Abstract
B-cell development in the bone marrow comprises proliferative and resting phases in different niches. We asked whether B-cell metabolism relates to these changes. Compared to pro B and small pre B cells, large pre B cells revealed the highest glucose uptake and ROS but not mitochondrial mass, whereas small pre B cells exhibited the lowest mitochondrial membrane potential. Small pre B cells from Rag1−/−;33.C9 μ heavy chain knock-in mice revealed decreased glycolysis (ECAR) and mitochondrial spare capacity compared to pro B cells from Rag1−/− mice. We were interested in the step regulating this metabolic switch from pro to pre B cells and uncovered that Swiprosin-2/EFhd1, a Ca2+-binding protein of the inner mitochondrial membrane involved in Ca2+-induced mitoflashes, is expressed in pro B cells, but downregulated by surface pre B-cell receptor expression. Knockdown and knockout of EFhd1 in 38B9 pro B cells decreased the oxidative phosphorylation/glycolysis (OCR/ECAR) ratio by increasing glycolysis, glycolytic capacity and reserve. Prolonged expression of EFhd1 in EFhd1 transgenic mice beyond the pro B cell stage increased expression of the mitochondrial co-activator PGC-1α in primary pre B cells, but reduced mitochondrial ATP production at the pro to pre B cell transition in IL-7 cultures. Transgenic EFhd1 expression caused a B-cell intrinsic developmental disadvantage for pro and pre B cells. Hence, coordinated expression of EFhd1 in pro B cells and by the pre BCR regulates metabolic changes and pro/pre B-cell development.
The hallmark of B-cell development is a continuous selection pressure imposed on pre B and B-cell receptors (BCRs), consisting of immunoglobulin (Ig) light chains (LCs) and heavy chains (HCs).1, 2 Ig μHC genes are rearranged in pro B cells and LC genes in small pre B cells.3, 4 Mice deficient in Rag genes show a developmental block at the pro B-cell stage and accumulate pro B cells in the BM.4 A rearranged μHC together with the surrogate LC, VpreB and λ5, forms the pre BCR in pre B cells. Hence, expression of a μHC on a Rag2−/− background in mice rescues development of phenotypic pre B cells.3 Expression of the pre BCR inhibits further rearrangements of V to D-J loci of the not yet rearranged μHC (allelic exclusion).1 The pre BCR also elicits an increase in the cytosolic Ca2+ concentration 5 and acts as an inducible proliferative signal of pre B cells with an expansion factor of 20–100 (about four to six cell divisions).6 IL-7 provides important signals for pro B-cell proliferation7 via stabilization of mitochondria.8 In vitro, IL-7 supports the proliferation of pro B cells expressing ckit (Hardy fraction B and C).9, 10 IL-7 also promotes glycolysis via Akt activation, 8, 11, 12 but from pro to immature B cells, the responsiveness to IL-7 decreases.7, 13 In parallel to the decreasing IL-7 responsiveness, the pre BCR initiates expression of SLP-65/BLNK. This allows large pre B differentiation into small pre B cells through inhibition of the PI3K/Akt pathway and upregulation of FOXO1.13, 14, 15 LC rearrangement in small pre B cells enables BCR expression on immature B cells that migrate to the spleen and mature to resting follicular and marginal zone B cells. Resting splenic B cells exhibit a low metabolic activity and consume fatty acids to produce ATP via oxidative phosphorylation (OxPhos).16 Upon activation through the BCR or through TLR4 by lipopolysaccharide, murine B cells upregulate both OxPhos and glycolysis, concomitant with the glucose transporter glut1, in a balanced ratio.16 Anabolic pathways, controlled by PI3K/Akt/mTor or Erk, favor proliferation and often rely on glycolysis.17 In particular, the PI3K and Erk pathways control proliferation during pre B cell development.12, 13, 18 Genes controlling mitochondria and glycolysis are targets of the pre BCR and the BCR.16, 19 This infers that the metabolic machinery of B cells integrates signals downstream of (pre) BCR receptors together with growth factors,.13, 16 The pre B cell checkpoint is subject to control by cellular metabolism: mice with a null mutation of the folliculin-interacting protein 1 (Fnip1) reveal a block at the large pre B-cell stage due to imbalanced metabolism.20 However, these data cannot explain the pro/pre B-cell transition. Swiprosin-2/EFhd1 (EFhd1)21 is a protein of the inner mitochondrial membrane.22, 23, 24 We revealed its expression only in the Bcr-Abl expressing pro B-cell line 38B9,25 but not in B-cell lines expressing a pre BCR or a BCR.26 EFhd1 is a target gene of two transcription factors involved in B-cell identity and control of early B-cell development, Foxo1 and Brg1, in pro B cells.27, 28 In very aggressive human melanomas, EFhd1 is co-expressed with PGC-1α, a co-activator of PPAR that is involved in mitochondrial biogenesis.29 EFhd1 becomes upregulated, together with PGC-1α, UCP2 and other proteins involved in mitochondrial function, in distal convoluted tubule cells of the kidney through deletion of the cytosolic Ca2+ buffer parvalbumin 30 and is a positive regulator of mitochondrial flashes (mitoflashes) via Ca2+ binding to its EF hands.31 Mitoflashes indicate stochastic and brief periods of alkalinization in individual mitochondria, possibly as a result of acceleration in proton pumping, triggered by mitochondrial fusion and/or a change in ion homeostasis.32 Here, we tested the hypothesis that metabolism is altered during early B-cell development in the BM. We reveal that EFhd1 is normally downregulated by the pre BCR. Prolonged expression of EFhd1 beyond the pro B-cell stage in transgenic mice (EFhd1tg) counterbalances metabolic changes induced through the pre BCR. Hence, regulation of mitochondrial genes and metabolism by the pre BCR is crucial for B-cell development.
Results
Metabolism changes at the pro B to small pre B transition
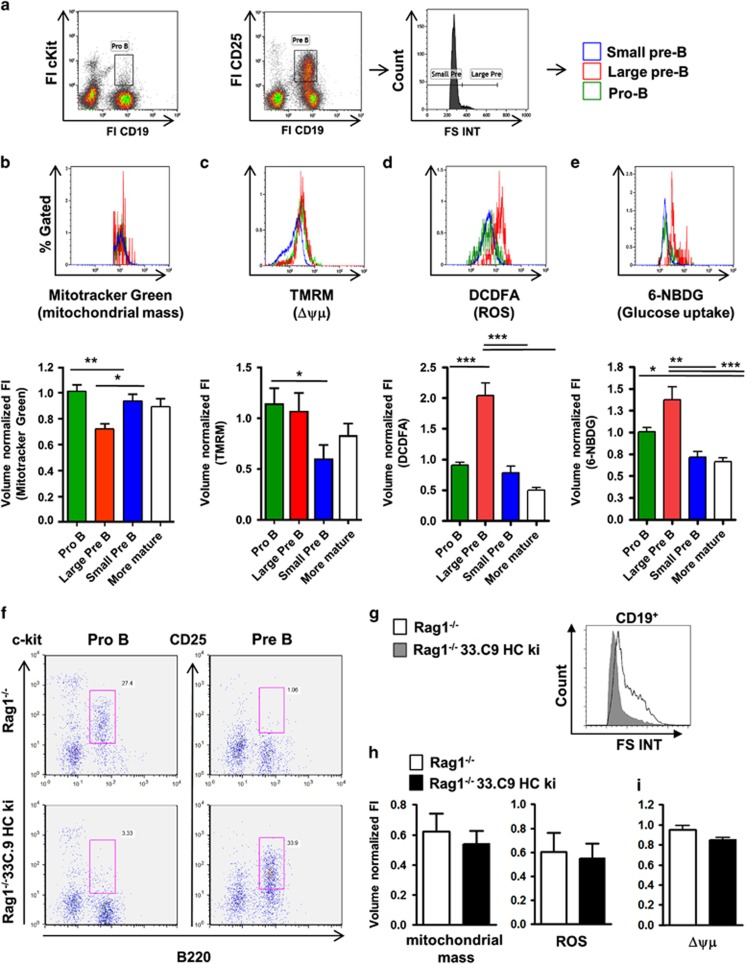
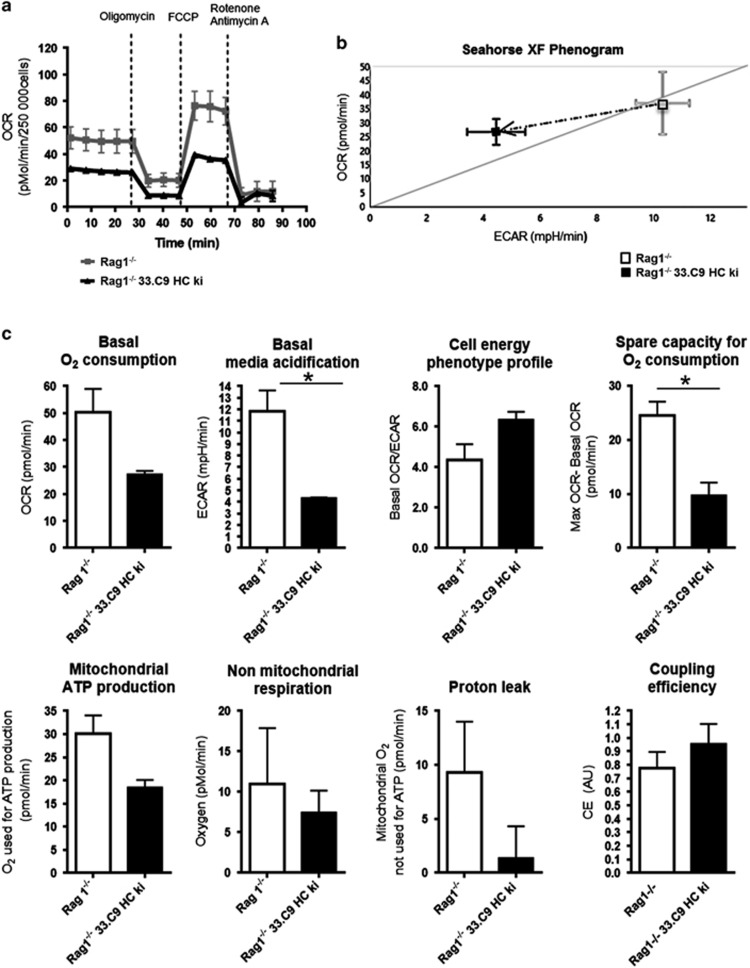
To characterize metabolic changes during early B-cell development in the BM, we used flow cytometry and assessed total mitochondrial mass (Mitotracker Green), mitochondrial membrane potential (Δψμ; TMRM), reactive oxygen species (ROS) (DCDFA) and glucose uptake (6-NBDG) normalized to cell size 33 (Figures 1a–e). Assessment of Δψμ with TMRM was calibrated with a protonophore (Supplementary Figure S1). Mitochondrial mass relative to cell size went down in large pre B cells, but remained constant during later B-cell development (Figure 1b). Pro B cells exhibited the highest Δψμ that decreased significantly in small pre B cells (Figure 1c). ROS production (Figure 1d) and glucose uptake (Figure 1e) were highest in large pre B cells and were reduced again in small pre B cells. We concluded that large pre B cells are metabolically more active than small pre B cells without having increased mitochondrial mass. These data correlate well with clonal expansion of large pre B cells. To functionally test for a metabolic transition of pro to small pre B cells, we established a μHC knock-in (ki) mouse model (33.C9μHCki) and crossed these mice to Rag1−/− mice4 (Rag1−/−; 33.C9μHCki). In accordance with other transgenic μHC crossed onto a Rag-deficient background,3 33.C9μHC expression established pre B-cell development (Figure 1f). Pre B cells from Rag1−/−;33.C9μHCki mice were mainly small (Figure 1g). In full accordance with data from wild-type (WT) pro and small pre B cells, Rag1−/−;33.C9μHCki pre B cells showed no difference in mitochondrial mass and ROS when compared to Rag1−/− pro B cells (Figure 1h), and a trend towards reduced Δψμ (P=0.055) as shown by flow cytometry and confocal microscopy (Figure 1h; Supplementary Figure S1). We then used a functional assay to assess metabolic activity of pro and small pre B cells. Extracellular flux analyses performed with a Seahorse device revealed in general a reduced oxygen consumption rate (OCR; an indicator for oxidative phosphorylation/OxPhos) and extracellular acidification rate (ECAR; an indicator for aerobic glycolysis) in Rag1−/−;33.C9μHCki pre B cells (Figures 2a–c). However, in relation, glycolysis was reduced significantly more; therefore, the OCR/ECAR ratio tended to increase (Figures 2b and c). In accordance, there were trends towards better mitochondrial coupling efficiency and less proton leak (Figure 2c). In contrast, mitochondrial spare capacity (maximal/basal O2 consumption) was significantly reduced in Rag1−/−;33.C9μHCki pre B cells, and there were tendencies towards less mitochondrial ATP production and respiration (Figure 2c). We conclude that expression of the pre BCR ultimately promotes metabolic quiescence, with a dominant reduction of glycolysis. A remaining question was how the metabolic transition of pro over highly active pre B cells towards rather inactive small pre B cells is handled.
