Abstract
Frequent KRAS mutations contribute to multiple cancers including ~40% of human colorectal cancers (CRCs). Systematic screening of 1255 microRNAs (miRNAs) identified miR-30a as a synthetic lethal in KRAS-mutant CRC cells. miR-30a was downregulated in CRCs and repressed by P65. miR-30a directly targeted malic enzyme 1 (ME1) and KRAS, and inhibited anchorage-independent growth and in vivo tumorigenesis by KRAS-mutant CRC cells. ME1 was significantly upregulated in KRAS-mutant CRCs. Eliminating ME1 by short hairpin RNA (shRNA) resulted in obviously decreased NADPH production, levels of triglyceride and fatty acid, and an inhibition of tumorigenicity of KRAS-mutant CRCs. miR-30a overexpression and ME1 suppression attenuated AOM/DSS-induced colorectal tumorigenesis. The critical roles of miR-30a and ME1 in the development of KRAS-mutant CRCs indicate therapy potentials for this subtype of cancer.
Colorectal cancer (CRC) is the third most prevalent cancer worldwide.1 Mutation of KRAS occurs in 42.4% of CRCs.2, 3, 4 Oncogenic KRAS mutations initiates and sustains colorectal tumorigenesis. However, directly targeted therapies to KRAS is still unsuccessful. The surface of this protein is too smooth for drugs to bind and a large family of related protein members share similar GTP-/GDP-binding domain, which makes KRAS therapeutic attack extremely challenging. KRAS protein has been regarded as an ‘undruggable’ target.5, 6 Thus it has been suggested that taking advantage of synthetic lethal interactions with KRAS mutation could be exploited as an effective therapeutic strategy in KRAS-mutant human cancers.7, 8, 9, 10, 11
MicroRNAs (miRNAs) are small non-coding RNAs, which inhibit the translation and/or stability of targeted mRNAs.12 Recently miRNAs have been implicated in the progression and development of varieties of cancers including CRCs.13, 14, 15, 16, 17, 18, 19 miR-206 and miR-342 specifically impair the growth of breast cancer cells with MYC addiction and BRCA1 mutations, respectively.15, 16 miR-17-92 cluster depletion interacts with p53 mutations in non-small-cell lung cancer.17 Some reports showed that miRNAs or their antagomirs might be effective therapeutic potentials.20, 21
In this study, we undertook a high-content screening to identify miRNAs that selectively impaired the growth of KRAS-mutant CRC cells. We found that miR-30a inhibited the growth and tumorigenicity of the KRAS-mutant CRC cells by directly inhibiting malic enzyme 1 (ME1) and KRAS. Furthermore, we investigated the effects of miR-30a and ME1 in KRAS-mutant CRC cells and AOM-/DSS-induced CRC mouse model. Manipulating the expression levels of miR-30a and ME1 might have therapeutic potentials in KRAS-mutant CRC patients.
Results
Identification of miR-30a as a specific attenuator of KRAS-mutant CRC cells by functional miRNA screening
CRC cells frequently harbor KRAS mutations. We investigated the KRAS status of several CRC cell lines. Results show that RKO, SW48 and HT29 are wild-type (WT) cells, while HCT116 and DLD1 cells carry G13D point mutations (Supplementary Figure S1A). Two distinct short hairpin RNAs (shRNAs) targeting KRAS were introduced into these cells to validate the growth dependency of KRAS (Supplementary Figure S1B). KRAS suppression attenuated both anchorage-dependent and -independent growth only in HCT116 and DLD1 KRAS-mutant cells (Supplementary Figures S1C–E). Thus, HCT116 and DLD1 cells clearly exhibit dependency on oncogenic KRAS mutations. We chose HCT116 and RKO cells to perform the primary screening.
We screened HCT116 and RKO CRC cells with the miRNA library comprised of 1255 individual miRNA expression vectors (miRBase release 18.0 (2012), the University of Manchester, Manchester, UK; Supplementary Table 1) generated by our laboratory.15, 22 MTT assay was applied to validate the effects of miRNAs on cell viability compared with the control. In the primary screening, 11 miRNAs showed marked inhibitory effects on cell viability only in HCT116 cells (Log2 relative growth ratio<−0.6, Figure 1a; Supplementary Table 1). After confirming the growth inhibitory effects of these miRNAs in HCT116 and RKO cells, we tested 11 candidates in three KRAS WT CRC cells (RKO, SW48 and HT29) and two KRAS-mutant CRC cells (HCT116 and DLD1).4, 8, 9, 10, 11 miR-30a significantly attenuates the growth of only KRAS-mutant cells (Figure 1b; Supplementary Figure S2).
