Abstract
Basal autophagy is tightly regulated by transcriptional and epigenetic factors to maintain cellular homeostasis. Dysregulation of cardiac autophagy is associated with heart diseases, including cardiac hypertrophy, but the mechanism governing cardiac autophagy is rarely identified. To analyze the in vivo function of miR-199a in cardiac autophagy and cardiac hypertrophy, we generated cardiac-specific miR-199a transgenic mice and showed that overexpression of miR-199a was sufficient to inhibit cardiomyocyte autophagy and induce cardiac hypertrophy in vivo. miR-199a impaired cardiomyocyte autophagy in a cell-autonomous manner by targeting glycogen synthase kinase 3β (GSK3β)/mammalian target of rapamycin (mTOR) complex signaling. Overexpression of autophagy related gene 5 (Atg5) attenuated the hypertrophic effects of miR-199a overexpression on cardiomyocytes, and activation of autophagy using rapamycin was sufficient to restore cardiac autophagy and decrease cardiac hypertrophy in miR-199a transgenic mice. These results reveal a novel role of miR-199a as a key regulator of cardiac autophagy, suggesting that targeting miRNAs controlling autophagy as a potential therapeutic strategy for cardiac disease.
Autophagy is a catabolic process initiated upon starvation, cellular stress, accumulation of damaged organelles or protein aggregates, which has a critical role in cardiac homeostasis and pathologies, including cardiac hypertrophy.1 There are controversial concepts regarding the function of autophagy in cardiac hypertrophy. Autophagy has been shown to have a protective role in cardiomyocytes.2, 3, 4, 5 Impaired autophagy induced by deficient autophagy-related genes (Atgs), Atg7 or Atg5, leads to cardiac hypertrophy and contractile dysfunction,2, 3 while sustained Atg7-induced autophagy ameliorates ventricular dysfunction and cardiac hypertrophy in a mouse model of desmin-related cardiomyopathy.4 Inhibition of autophagy caused by Mst1 (mammalian Ste20-like kinase 1) also promotes cardiac dysfunction in mice subjected to myocardial infarction.5 On the other hand, autophagic activity has been shown to be harmful for cardiac homeostasis.6, 7 Heterozygous deletion of Beclin1 (Atg6), a gene required for the initiation of autophagosome formation, reduces autophagic activity and improves cardiac function upon pressure overload.6 Autophagy suppression by histone deacetylase inhibitors decreases pressure overload-induced cardiac hypertrophy in mice.7 A possible explanation for this contradiction is that the cardiac homeostasis is maintained by sophisticatedly balanced autophagy. However, mechanisms governing cardiac autophagy are still largely unknown.
MicroRNAs (miRNAs) are small, endogenous noncoding RNAs, which inhibit the expression of specific genes at the posttranscriptional level by binding with the 3′-untranslated region (3′-UTR) of target mRNAs.8 miRNAs have been shown to have important roles in regulating cardiac hypertrophy and heart failure.9, 10, 11, 12 Although a considerable number of miRNAs are shown to be key regulators of autophagy,13 only a few miRNAs participating in cardiac autophagy have been discovered until recently.14, 15 Nevertheless, how miRNAs regulate cardiac autophagy and cardiac hypertrophy is underexplored.
miR-199a is frequently upregulated in pressure-overloaded hypertrophic hearts.16, 17, 18 In vitro studies have shown that miR-199a regulates cell size of cardiomyocyte.19 It has been shown that antagomir-mediated knockdown of miR-199a and miR-214 improved cardiac function of transverse aortic constriction pressure overload mice by regulating fatty acid metabolism.18 However, the in vivo causal link between miR-199a overexpression and cardiac hypertrophy is still lacking. Here we generated cardiomyocyte-specific miR-199a transgenic mice to show that miR-199a overexpression induced cardiac hypertrophy and heart failure. We demonstrated that miR-199a could inhibit cardiomyocyte autophagy in a cell-autonomous manner by activating mammalian target of rapamycin (mTOR) complex signaling. Moreover, activation of autophagy by overexpression of Atg5 or rapamycin treatment increased cardiomyocyte autophagy and attenuated cardiac hypertrophy induced by miR-199a overexpression.
Results
Cardiomyocyte-specific overexpression of miR-199a induces cardiac hypertrophy and cardiac dysfunction in mice
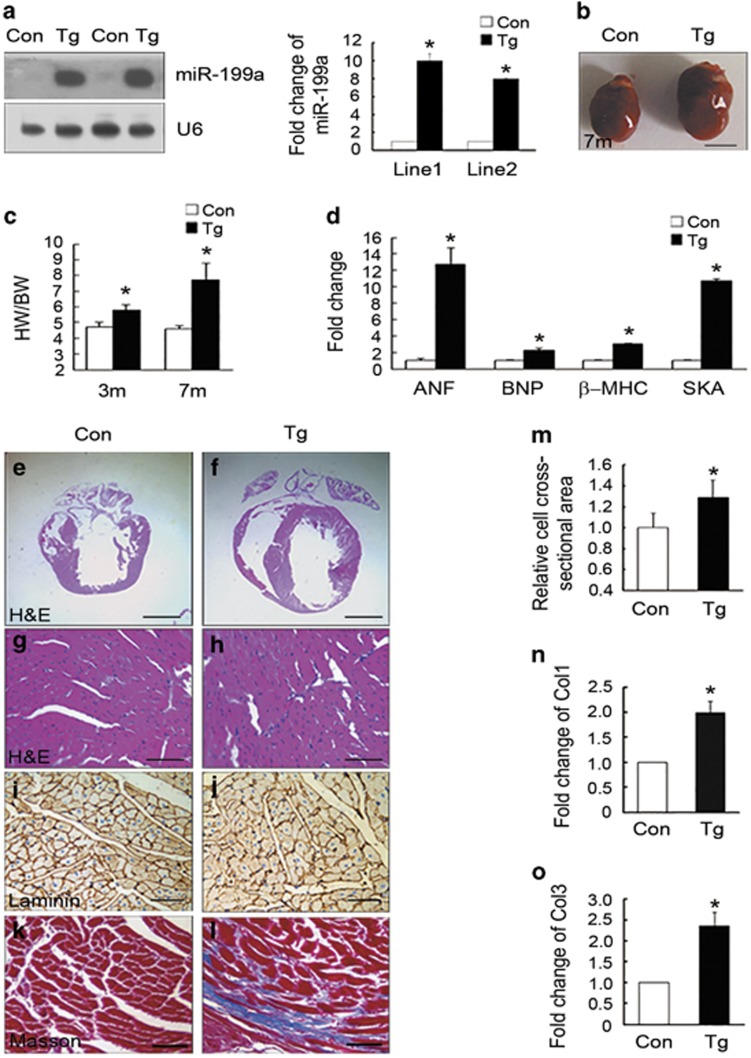
miR-199a was upregulated in heart samples from mouse models of pressure overload-induced cardiac hypertrophy and human heart failure (Supplementary Figures S1A and B). To study the consequences of miR-199a overexpression in vivo, we generated two cardiomyocyte-specific miR-199a overexpression transgenic mouse lines. Real-time PCR and Northern blotting results revealed that miR-199a was successfully overexpressed specifically in the transgenic hearts (Figure 1a and data not shown). The miR-199a transgenic mice developed obvious cardiac hypertrophy at 3 months of age and processed to heart failure at about 7 months of age. The transgenic mice exhibited enlarged hearts (Figure 1b) and higher heart weight/body weight (HW/BW) ratios (Figure 1c) than sex-matched littermate wild-type controls. The fetal genes such as atrial natriuretic factor (ANF), brain natriuretic peptide (BNP), myosin heavy chain β (β-MHC) and skeletal muscle and cardiac actin (SKA) were significantly upregulated in the heart tissues from miR-199a transgenic mice (Figure 1d).
