32-H39 integrase variant, no cassette expression) | This study |
| 13 | pS*-4GC | pS-4GC with inactivated PcS promoter (silent mutation, IntI1R32-N39 integrase variant, no cassette expression) | This study |
| 14 | pW*lexA*-4GC | pW-4GC with inactivated PcW promoter (no cassette expression) and IntI1R32-H39 constitutive expression (mutated LexA box) | This study |
| 15 | pWTGN-10*lexA*-4GC | pWTGN-10-4GC with inactivated PcWTGN-10 promoter (no cassette expression) and IntI1P32-H39 constitutive expression (mutated LexA box) | This study |
| 16 | pS*lexA*-4GC | pS-4GC with inactivated PcS promoter (no cassette expression) and IntI1R32-N39 constitutive expression (mutated LexA box) | This study |
| 17 | pWY312F*lexA*-4GC | pW* lexA*-4GC encoding an inactive integrase (catalytic amino acid Y312 mutated) | This study |
| 18 | pS-2GC | integron intI1-aadA1-catB9 with PcS promoter cloned into pZA2 | This study |
| 19 | pW*PintI*-2GC | pS-2GC with inactivated Pc promoter and inactivated PintI promoter (no intI1 expression nor GCs expression) | This study |
| 20 | pS-aadA1 | integron intI1-aadA1 with PcS promoter cloned into pZA2 | This study |
| 21 | pS-catB9 | integron intI1- catB9 with PcS promoter cloned into pZA2 | This study |
| 22 | pS-aac(6′)-Ib | integron intI1- aac(6′)-Ib with PcS promoter cloned into pZA2 | This study |
| 23 | pS-dfrA15 | integron intI1- dfrA15 with PcS promoter cloned into pZA2 | This study |
| 24 | pW*PintI*-aadA1 | pW-aad with inactivated Pc promoter and inactivated PintI promoter (no intI1 expression nor GCs expression) | This study |
| 25 | pW-aac(6′)-Ib | integron intI1- aac(6′)-Ib with PcW promoter cloned into pZA2 | This study |
Abbreviation: GC, gene cassette. Bold was used to highlight the change of Pc variant between constructions.
All integrons were generated by assembly PCR, as described in Supplementary Figure S1, and cloned into a pZA2 backbone via the XhoI and BamHI restriction sites.
Construction of the MG1656 λatt::gfp strain
The MG1656 λatt::gfp strain, expressing green fluorescent protein (GFP), was constructed by integrating a km-frt-gfpmut3 cassette at the E. coli λatt site in a three-step PCR-based approach as previously described (Cherepanov and Wackernagel, 1995). Primers used are available in Supplementary Table S1.
Determination of minimal inhibitory concentrations
MICs were determined by broth dilution with an inoculum of 105 CFU ml−1 in DM25 medium. Each test was done at least three times.
Competition experiments
The fitness of the MG1656 strain carrying a given plasmid, relative to the MG1656 λatt::gfp strain containing another plasmid, was estimated in pairwise competition experiments (Lenski et al., 1991). Briefly, independent pre-cultures of both competitors were grown in DM25 overnight and mixed at a 1:1 volume ratio the following day. Fresh DM25 cultures were then inoculated with this mix performing a 100-fold dilution. Cultures were then propagated for 4 days with a daily 100-fold dilution in fresh medium. Samples were taken each day to follow the relative frequency of the two competitors by flow cytometry.
Flow cytometry
The relative frequency of GFP bacteria was determined with a BD FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). Measurements were performed directly on bacterial cultures. Bacterial cells were gated on the basis of their distribution on forward vs side scatter plots. GFP bacteria were then separated from non-GFP bacteria by using the fluorescent marker. Green fluorescence was collected in the FL1 channel (530±15 nm). Thresholds were applied manually to determine the boundaries between the two populations, and remained constant throughout all the experiments. The relative frequency of the two competitors was calculated by counting 10 000 bacterial cells. All measurements were made in triplicate. Controls were performed to assess the level of false positives and false negatives in GFP quantification. Both were shown to be negligible and constant throughout independent daily measurements (Supplementary Data and Supplementary Figure S2).
Calculating the selection coefficient
For each competition, the selection coefficient of the non-GFP strain was determined from the slope of the regression ln [freqnonGFP/freqGFP] plotted against the time course in generations (Lenski et al., 1991). Experiments were performed in at least 12 replicates. A single regression line was then fitted through the points from all replicates (Schrag et al., 1997; Rodríguez-Verdugo et al., 2013). Reported standard errors and P-values were calculated for the regression coefficient. Error bars correspond to the 99% confidence interval. The fluorescent reporter used in the MG1656 λatt::gfp strain had no cost when compared with the MG1656 parental strain (Supplementary Data and Supplementary Table S2).
Results
Effect of Pc polymorphism on the cost of class 1 integrons in E. coli
We first assessed the impact of Pc polymorphism on the cost of class 1 integrons in E. coli by performing pairwise competition experiments between strains carrying and lacking the integron (Supplementary Figure S3A). Integrons differing only by their Pc sequence were used (plasmids 2–5; Table 1).