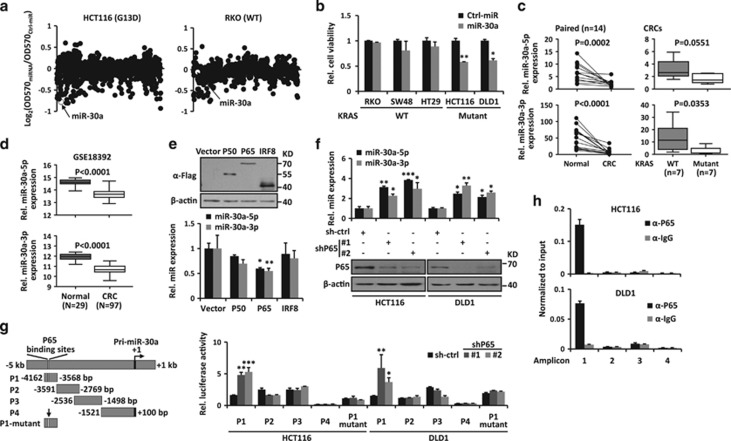
Figure 1.
miR-30a is downregulated and repressed by P65 in CRC cells. (a) High-content functional library screening results of miRNAs in HCT116 and RKO cells. miR-30a is indicated in hollow dot. (b) The effects of miR-30a on growth of indicated KRAS WT and -mutant cancer cells. (c) Expression levels of miR-30a-5p/3p were measured by RT-qPCR in paired colorectal tissues (left). Data from CRC tissues was showed as WT versus mutant KRAS group (Mut) according to their KRAS status (right). (d) Expression levels of miR-30a-5p/3p were analyzed from public available GSE18392 data set. (e) Upper: overexpression of P50, P65 and IRF8 was detected by immunoblot in HEK-293 cells. Lower: expression levels of miR-30a-5p/3p were determined by RT-qPCR. (f) Upper: P65 suppression elevated miR-30a-5p/3p expression. Lower: P65 knockdown by shRNA was confirmed by immunoblot in HCT116 and DLD1 cells. β-actin was used as the loading control. (g) Left: schematic diagram of miR-30a promoter. miR-30a promoter is divided into four fragments and the first fragment (P1) contains predicted P65-binding sites. The mutant P65-binding sites were indicated by a vertical arrow. Right: luciferase activity in HCT116 and DLD1 cells transfected with miR-30a promoter reporter and P65 shRNA-expressing vector or control vector. (h) ChIP assay was performed in HCT116 and DLD1 cells. Enrichment of each amplicon was detected by qPCR. Data are shown as the means±S.D. *P<0.05, **P<0.01, ***P<0.001
miR-30a is downregulated in human CRC specimens
The miR-30a locus encodes two miRNAs, miR-30a-5p/3p. We analyzed their expression in 14 human CRCs and paired normal colon tissues. These studies showed that both miR-30a-5p and 3p were downregulated in CRCs compared with their adjacent non-tumor tissues (n=14, P=0.0002 for miR-30a-5p and P<0.0001 for miR-30a-3p) (Figure 1c). A similar downregulation of miR-30a was observed in a published non-coding RNA profiling array, GSE18392 (P<0.0001) (Figure 1d).23 To investigate the correlation between downregulation of miR-30a and KRAS mutations in CRCs, we sequenced the first exon of KRAS genomic DNA extracted from CRC tissues (Supplementary Figure S3). About 50% (7 of 14) of the CRC tissues harbor KRAS mutations, convinced with previous report.24 miR-30a-5p/3p were downregulated in KRAS-mutant CRC specimens compared with WT ones (Figure 1c). These data suggest that miR-30a might play critical roles in the development of KRAS-mutant CRCs.