Figure 1.
Overexpression of miR-199a induced cardiac hypertrophy in vivo. (a) Northern blottings and quantification data showed approximately 8–11-fold increases of miR-199a expression in the hearts of miR-199a transgenic mice (Tg) compared with control wild-type littermates (Con). Loading control was performed with a RNA probe against U6. (b) Gross morphology showing that the miR-199a Tg heart was enlarged compared with the control heart at 7 months. Scale bar represents 5 mm. (c) Heart weight to body weight (HW/BW) ratios of 3- and 7-month-old miR-199a Tg mice (n=12) were significantly higher than controls (n=8). (d) Analyses of ANF, brain natriuretic peptide (BNP), β-MHC and skeletal muscle and cardiac actin (SKA) by real-time PCR in the hearts from miR-199a Tg and control mice (n=3 per genotype). *P<0.05. (e–h) Macroscopic view of H&E-stained transverse sections from the control (e and g) and miR-199a Tg hearts (f and h). Scale bars represent 1 mm (e and f) and 25 μm (g and h). (i and j) Immunostaining for laminin on transverse sections of myocardium from control (i) and miR-199a Tg mice (j). Scale bar represents 25 μm. (k and l) Representative images of Masson staining from control (k) and miR-199a Tg mice (l). Scale bar represents 25 μm. (m) Quantification of cell cross-sectional areas from histological sections stained with laminin. Mean cardiomyocyte cross-sectional area of the miR-199a Tg hearts was significantly larger than that of the control hearts (388 cells from transgenic hearts, 350 cells from control hearts), *P<0.001. (n and o) Detection of fibrosis marker genes, Col1 and Col3, in the hearts from control mice and miR-199a Tg mice. Data represent means±S.E.M. *P<0.05
To ascertain whether the increased heart size was due to cell growth (increase in cell size) or cell proliferation (increase in cell number), we first assessed cardiomyocyte proliferation by BrdU labeling experiment. The cardiomyocyte proliferation ratio of the miR-199a transgenic mice was not significantly different from the littermate controls (Supplementary Figure S1C). Cross-sections of the hearts confirmed that left ventricle (LV) was enlarged (Figures 1e and f) and the size of cardiomyocytes was increased in the transgenic mice (Figures 1g and j). Masson staining of cardiac section demonstrated increased cardiac fibrosis in the transgenic hearts (Figures 1k and l). Quantitative measurement of miR-199a transgenic cardiomyocytes revealed a 35% increase in cell cross-sectional area relative to controls (Figure 1m). The fibrosis markers, type I collagen (Col1) and Col3, were also significantly upregulated in the transgenic hearts (Figures 1n and o).
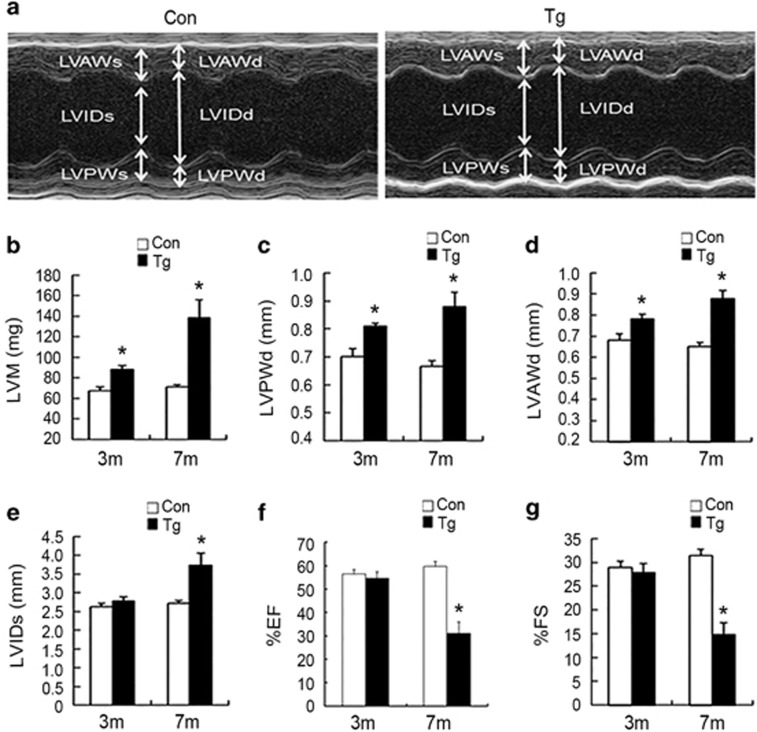
To evaluate the effect of miR-199a overexpression on cardiac function, we assessed the LV dimensions and the systolic functions of 3- and 7-month-old miR-199a transgenic mice by using M-mode echocardiography (Figure 2a and Supplementary Table S1). The miR-199a transgenic mice showed increased LV mass (Figure 2b). The thickened LV walls (LVPWd and LVAWd) (Figures 2c and d) and an increase in the LV internal diameter in systole (LVIDs) were observed in miR-199a transgenic mice (Figure 2e). In addition, the ejection fraction and fractional shortening, two echocardiographic indexes of LV contractile function, were found to be decreased in the miR-199a transgenic mice (Figures 2f and g). These observations indicate that miR-199a overexpression is sufficient to induce cardiac hypertrophy and the impairment of cardiac function.