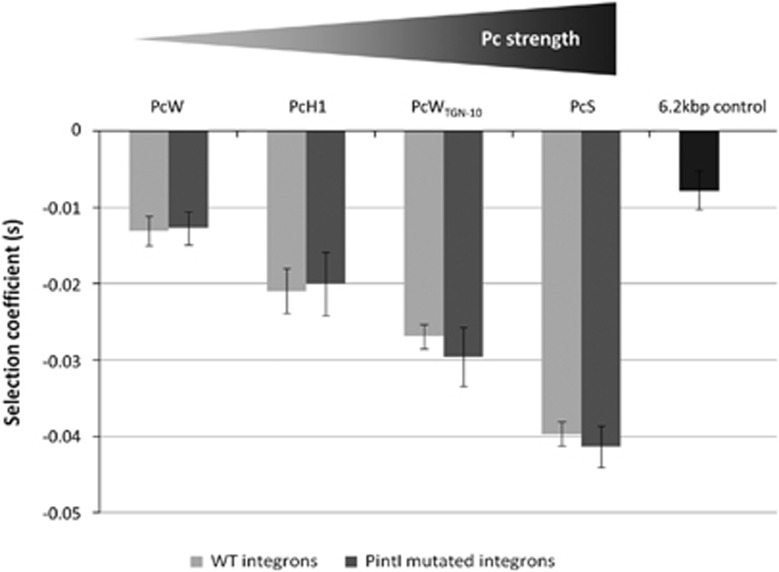
Whatever the Pc variant, integrons appeared to be costly for E. coli (Figure 1, light gray). The selection coefficients of the integron-carrying strains were all negative, ranging from −0.013±0.002 to −0.04±0.003 (all P-values <1 × 10−10), meaning that the integrons slowed bacterial growth by 1.3–4%. Interestingly, the cost of class 1 integrons appeared to correlate with the Pc strength: the stronger the Pc, the costlier the integron (Figure 1). This correlation was associated with higher resistance level as we observed that stronger Pc variants (PcS and PcWTGN-10) exhibit higher resistance levels (Table 2).
Figure 1.
Effect of Pc polymorphism on the cost of a class 1 integron in E. coli. MG1656 strains containing plasmid-borne integrons varying only by their Pc sequence were placed in competition with the MG1656 λatt::gfp+pZA2 strain (Supplementary Figure S3A). Experiments were performed with either wild-type (WT) integrons (plasmids 2–5 in Table 1) or PintI-mutated integrons (no intI1 expression, plasmids 6–9 in Table 1). The 6.2 kbp control plasmid (no intI1 expression or cassette expression, plasmid 10 in Table 1) was used to evaluate the effect of plasmid size between plasmid pZA2 (2.1 kbp) and the integron-carrying plasmids (6.2 kbp).
Table 2. MICs of MG1656 strains containing the four gene cassettes with different Pc variants.
|
MIC (μg ml−1) |
||||
|---|---|---|---|---|
| SPT | CHL | TOB | TMP | |
| MG1656+pZA2 | 2 | 4 | 0.25 | − |
| MG1656+pW-4GC | 128 | 16 | 4 | − |
| MG1656+pH1-4GC | 256 | 32 | 8 | − |
| MG1656+pWTGN-10-4GC | 512 | 64 | 16 | − |
| MG1656+pS-4GC | 512 | 128 | 16 | − |
Abbreviations: CHL, chloramphenicol; SPT, streptomycin; TMP, trimethoprim; TOB, tobramycin. Trimethoprim MICs were not determined due to solubility issues in DM25 medium.
We further examined whether or not the observed cost was due solely to gene cassette expression. We therefore performed competition experiments using the same above-mentioned plasmids but with mutations in the PintI promoter which prevented intI1 expression (plasmids 6–9; Table 1) (Guérin et al., 2011). The overall cost of the integron was similar whether PintI was mutated or not, suggesting that, in wild-type conditions in which intI1 is repressed, the fitness cost of an integron is due to gene cassettes expression (Figure 1).
The strain with plasmid 10 was used as a control to assess the contribution of the 6.2 kpb plasmid to host fitness. In this plasmid, the PintI and Pc promoters were both mutated in order to relate its cost to its size alone (no expression of the intI1 gene and gene cassettes; Guérin et al., 2011). The selection coefficient for this plasmid (s=−0.008±0.003, P<1 × 10-8) showed that the 6.2 kbp plasmidic backbone itself slowed bacterial growth by 0.8% (Figure 1).