P65 mediates miR-30a downregulation
To study the underlying mechanism behind the downregulation of miR-30a, we analyzed the transcription factors binding to the miR-30a promoter region (−5 to +2 kb) in MotifMap website.25 These showed three candidate transcription factors, P50, P65 and IRF8 (Supplementary Figure S4A). However, only P65 overexpression significantly suppressed miR-30a-5p/3p expression (Figure 1e). On the other side, P65 knockdown in HCT116 and DLD1 CRC cells markedly increased miR-30a-5p/3p expression (Figure 1f). To determine whether P65 binds to the miR-30a promoter region, luciferase reporter assays were conducted to examine whether P65 inhibits miR-30a-5p/3p expression. Indeed, suppression of P65 using its specific shRNA enhanced the activity of P1 reporter vector containing the predicted P65-binding site in dual luciferase reporter assays, while mutation in P65-binding sites abrogated this upregulation (Figure 1g). The binding of P65 to the miR-30a promoter was confirmed by chromatin immunoprecipitation (ChIP) in HCT116 and DLD1 cells (Figure 1h). ChIP assay in HEK-293 cells with ectopic expression of P65 also showed a direct binding in the first amplicon (Supplementary Figure S4B). P65 is a critical subunit of NF-κB and commonly upregulated in CRC cells. Thus, downregulation of miR-30a might result from elevated P65 in CRC cells.
miR-30a attenuates tumor growth, migration and invasion of KRAS-mutant CRC cells
We evaluated the effects of miR-30a in KRAS-mutant cancer cells by gain- and loss-of-function experiments (Figure 2). Indeed, miR-30a overexpression inhibited cell growth, focus formation and anchorage-independent soft agar growth of only KRAS-mutant CRC cells (HCT116 and DLD1), while it showed no significant effect in KRAS WT CRC cells (RKO, SW48 and HT29) (Figures 2a–d; Supplementary Figure S5). To better elucidate the selective tumor suppressor activity on KRAS-mutant CRC cells, we performed the wound-healing assay, cell migration and invasion assays (Supplementary Figure S6). miR-30a overexpression inhibited wound-healing closure, cell migration and invasion of only KRAS-mutant CRC cells, while it showed no significant effect in KRAS WT CRC cells. Also miR-30a overexpression significantly increased cleaved PARP-1, indicating induction of apoptosis in KRAS-mutant CRC cells, while no obvious effect in WT CRC cells (Figure 2e). Flow cytometry analysis of Sub-G1 DNA content in those cells also revealed a similar result (Supplementary Figure S7).
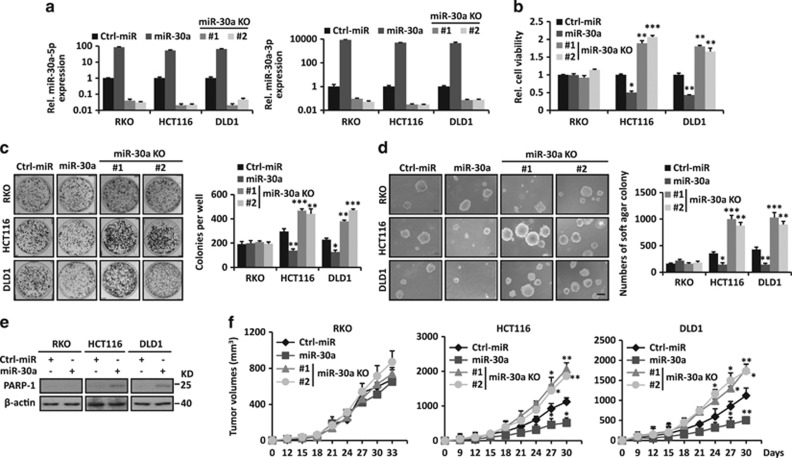
Figure 2.