Figure 1.
Analysis of mitochondrial activity and glucose uptake of pro and pre B cells by flow cytometry. (a) BM cells from C57BL/6 mice were incubated with fluorescent probes and then simultaneously stained for surface antigens (Pro B cells: CD19+, cKit+, large pre B cells: CD19+, CD25+, FS INThi, small pre B cells: CD19+, CD25+, FS INTlow and more differentiated B-cell populations (CD19+, cKit−, CD25−) and analyzed by flow cytometry. Representative plots of the median fluorescence intensity for Mitotracker Green (b), TMRM (c), DCFDA (d) and 6-NBDG (e) are shown. Fluorescence was normalized to spheroid cell volume by FS TOF signal as an indicator for cell size. TMRM fluorescence was normalized with CCCP. Data were normalized to pro B cells and are represented as median±S.E.M., n=10 C57BL/6 mice from two experiments (b, d and e), n=5 mice from four experiments (c). Significance was tested by ANOVA with Bonferroni correction, except for C (two-tailed Student’s t-test; only pro and small pre B cells were compared). (f) BM of Rag1−/− and Rag1−/− 33.C9μHCki mice was analyzed by flow cytometry (Pro B cells: CD19+, cKit+, pre B cells: CD19+, CD25+), (g) Histograms representing FS Int of pro and pre B cells of Rag1−/− and Rag1−/− 33.C9μHC ki mice, (h) CD19-positive BM cells of Rag1−/− and Rag1−/−;33.C9μHCki mice were stained with Mitotracker Green and DCDFA, analyzed by flow cytometry and normalized to cell volume and pro B cells. Results are represented as mean±S.D., n=5, (i) CD19-positive BM cells of 5 Rag1−/− and Rag1−/−;33.C9μHCki mice were stained with TMRM, duplicate CCCP-calibrated samples were analyzed by flow cytometry and normalized to cell volume and pro B cells. Results are represented as mean±S.E.
Figure 2.
Influence of μ heavy chain expression on metabolic activity of BM B lymphocytes. (a) BM cells of Rag1−/− and Rag1−/−;33.C9μHCki mice were pooled and pro and pre B lymphocytes were isolated by positive selection (anti-CD19) from Rag1−/− and Rag1−/− 33.C9 HC ki mice. Mitochondrial activity of pro and pre B cells was directly analyzed by Mito Stress Test with a Seahorse XFe96 extracellular flux analyzer. Five readings were taken for basal oxygen consumption rate (OCR) before subsequent addition of oligomycin, FCCP and antimycin+rotenone; summary of three experiments with each two to four pooled mice, (b) Representative cellular energy phenotype profiles (basal OCR/ basal extracellular acidification (ECAR) of Rag1−/− and Rag1−/−;33.C9μHCki cells from one experiment are compared in a 2D plot, (c) The basal OCR, ECAR and the cell energy phenotype profile (OCR/ECAR) are shown as bar diagrams. Also shown are the mitochondrial spare capacity (the increase from basal to maximum OCR when uncoupling mitochondrial proton flow by FCCP), the mitochondrial ATP production rate (OCR, which can be blocked by adding the mitochondrial ATPase inhibitor oligomycin), the non-mitochondrial respiration (O2 consumption after inhibition of mitochondria by oligomycin, (FCCP), antimycin and rotenone), proton leak (difference in O2 consumption after oligomycin and full mitochondrial inhibition by additional antimycin A and rotenone) and coupling efficiency (ratio of mitochondrial ATP production and proton leak). Summary of n=3 with each two to four pooled mice
The pre BCR downregulates EFhd1
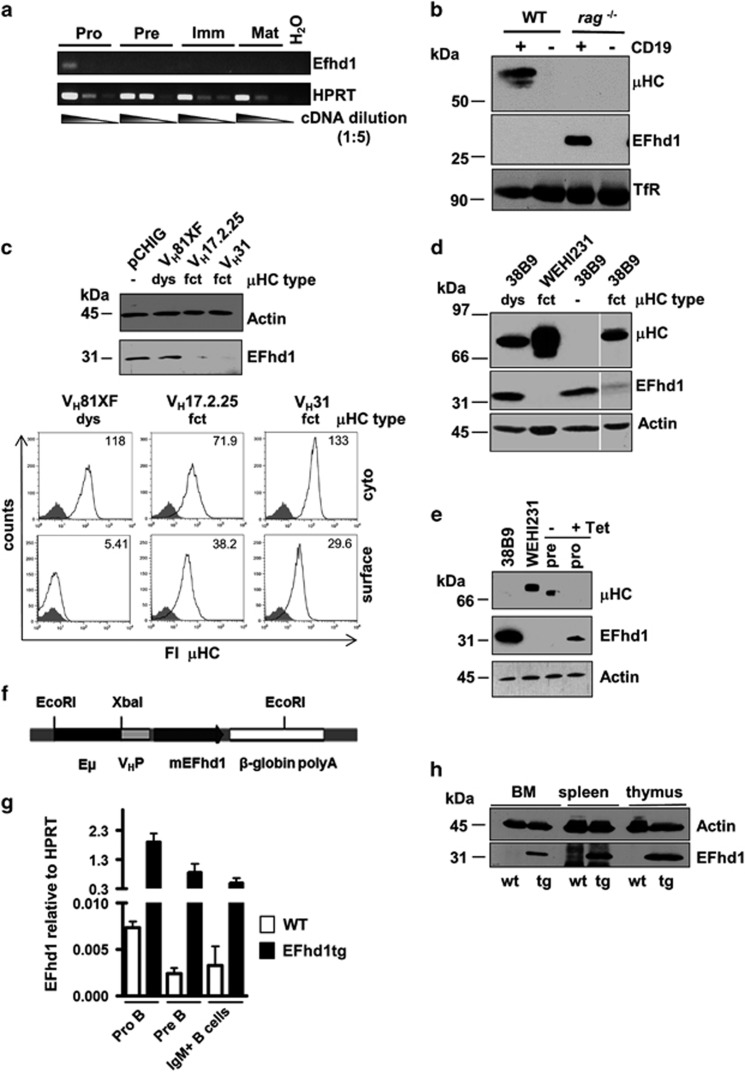
To assess the relevance of the metabolic switch at the pro/pre B-cell transition, we made use of the EF hand containing Ca2+-binding mitochondrial protein Swiprosin-2/EFhd1. We had detected its expression only in the pro B-cell line 38B9.26 RT-PCR analysis of sorted BM B cells34 revealed the strongest efhd1 mRNA expression in pro B cells (Figure 3a). Western blot analysis of pro B cells from Rag1−/− mice confirmed EFhd1 protein expression in pro B cells. EFhd1 was neither detectable in total BM IgM+ CD19+ B cells because pro B cells represent only 1% within CD19+ cells, nor in CD19− cells (Figure 3b). Retroviral transduction of the Rag2−/− IL-7-dependent pro B cell line R5B35 with recombined μHC (functional μHC, μ fct) showed that surface pre BCR formation led to downregulation of EFhd1 at the protein level (Figure 3c). Cytoplasmic μHC expression, which was achieved by expression of the dysfunctional μHC VH81.X (μ dys),36 could not downregulate EFhd1 (Figure 3c). Similar results were obtained with 38B9 cells (Figure 3d). Inducible expression of the pre BCR by removal of tetracycline of pro B-cell cultures from Rag2−/− dTg (Ig-tTA/tet-μ) mice6 led to downregulation of EFhd1 on the protein level in those primary cells (Figure 3e). Retrospective analysis of microarray data of pre BCR-regulated genes19 confirmed these results (efhd1 signal in pro B cells: array 1/2, raw data 625.5/723.9, efhd1 signal in pre B cells: array 1/2, raw data 90.6/147.2). We reasoned that the downregulation of efhd1 by the pre BCR could support a hitherto unknown role of the pre BCR in regulating metabolic function.