Figure 2.
Cardiac dysfunction in miR-199a transgenic mice. (a) Representative M-mode images of a miR-199a Tg mouse (right) and the wild-type littermate control (left). (b) Quantification of LV mass. (c and d) Measurements of the LV wall thickness in diastole (LVPWd and LVAWd). (e) Measurements of the LV internal diameters in systole (LVIDs). (f and g) Quantification of ejection fraction (EF) and fractional shortening (FS) in the hearts from miR-199a Tg mice and control mice at 3 and 7 months. Mean values±SEM were determined by echocardiography. 3-Month-old control mice: n=5; 3-month-old Tg mice: n=6; 7-month-old control mice: n=7; 7-month-old Tg mice: n=8; *P<0.05
miR-199a inhibits cardiac autophagy and activates mTOR signaling in mice
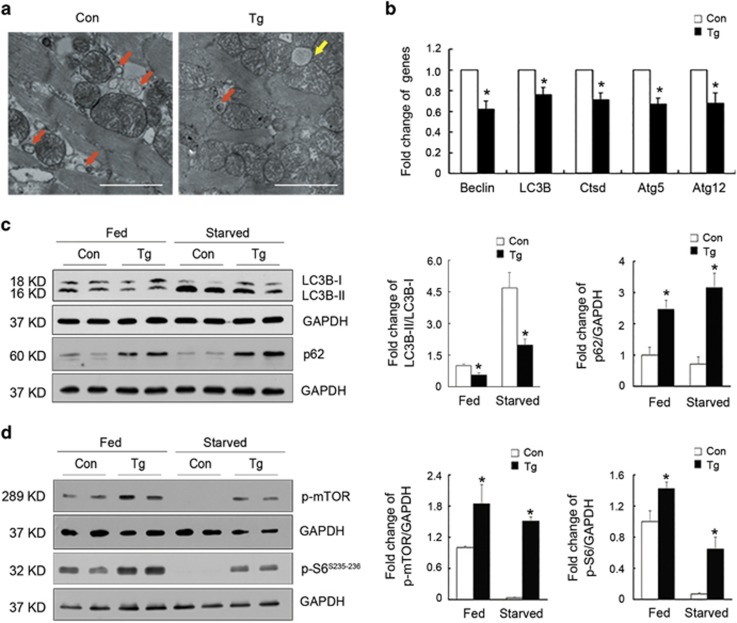
To examine whether the ultrastructural architecture was influenced by miR-199a overexpression, LV tissues obtained from transgenic and control mice were subjected to electron microscopy. We found that the number of autophagic vacuoles was decreased in the miR-199a transgenic hearts compared with that of control hearts (Figure 3a). We next determined to identify whether cardiac autophagy was impaired in miR-199a transgenic mice. The autophagic marker genes, such as Atg5, Atg12, Beclin1 and LC3B, were dramatically decreased in the transgenic hearts (Figure 3b). We then explored the alteration of LC3B and p62 in the control and Tg hearts under fed and starved conditions. We found that the LC3B lipidation (LC3B-II/LC3B-I ratios) and transcriptional levels were lower, whereas autophagy substrate p62 levels were higher in the miR-199a-overexpressing transgenic hearts compared with that of the control hearts under both fed and starved conditions (Figure 3c and Supplementary Figure S2A), indicating an inhibitory effect of miR-199a on cardiomyocyte autophagy. Starvation induced LC3B lipidation and transcript level no matter whether miR-199a was overexpressed (Figure 3c and Supplementary Figure S2A), indicating that the Tg hearts did not completely lose the response to autophagic stimuli. However, starvation failed to significantly decrease p62 level in the control hearts and rather induced a slight increase of p62 in the Tg hearts (Figure 3c). So we detected the transcriptional level of p62, which was upregulated under starved condition (Supplementary Figure S2B), suggesting a transcriptional upregulation of p62 in response to starvation and some unknown mechanism of p62 regulated by miR-199a. Because decreased levels of LC3B-II could be caused by either decreased LC3B synthesis or enhanced LC3B degradation, we then performed autophagic flux assay using lysosomal inhibitor chloroquine (CQ) to block lysosomal function and thus LC3B degradation. CQ treatment effectively increased the LC3B-II and p62 levels both in the control and Tg hearts, while the Tg hearts displayed inhibited autophagy under both conditions, indicating that miR-199a inhibited autophagy by suppressing the formation of autophagic vesicles (Supplementary Figure S2C). Furthermore, mTOR and its downstream S6 ribosomal protein (S6), a classical signaling pathway involved in the inhibition of autophagy, were found to be activated in the miR-199a transgenic hearts under fed and starved conditions (Figure 3d).
Figure 3.