The IntI1 fitness cost is related to integrase catalytic activity
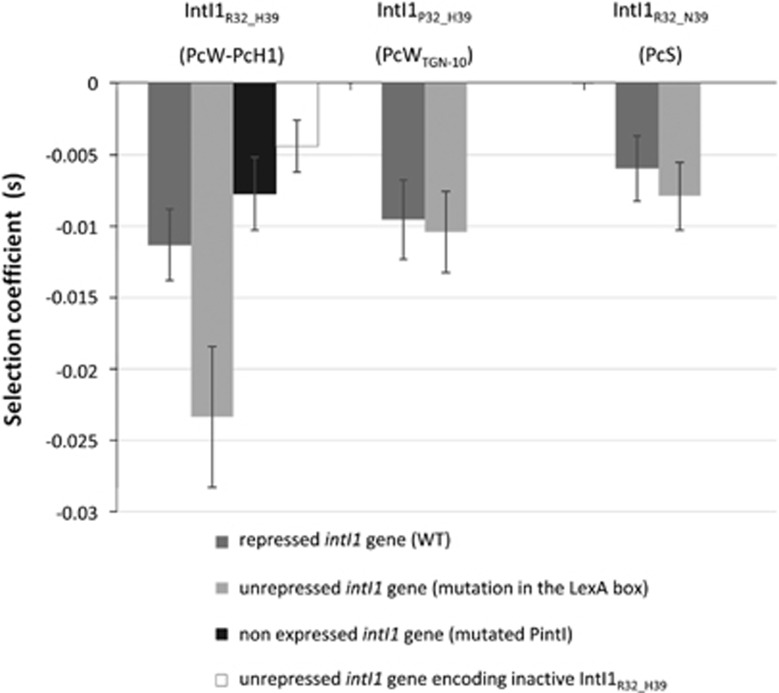
To assess the fitness cost of integrase expression, we used integron-carrying MG1656 strains in which the Pc promoter was mutated, thus preventing gene cassette expression without affecting the IntI1 protein sequence (plasmids 11–16; Table 1) (Guérin et al., 2011). The integrons contained either the normally repressed intI1 gene (plasmids 11–13) or the unrepressed intI1 gene (mutated LexA binding site) (plasmids 14–16). Competition experiments were performed with each IntI1 protein variant: IntI1R32_H39 (associated with the PcW or PcH1 promoter), IntI1P32_H39 (associated with PcWTGN-10) and IntI1R32_N39 (associated with PcS).
Whatever the Pc variant, when the intI1 gene was repressed, its cost was not different from the control in which the PintI promoter was mutated (Figure 2). This confirmed that when intI1 is repressed, the observed cost of an integron is only due to gene cassettes expression. When unrepressed (mutated LexA binding site), a twofold increase in fitness cost was observed with the integron encoding IntI1R32_H39 integrase (the integrase associated with the PcW sequence), rising from −0.011±0.002 when repressed to −0.023±0.004 when unrepressed (P<1 × 10−10; Figure 2). Interestingly, IntI1R32_H39 is also the most active integrase (Jové et al., 2010). To determine whether the cost of this integrase was related to its catalytic activity, we used the MG1656 strain carrying an integron with a mutated intI1 gene encoding a non-functional IntI1R32_H39 variant (IntI1Y312F) (plasmid 17; Table 1). The Y312F mutation was sufficient to abrogate the cost associated with the IntI1R32_H39 integrase (Figure 2).
Figure 2.
The cost of integrase expression. The MG1656 λatt::gfp+pZA2 strain was placed in competition with MG1656 strains carrying plasmidic integrons with a mutated Pc promoter (no gene cassette expression, plasmids 11–17; Table 1). Mutations were silent, so as not to affect the IntI1 protein variant encoded by the intI1 gene. The intI1 gene was either repressed (plasmids 11–13) or unrepressed (plasmids 14–17). Derepression of the intI1 gene was achieved by mutating the LexA box and thus preventing repressor binding. Plasmid 17 encoded an inactive integrase. Plasmid 10, with neither intI1 nor gene cassette expression (Table 1), was used as a control.
Influence of the cassette array contents
As cassette arrays can vary by their size and gene content, we wondered whether these two parameters might impact the global cost of integrons.
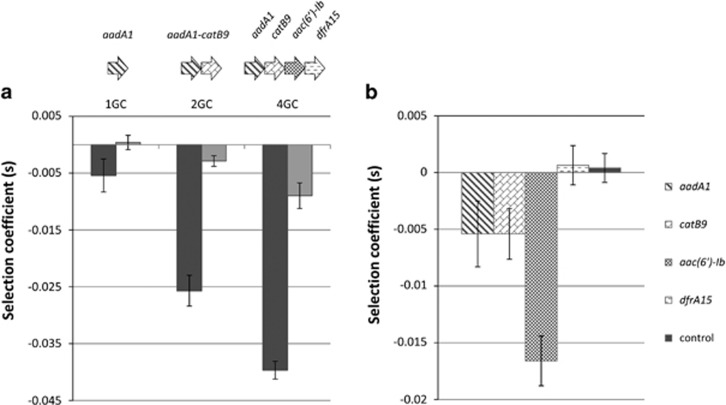
We first analyzed the fitness cost in MG1656 strains carrying integrons with one, two or four gene cassettes under the control of the strong promoter variant PcS (plasmids 20, 18 and 5). As controls, plasmids 10, 19 and 24, in which the Pc and PintI promoters were both mutated, were used to evaluate the inherent cost related to plasmid size. We found that the cost of an integron increased with the number of gene cassettes (Figure 3a), rising eightfold from one to four gene cassettes. A minor part of this cost was related to plasmid size (Figure 3a, light gray).
Figure 3.
Effect of the gene cassettes array content. (a) Effect of cassettes network size. MG1656 strains containing plasmidic integrons, varying only by the number of gene cassettes (GC) they carried (plasmids 5, 18 and 20; Table 1), were placed in competition with the MG1656 λatt::gfp+pZA2 strain (Supplementary Figure S3B). For each condition (1, 2 or 4 gene cassettes), control plasmids with neither intI1 nor cassette expression (plasmids 10, 19 and 24; Supplementary Table S1) were used to evaluate the inherent cost of carrying this plasmid size (light gray). (b) Influence of the nature of the gene cassettes. Competition experiments were performed between the MG1656 λatt::gfp+pZA2 strain and MG1656 strains carrying plasmids with a one-cassette integron (plasmids 20–23; Table 1) (Supplementary Figure S3C). The cassette was aadA1, catB9, aac(6′)-Ib or dfrA15. Plasmid 24 was used as a control to evaluate the carriage cost of this plasmid size.