miR-30a impairs growth and tumorigenicity of KRAS-mutant CRC cells. (a) miR-30a-5p/3p levels were measured in CRC cells with miR-30a overexpression or KO. (b–d) miR-30a overexpression inhibits cell viability (b), focus formation (c) and anchorage-independent soft agar growth (d) in HCT116, DLD1 KRAS-mutant CRC cells. MiR-30a KO showed opposite effect in KRAS-mutant CRC cells. Scale bar, 200 μm. Colonies of focus formation and anchorage-independent soft agar were counted and showed in bar graphs. (e) Overexpression of miR-30a promotes apoptosis of KRAS-mutant CRC cells as indicated by immunoblot using PARP-1 antibody. β-actin was used as the loading control. (f) miR-30a selectively slows tumor growth in HCT116 and DLD1 xenograft (n=5). Data are shown as the means±S.D. *P<0.05, **P<0.01, ***P<0.001
miR-30a knockout (KO) CRC cells were obtained by CRISPR/Cas9 systems with two distinct guide RNAs (Supplementary Figures S8A and B). miR-30a KO obviously promoted the anchorage-dependent and -independent growth of KRAS-mutant CRC cells, while it showed no significant effect in KRAS WT RKO cells (Figures 2a–d). To evaluate the roles of miR-30a in vivo, indicated CRC cells were injected into the flanks of nude mice. Stable overexpression of miR-30a in KRAS-mutant CRC cells suppressed the tumor growth, while miR-30a KO in these cells showed the opposite effect (Figure 2f). However, manipulation of miR-30a expression levels had no significant effect in KRAS WT CRC cells (Figure 2f; Supplementary Figure S5). Immunohistochemistry of HCT116 and DLD1 xenografts overexpressing miR-30a showed inhibition of cell proliferation and induction of apoptosis, as indicated by the decreased Ki-67 staining and increased caspase-3 cleavage (Supplementary Figure S8C). These data indicate miR-30a impairs the growth, motility and tumorigenicity of KRAS-mutant CRC cells.
miR-30a directly suppresses ME1 and KRAS
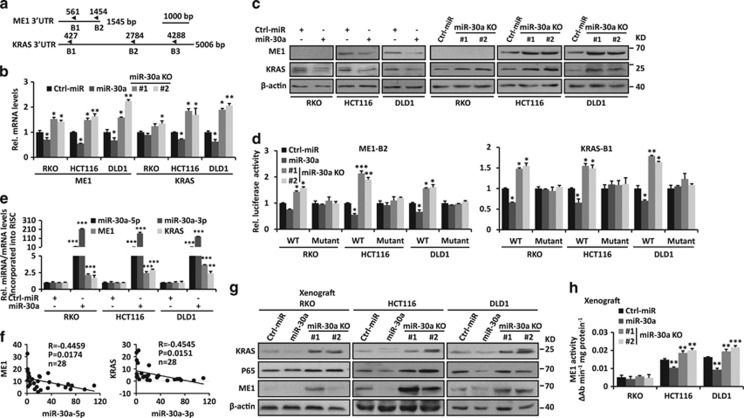
As miR-30a has critical effects on KRAS-mutant CRC cells, we performed prediction analyses to identify its targets. We found that the 3′-untranslated regions (UTR) of ME1 and KRAS contain two miR-30a-5p and three miR-30a-3p-binding sites, respectively (Figure 3a; Supplementary Figures S9A and B). ME1 is a cytosolic NADP+-dependent enzyme, which is responsible for NADPH production and lipogenesis.26 miR-30a overexpression inhibited ME1 and KRAS at the mRNA and protein levels in CRC cells, while miR-30a KO showed the opposite effects (Figures 3b and c). To validate the direct miRNA–mRNA interactions, we performed luciferase reporter assays in CRC cells with stable miR-30a overexpression or deletion. Results showed that miR-30a overexpression inhibited both ME1 and KRAS 3′UTR (only ME1-B2 and KRAS-B1) luciferase activities, while miR-30a deletion increased these activities (Figure 3d; Supplementary Figure S9C). Mutations in miR-30a-binding sites abrogated these effects (Figure 3d). We further used co-immunoprecipitation with an antibody AGO-2 to precipitate RNAs associated with the RNA-induced silencing complex. As shown in Figure 3e, the above target RNAs were significantly enriched in miR-30a overexpressed CRC cells versus control ones. Furthermore, negative correlations were observed in the RNA levels between of miR-30a-5p, 3p and ME1, KRAS in CRCs (n=28, P=0.0174 for ME1 and P=0.0151 for KRAS) (Figure 3f), respectively. Similar results were also found in mouse xenografts. miR-30a overexpression significantly decreased ME1 and KRAS protein levels, and ME1 activities, while miR-30a KO had the opposite effects in KRAS-mutant CRC cells (Figures 3g and h). However, miR-30a overexpression or KO had no significant effect in WT RKO CRC cells. Taken together, these data indicated that miR-30a can directly inhibit ME1 and KRAS.