Cardiomyocyte autophagy was inhibited in miR-199a transgenic mice. (a) Representative electron microscopy of autophagosomes in the hearts from control and miR-199a transgenic mice. Red arrows indicate autophagosomes, and the yellow arrow indicates a lipid drop. Scale bars represent 2 μm. (b) mRNA expression levels of autophagic marker genes in the hearts of control and miR-199a Tg mice (n=3 per genotype). (c) Western blotting analyses of LC3B-I, LC3B-II and p62 levels in control and miR-199a Tg mice under fed and starved conditions. Quantification of the results is shown right (n=6). *P<0.01. (d) Western blotting analyses of p-mTOR and its downstream p-S6 in the hearts of control and miR-199a Tg mice under fed and starved conditions. Quantification of the results is shown right (n=4). *P<0.05. GAPDH, glyceraldehyde 3-phosphate dehydrogenase
miR-199a-mediated autophagy inhibition is cell autonomous and indispensable for cardiomyocyte hypertrophy
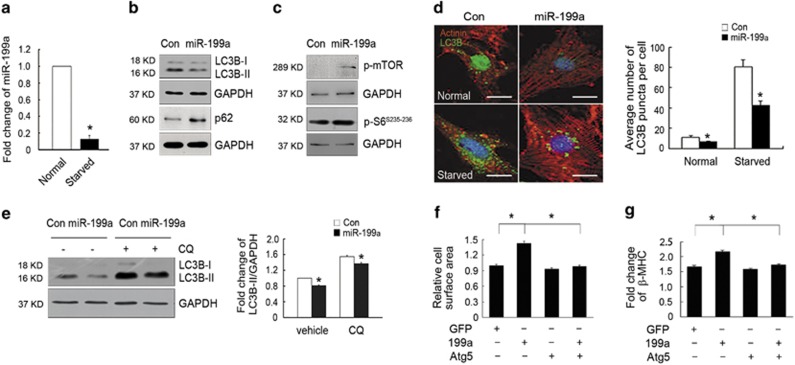
To further verify the direct role of miR-199a in the inhibition of cardiomyocyte autophagy, we overexpressed miR-199a in cardiomyocytes and induced the autophagic condition by starving the cardiomyocytes in serum/glucose-free media. As expected, the autophagic genes, Beclin1, LC3B, Atg5 and Atg12, were significantly upregulated upon starvation (Supplementary Figure S3A). The expression of miR-199a was significantly decreased under starved condition (Figure 4a). miR-199a overexpression downregulated the autophagic marker genes (Supplementary Figures S3B and C), decreased the LC3B lipidation level (LC3B-II/LC3B-I ratios) and increased autophagy substrate p62 level (Figure 4b). Consistently, mTOR/S6 signaling was activated upon miR-199a overexpression (Figure 4c). We then infected primary cardiomyocytes with an adenovirus carrying GFP-LC3B fusion protein, which allows the visualization of autophagic structures as fluorescent puncta. The average number of puncta was reduced in miR-199a overexpression cardiomyocytes compared with that of control cells under both normal and starved condition (Figure 4d). We also performed autophagic flux assay in vitro and found that miR-199a overexpression resulted in reduced LC3B-II levels and increased p62 accumulation under both basal and lysosome-inhibited conditions (Figure 4e and Supplementary Figure S3D), indicating a significant reduction in LC3B synthesis relative to controls. These data demonstrate that miR-199a overexpression inhibits cardiomyocyte autophagy in a cell-autonomous manner.
Figure 4.
miR-199a inhibit cardiomyocyte autophagy in a cell-autonomous manner. (a) Real-time PCR analyses of miR-199a expression under normal or serum/glucose-deprivation (starved) conditions. (b and c) Western blotting analyses of LC3B-I, LC3B-II, p62 and mTOR/S6 protein levels in cardiomyocytes infected with adenoviruses expressing control or miR-199a miRNAs. (d) Representative images and quantification of GFP-LC3B puncta (green) in control and miR-199a-overexpressing cardiomyocytes under normal or starved conditions (n=30 cells/per condition). Scale bars represent 5 μm. (e) Representative western blotting and quantification showing reduced LC3B-II levels in miR-199a overexpression cardiomyocytes treated with or without CQ. (f) Atg5 overexpression rescued the miR-199a-induced hypertrophic growth of cardiomyocytes. Fold change in mean cell surface area of α-actinin-immunostained cardiomyocytes infected with adenoviruses (n=160). *P<0.01. (g) Real-time PCR showing that Atg5 overexpression rescued β-MHC upregulation induced by miR-199a overexpression (n=3). *P<0.05. GAPDH, glyceraldehyde 3-phosphate dehydrogenase
We further tested whether reactivation of autophagy could attenuate the detrimental effects of miR-199a overexpression. We infected neonatal cardiomyocytes with an adenovirus expressing Atg5, which was essential in the early stages of autophagosome formation, and found that overexpression of Atg5 (Supplementary Figure S4A) could rescue the miR-199a-induced hypertrophic growth of cardiomyocytes and upregulation of β-MHC (Figures 4f and g, and Supplementary Figure S4B). All these results indicate that inhibited cardiomyocyte autophagy might be one of the causes of cardiac hypertrophy induced by miR-199a overexpression.