To estimate the influence of the nature of the gene cassettes, we performed competition experiments with MG1656 strains carrying an integron in which only one cassette was fully expressed from the strong Pc variant. Four gene cassettes were tested: aadA1, catB9, aac(6′)-Ib and dfrA15 (plasmids 20–23; Table 1). As shown in Figure 3b, the cost of an integron was found to be dependent on the gene cassette it contained. The aac(6′)-Ib gene cassette was the costliest, with a selection coefficient of −0.017±0.002 (P<1 × 10−10). Cassettes catB9 and aadA1 were slightly detrimental, with respective costs of −0.005±0.002 (P<1 × 10-8) and −0.005±0.003 (P<1 × 10−10). Given the detection limit of our approach (detection of fitness differences ⩾|2 × 10−3|), the dfrA15 gene cassette appeared to bear no cost.
Impact of subinhibitory antibiotic concentrations
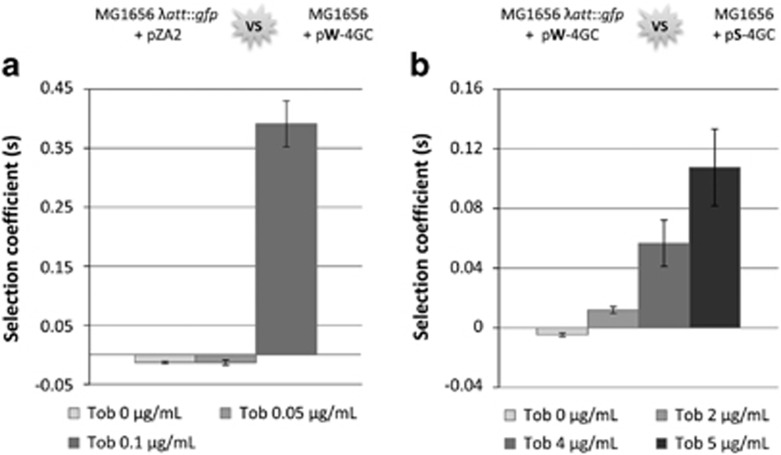
To study whether subinhibitory antibiotic concentrations favor bacteria carrying integrons with appropriate resistance gene cassettes, we analyzed the impact of a subinhibitory tobramycin (Tob) concentration on the cost of a four-cassettes integron containing the aac(6′)-Ib cassette in third position within the array (MIC 4 μg ml−1). Competition experiments between resistant bacteria carrying the integron and susceptible strain lacking the integron were performed in the presence of various subinhibitory Tob concentrations. The chosen concentrations were 2.5 (0.1 μg ml−1) and 5 times (0.05 μg ml−1) lower than the MIC of the strain without the integron (MIC 0.25 μg ml−1).
No effect was observed at 0.05 μg ml−1 Tob but, at 0.1 μg ml−1, bacteria carrying the integron appeared superfit (s=0.39±0.04, P<1 × 10−10; Figure 4a). They grew 1.39 times faster than their integron-less counterparts, showing that subinhibitory antibiotic concentrations can favor the selection of bacteria containing an integron with an appropriate resistance gene and thereby promote the maintenance and dissemination of integrons.
Figure 4.
Effect of subinhibitory antibiotic concentrations on integron carriage. (a) Integron with PcW. Competition experiments were performed between GFP bacteria without an integron (MG1656 λatt::gfp+pZA2) and non-GFP bacteria carrying an integron (MG1656+plasmid 2; Table 1). Different subinhibitory tobramycin (Tob) concentrations were added to the growth media during competition. These concentrations were chosen to be below the MIC of the most susceptible competitor (0.25 μg ml−1). (b) Integrons with different Pc. Competition experiments were performed between two bacteria carrying the same integron except for the Pc variant, one with a weak Pc promoter (GFP bacteria with plasmid 25; Table 1) and the other with a strong Pc promoter (non-GFP bacteria with plasmid 22; Table 1) (Supplementary Figure S3D). Subinhibitory Tob concentrations below the MIC of the more susceptible competitor (8 μg ml−1) were added to the growth media during competition.
To determine whether subinhibitory antibiotic concentrations could favor the maintenance of integrons with given promoter variants, we performed direct competition experiments between bacteria carrying integrons with either a weak Pc (PcW; plasmid 25, Table 1) or a strong Pc (PcS; plasmid 22, Table 1 and Supplementary Figure S3D), in the absence and presence of subinhibitory Tob concentrations (Figure 4b). Again, the chosen Tob concentrations were below the MIC of the most susceptible bacteria (8 μg ml−1).
In the absence of tobramycin, bacteria carrying the integron with the weak Pc were fitter than those carrying the integron with the strong Pc. However, when tobramycin was added, the selection coefficient became positive, showing that the bacteria carrying the integron with the strong Pc became fitter than those with the weak Pc. Tob concentrations from 2 to 5 μg ml−1 were tested. Selection coefficients rose from 0.012±0.002 (P<1 × 10−10) at the lowest Tob concentration to 0.107±0.003 (P<1 × 10−10) at the highest Tob concentration, meaning that bacteria carrying the integron with the strong Pc grew 1.2–10.7% faster, depending on the antibiotic concentration (Figure 4b). These results show how integrons associated with strong Pc variants confer a major fitness advantage in the presence of subinhibitory antibiotic concentrations.