Figure 3.
Downregulation of EFhd1 by the pre BCR and establishment of EFhd1tg mice. (a) Pro, pre and immature B cells of the BM were isolated through FACS and mature B cells of the spleen by MACS. Total RNA from the indicated cells was isolated, reversely transcribed to cDNA and amplified with efhd1 or hprt-specific primers in fivefold serial dilutions, (b) BM cells of WT and Rag1−/− mice positively purified via anti-CD19 and protein lysates of CD19+ and CD19− cells were subjected to western blot analysis using anti transferrin receptor (TfR), anti μHC and anti EFhd1 antibodies, (c) IL-7-dependent μHC-negative R5B pro B cells were infected with retroviral particles derived from pCHIG or various functional (fct) and dysfunctional (dys) μHC idiotypes cloned into pCHIG, selected and analyzed by western blot using anti Actin and anti EFhd1 antibodies (upper panel). Cells were either analyzed for surface μHC or cytoplasmic μHC expression by flow cytometry (lower panel). MFI is indicated, (d) Lysates of WEHI231 cells, 38B9 cells expressing no μHC, a dysfunctional or a functional μHC were subjected to western blot analysis using anti μHC, anti Actin and anti EFhd1 antibodies. Alle lanes are from the same western blot and exposure, (e) Lysates of 38B9, WEHI231 and pro and pre B cells obtained from a double transgenic tetracycline (tet)-off controlled SP6 μHC expression system were subjected to western blot analysis using anti μHC, anti Actin and anti EFhd1 antibodies. In this experiment, 73.6% of induced cells (- tet) expressed the SP6 μHC, (f) Schematic of the transgenic expression cassette, Eμ: μ enhancer, VHP: VH promoter, (g) Pro, pre and immature B cells isolated through FACS from wild-type or EFhd1tg mice were analyzed by qPCR for EFhd1 expression, (h) Protein lysates of total BM, spleen and thymus were analyzed by western blot using anti Actin and anti EFhd1 antibodies. Molecular mass standards are indicated on the left (kDa)
Ectopic EFhd1 expression distorts early B-cell development
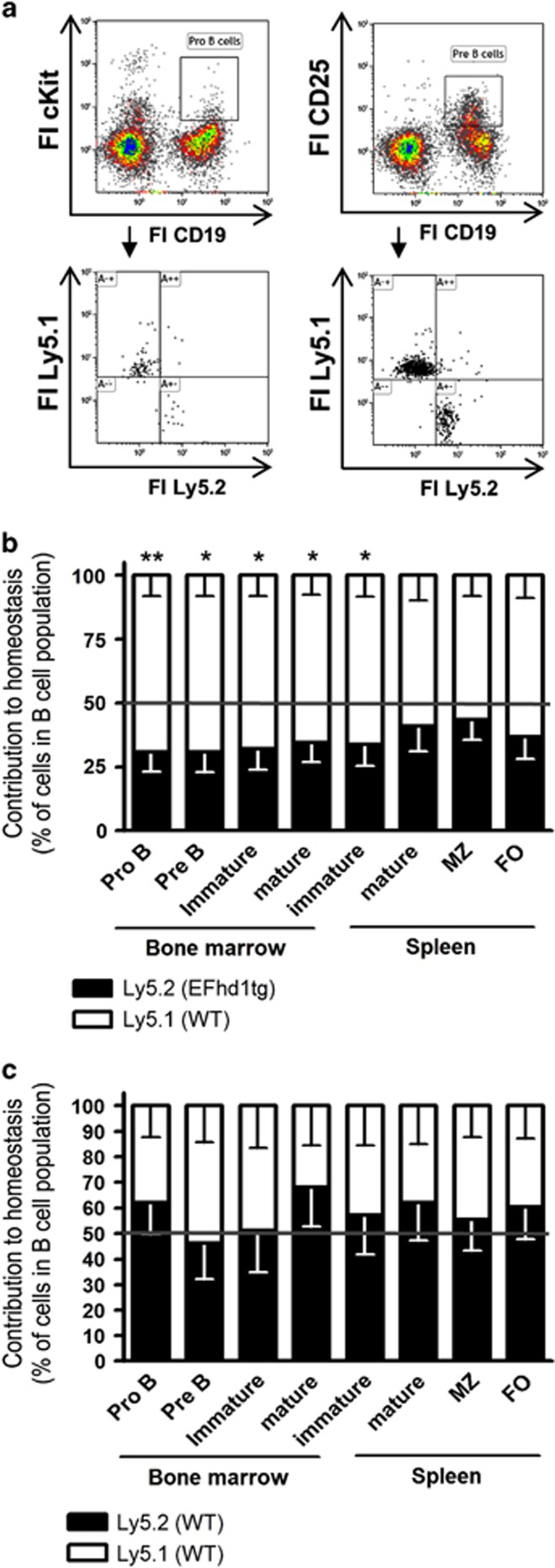
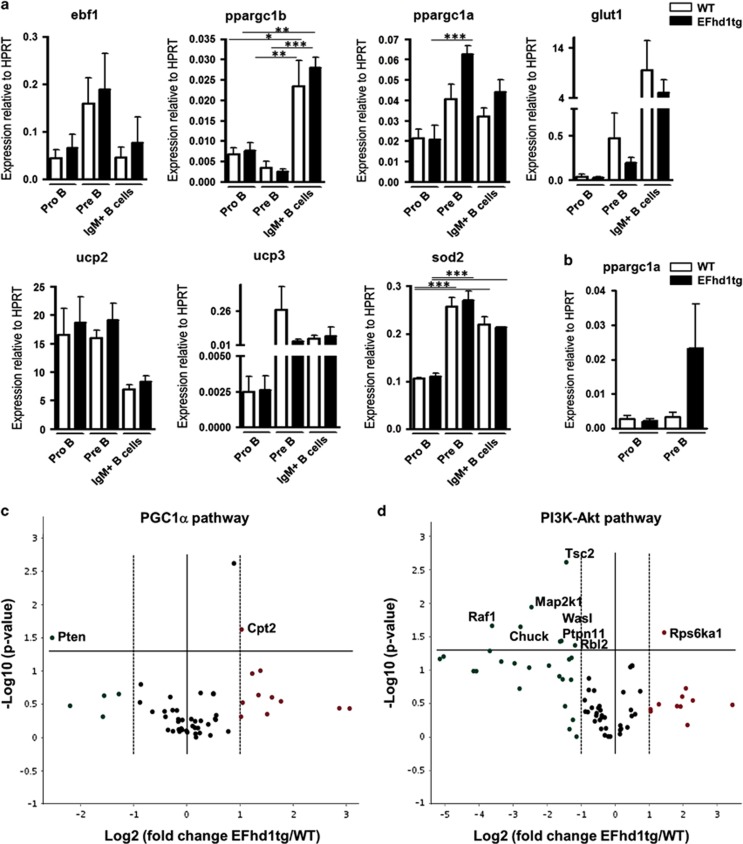
To test whether pre-BCR-induced downregulation of EFhd1 serves as a metabolic checkpoint, we generated EFhd1 transgenic mice (EFhd1tg) expressing EFhd1 under control of the Eμ enhancer and VH promoter (Figure 3f). Southern blot and circle PCR analysis revealed that seven copies of the transgene had inserted in a non-coding region of the gene ms4a12 on chromosome 19 (data not shown). Ms4a12 is a colon-specific protein,37 and thus, integration of the EFhd1 transgene per se should not interfere with B cell development. Quantitative PCR (qPCR) analysis confirmed ectopic expression of EFhd1 in pre B cells and IgM+ B cells, and once more its downregulation in WT pre B cells (Figure 3g). Ectopic EFhd1 protein was detected in BM, spleen and thymus, because the Eμ enhancer is also active in T cells (Figure 3h).38 Interestingly, we also noted reduced expression of ectopic EFhd1 in pre B cells (Figure 3g), suggesting that only a certain expression level of EFhd1 is tolerated in pre B cells. Hence, we tested whether prolonged EFhd1 expression intrinsically affects early B-cell development. Mixed BM chimeras (EFhd1tg; Ly5.2/WT; Ly5.1 or WT; Ly5.2/WT; Ly5.1) (Figure 4a) showed a competitive disadvantage of EFhd1tg B cells in the pro B, pre B, immature B-cell compartments in BM and spleen, as well as in recirculating BM B cells (Figure 4b). This was not due to differential homing (Supplementary Figure S2). WT/WT chimeras did not show significant differences (Figure 4c). Thus, prolonged expression of EFhd1 beyond the pro B-cell stage and ectopic expression of EFhd1 in pro B cells distorts early B-cell development. We next sorted pro, pre (large and small) and IgM+ B cells from BM and assessed expression of genes involved in glucose uptake (glut1), mitochondrial function, biogenesis and ROS detoxification (ppargc1a, ppargc1b, sod2, ucp2, ucp3).39, 40 Pre B cells expressed more ebf1, as expected,41 and upregulated ppargc1a (encoding PGC-1α), glut1, ucp3 and sod2 (Figure 5a). Upregulation of sod2 in pre B cells correlates well with its function in suppressing EFhd1 expression.42 Ppargc1b became strongly upregulated in IgM+ B cells (Figure 5a). EFhd1 expression significantly enhanced ppargc1a mRNA in pre B cells. Interestingly, upregulation of ppargc1a by EFhd1 was also visible in in pre B cells of IL-7 cultures (Figure 5b). Whereas IgM+ B cells downregulated ucp2, pre B cells, and IgM+ B cells upregulated ucp3, pointing to a change in mitochondrial coupling. As EFhd1 upregulated PGC-1α but rather prevented upregulation of glut1, an Akt target gene43 in pre B cells (Figure 5), we next evaluated expression of genes involved in PGC-1α and Akt pathways through qPCR arrays (RT2 profiler) using cDNA of sorted pre B cells (pool of large and small) (Figures 5c and d). These experiments uncovered a significant EFhd1-mediated upregulation of the PGC-1α target gene carnitine palmitoyl transferase 2 (cpt2)44 and downregulation of pten (Figure 5c). Most of the other genes upregulated by EFhd1, although not significantly in this assay, are PGC-1α target genes44 (Figure 5c). Our analysis also unraveled dysregulation of the Akt pathway (Figure 5d). We observed a downregulation of genes involved in canonical Erk signaling (ptpn11, raf1, map2k1), downregulation of tsc2 and upregulation of rps6ka, indicating activation of the mTOR pathway.20 These data and the fact that EFhd1 increased ppargc1a predicted that EFhd1 expression favors mitochondria as the preferential source of energy, leading to an increase in the OCR/ECAR ratio, but may simultaneously induce an anabolic phenotype, that is, increased ATP consumption and finally exhaustion.20
Figure 4.