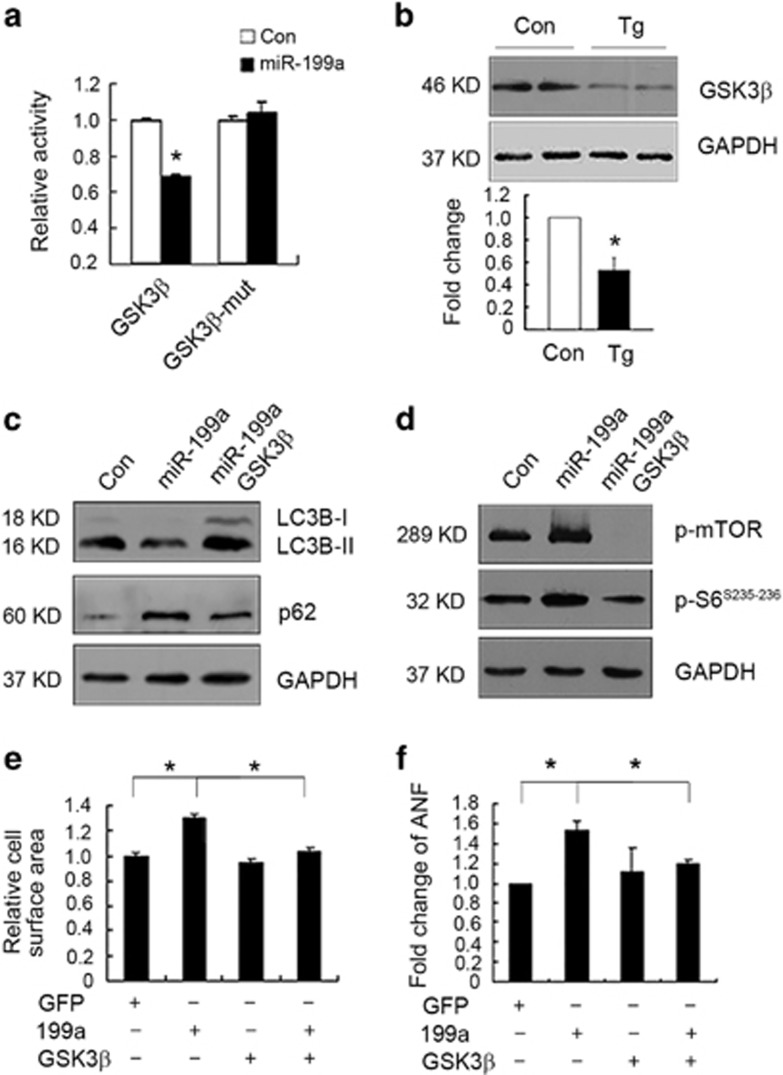
miR-199a targets pro-autophagic and antihypertrophic glycogen synthase kinase 3β (GSK3β) to activate mTOR signaling
As mTOR signaling was found to be significantly activated in the miR-199a transgenic hearts, we then determined the potential mechanism of how mTOR signaling was activated by miR-199a. We screened the candidate miR-199a target mRNAs that have been reported to regulate mTOR signaling using TargetScan and identified GSK3β, whose mRNA 3′-UTR region comprised the seed sequences and flanking nucleotides matching miR-199a which is highly conserved among the human, mouse and rat (Supplementary Figures S5A and B). To confirm whether GSK3β was a direct target of miR-199a, we performed luciferase reporter assays in HEK293 cells. Cotransfection of miR-199a with the luciferase reporter gene linked to the wild-type 3′-UTR of GSK3β strongly inhibited the luciferase activity, while no effect was observed with the construct harboring a mutant fragment of GSK3β 3′-UTR (Figure 5a). Moreover, the GSK3β protein levels were significantly decreased in heart tissues of miR-199a transgenic mice (Figure 5b).
Figure 5.
GSK3β was the target of miR-199a in cardiomyocytes. (a) Dual luciferase activity assay of HEK293 cells cotransfected with a luciferase reporter plasmid containing the naive or mutant GSK3β 3′-UTR. n=4, *P<0.05. (b) Western blotting analyses of GSK3β protein levels in the control and miR-199a Tg hearts. Quantification is shown lower. (c and d) Western blotting analyses of LC3B-I, LC3B-II, p62 and mTOR/S6 protein levels in cardiomyocytes infected with adenoviruses expressing control, miR-199a miRNAs with or without GSK3β. (e) GSK3β overexpression rescued the hypertrophic growth of cardiomyocytes induced by miR-199a overexpression. Fold change in mean cell surface area of α-actinin-immunostained cardiomyocytes infected with adenoviruses (n=120). *P<0.05. (f) Real-time PCR showing that GSK3β overexpression rescued ANF upregulation induced by miR-199a overexpression (n=3). *P<0.05. GAPDH, glyceraldehyde 3-phosphate dehydrogenase
To further test whether GSK3β mediated the effect of miR-199a on the regulation of mTOR signaling, cardiomyocyte autophagy and cardiac hypertrophy, we developed Ad-GSK3β to overexpress GSK3β in cardiomyocytes (Supplementary Figure S5C). We found that GSK3β overexpression could induce the expression of autophagic marker genes (Supplementary Figure S5D), attenuate miR-199a-induced reduction of LC3B lipidation level and upregulation of autophagy substrate p62 level (Figure 5c). miR-199a-activated mTOR signaling was significantly suppressed by GSK3β overexpression (Figure 5d). We also found that GSK3β overexpression could rescue miR-199a-induced enlargement of cardiomyocyte cell surface area and upregulation of ANF (Figures 5e and f and Supplementary Figure S5E). All these observations suggest that GSK3β is the target of miR-199a and mediates the effects of miR-199a on mTOR signaling activation, cardiomyocyte autophagy and hypertrophy.
Rapamycin induces cardiac autophagy and attenuates cardiac hypertrophy in miR-199a transgenic mice
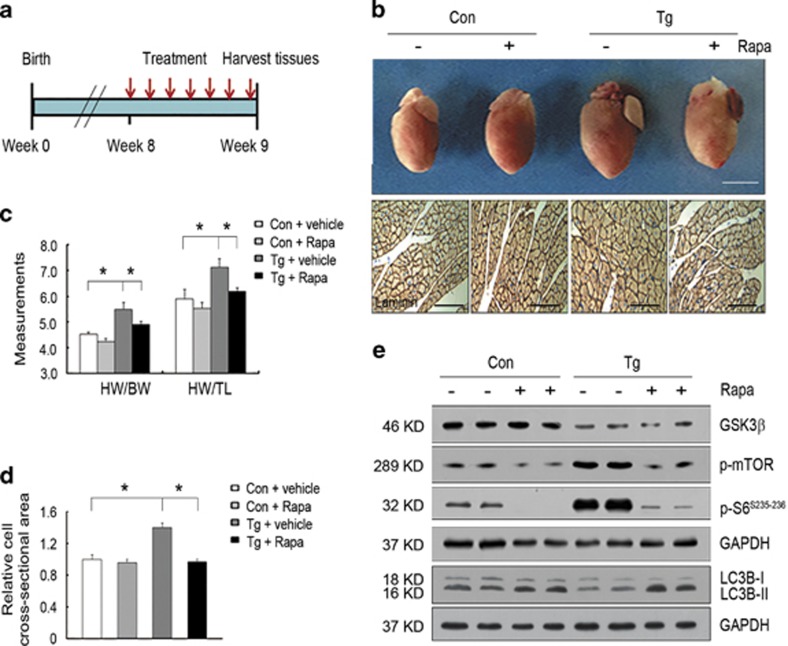
We next investigated whether inhibition of mTOR could induce cardiac autophagy and rescue the cardiac phenotypes of miR-199a transgenic mice. As rapamycin could inhibit mTOR and induce autophagy,20, 21, 22 we treated 8-week-old miR-199a transgenic and control mice with rapamycin or vehicle daily for 1 week (Figure 6a). Morphological and histological analyses of the hearts revealed that miR-199a transgenic mice treated with vehicle developed significant cardiac hypertrophy, whereas an attenuation of the cardiac hypertrophy was observed in miR-199a transgenic mice treated with rapamycin (Figure 6b). The ratio of heart weight to body weight or tibia length was increased in miR-199a transgenic mice treated with vehicles compared with vehicle-treated control mice, whereas these two ratios were decreased in miR-199a transgenic mice treated with rapamycin (Figure 6c). The cell cross-section area was significantly reduced in rapamycin-treated miR-199a transgenic mice compared with vehicle-treated transgenic animals (Figure 6d). Expectedly, the activity of mTOR signaling was reduced in the miR-199a transgenic hearts treated with rapamycin, as demonstrated by decreased p-mTOR and p-S6 (Figure 6e). Importantly, the conversion of LC3B-I to LC3B-II was enhanced in the miR-199a transgenic hearts treated with rapamycin compared with that of the vehicle-treated groups, suggesting that rapamycin was sufficient to rescue autophagy reduction caused by miR-199a overexpression (Figure 6e). Collectively, these data demonstrated that rapamycin could induce cardiac autophagy and attenuate cardiac hypertrophy in miR-199a transgenic mice.