Discussion
This study shows that class 1 integrons are low-cost structures in E. coli. A previous meta-analysis involving 760 chromosomal resistance mutations and 49 resistance plasmids suggested that the average cost of chromosomal and plasmid-borne resistance was equivalent to a 21–9% decline in host fitness, respectively (Vogwill and MacLean, 2015). Thus with a 1.3–4% decrease in the host fitness, integron-mediated resistance would be 2–7 times less costly than plasmidic resistance, and 5–15 times less costly than chromosomal resistance. Furthermore, in this study we used a p15a replicon yielding approximately 15 plasmid copies per bacterial cell. In clinical and environmental settings, class 1 integrons are found mainly in low copy plasmids. Roughly, their cost might even be five times smaller for a natural integron-bearing plasmid with three copies such as the R388 plasmid (Avila and de la Cruz, 1988). In return for this very low fitness cost, integrons would allow E. coli to adapt more readily to antibiotic pressure.
This very low cost could explain why integrons are highly prevalent in E. coli and, probably, in many other Enterobacteriaceae (Ahmed et al., 2005; Guerra et al., 2006; Rao et al., 2006). Dissemination alone cannot explain the success of integrons. Indeed, while dissemination has contributed to expanding the host range of integrons, newly acquired genetic elements are not necessarily maintained in bacterial populations. For instance, a previous study showed that various IncA/C plasmids harboring the blaCMY-2 resistance gene were lost in the absence of antibiotic selective pressure (Subbiah et al., 2011). Likewise, selective pressure alone cannot explain integron maintenance in bacterial populations. Indeed, integrons have been found in environments preserved from the effects of human activity (Wright et al., 2008; Gaze et al., 2011). The very low cost of integrons might therefore have favored their maintenance and spread in bacterial populations, notably in antibiotic-free environments. Moreover, this would help bacteria to retain resistance cassettes in the absence of antibiotic pressure, keeping them readily available in the event of antibiotic stress.
The low cost of class 1 integrons in E. coli could be explained by the tight regulation of integrase expression. Indeed, we found that LexA-mediated repression of intI1 expression significantly lowered the cost of integrons (Figure 2). Regulatory systems that reduce the cost of resistance determinants have already been described. In Enterococcus for instance, the cost of the vancomycin resistance operon is lowered by a tight regulatory mechanism (Foucault et al., 2010). Previously, the regulation of a tetracycline resistance operon derived from a Tn10 transposon was found to abrogate the cost of the structure in E. coli (Nguyen et al., 1989). LexA regulation of integrase expression could therefore be an adaptive response designed to lower the cost of integrons and thereby to favor their maintenance and dissemination.
LexA-mediated repression of integrase expression might also be an adaptive response designed to prevent the loss of a costly protein. Truncated and inactivated intI1 genes are commonly found in bacteria (Gillings et al., 2005; Nemergut et al., 2008). This loss of integrase functionality could be due to the burden they represent for the host. Yet, in silico evolutionary studies have shown the importance of integrase integrity and functionality for adaptation to environmental challenges (Engelstadter et al., 2016). Interestingly, an in silico study correlated the loss of integrase functionality with the absence of LexA-mediated regulation in bacteria, suggesting that the integrase would be lost when intI1 gene expression was not regulated by the SOS response (Cambray et al., 2011). Further evidence of the protective role of LexA regulation came from evolutionary experiments with integron-containing A. baylyi, a species lacking the LexA repressor, in which frameshift mutations rapidly emerge to inactivate IntI1 protein and to restore host fitness (Starikova et al., 2012). LexA repression could therefore have been needed to prevent the loss of a costly but adaptively important gene.
Interestingly, we found that the cost of a given integrase was related to its catalytic activity (Figure 2): integrase with high activity (IntI1R32_H39) was costlier than those with low activity (IntI1P32_H39 and IntI1R32_N39). Furthermore, a single mutation in the catalytic amino acid codon of this highly active integrase was sufficient to abolish its cost. It has been proposed that the cost of an integrase may be related to the genomic instability it confers (Starikova et al., 2012). Even though the IntI1 integrase mostly catalyzes recombination reactions between attI and attC sites carried by integrons and cassettes, it can also recognize non-canonical sites in the host genome and thereby create genomic instability (Recchia et al., 1994). Recent work has notably highlighted the importance of attI × attI recombination in vivo. Despite having a lower recombination rate when compared with attI × attC reaction, attI × attI recombination involves sites that do not require a proper folding to become recombinogenic (in contrast with attC sites; Escudero et al., 2016). Thus, attI × attI reactions could be more prevalent than previously thought. As a GNT is sufficient to become a secondary attI site (Recchia et al., 1994), such recombination events are likely to be very important in integrase-mediated genome instability. In this context, a more active integrase would generate more recombination events and create more instability, thus being more deleterious for the host.