Contribution of EFhd1tg precursors to B-cell development in mixed bone marrow chimeras. CD45.1 BL6 (Ly5.1) recipient mice were lethally irradiated with 11 Gy and reconstituted with 1 × 106 WT CD45.1 (Ly5.1) and 1 × 106 CD45.2 (Ly5.2) EFhd1tg or wild-type liitermate BM cells. After 6 weeks repopulation, the contribution of CD45.1 and CD45.2 to B-cell populations in the BM and spleen was analyzed by flow cytometry for pro B (CD19+, cKit+), pre B (CD19+, CD25+), immature (CD19+, IgM+, IgD−), mature (CD19+, IgM+, IgD+), marginal zone (CD19+, CD21high, CD23−) and follicular (CD19+, CD21low, CD23+) B cells. (a) Representative analysis of WT/EFhd1tg chimeras. (b) Summary of WT/EFhd1tg chimeras. (c) Summary of WT/WT chimeras. Data are represented as mean+S.E.M. of 7–11 mice from four experiments. Statistics was done by Mann–Whitney-U test
Figure 5.
Expression of metabolic and mitochondrial genes in BM B lymphocyte subsets. (a) BM was isolated from EFhd1tg and WT mice and pro B (CD19+, cKit+, CD25−, sIgM−), pre B cells (CD19+, cKit−, CD25+, sIgM−) and BCR+ B cells (CD19+, CD25−, cKit−, sIgM+) were sorted by FACS. RNA was isolated, and converted into cDNA. Expression of ebf1, ppargc1a, ppargc1b, glut1, ucp2, ucp3 and sod2 was evaluated by SYBR Green qPCR, analyzed with the ΔΔCT method and normalized to hprt. mRNA expression of is represented as mean±S.E.M. of three independent experiments and was tested for statistical significance by ANOVA with Bonferroni correction for multiple testing, (b) RNA was isolated from sorted pro (CD19+, cKit+, sIgM−) and pre B (CD19+, cKit−, sIgM−) of EFhd1tg and WT mice after 7 days culture with 5 ng/ml IL-7. RNA was converted to cDNA and ppargc1a (PGC-1α) expression was analyzed as described in (a). Data is represented as mean+S.E.M. of four independent cultures and sorts per genotype and statistical analysis was carried out by Mann–Whitney-U test. (c and d) RNA was isolated from pre B cells (CD19+, cKit−, CD25+, sIgM+) sorted from EFhd1tg and WT BM. cDNA conversion and expression profile analysis were carried out by Qiagen Mouse PPAR Targets (a) or (b) PI3K-Akt pathway RT2 Profiler PCR Array. Data was analyzed with the available online software (www.SABiosciences.com/pcrarraydataanalysis.php). Normalization was carried out by an automatic algorithm against suitable genes of the whole plate. The data are represented as heat map and volcano plot of Log2 fold change in gene expression of EFhd1tg to WT pre B cells against −Log10 of the P-value. Values represent the mean of three EFhd1tg against three WT mice
EFhd1 decreases mitochondrial ATP production in pre B cells
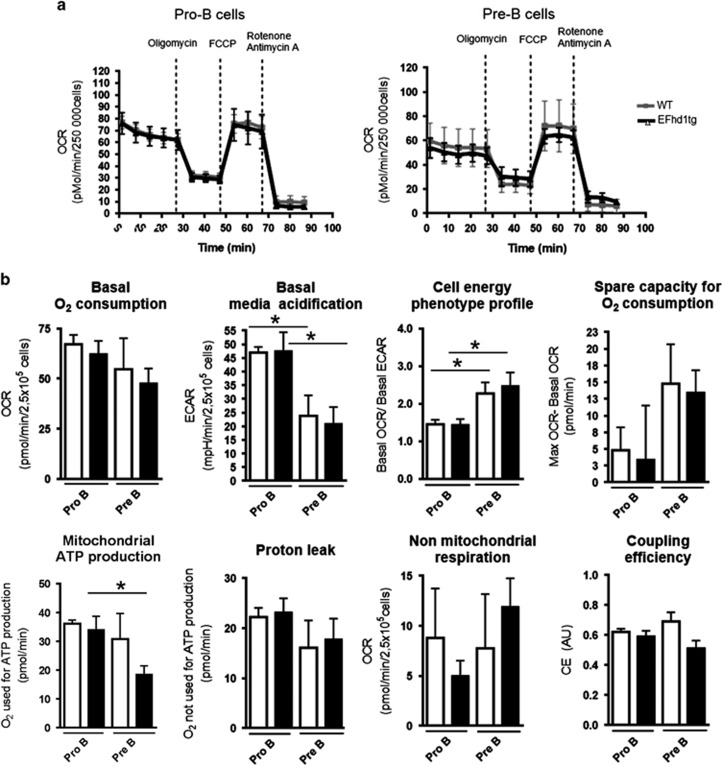
To test the hypothesis that EFhd1 alters mitochondrial activity or glycolysis, we performed extracellular flux analyses with a Seahorse device, using WT and EFhd1tg pro and pre B cells. Because of the very limited cell numbers obtainable by cell sorting (~105 pro B cells per mouse, with 1.6 × 106 cells needed per mouse per experiment), we had to expand pro B cells using IL-7 ex vivo cultures. We chose an IL-7 concentration where pro B cells do not outnumber pre B cells (Supplementary Figures S4 and S5, related to Figure 6). IL-7 derived pro and pre B cells behaved very similar to directly ex vivo assayed pro and pre B cells of the Rag1−/−;33.C9 μHCki system (Figure 6; compare Figure 2) since pre B cells were smaller than pro B cells (Figure 1; Supplementary Figure S3). However, pro and pre B cells from IL-7 cultures were in general larger than their freshly isolated counterparts (Supplementary Figure S3). Pre B cells from IL-7 cultures showed also a significant downregulation of glycolysis (ECAR) when compared to pro B cells, yielding a significantly increased OCR/ECAR ratio (Figures 6a and b). In contrast to the Rag1−/−;33.C9 μHCki system, pre B cells from IL-7 cultures exhibit a higher mitochondrial spare capacity (ratio of maximal to basal O2 consumption) than pro B cells from IL-7 cultures (Figure 6b). Thus, IL-7 does exert major effects on pro and pre B cell metabolism. IL-7 cultures from WT and EFhd1tg mice were comparable in proliferation and differentiation (Supplementary Figures S3 and S4), but freshly isolated WT pro B cells briefly expanded in the absence of IL-7, whereas EFhd1tg pro B cells did not do so at all (Supplementary Figure S4). Ectopic expression of EFhd1 had only minor effects on ECAR and OCR (Figures 6a and b). Unexpectedly, but in line with a competitive disadvantage of EFhd1tg pre B cells, EFhd1 expression resulted in significantly reduced mitochondrial ATP production (Figure 6b). Like in the Rag1−/−;33.C9 μHCki system, albeit not to the same extent, mitochondrial coupling efficacy increased slightly from WT pro to pre B cells. However, EFhd1 expression in pre B cells reduced mitochondrial coupling efficacy at the pro to pre B cell transition (P=0.057) (Figures 6d and e). Taken together, prolonged expression of EFhd1 in pre B cells distorts mitochondrial ATP production. This was paralleled by a less efficient induction of mitochondrial coupling at the pro to pre B-cell transition.
Figure 6.
Metabolic profile of EFhd1tg and WT IL-7 pro B cell cultures. (a) Sorted pro B cells were cultured with 5 ng/ml IL-7. After 7 days, pro (CD19+, cKit+, sIgM−) and pre B (CD19+, cKit−, sIgM−) cells were sorted and metabolic activity of pro and pre B cells from the IL-7 culture was directly determined by Mito Stress Test with a Seahorse XFe96 analyzer. Five basal readings were taken for oxygen consumption rate (OCR) and basal extracellular acidification rate (ECAR). The results are represented as mean±S.E.M. Three readings were taken after complex V (ATPase) inhibition by oligomycin and three readings for maximum OCR were taken after addition of the mitochondrial uncoupler FCCP. (b) An energy phenotype of the cells is calculated by dividing OCR by ECAR. Spare capacity (the increase from basal to maximum OCR) and the mitochondrial ATP production rate (the OCR which can be blocked by adding ATPase inhibitor oligomycin) are shown as bar diagrams. Mitochondrial proton leakage, (OCR which can be further blocked by antimycin A and rotenone after oligomycin administration) and non-mitochondrial respiration (OCR remaining after fully blocking the mitochondria with oligomycin, antimycin A and rotenone) were calculated. The coupling efficiency represents the ratio of mitochondrial ATP production and proton leak. Data are represented as mean±S.E.M. of four independent sorts and seahorse experiments of each time three pooled EFhd1tg and WT mice. Statistical analysis was done by ANOVA with Bonferroni correction for multiple testing
EFhd1 knockdown decreases the OCR/ECAR ratio in 38B9 cells
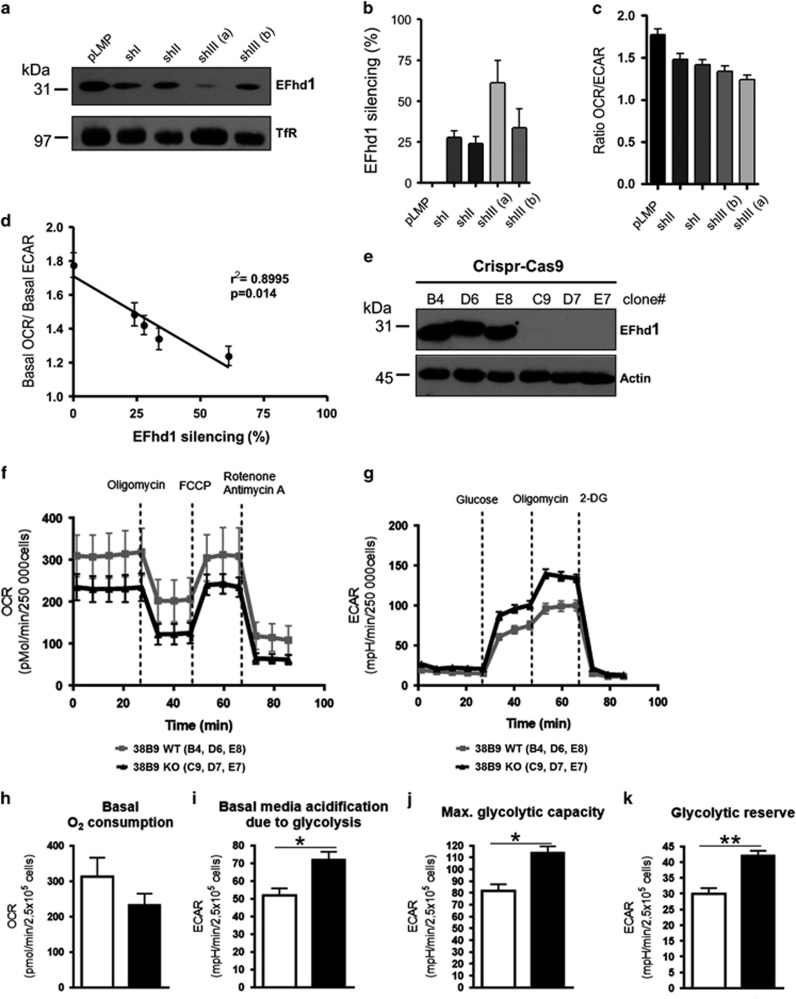
Next, we wished to assess the mitochondrial function of EFhd1 in pro B cells independent of IL-7. To this end, we knocked down EFhd1 by an shRNA approach using three different shRNAs that resulted in different knockdown efficacies in the 38B9 pro B-cell line (Figures 7a and b; knockdown efficacy is indicated by a gray color code). We then tested mitochondrial activity and glycolysis by measuring OCR and ECAR. The observed reduction of the OCR/ECAR ratio correlated directly and in a linear manner with reduction of EFhd1 expression (Figures 7b and c) and suggested increased glycolytic rates. As knockdown efficacies were low, we established Crispr-Cas9 mediated knockout of EFhd1 (Figure 7e). 38B9 EFhd1 knockout cells revealed trends towards decreased OCR, but significantly increased basal glycolysis, maximal glycolytic capacity and glycolytic reserve (Figures 7f and g). Consequently, the OCR/ECAR ratio was reduced (P=0.052, Student’s t-test). Taken together, we have unraveled for the first time metabolic adaption during early B-cell development and pre-BCR-induced regulatory signals controlling mitochondrial metabolism, ATP synthesis and glycolysis through regulation of the Ca2+-binding mitochondrial protein EFhd1.