Figure 6.
Administration of rapamycin induced autophagy and attenuated the cardiac hypertrophy in miR-199a transgenic mice. Eight control mice were treated with vehicle or rapamycin (Rapa), five miR-199a Tg mice were treated with vehicle, and seven miR-199a Tg mice were treated with rapamycin. (a) The strategy of rapamycin administration. Rapamycin or vehicle treatment was started at 8 weeks of age and injected daily for 1 week. (b) Representative gross morphology (upper) and laminin immunostaining analyses (lower) of the hearts from control and miR-199a Tg mice treated with vehicle or rapamycin. The upper scale bar represents 5 mm, the lower scale bar represents 50 μm. (c) Heart weight to body weight or tibia length ratios of control and miR-199a Tg mice treated with vehicle or rapamycin. *P<0.05. (d) Quantification of relative cell cross-sectional area of cardiomyocytes of control and miR-199a Tg mice treated with vehicle or rapamycin (n=300). *P<0.05. (e) Western blotting analyses of GSK3β, p-mTOR, p-S6 and LC3B in control and miR-199a Tg mice treated with vehicle or rapamycin. GAPDH, glyceraldehyde 3-phosphate dehydrogenase
Discussion
Our study provided the first in vivo data to demonstrate that cardiomyocyte-specific overexpression of miR-199a is sufficient to impair cardiomyocyte autophagy and induce cardiac hypertrophy, uncovering a new insight into the mechanism of miRNA-regulated cardiac hypertrophy. Most of our findings were carried out using the cardiomyocyte-specific miR-199a transgenic mice, which allowed dissecting the function and mechanism of miR-199a in a pathophysiological setting, as miR-199a overexpression has been observed in the mouse and human hypertrophic hearts (Supplementary Figures S1A and B).16, 17, 18 Consistent with the previous in vitro studies,19 we demonstrated that overexpression of miR-199a was able to trigger cardiac hypertrophy and cause heart failure in vivo. It has been shown that miR-199a regulates cardiomyocyte size and mitochondrial fatty acid oxidation,18 while our current study revealed that miR-199a inhibited cardiac autophagy in a cell-autonomous manner, suggesting a novel mechanism underlying miR-199a-induced cardiac hypertrophy. Supportively, rescue experiments using rapamycin administration or Atg5 overexpression proved that increased autophagy activity significantly attenuated miR-199a-induced cardiac hypertrophy, indicating a possible causal link between impaired autophagy and cardiac hypertrophy.
Our experiments identified miR-199a as a negative regulator of cardiomyocyte autophagy, demonstrated by the inhibited LC3B lipidation and enhanced p62 accumulation under both normal and starved conditions. However, there seemed to be some discrepancies regarding the LC3B and p62 alterations in response to starvation. Both the control and Tg hearts displayed enhanced autophagy under starvation indicated by a higher LC3B lipidation, but starvation failed to significantly decrease p62 level in the control hearts and rather induced a slight increase of p62 in the Tg hearts. With respect to the p62 level under starvation, we found that the p62 mRNA level was elevated in starved heart tissues (Supplementary Figure S2B), suggesting a significant transcriptional upregulation of p62. which is consistent with previous studies in skeletal muscle.23, 24 Moreover, a recent study indicates that the level of p62 in starved cells is determined by at least three factors: autophagic degradation, transcriptional upregulation, and availability of lysosomal-derived amino acids, which suggests a complex effect of starvation on p62 level and implicates that the expression level of p62 does not always inversely correlate with autophagic activity.25 On the other hand, as p62 level can also be modulated by a number of factors,26, 27 miR-199a may affect these factors to induce p62 expression that may be even pronounced under starvation condition. Nevertheless, our study proposed that cardiac hypertrophy induced by miR-199a was at least partially due to inhibited cardiac autophagy, which was in favor of the concept that basal level of autophagy had a protective role in the maintenance of cardiac homeostasis.
Searching for molecular mechanisms underlying the role of miR-199a in suppressing cardiomyocyte autophagy and inducing cardiac hypertrophy, we found that mTOR signaling was significantly activated in transgenic hearts. Mechanistically, miR-199a targets GSK3β, a pro-autophagic and antihypertrophic gene, which has been shown to mediate mTOR suppression.28, 29 Consistently, GSK3β expression was significantly downregulated and mTOR was activated in the hypertrophic hearts of miR-199a transgenic mice. Most importantly, we performed rescue experiments to confirm that GSK3β overexpression could inhibit the activity of mTOR, induce the autophagy and decrease the hypertrophic growth of cardiomyocytes induced by miR-199a. It has been established that mTOR signaling is a major negative regulatory axis of autophagy, and direct inhibition of mTOR is essential and sufficient to trigger autophagy induction.20, 21, 22 mTOR pathway has also been shown to have a key regulatory function in cardiac hypertrophy.30, 31, 32, 33, 34 mTOR activity is increased during the cardiomyocyte hypertrophic response to multiple pathological stimuli30, 31 and mTOR inhibition significantly reduces cardiac hypertrophy.31, 32, 33, 34 In our study, the rescue experiment using rapamycin suggested that mTOR activation had important roles in miR-199a-regulated cardiac autophagy and cardiac hypertrophy. However, we do not exclude the possible parallel autophagy independent roles of mTOR activation in miR-199a-induced cardiac hypertrophy.