Our results also demonstrate that the cost of class 1 integrons correlates with the strength of the Pc promoter (Figure 1): the stronger the Pc (and thus the expression level of antibiotic resistance), the costlier the integron. Epidemiological studies revealed a high prevalence of integrons associated with weak Pc variants (PcW and PcH1) in Enterobacteriaceae (Vinué et al., 2011; Wei et al., 2013). This high prevalence could be explained by the observation that integrons with a weak Pc, encoding high-activity integrases (Jové et al., 2010), are adaptively advantageous. Indeed, more efficient integrases would allow bacteria to adapt more efficiently to challenging environments by acquiring new gene cassettes. Besides, we show that integrons associated with weak Pc variants are also the least costly. Thus, weak Pc promoters would be doubly advantageous, favouring both integron maintenance and environmental adaptation.
Furthermore, the gene cassette contents also affect the cost of class 1 integrons: the higher the number of cassettes in the array, the costlier the integron (Figure 3a). This observation is consistent with previous reports on the cost of plasmidic resistance, showing that the average cost of a plasmid increases with the number of resistance genes it carries, independently of the size of the plasmid (Vogwill and MacLean, 2015). This could explain why class 1 integrons usually carry a limited number of gene cassettes (2 or 3 on average, and up to 10; Integrall database, http://integrall.bio.ua.pt/). However, this correlation between the cost of an integron and its size has to be balanced. Indeed, it has been shown that the level of gene cassette expression is directly dependent on its distance from the Pc promoter (Collis and Hall, 1995) and that the expression level is reduced by the action of attC secondary structure which impedes translation activity (Jacquier et al., 2009). Thus, beyond a certain number of cassettes, host fitness might increase only slightly as further cassettes are acquired.
The gene cassette content also seems to impact host fitness. In a model of a one-cassette integron, we found that different gene cassettes had different costs, aadA1 and dfrA15 being the least costly and aac(6′)-Ib the costliest (Figure 3b). Several explanations for these differences can be proposed. This could be linked to the recombinogenic activity of the attC sites. The low-cost dfrA15 and aadA1 gene cassettes, with their respective T-N6-C and T-N6-G extrahelical base motifs, are supposed to be highly recombinogenic (Larouche and Roy, 2011). In contrast, the costly aac(6′)-Ib gene cassette with the TC-N6-G attC site exhibiting low affinity for IntI1 might have the lowest recombination rate. Furthermore, previous studies have shown that the length of an attC site and the structure of its VTS (variable terminal sequence) can prevent proper folding of an attC site (Loot et al., 2014). Thus, in our study, the following order of gene cassette recombination rates should be observed: aadA1 (favorable extrahelical base motifs and short attC site)>dfrA15 (favorable extrahelical base motifs but longer attC site) >>> catB9 (extrahelical base motifs and long attC site)≈aac(6′)-Ib (very unfavorable extrahelical base motifs and moderate attC size). However, given the cost of each cassette (aac(6′)-Ib>catB9=aadA1>dfrA15), no clear correlation emerges between the recombinogenic activity of the attC site and the cost of the gene cassette. Another possibility is that the observed differences are related to the nature of the encoded resistance mechanisms. Indeed, a meta-analysis of chromosomal resistance mutations showed that the average cost of various resistance mechanisms could differ significantly (Vogwill and MacLean, 2015). It has been reported that the prevalence of dfr genes in clinical isolates remained unchanged for 2 years despite an 85% reduction in trimethoprim prescriptions, suggesting that these genes are highly stable even in the absence of selective pressure (Brolund et al., 2010). We infer that the family of dfr genes might have particularly low cost. On the other hand, we found that the catB9 and aac(6′)-Ib gene cassettes, which both encode acetyltransferases and have similar attC recombinogenic activities, had different costs (Figure 3b). Some acetyltransferases involved in aminoglycoside acetylation are able to acetylate peptidoglycans in vitro (Payie and Clarke, 1997). Thus, other putative targets of AAC(6′)-Ib protein might affect bacterial metabolism and thereby entail a higher cost.
We found that the dfrA15 and aadA1 gene cassettes entailed a negligible fitness cost. Interestingly, the aadA and dfr gene cassette families are highly prevalent in class 1 integrons (Vasilakopoulou et al., 2009; Vinué et al., 2011; Kheiri and Akhtari, 2016), reaching 63% in E. coli class 1 integrons (data from the Integrall database), suggesting that their low cost could favor their maintenance in cassette networks. By contrast, aac gene cassettes, including the costly aac(6′)-Ib cassette, are only found in 11% of E. coli class 1 integrons (Integrall database).
The different costs of gene cassettes also raise the question of the impact of the order of the cassettes in a given array. A costly cassette located in first position, close to the Pc and therefore strongly expressed, would be highly deleterious for the host bacterium. By contrast, transfer of a low-cost cassette to first position might reduce the overall cost of the gene cassette array. An integron could therefore reduce its overall cost by reshuffling the gene cassettes into a more cost-effective order. This hypothesis is supported by the fact that, when present, low-cost dfr cassettes are often found in first position (70% of the time according to the Integrall database). However, other factors are also known to influence the position of gene cassettes. For example, some cassettes lack an efficient translation initiation region (TIR) (Hanau-Berçot et al., 2002) and would need to be in first position in order to be properly expressed, relying on the translation of a short ORF11 peptide encoded by the attI site (Hanau-Berçot et al., 2002). This would explain why, despite their cost, aac(6′)-Ib cassettes lacking an efficient TIR are often found in first position (60% of the time when present, Integrall).