Figure 7.
Metabolic profile of 38B9 pro B-cell lines with shRNA-mediated EFhd1 knockdown. (a) 38B9 cells stably transfected with pLMP and four stable knockdown cell lines were analyzed for EFhd1 protein expression by western blot, Molecular mass standards are indicated on the left (kDa). (b) EFhd1 expression was normalized to TfR protein expression and represented as mean percentage±S.E.M. in comparison to pLMP, n=3, (c) The metabolic activity of the 38B9 cell lines was analyzed by Mito Stress Test with a Seahorse XF analyzer. Basal OCR, as well as the basal ECAR and the cell energy phenotype profiles (OCR/ECAR) are represented as mean±S.D. of three to five measurements, (d) Correlation of the percentage of EFhd1 silencing with the cells’ energy phenotype profile (OCR/ECAR), (e) Western blot analysis of Crispr-Cas9 engineered 38B9 EFhd1 knockout and control clones, (f) Mitochondrial activity of of randomly chosen 38B9 WT and EFhd1 knockout clones was analyzed by Mito Stress Test with a Seahorse XFe96 extracellular flux analyzer. Five readings were taken for basal oxygen consumption rate (OCR) before subsequent addition of oligomycin, FCCP and antimycin+rotenone, (g) Glycolytic activity of randomly chosen 38B9 WT and EFhd1 knockout clones was analyzed by Glyco Stress Test with a Seahorse XFe96 extracellular flux analyzer. Five readings were taken for basal ECAR in glucose-free medium before subsequent addition of glucose, oligomycin and 2-deoxyglucose (2-DG), (h) Basal O2 consumption from Mito stress test, (i) Basal media acidification (glycolysis stress test), (j) Maximal glycoloytic capacity after addition of glucose (glycolysis stress test), (k) Maximal glycoloytic capacity after addition of oligomycin (glycolysis stress test). Statistical analysis was done with Student’s t-test
Discussion
Previously, IL-7/CXCL12 cultures of mixed pro/pre B cells from WT and Fnip1−/− mice had been analyzed by extracellular flux analysis.20 Yet, to our best knowledge, neither a comprehensive comparative metabolic analysis during B-cell development in the BM nor a metabolic analysis of separated primary pro and pre B cells in the absence or presence of IL-7 has been performed so far. We noted that IL-7 has specific effects: It increases mitochondrial spare capacity and it maintains mitochondrial ATP production in cultured small pre B cells. Nevertheless, several results obtained with the Rag1−/−;33.C9μHCki system in the absence of IL-7 were corroborated in IL-7 cultures of pro and pre B cells (decreased ECAR, increased OCR/ECAR ratio in small pre B cells). We demonstrate that small pre B cells downregulate their metabolic activity even when compared to pro B cells, and that large pre B cells are superior in glucose uptake and ROS production. The fact that we observed the most glucose uptake by large pre B cells matches the higher energy demand certainly required in large pre B cells. Increased metabolic activity in large pre B cells might be at least partially driven by the higher glucose consumption. A finding that supports our interpretation of increased metabolic activity in large pre B cells is the increased ROS production in large pre B cells, as ROS is a byproduct of a highly active mitochondrial respiratory chain. It is relevant in this respect that downregulation of EFhd1 in 38B9 pro B cells, normally achieved by surface pre BCR expression, increases glycolytic parameters. The downregulation of EFhd1 by the pre BCR represents therefore one possible mechanism of how the pre BCR increases metabolic activity in large pre B cells. Our extracellular flux analysis of primary pro and small pre B cells missed large pre B cells due to technical reasons. Large pre B cells may in the future be enriched from Irf4/8 double or BLNK/SLP-65 knockout mice. To sum up, it is tempting to speculate that increased aerobic glycolysis occurs in large pre B cells through downregulation of EFhd1, providing more energy and more anaplerotic reactions for macromolecules and intermediate products protecting from ROS.45 Glucose uptake dropped in small pre B cells, even below the level of pro B cells. In full accordance, we measured a decline of ECAR in small pre B cells as compared to pro B cells. The PI3K/Akt pathway regulates glycolysis positively and is an important regulator of B-cell differentiation.12 Thus, the decrease of glycolysis (as defined by ECAR) in small pre B cells compared to pro B cells is in accordance with inactivation of the Akt pathway and may implement nuclear commands signaling transient quiescence.13, 46 Vice versa, increased glycolysis in 38B9 EFhd1 knockout pro B cells may comprise a mechanism of early pre BCR signaling, leading normally to downregulation of EFhd1. We observed downregulation of glucose uptake, Δψμ and ROS generation in small pre B cells. Together with Pax5, FOXO1 transactivates Rag1/2, IRF4 and p27 that induces cell cycle arrest while LC rearrangement takes place in small pre B cells.28, 39, 47 FOXO1 is also activated by ROS and upregulates superoxide dismutase 2 (sod2).40 The increase in the FOXO1 target gene, sod2, in pre B cells is compatible with an induction of FOXO1 through inactivation of Akt40 and might limit ROS. Although small pre B cells reduced both glycolysis (ECAR) and OxPhos (OCR), ECAR was in relation reduced more. These observations were congruent in pre B cells from both the 33.C9 μHC system and from WT IL-7 cultures. Our interpretation is that pro B cells rely more on aerobic glycolysis for energy production than small pre B cells. OxPhos often takes over in cells differentiating away from stem cells.48 The shift towards a more mitochondrial metabolism in small pre B cells is therefore in line with their advanced differentiation status. In line, we uncovered that the mitochondrial co-activator PGC-1α 49 is increased in pre B cells. We have uncovered a causative relation between EFhd1 and PGC-1α: EFhd1tg pre B cells show significantly increased PGC-1α expression. In accordance, they revealed a slightly increased OCR/ECAR ratio, with knockout of EFhd1 in the pro B-cell line 38B9 showing the opposite. Although PGC-1α stimulates mitochondrial biogenesis,49 we did not observe increased mitochondrial mass in EFhd1 expressing pre B cells (unpublished data). It is possible that EFhd1 blocks another co-factor required for this function of PGC-1α, which might also explain why not many PGC-1-α target genes are upregulated by EFhd1. Interestingly, EFhd1 interacts in mitochondria with the estrogen receptor alpha chain (esr1, nr3a1) and there are numerous studies about the bidirectional interdependence of Esr1 and PGC-1α. 50 EFhd1 could therefore impact expression of PGC-1α and its target genes via Esr. Given that PGC-1α upregulation is expected to support mitochondrial metabolism, why then does EFhd1 counteract mitochondrial ATP production in pre B cells? One interpretation could be that the normal downregulation of EFhd1 in pre B cells prevents ATP exhaustion perhaps by generation of ATP via glycolysis. Increased rps6ka1 expression induces anabolic ATP exhaustion in Fnip1−/− pro/pre B cells and also in PTEN−/+ and PTEN−/− pre B ALL cells.20, 51 Hence, the regulation of the pten/tsc2/rps6ka1 axis may exaggerate the disadvantage of EFhd1tg pro and pre B cells in vivo. Pre B cells face heavy selection pressure for an optimal pre BCR signaling strength.1 EFhd1tg pre B cells would therefore represent pre B cells where the pre BCR has induced only insufficient signals to mediate downregulation of EFhd1, such as intracellular μHC that are unable to pair with the surrogate LC, do not provide optimal signals52 and cannot downregulate EFhd1. Through EF hand mediated Ca2+ binding,31 EFhd1 positively regulates mitoflashes under conditions of high intracellular Ca2+. The alkalinization of the mitochondrial matrix during a mitoflash may reduce the efficacy of mitochondrial ATP production.32 EFhd1 might therefore translate the pre BCR-elicited increase in Ca2+ into mitoflashes,5 compromising ATP generation. We propose that downregulation of EFhd1 represents a sensor of optimal pre BCR Ca2+-signaling strength governing μHC selection in the BM. Taken together, an intimate interplay of pre BCR controlled genes involved in metabolism and Ca2+ sensing, such as efhd1, appears to be important for fitness of early B cells.