In conclusion, we demonstrated that cardiomyocyte-specific miR-199a overexpression evoked cardiac hypertrophy by suppressing cardiac autophagy through activation of mTOR signaling. Further characterization of miRNAs regulating cardiac autophagy and hypertrophy might reveal more potential therapeutic targets for heart failure.
Materials and Methods
Cloning and expression of miRNAs
A 438-bp genomic fragment containing the miR-199a gene was amplified by PCR and ligated into adenoviral vectors to generate Ad-miR-199a using 293 A cells. PCR primers for miR-199a were as following: miR-199a sense: 5′-TTTCTCGAGGAAGATGCTCACCAGCCCTTTA-3′, and miR-199a antisense: 5′-TTTTCTAGAGCATCATCTTGCCAGCGACT-3′. miR-199a mimic oligonucleotides were synthesized and purified with high-performance liquid chromatography by GenePharma Co. Ltd (Shanghai, China). The mimic sequence is 5′-CCCAGUGUUCAGACUACCUGUUC-3′.
Isolation and culture of neonatal cardiomyocytes
Neonatal rat ventricular myocytes were isolated from 2-day-old pups, infected and stained as described.35 Briefly, hearts from 1–2-day-old rats were aseptically removed. Their ventricles were dissected, minced and trypsinized overnight at 4 °C. The next day, cells were dissociated with collagenase and preplated for 2 h at 37 °C. The non-adherent cardiomyocytes were removed and plated in 24-well plates in DMEM/F-12 medium containing 10% FBS and 0.1 mM bromodeoxyuridine (Sigma, St. Louis, MO, USA). A total of 1 × 105 cells/cm2 were seeded in 24-well plate for further experiments. This procedure yielded cultures with 90–95% cardiomyocytes, as assessed by microscopic observation of cell beating.
Generation of miR-199a transgenic mice
A mouse genomic fragment containing the miR-199a gene was amplified using primers 5′-TTTGTCGACCTGCAAATGTGCCACGTCAA-3′ and 5′-TTTGAATTCCTTACCCGCTGGTCTTCCCT-3′ and then sub-cloned into a cardiac-specific expression plasmid containing the α-MHC promoter and human growth hormone (GH) poly (A) signal.35 To genotype the mouse, genomic DNA was isolated from mouse tail biopsy and analyzed by PCR using specific primers: 5′-ATGACAGACAGATCCCTCCTATCTCC-3′ and 5′-GAATTCCTTACCCGCTGGTC TTCCCT-3′. miR-199a transgenic mice at 3 and 7 months of age were used for functional and histological studies and were compared with their sex-matched wild-type littermates.
Western blotting analysis
Western blottings were carried out on myocardial extracts as described.12 In all, 30 μg of proteins were electrophoresed on 12% SDS-PAGE and transferred onto PVDF membranes. Immunoblotting was performed according to the manufacturer’s instructions using the following antibodies: GSK3β, p62, LC3B, p-mTOR, p-S6S235-236 (Cell Signaling, Beverly, MA, USA), and GAPDH (Santa Cruz, Dallas, TX, USA).
Luciferase assay
For target assay, we performed luciferase reporter experiments in the HEK293 cells. The 3′-UTR fragment of GSK3β was amplified using primers 5′-TTTAGATCTCCTTCTGCATCAGCTTCCAACTC-3′ and 5′-TTTACGCGTTCTTTCCAAACGTGACCAGTGT-3′ and cloned into pGL3-CM luciferase reporter vector (Ambion, Grand Island, NY, USA). The construct (Luc-GSK3β mutant) containing a mutated GSK3β-UTR (ACACUGG was mutated to ACGCGU) was also constructed. For luciferase assay, reporter plasmids were co-transfected using lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). The phRG-TK vector (Promega, Madison, WI, USA), which expresses a synthetic renilla luciferase, was used for normalizing transfection efficiency. After transfection, luciferase activities were measured with a Dual Luciferase Reporter Assay Kit (Invitrogen) using LB 960 Centro XS3 luminometer (Berthold Technologies, Bad Wildbad, Germany).36
Transthoracic echocardiography measurement
Cardiac function and heart dimensions were evaluated by echocardiography as described.35 Briefly, mice were anesthetized with tribromoethanol and analyzed for anatomy and function on a Vivid 7 Dimension cardiovascular ultrasound system with a 12-MHz microprobe (GE Healthcare, Buckinghamshire, UK). Echocardiographic measurements were taken on M-mode in triplicate from more than five separate mice per group.
Autophagic flux assays
To measure autophagic synthesis versus degradation, LC3B-II protein levels were quantified with and without lysosomal inhibitor under different conditions. To inhibit lysosomal function, media containing 50 μM chloroquine (Sigma) was added to the cardiomyocytes for 4 h.
Administration of rapamycin
Rapamycin (2 mg/kg/day) or vehicle was administered intraperitoneally to miR-199a transgenic mice and control mice for 1 week as previously described.37
Statistical analysis
All statistical analyses were performed using the SPSS software (SPSS Inc., Chicago, IL, USA). Results are means±S.E.M. Statistical differences were determined by Student’s t-test. P-values<0.05 were considered significant.
Acknowledgments
This work was supported by the Chinese National Key Program on Basic Research (2012CB945103) and the National Natural Science Foundation of China (31430057, 31071992, 30971161 and 81030001).
Glossary
- LV
left ventricle
- ANF
atrial natriuretic factor
- α-MHC
myosin heavy chain α
- GSK3β
glycogen synthase kinase 3β
- mTOR
mammalian target of rapamycin
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by H-U Simon
The authors declare no conflict of interest.