Lastly, our experiments indicate that subinhibitory tobramycin concentrations favor the maintenance and enrichment of bacterial populations carrying an integron with the appropriate resistance gene cassette (Figure 4a). Furthermore, we found that integrons associated with strong Pc variants were fitter than those associated with weak Pc variants (Figure 4b). These findings are in agreement with a previous study showing that antibiotic concentrations up to several hundred times below the MIC can led to enrichment in resistant bacteria (Gullberg et al., 2011).
Altogether, these results reveal the cost-effectiveness of integrons in E. coli. In return for a low fitness cost, they allow their host bacteria to adapt readily to antibiotic pressure. This low cost would be due to the SOS repression system, which prevents the expression of a costly integrase. The low cost of integrons would also explain their success and prevalence in many bacterial species, and would make them easily selected in the presence of subinhibitory antibiotic concentrations.
Acknowledgments
We thank C Ouk from the CIM technical platform (University of Limoges) for technical support with flow cytometry, M Baltazar for the MG1656 λatt::gfp strain construct, C Guet and D Mazel for respectively providing us with the pZE1-gfpmut3 and p8741 plasmids. This work was supported by grants from Ministère de l’Enseignement Supérieur et de la Recherche (MESR), Institut National de la Santé et de la Recherche Médicale (INSERM) and Fondation pour la recherche Médicale (FRM grant DEQ20150331742).
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Ahmed AM, Nakano H, Shimamoto T. (2005). Molecular characterization of integrons in non-typhoid Salmonella serovars isolated in Japan: description of an unusual class 2 integron. J Antimicrob Chemother 55: 371–374. [DOI] [PubMed] [Google Scholar]
- Avila P, de la Cruz F. (1988). Physical and genetic map of the IncW plasmid R388. Plasmid 20: 155–157. [DOI] [PubMed] [Google Scholar]
- Baharoglu Z, Mazel D. (2011). Vibrio cholerae triggers SOS and mutagenesis in response to a wide range of antibiotics: a route towards multiresistance. Antimicrob Agents Chemother 55: 2438–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud O, Ploy M-C. (2015). Diversity of class 1 integron gene cassette rearrangements selected under antibiotic pressure. J Bacteriol 197: 2171–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloin C, Valle J, Latour-Lambert P, Faure P, Kzreminski M, Balestrino D et al. (2004). Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol Microbiol 51: 659–674. [DOI] [PubMed] [Google Scholar]
- Brolund A, Sundqvist M, Kahlmeter G, Grape M. (2010). Molecular characterisation of trimethoprim resistance in Escherichia coli and Klebsiella pneumoniae during a two year intervention on trimethoprim use. PLoS ONE 5: e9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambray G, Guerout, Mazel D. (2010). Integrons. Annu Rev Genet 44: 141–166. [DOI] [PubMed] [Google Scholar]
- Cambray G, Sanchez-Alberola N, Campoy S, Guerin É, Da Re S, González-Zorn B et al. (2011). Prevalence of SOS-mediated control of integron integrase expression as an adaptive trait of chromosomal and mobile integrons. Mob DNA 2: 6–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaveroche M-K, Ghigo J-M, d’Enfert C. (2000). A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res 28: e97–e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158: 9–14. [DOI] [PubMed] [Google Scholar]
- Collis CM, Hall RM. (1995). Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother 39: 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelstadter J, Harms K, Johnsen PJ. (2016). The evolutionary dynamics of integrons in changing environments. ISME J 10: 1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero JA, Loot C, Nivina A, Mazel D. (2015). The integron: adaptation on demand. Microbiol Spectr 3: MDNA3-0019-2014. [DOI] [PubMed] [Google Scholar]
- Escudero JA, Loot C, Parissi V, Nivina A, Bouchier C, Mazel D. (2016). Unmasking the ancestral activity of integron integrases reveals a smooth evolutionary transition during functional innovation. Nat Commun 7: 10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espéli O, Moulin L, Boccard F. (2001). Transcription attenuation associated with bacterial repetitive extragenic BIME elements1. J Mol Biol 314: 375–386. [DOI] [PubMed] [Google Scholar]
- Foucault M-L, Depardieu F, Courvalin P, Grillot-Courvalin C. (2010). Inducible expression eliminates the fitness cost of vancomycin resistance in enterococci. Proc Natl Acad Sci USA 107: 16964–16969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaze WH, Zhang L, Abdouslam NA, Hawkey PM, Calvo-Bado L, Royle J et al. (2011). Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J 5: 1253–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings MR, Holley MP, Stokes HW, Holmes AJ. (2005). Integrons in Xanthomonas: A source of species genome diversity. Proc Natl Acad Sci USA 102: 4419–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin É, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, Da Re S et al. (2009). The SOS Response Controls Integron Recombination. Science 324: 1034–1034. [DOI] [PubMed] [Google Scholar]
- Guerra B, Junker E, Schroeter A, Helmuth R, Guth BEC, Beutin L. (2006). Phenotypic and genotypic characterization of antimicrobial resistance in Escherichia coli O111 isolates. J Antimicrob Chemother 57: 1210–1214. [DOI] [PubMed] [Google Scholar]
- Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D et al. (2011). Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog 7: e1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérin E, Jové T, Tabesse A, Mazel D, Ploy M-C. (2011). High-level gene cassette transcription prevents integrase expression in class 1 integrons. J Bacteriol 193: 5675–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanau-Berçot B, Podglajen I, Casin I, Collatz E. (2002). An intrinsic control element for translational initiation in class 1 integrons. Mol Microbiol 44: 119–130. [DOI] [PubMed] [Google Scholar]
- Hare JM, Adhikari S, Lambert KV, Hare AE, Grice AN. (2012). The Acinetobacter regulatory UmuDAb protein cleaves in response to DNA damage with chimeric LexA/UmuD characteristics. FEMS Microbiol Lett 334: 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier H, Zaoui C, Sanson-le Pors M-J, Mazel D, Berçot B. (2009). Translation regulation of integrons gene cassette expression by the attC sites. Mol Microbiol 72: 1475–1486. [DOI] [PubMed] [Google Scholar]
- Jové T, Da Re S, Denis F, Mazel D, Ploy M-C. (2010). Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet 6: e1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheiri R, Akhtari L. (2016). Antimicrobial resistance and integron gene cassette arrays in commensal Escherichia coli from human and animal sources in IRI. Gut Pathog 8: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larouche A, Roy PH. (2011). Effect of attC structure on cassette excision by integron integrases. Mob DNA 2: 3–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC. (1991). Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat 138: 1315–1341. [Google Scholar]
- Levy SB, Marshall B. (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 10: S122–S129. [DOI] [PubMed] [Google Scholar]
- Loot C, Parissi V, Escudero JA, Amarir-Bouhram J, Bikard D, Mazel D. (2014). The integron integrase efficiently prevents the melting effect of Escherichia coli single-stranded DNA-binding protein on folded attC sites. J Bacteriol 196: 762–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, Bujard H. (1997). Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res 25: 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel D. (2006). Integrons: agents of bacterial evolution. Nat Rev Microbiol 4: 608–620. [DOI] [PubMed] [Google Scholar]
- Nemergut DR, Robeson MS, Kysela RF, Martin AP, Schmidt SK, Knight R. (2008). Insights and inferences about integron evolution from genomic data. BMC Genomics 9: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TN, Phan QG, Duong LP, Bertrand KP, Lenski RE. (1989). Effects of carriage and expression of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli K12. Mol Biol Evol 6: 213–225. [DOI] [PubMed] [Google Scholar]
- Payie KG, Clarke AJ. (1997). Characterization of gentamicin 2’-N-acetyltransferase from Providencia stuartii: its use of peptidoglycan metabolites for acetylation of both aminoglycosides and peptidoglycan. J Bacteriol 179: 4106–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Valdespino A, Lazarini-Martínez A, Rivera-González AX, García-Hernández N, Curiel-Quesada E. (2016). Dynamics of a class 1 integron located on plasmid or chromosome in two Aeromonas spp. strains. Front Microbiol 7: 1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AN, Barlow M, Clark LA, Boring JR, Tenover FC, McGowan JE. (2006). Class 1 integrons in resistant Escherichia coli and Klebsiella spp., US hospitals. Emerg Infect Dis 12: 1011–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recchia GD, Stokes HW, Hall RM. (1994). Characterisation of specific and secondary recombination sites recognised by the integron DNA integrase. Nucleic Acids Res 22: 2071–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Verdugo A, Gaut BS, Tenaillon O. (2013). Evolution of Escherichia coli rifampicin resistance in an antibiotic-free environment during thermal stress. BMC Evol Biol 13: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrag SJ, Perrot V, Levin BR. (1997). Adaptation to the fitness costs of antibiotic resistance in Escherichia coli. Proc R Soc B Biol Sci 264: 1287–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starikova I, Harms K, Haugen P, Lunde TTM, Primicerio R, Samuelsen Ø et al. (2012). A trade-off between the fitness cost of functional integrases and long-term stability of integrons. PLoS Pathog 8: e1003043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes HW, Hall RM. (1989). A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol 3: 1669–1683. [DOI] [PubMed] [Google Scholar]
- Subbiah M, Top EM, Shah DH, Call DR. (2011). Selection pressure required for long-term persistence of blaCMY-2-positive IncA/C Ppasmids. Appl Environ Microbiol 77: 4486–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakopoulou A, Psichogiou M, Tzouvelekis L, Tassios PT, Kosmidis C, Petrikkos G et al. (2009). Prevalence and characterization of class 1 integrons in Escherichia coli of poultry and human origin. Foodborne Pathog Dis 6: 1211–1218. [DOI] [PubMed] [Google Scholar]
- Vinué L, Jové T, Torres C, Ploy M-C. (2011). Diversity of class 1 integron gene cassette Pc promoter variants in clinical Escherichia coli strains and description of a new P2 promoter variant. Int J Antimicrob Agents 38: 526–529. [DOI] [PubMed] [Google Scholar]
- Vogwill T, MacLean RC. (2015). The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol Appl 8: 284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Jiang X, Li M, Li G, Hu Q, Lu H et al. (2013). Diversity of gene Cassette promoter variants of class 1 integrons in uropathogenic Escherichia coli. Curr Microbiol 67: 543–549. [DOI] [PubMed] [Google Scholar]
- Wright MS, Baker-Austin C, Lindell AH, Stepanauskas R, Stokes HW, McArthur JV. (2008). Influence of industrial contamination on mobile genetic elements: class 1 integron abundance and gene cassette structure in aquatic bacterial communities. ISME J 2: 417–428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.