Materials and methods
Mice, BM transfer and B cell homing
A transgene expression cassette, pBSVE6BK (kind gift of Dr. Carlo Croce) with blunt-end EcoRV inserted murine EFhd1 cDNA under control of VH promoter (V186.2), and Eμ-enhancer53 was linearized and injected into the pronucleus of C57Bl/6 zygotes. Offspring was characterized with the following PCR primers and conditions (fwd 5′-GAACTTCTTCGAAGCCAA-3′ and rev 5′-ATGACATGAACTTAACCATA-3′ 94 °C for 5 min; 35 cycles (94 °C, 30 s/60 °C, 30 s/72 °C, 1 min). Southern blot analysis was used to quantify integrated transgene copies using a 292 bp probe representing complete efhd1 exon1. The insertion site of the transgene was mapped by digestion of genomic DNA with Msp1, religation and PCR with primers Eμfwd (5′-ttggtggggctggacagag-3′), Eμrev (5′-ACCGAAAGTCCAGGCTGAGC-3′), βGlobfwd (5′-TTTTATGGTTGGGATAAGGCTG-3′) and βGlobrev (5′-GCAGAATGGTAGCTGGATTG-3′). PCR products were cloned, sequenced and insertion points were identified by BLAST. Mice were maintained in IVC cages. Animal experiments were conducted according to institutional ethical guidelines for animal of the Friedrich-Alexander-University (FAU) Erlangen-Nuremberg and approved by the state of Bavaria. Unless stated otherwise, 6–20-week-old littermates of both sexes were used. C57Bl/6/Ly5.1 (B6.SJL-Ptprca Pep3b/BoyJ) mice were originally obtained from the Jackson Laboratory. 33.C9μHCki mice were generated by homologous recombination in V6.5 embryonic stem cells. The HC variable region of the hybridoma 33.C9 backmutated to its germ line sequence54 was inserted into the targeting vector pIVHL2neo kindly provided by Roberta Pelanda.55 Heterozygous knock-in mice were backcrossed to C57Bl/6 für 12 generations and mated with Rag1−/− mice obtained from Irmgard Förster (University Munich). Rag2−/− dTg (Ig-tTA/tet-μ) mice were treated for 7 days with tetracycline in the drinking water as described previously.6 For mixed BM chimeras, C57Bl/6/Ly5.1 or C57Bl/6/L5.2 mice were irradiated with one dose of 6 Gy and one dose of 5 Gy given 4 h apart, followed by i.v. reconstitution with a 1 : 1 mix of 2 × 106 BM cells in PBS prepared from the femurs of C57Bl/6 Ly5.1 and C57BL76 EFhd1tg/Ly5.2, or C57Bl/6/Ly5.1 and C57Bl/6 WT Ly5.2 mice. Recipients were analyzed after 6 weeks reconstitution. For B-cell homing, non-irradiated recipient mice were injected i.v. with a 1 : 1 mix of 2 × 07 WT B cells labeled with CSFE (eBioScience)/EFhd1tg B cells labeled with eF670 (eBioScience) l (or vice versa) in PBS. B cells were isolated by CD43 depletion (Milteny, Bergisch-Gladbach, Germany). BM, spleen and inguinal LN were analyzed either after 3 h or 24 h post injection for CSFE and eF670 positive viable lymphocytes.
Antibodies and chemicals
All chemicals were purchased from Sigma-Aldrich (Taufkirchen, Germany), Merck (Darmstadt, Germany) or Roth (Karlsruhe, Germany) unless stated otherwise. Cell culture medium and medium supplements were obtained from Invitrogen LifeTechnologies. Antibodies were purchased from BD Pharmingen (Heidelberg, Germany), BioLegend (London, England), eBioscience (Dreieich, Germany), Jackson ImmunoResearch Laboratories (distributed by Dianova, Hamburg, Germany) unless stated otherwise. The following antibodies were used for flow cytometry and cell sorting: unconjugated anti-CD16/32 (FC Block; 93; eBioscience), Alexa fluor 647 (AF647)-conjugated CD19 (eBio1D3; eBioscience), CD2 phycoerythrin (PE)-conjugated anti-CD19 (eBio1D3; eBioscience), Brilliant Violet 421 (BV)-conjugated anti-CD19 (BioLegend), fluorescein (FITC)-conjugated anti-CD21/35 (CR2/CR1; eBio8D9; eBioscience), FITC-conjugated anti-CD23 (eBioscience), PE-conjugated anti-CD23 (FcεRII; B3B4; eBioscience), APC-conjugated anti-CD25 (PC61; BioLegend), FITC-conjugated anti-CD43 (Ly-48, S7; BD), peridinin-chlorophyll protein-cyanine5.5 (PerCP)-conjugated anti-CD45R (B220; eBioscience), PE-conjugated anti-CD117 (c-kit; ACK45; BD), APC-conjugated anti-CD117 (c-kit, eBioscience), BV 421-conjugated anti-CD117 (c-kit, eBioscience), PerCP-Cy5.5-conjugated antiCD45.2 (eBioscience), Pacific Blue (PB)-conjugated anti-CD45.1 (BioLegend), Biotin-conjugated anti-IgD (Southern Biotech, distributed by Biozol, Eching, Germany), FITC-conjugated anti-IgM (Southern Biotech, distributed by Biozol, Eching, Germany), and Cyanine (Cy)5-conjugated anti-IgM (Southern Biotech); to detect biotinylated antibodies, PerCP-conjugated streptavidin (BD) were used.
FACS analysis
A total of 0.5–2 × 106 cells were washed with 500 μl FACS buffer (2% FCS/0.01% NaN3/PBS), resuspended in 100 μl antibody staining solution (FACS buffer with primary antibodies) for 20 min at 4 °C in the dark, washed and if required stained with secondary antibodies and kept on ice until measurement. For intracellular analysis, cells were fixed and permeabilized according to manufacturer’s instructions (von der Grub). Data were acquired on a FACSCalibur (BD) or Gallios flow cytometer (Beckman Coulter, Krefeld, Germany), and analyzed with the Kaluza (Beckman coulter, Version 1.2) or FlowJo software (Version 8.8.7, Ashland, OR, USA). To determine cell numbers by flow, cells were resuspended in 250 μl FACS buffer and 10 μl Coulter CC size standard L10 (Beckman Coulter, #B01828-AA) in PBS with a known concentration of beads (3–5 × 105 beads per ml).
Mitochondrial and metabolic stains
Cells were washed once in unsupplemented OptiMEM medium and then incubated for 30 min at 37 °C, 5% CO2 in the same medium with either 10 nM MitoTracker Green FM (Cell signalling #9074S, Frankfurt, Germany), 25 nM tetramethylrohdamine (TMRM) (Life Technologies, Dreieich, Germany, #T668) in the absence or presence of 100 μM carbonyl cyanide 3-chlorophenlyhydrazon (CCCP) or 1 μM DCFDA (Sigma, Taufkirchen, Germany). Fluorescence of CCCP-treated cells was subtracted to obtain specific TMRM signals. Cells were washed with unsupplemented OptiMEM and afterwards stained for surface markers as described in the previous section, but in OptiMEM medium. To measure glucose uptake, the cells were stained with 300 μM 6-NBDG (Life Technologies, #N23106) in glucose-free medium and washed in the same.
Cell isolation and cell sorting
Splenic cells were isolated by passing the organ through a 70 μm nylon filter. Splenic B cells were isolated to a purity of routinely >92% with an EasySep Mouse B cell isolation negative selection kit (EasySep #19854A) according to the manufacturer’s instructions. The cells were cultured in R10+ medium (RPMI 1640 medium (Gibco-Invitrogen, Dreieich, Germany) supplemented with 10 % v/v fetal calf serum (FCS; Gibco-Invitrogen, heat inactivated at 56 °C for 45 min), 2 mM glutamate, 1 mM sodium pyruvate, 50 U/ml penicillin G, 50 μg/ml streptomycin and 50 μM β-mercaptoethanol) at 37 °C, 5 % v/v CO2) with further additions as indicated. BM cells were isolated by flushing the bones with a G23 needle with cold OptiMEM10 medium (OptiMEM medium (Gibco-Invitrogen) with the same additions as R10+ medium). Rag1−/− and Rag1−/−;33.C9μHCki B cells were isolated by Mouse CD19-positive selection kit (EasySep, Stem cell technologies, Köln, Germany, #18754). Pro B cells (CD19+, c-kit+, sIgM−), Pre B cells (CD19+, CD25+, sIgM−), and immature/mature B cells (CD19+, IgM+) were sorted by MoFlo (Beckman Coulter, Krefeld, Germany) or Astrios (Beckman Coulter, Krefeld, Germany) to a purity >95% into medium or directly into RLT buffer (Qiagen kit) for RNA extraction. Pro B cells from IL-7 cultures were sorted as CD19+, cKit+, sIgM−, pre B cells CD19+, cKit−, sIgM− and immature B cells as CD19+, cKit−, sIgM+.
Confocal microscopy
B cells were positively sorted from the BM of Rag1−/− or Rag1−/−;33.C9HCki mice with anti-CD19 magnetic beads and attached to slides. Cells were stained with TMRM (100 nM) and anti-CD19 antibody coupled to FITC in the absence or presence of CCCP (100 μM), washed in OptiMEM and covered with cover slips. Images were taken along the central plane of cells. (Zeiss LSM700 (Oberkochen, Germany), 63 × oil objective, 90 nm voxel size, eightfold averaging, 16 bit image depth).
Pro B-cell culture
Pro B cells were seeded at 105 cells per ml and cultured in OptiMEM medium supplemented with 10 % v/v FCS (Gibco-Invitrogen, heat inactivated at 56 °C for 45 min), with IL-7 (Peprotech, Hamburg, Germany, #217-17) as indicated. IL-7 was added freshly every 3 days, new medium added on day 4 and the cells split to original density on day 5. To analyze proliferation, freshly sorted pro B cells were labeled with eFlour 670 (eBioscience) according to manufacturer’s instructions.