Supplementary Material
References
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147: 728–741. [DOI] [PubMed] [Google Scholar]
- Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med 2007; 13: 619–624. [DOI] [PubMed] [Google Scholar]
- Taneike M, Yamaguchi O, Nakai A, Hikoso S, Takeda T, Mizote I et al. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy 2010; 6: 600–606. [DOI] [PubMed] [Google Scholar]
- Bhuiyan MS, Pattison JS, Osinska H, James J, Gulick J, McLendon PM et al. Enhanced autophagy ameliorates cardiac proteinopathy. J Clin Invest 2013; 123: 5284–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima Y, Kyoi S, Zhai P, Liu T, Li H, Ivessa A et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med 2013; 19: 1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest 2007; 117: 1782–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao DJ, Wang ZV, Battiprolu PK, Jiang N, Morales CR, Kong Y et al. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci USA 2011; 108: 4123–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 2007; 316: 575–579. [DOI] [PubMed] [Google Scholar]
- da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, van Oort RJ, Pinto YM et al. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation 2008; 118: 1567–1576. [DOI] [PubMed] [Google Scholar]
- Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest 2009; 119: 2772–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Song Y, Zhang Y, Xiao H, Sun Q, Hou N et al. Cardiomyocyte overexpression of miR-27b induces cardiac hypertrophy and dysfunction in mice. Cell Res 2012; 22: 516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wang Y, Tan X, Jing H. MicroRNAs in autophagy and their emerging roles in crosstalk with apoptosis. Autophagy 2012; 8: 873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun 2012; 3: 1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M, Wang J, Wang C, Wang X, Dong W, Qiu W et al. MicroRNA-221 inhibits autophagy and promotes heart failure by modulating the p27/CDK2/mTOR axis. Cell Death Differ 2015; 22: 986–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA 2006; 103: 18255–18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res 2007; 100: 416–424. [DOI] [PubMed] [Google Scholar]
- el Azzouzi H, Leptidis S, Dirkx E, Hoeks J, van Bree B, Brand K et al. The hypoxia-inducible microRNA cluster miR-199a approximately 214 targets myocardial PPARdelta and impairs mitochondrial fatty acid oxidation. Cell Metab 2013; 18: 341–354. [DOI] [PubMed] [Google Scholar]
- Song XW, Li Q, Lin L, Wang XC, Li DF, Wang GK et al. MicroRNAs are dynamically regulated in hypertrophic hearts, and miR-199a is essential for the maintenance of cell size in cardiomyocytes. J Cell Physiol 2010; 225: 437–443. [DOI] [PubMed] [Google Scholar]
- Castedo M, Ferri KF, Kroemer G. Mammalian target of rapamycin (mTOR): pro- and anti-apoptotic. Cell Death Differ 2002; 9: 99–100. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet 2004; 36: 585–595. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol 2005; 17: 596–603. [DOI] [PubMed] [Google Scholar]
- Castets P, Lin S, Rion N, Di Fulvio S, Romanino K, Guridi M et al. Sustained activation of mTORC1 in skeletal muscle inhibits constitutive and starvation-induced autophagy and causes a severe, late-onset myopathy. Cell Metab 2013; 17: 731–744. [DOI] [PubMed] [Google Scholar]
- Paul PK, Bhatnagar S, Mishra V, Srivastava S, Darnay BG, Choi Y et al. The E3 ubiquitin ligase TRAF6 intercedes in starvation-induced skeletal muscle atrophy through multiple mechanisms. Mol Cell Biol 2012; 32: 1248–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahani MH, Itakura E, Mizushima N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy 2014; 10: 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S et al. TFEB links autophagy to lysosomal biogenesis. Science 2011; 332: 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Hong SK, Wu PK, Richards AL, Jackson WT, Park JI. Raf/MEK/ERK can regulate cellular levels of LC3B and SQSTM1/p62 at expression levels. Exp Cell Res 2014; 327: 340–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 2006; 126: 955–968. [DOI] [PubMed] [Google Scholar]
- Zhai P, Sadoshima J. Glycogen synthase kinase-3beta controls autophagy during myocardial ischemia and reperfusion. Autophagy 2012; 8: 138–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciarretta S, Volpe M, Sadoshima J. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circ Res 2014; 114: 549–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioi T, McMullen JR, Tarnavski O, Converso K, Sherwood MC, Manning WJ et al. Rapamycin attenuates load-induced cardiac hypertrophy in mice. Circulation 2003; 107: 1664–1670. [DOI] [PubMed] [Google Scholar]
- Marin TM, Keith K, Davies B, Conner DA, Guha P, Kalaitzidis D et al. Rapamycin reverses hypertrophic cardiomyopathy in a mouse model of LEOPARD syndrome-associated PTPN11 mutation. J Clin Invest 2011; 121: 1026–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkers M, Toko H, Doroudgar S, Din S, Quijada P, Joyo AY et al. Pathological hypertrophy amelioration by PRAS40-mediated inhibition of mTORC1. Proc Natl Acad Sci USA 2013; 110: 12661–12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Cao Y, Nie J, Liu H, Lu S, Hu X et al. Genetic and pharmacological inhibition of Rheb1-mTORC1 signaling exerts cardioprotection against adverse cardiac remodeling in mice. Am J Pathol 2013; 182: 2005–2014. [DOI] [PubMed] [Google Scholar]
- Wang J, Xu N, Feng X, Hou N, Zhang J, Cheng X et al. Targeted disruption of Smad4 in cardiomyocytes results in cardiac hypertrophy and heart failure. Circ Res 2005; 97: 821–828. [DOI] [PubMed] [Google Scholar]
- Guo SL, Ye H, Teng Y, Wang YL, Yang G, Li XB et al. Akt-p53-miR-365-cyclin D1/cdc25A axis contributes to gastric tumorigenesis induced by PTEN deficiency. Nat Commun 2013; 4: 2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen JR, Sherwood MC, Tarnavski O, Zhang L, Dorfman AL, Shioi T et al. Inhibition of mTOR signaling with rapamycin regresses established cardiac hypertrophy induced by pressure overload. Circulation 2004; 109: 3050–3055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.