Retroviral infection of cells
Three commercially available shRNAS directed against murine EFhd1 and a control (Biocat, Heidelberg, Germany) were cloned into the retroviral vector pLMP (GE healthcare, Dornstadt, Germany; please note that EFhd1 shRNA-targeting sequences were not published and not available on demand from the distributor; shRNAS directed against EFhd1 are therefore numbered 1–3). Retroviral particles from the pLMP constructs were produced as described previously,34 38B9 cells25 were infected according to standard procedures34 and selected with 5μg/ml puromycin. The pCHIG (CMV promoter μH chain IRES-GFP) retroviral vectors expressing μHC were constructed as follows: VH fragments were generated by PCR from cDNA of C57Bl/6 splenic B cells using degenerate primers (VHdeg fwd: 5′-aggtsmarctgcagsagtcwgg-3′, JHdeg rev: 5′-TTTGGGAAGCTTTGACTCTCTGAGGARACGGTG-3′; M: A/C, R: A/G, S: C/G, W: A/T), cloned into pCR2.1 (Invitrogen, Dreieich, Germany) and sequenced. Specific VH fragments (17.2.25, 31, 81X) were then cloned into SalI/HindIII digested LXSPstar-81XBC2μm.56 The resulting vector containing a complete μHC including VH leader was then used as PCR template to generate a 181 bp fragment of the VH region containing a 5′ XhoI and a 3′ EcoRI site, and to generate an EcoRI/NlotI fragment. The XhoI/EcoRI digested PCR product and the EcoRI/NotI fragment were then co-ligated with the XhoI/NheI digested retroviral vector pCIG/pCru5 containing an IRES-GFP cassette.34 Retroviral particles were produced as described previously.34 IL-7-dependent R5B cells (kind gift of Dr. Chris Paige)57 and 38B9 cells25 were infected according to standard procedures34 and sorted by means of GFP expression.
Western blot
Western blot analyses were performed according to standard procedures as previously described using rabbit anti EFhd1,26 rabbit anti Actin (Sigma) and mouse anti Transferrin receptor antibodies (Thermo Scientific, Dreieich, Germany), followed by horseradish-peroxidase coupled secondary antibodies and development by enhanced chemiluminescence. Goat anti μ HC coupled to horseradish-peroxidase was purchased from Southern Biotech.
RT-PCR
RNA was isolated with the help of a QIAshredder (Qiagen, #79654) and RNeasy Mini kit (Qiagen, #74104). 50–100 ng cDNA was transcribed from isolated RNA with the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, #K1622) using oligo(dT)18 primers according to the manufacturer’s instructions or the QuantiTect Reverse Transcription kit (Qiagen, #205311) according to the manufacturer’s instructions. Primers were: efhd1: fwd 5′-CGGACTCCGAACTGAACCTC-3′, rev 5′-AACTCCGGGAACTCGGTGTA-3′, hprt fwd 5′-TCAGTCAACGGGGGACATAAA-3′, rev 5′-GGGGCTGTACTGCTTAACCAG-3′, sod2: fdw 5′-CAGACCTGCCTTACGACTATGG-3′, rev 5′-CTCGGTGGCGTTGAGATTGTT-3′, ppargc1a: fwd 5′-TATGGAGTGACATAGAGTGTGCT-3′, rev 5′-CACTTCAATCCACCCAGAAAG-3′, ppargc1b: fwd 5′-CGCTCCAGGAGACTGAATCCAG-3′, rev 5′-CTTGACTACTGTCTGTGAGGC-3′, upc2: fwd 5′-GCATCCAACGGAGTGGAAG-3′, rev 5′-GATTTCCGCAGGTTAGAAGGC-3′, ucp3: fwd 5′-CTGCACCGCCAGATGAGTTT-3′, rev 5′-ATCATGGCTTGAAATCGGACC-3′, ebf1 gene: fwd: 5′-GCATCCAACGGAGTGGAAG-3′, rev 5′-GATTTCCGCAGGTTAGAAGGC-3′, glut1: fwd: 5′-CAGTTCGGCTATAACACTGGTG-3′, rev 5′-GCCCCCGACAGAGAAGATG-3′ (Harvard Primer Bank58). The qRT-PCR was carried out in triplicate samples with 7.5 μl SYBRgreen MIX (Applied Biosystems, Life Technologies #4309155) 1 μl (1–5 ng) cDNA/RNA template, 0.25 μl (5 μM) fwd/rev primer and 6 μl ultra-pure H2O (Sigma-Aldrich, #HN68.2) per reaction. The DNA was melted at 95 °C for 15 min and heated for 30 s at 95 °C and 30 s at 60 °C for 40 cycles in an Applied Biosystems 7300 cycler and analyzed by ΔΔCT method. Non reversely transcribed RNA with HPRT primers and cDNA with H2O were included as quality control for each plate. RT2 profiler PCR arrays (Qiagen, Mouse PI3K-AKT, #PAMM-0582F-6 or Mouse PPAR Targets, #PAMM-149ZA-6) were carried out according to the manufacturer’s instructions with RT2 SYBR Green qPCR Mastermix (Qiagen, Hilden, Germany, # 330510) in an Applied Biosystems (Dreieich, Germany) 7300 cycler after generation of cDNA from 100 ng RNA per 96-well plate prepared by RNeasy kit (Qiagen, #74104) and converted with the RT2 First Strand kit (Qiagen # 330401).
Extracellular flux analysis
Extracellular flux analyses were carried out with a Seahorse XF-96 analyzer as previously published.59 Cells were isolated and cultured as indicated and washed three times in seahorse assay medium. Cells were analyzed for cell count and viability and seeded at 8.9 × 104–2.5 × 105 cells per well (same number of viable cells used for compared data sets). Mitochondrial metabolism (OCRs) and glycolysis (ECAR) were analyzed with Seahorse Mito and Glycolysis Stress Test kits (Agilent, Waldbronn, Germany) according to the manufacturer’s instructions.
CRISPR-Cas9 mediated knockout of EFhd1 in 38B9 pro B cells
The guide RNA targeting the EFhd1 gene was designed to bind within the first exon using the Zhang Lab online tool (http://crispr.mit.edu/) (sgRNA EFhd1 (1): 5′-CAACGACATCCACCAGGGTACAGCG-3′, sgRNA EFhd1 (2): 5′-AAACCGCTGTACCCTGGTGGATGTC-3′). Oligos of 10 μM were incubated in 10 μl H2O at 95 °C for 5 min and ligated into the BbSI digested px458 vector (pSpCas9(BB)-2A-GFP)60 with T4 DNA ligase (New England Biolabs, Frankfurt, Germany) according to the manufacturer’s instructions. The pro B-cell line 38B925 was transfected with the px458 vector containing the guide RNA using the Amaxa Cell Line Nucleofector Kit L (Lonza, Köln, Germany) according to the manufacturer’s instructions. Twenty-four hours after transfection GFP-positive cells were sorted individually into 96-well plates and cultivated until further analysis. Out of 53 clones grown originally, 11 survived, 2 had a shortened protein, 4 contained a full-length EFhd1 protein with normal abundance and served as controls, and no EFhd1 protein was detectable anymore in 5 clones.
Data and statistical analysis
GraphPad Prism 5 (GraphPad Software, LaJolla, CA, USA) was used for all statistical analyses. Mann–Whitney-U tests and unpaired Student’s t-tests were used for statistical comparison and calculation of statistical significance depending on whether data passed Kolmogorov–Smirnov normal distribution tests. For comparison of more than four groups, ANOVA with Bonferroni correction was used. Significance is shown as *P≤0.05, **P≤0.01 and ***P≤0.001.
Acknowledgments
We thank Dr. Carlo Croce for the VHP-EμH vector used to generate the transgene cassette, Dr. Roberta Pelanda for pIVHL2neo, Dr. Irmgard Förster for Rag1−/− mice, Dr. Chris Paige for R5B cells and Dr. Jan Tuckermann for support. We also thank Drs. Mark Shlomcik and Florian Weisel for helpful comments during preparation of this manuscript. We thank Uwe Appelt and Markus Mroz for cell sorting and Stefan Hofmann as well as Domenica Saul for technical assistance. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) (TRR130, to DM, H-MJ, THW; SPP1468, to DM and AB, IRTG1660, to DM, H-MJ, THW).
Author contributions
MS, SD and DM designed experiments, performed experiments, analyzed data and wrote the paper. DM designed experiments and wrote the paper. MB and THW designed and performed the experiments. KF, DR, SU and TS performed experiments. AB, WS and H-MJ provided important conceptual input and wrote the paper.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by L Scorrano
The authors declare no conflict of interest.
Supplementary Material
References
- Melchers F. The pre-B-cell receptor: selector of fitting immunoglobulin heavy chains for the B-cell repertoire. Nat Rev Immunol 2005; 5: 578–584. [DOI] [PubMed] [Google Scholar]
- Vettermann C, Jäck HM. The pre-B cell receptor: turning autoreactivity into self-defense. Trends Immunol 2010; 31: 176–183. [DOI] [PubMed] [Google Scholar]
- Young F, Ardman B, Shinkai Y, Lansford R, Blackwell TK, Mendelsohn M et al. Influence of immunoglobulin heavy- and light-chain expression on B-cell differentiation. Genes Dev 1994; 8: 1043–1057. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Johnson RS, Herrup K, Tonegawa S, Papaioannouo VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 1992; 68: 869–877. [DOI] [PubMed] [Google Scholar]
- Guo B, Kato RM, Garcia-Lloret M, Wahl MI, Rawlings DJ. Engagement of the human pre-B cell receptor generates a lipid raft-dependent calcium signaling complex. Immunity 2000; 13: 243–253. [DOI] [PubMed] [Google Scholar]
- Hess J, Werner A, Wirth T, Melchers F, Jäck HM, Winkler TH. Induction of pre-B cell proliferation after de novo synthesis of the pre-B cell receptor. Proc Natl Acad Sci USA 2001; 98: 1745–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne CD, Paige CJ. IL-7: a key regulator of B lymphopoiesis. Semin Immunol 2006; 18: 20–30. [DOI] [PubMed] [Google Scholar]
- Clark MR, Mandal M, Ochiai K, Singh H. Orchestrating B cell lymphopoiesis. Nat Rev Immunol 2013; 14: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med 1991; 173: 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Kasai H, Watanabe A, Lai AY, Kondo M. IL-7 specifies B cell fate at the common lymphoid progenitor to pre-ProB transition stage by maintaining early B cell factor expression. J Immunol 2008; 181: 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wofford JA, Wieman HL, Jacobs SR, Zhao Y, Rathmell JC, Jeffrey C. IL-7 promotes Glut1 trafficking and glucose uptake via STAT5-mediated activation of Akt to support T cell survival IL-7 promotes Glut1 trafficking and glucose uptake via STAT5-mediated activation of Akt to support T cell survival. Blood 2007; 111: 2101–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baracho GV, Miletic AV, Omori SA, Cato MH, Rickert RC. Emergence of the PI3-kinase pathway as a central modulator of normal and aberrant B cell differentiation. Curr Opin Immunol 2011; 23: 178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai K, Maienschein-Cline M, Mandal M, Triggs JR, Bertolino E, Sciammas R et al. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat Immunol 2012; 13: 300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming HE, Paige CJ. Cooperation between IL-7 and the pre-B cell receptor: a key to B cell selection. Semin Immunol 2002; 14: 423–430. [DOI] [PubMed] [Google Scholar]
- Flemming A, Brummer T, Reth M, Jumaa H. The adaptor protein SLP-65 acts as a tumor suppressor that limits pre-B cell expansion. Nat Immunol 2003; 4: 38–43. [DOI] [PubMed] [Google Scholar]
- Caro-Maldonado A, Wang R, Nichols AG, Kuraoka M, Milasta S, Sun LD et al. Metabolic reprogramming is required for antibody production that is suppressed in anergic but exaggerated in chronically BAFF-exposed B cells. J Immunol 2014; 192: 3626–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agathocleous M, Harris WA. Metabolism in physiological cell proliferation and differentiation. Trends Cell Biol 2013; 23: 484–492. [DOI] [PubMed] [Google Scholar]
- Yasuda T, Sanjo H, Pagès G, Kawano Y, Karasuyama H, Pouysségur J et al. Erk kinases link pre-B cell receptor signaling to transcriptional events required for early B cell expansion. Immunity 2008; 28: 499–508. [DOI] [PubMed] [Google Scholar]
- Schuh W, Meister S, Herrmann K, Bradl H, Jäck HM. Transcriptome analysis in primary B lymphoid precursors following induction of the pre-B cell receptor. Mol Immunol 2008; 45: 362–375. [DOI] [PubMed] [Google Scholar]
- Park H, Staehling K, Tsang M, Appleby MW, Brunkow ME, Margineantu D et al. Disruption of Fnip1 reveals a metabolic checkpoint controlling B lymphocyte development. Immunity 2012; 36: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dütting S, Brachs S, Mielenz D. Fraternal twins: Swiprosin-1/EFhd2 and Swiprosin-2/EFhd1, two homologous EF-hand containing calcium binding adaptor proteins with distinct functions. Cell Commun Signal 2011; 9: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnoczky G, Booth D, Csordás G, Debattisti V, Golenár T, Naghdi S et al. Reliance of ER-mitochondrial calcium signaling on mitochondrial EF-hand Ca2+ binding proteins: Miros, MICUs, LETM1 and solute carriers. Curr Opin Cell Biol 2014; 29: 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 2003; 115: 629–640. [DOI] [PubMed] [Google Scholar]
- Tominaga M, Kurihara H, Honda S, Amakawa G, Sakai T, Tomooka Y. Molecular characterization of mitocalcin, a novel mitochondrial Ca2+-binding protein with EF-hand and coiled-coil domains. J Neurochem 2006; 96: 292–304. [DOI] [PubMed] [Google Scholar]
- Alt F, Rosenberg N, Lewis S, Thomas E, Baltimore D. Organization and reorganization of immunoglobulin genes in A-MULV- transformed cells: rearrangement of heavy but not light chain genes. Cell 1981; 27: 381–390. [DOI] [PubMed] [Google Scholar]
- Brachs S, Lang C, Buslei R, Purohit P, Fürnrohr B, Kalbacher H et al. Monoclonal antibodies to discriminate the EF hand containing calcium binding adaptor proteins EFhd1 and EFhd2. Monoclon Antib Immunodiagn Immunother 2013; 32: 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossen C, Murre CS, Chang AN, Mansson R, Rodewald H-R, Murre C. The chromatin remodeler Brg1 activates enhancer repertoires to establish B cell identity and modulate cell growth. Nat Immunol 2015; 16: 775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow KT, Timblin GA, McWhirter SM, Schlissel MS. MK5 activates Rag transcription via Foxo1 in developing B cells. J Exp Med 2013; 210: 1621–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Lim J-H, Chim H, Bhalla K, Girnun G, Pierce K et al. P. PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell 2013; 23: 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzi T, Schwaller B. Antagonistic regulation of parvalbumin expression and mitochondrial calcium handling capacity in renal epithelial cells. PLoS ONE 2015; 10: e0142005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T, Jian C, Xu J, Huang AY, Xi J, Hu K et al. Identification of EFHD1 as a novel Ca2+ sensor for mitoflash activation. Cell Calcium 2016; 59: 262–270. [DOI] [PubMed] [Google Scholar]
- Hou T, Wang X, Ma Q, Cheng H. Mitochondrial flashes: new insights into mitochondrial ROS signalling and beyond. J Physiol 2014; 17: 3703–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur A, Moore JK, Jorgensen P, Shapiro HM, Kirschner MW. Optimizing optical flow cytometry for cell volume-based sorting and analysis. PLoS ONE 2011; 6: e16053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramidou A, Kroczek C, Lang C, Schuh W, Jäck H-M, Mielenz D. The novel adaptor protein Swiprosin-1 enhances BCR signals and contributes to BCR-induced apoptosis. Cell Death Differ 2007; 14: 1936–1947. [DOI] [PubMed] [Google Scholar]
- Milne CD, Corfe SA, Paige CJ. Heparan sulfate and heparin enhance ERK phosphorylation and mediate preBCR-dependent events during B lymphopoiesis. J Immunol 2008; 180: 2839–2847. [DOI] [PubMed] [Google Scholar]
- Martin F, Won WJ, Kearney JF. Generation of the germline peripheral B cell repertoire: VH81X-lambda B cells are unable to complete all developmental programs. J Immunol 1998; 160: 3748–3758. [PubMed] [Google Scholar]
- Koslowski M, Sahin U, Dhaene K, Huber C, Türeci Ö. MS4A12 is a colon-selective store-operated calcium channel promoting malignant cell processes. Cancer Res 2008; 68: 3458–3466. [DOI] [PubMed] [Google Scholar]
- Möröy T, Fisher P, Guidos C, Ma A, Zimmerman K, Tesfaye A et al. IgH enhancer deregulated expression of L-myc: abnormal T lymphocyte development and T cell lymphomagenesis. EMBO J 1990; 9: 3659–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MR, Mandal M, Ochiai K, Singh H. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat Rev Immunol 2014; 14: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick SM. The cunning little vixen: Foxo and the cycle of life and death. Nat Immunol 2009; 10: 1057–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber T, Mandel EM, Pott S, Györy I, Firner S, Liu ET et al. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity 2010; 32: 714–725. [DOI] [PubMed] [Google Scholar]
- Hurt EM, Thomas SB, Peng B, Farrar WL. Molecular consequences of SOD2 expression in epigenetically silenced pancreatic carcinoma cell lines. Br J Cancer 2007; 97: 1116–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel A, Okino ST, Liao J, Nakatani K, Li J, Whitlock JP et al. Regulation of GLUT1 gene transcription by the serine/threonine kinase Akt1. J Biol Chem 1999; 274: 20281–20286. [DOI] [PubMed] [Google Scholar]
- Rakhshandehroo M, Knoch B, Müller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res 2010; 2010: 612089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Yang C, Yang Z, Zhang D, Ma X, Mills G et al. Homeostasis of redox status derived from glucose metabolic pathway could be the key to understanding the Warburg effect. Am J Cancer Res 2015; 5: 1265–1280. [PMC free article] [PubMed] [Google Scholar]
- Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol 2009; 9: 195–205. [DOI] [PubMed] [Google Scholar]
- Amin RH, Schlissel MS. Foxo1 directly regulates the trancription of recombination activating genes during B cell development. Nat Immunol 2008; 9: 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann AD, Beyer M, Krause-Buchholz U, Wobus M, Bornhäuser M, Rödel G. Oxphos supercomplexes as a hallmark of the mitochondrial phenotype of adipogenic differentiated human MSCS. PLoS ONE 2012; 7: e35160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villena JA. New insights into PGC-1 coactivators: redefining their role in the regulation of mitochondrial function and beyond. FEBS J 2015; 282: 647–672. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Zhou J, Du Y. Estrogen receptor alpha interacts with mitochondrial protein HADHB and affects beta-oxidation activity. Mol Cell Proteomics 2012; 11: M111.011056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shojaee S, Chan LN, Buchner M, Cazzaniga V, Cosgun KN, Geng H et al. PTEN opposes negative selection and enables oncogenic transformation of pre-B cells. Nat Med 2016; 4: 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielenz D, Ruschel A, Vettermann C, Jäck H-M. Immunoglobulin mu heavy chains do not mediate tyrosine phosphorylation of Ig alpha from the ER-cis-Golgi. J Immunol 2003; 171: 3091–3101. [DOI] [PubMed] [Google Scholar]
- Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci USA 2002; 99: 6955–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmann U, Letz M, Herrmann M, Angermüller S, Kalden JR, Winkler TH. The evolution of human anti-double-stranded DNA autoantibodies. Proc Natl Acad Sci USA 2005; 102: 9258–9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelanda R, Schaal S, Torres RM, Rajewsky K. A prematurely expressed Ig(kappa) transgene, but not a V(kappa)J(kappa) gene segment targeted into the Ig(kappa) locus, can rescue B cell development in lambda5-deficient mice. Immunity 1996; 5: 229–239. [DOI] [PubMed] [Google Scholar]
- Martin DA, Bradl H, Collins TJ, Roth E, Jäck H-M, Wu GE. Selection of Ig mu heavy chains by complementarity-determining region 3 length and amino acid composition. J Immunol 2003; 171: 4663–4671. [DOI] [PubMed] [Google Scholar]
- Milne CD, Fleming HE, Paige CJ. IL-7 does not prevent pro-B/pre-B cell maturation to the immature/sIgM(+) stage. Eur J Immunol 2004; 34: 2647–2655. [DOI] [PubMed] [Google Scholar]
- Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res 2010; 38: D792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitschin R, Hofmann AD, Bruns H, Gießl A, Bricks J, Berger J et al. Mitochondrial metabolism contributes to oxidative stress and reveals therapeutic targets in chronic lymphocytic leukemia. Blood 2014; 123: 2663–2672. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Prot 2013; 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.