Figure 3.
miR-30a directly targets ME1 and KRAS. (a) Schematic representation of predicted miR-30a-binding sites of ME1 and KRAS 3′UTR. (b–c) ME1 and KRAS mRNA and protein levels were detected in miR-30a-overexpressing or miR-30a KO CRC cells. (d) miR-30a-overexpressing or miR-30a KO CRC cells were transiently transfected with reporter vector (ME1-B2 or KRAS-B1) containing WT or mutant predicted miR-30a-binding site. Luciferase activity was measured. (e) RT-qPCR analysis was performed to measure miR-30a-5p/3p, ME1 and KRAS levels incorporated into RNA-induced silencing complex in miR-30a-overexpressing CRC cells compared with those of the control. (f) Negative correlation between miR-30a expression levels and KRAS/ME1 mRNA levels in colorectal tissues (n=28). (g) Protein levels of ME1, KRAS and P65 in xenograft from Figure 2f were detected by immunoblot. β-actin was used as the loading control. (h) ME1 activity was measured in xenograft from Figure 2f. Data are shown as the means±S.D. *P<0.05, **P<0.01, ***P<0.001
ME1 is upregulated in human KRAS-mutant CRCs
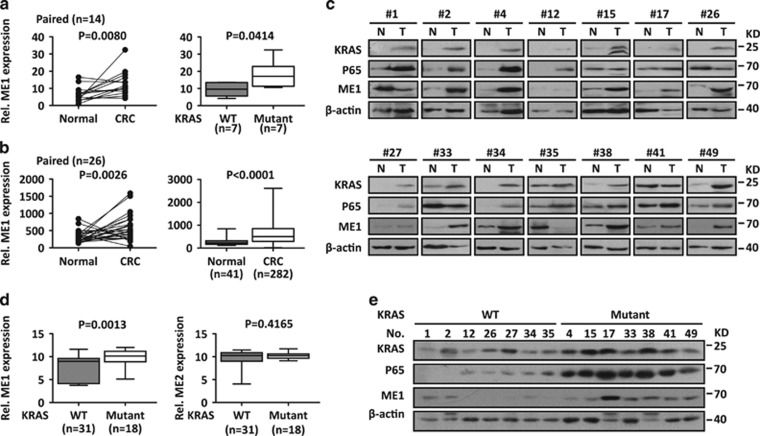
Next ME1 expression was determined in human CRCs. Results showed that ME1 was upregulated in CRCs compared with adjacent non-tumor tissues (n=14, P=0.008, Figure 4a). ME1 upregulation in CRCs was also observed both in paired (n=26, P=0.0026) and unpaired (n=41 for non-tumor tissues and n=282 for CRCs, P<0.0001) CRC samples (Figure 4b) in the Cancer Genome Atlas (TCGA) data set. ME1 upregulation at the protein levels was also confirmed by immunoblot analysis (Figure 4c).
Figure 4.
ME1 is upregulated in human KRAS-mutant CRCs. (a) Left: expression of ME1 normalized to β-actin was measured by RT-qPCR in paired colorectal tissues. Right: data from CRC tissues were showed as WT versus KRAS-mutant group (Mut) according to KRAS status. (b) ME1 is upregulated in CRC tissues from TCGA database. Left: paired tissues; Right: unpaired tissues. (c) Protein levels of ME1, KRAS and P65 were detected by immunoblot in colorectal tissues. β-actin was used as the loading control. (d) ME1, ME2 mRNA expression in KRAS WT and Mut cancer cells from CCLE database. (e) Protein levels of ME1, KRAS and P65 in CRC tissues. β-actin was used as the loading control. N, normal tissues; T, CRC tissues
Interestingly, the high expression of ME1 was noticed in KRAS-mutant HCT116, DLD1 CRC cells compared with KRAS WT RKO ones (Figure 3c). Further studies showed that ME1 mRNA increased in KRAS-mutant cancer cells (n=18) compared with WT ones (n=31), while ME2 expression showed no significant difference (Figure 4d) from the Cancer Cell Line Encyclopedia (CCLE, the Broad Institute, Cambridge, MA, USA) data set.10, 11, 27 Elevated ME1 was also detected in primary CRC specimens in mRNA (Figure 4a, right) and protein levels (Figure 4e). KRAS suppression decreased ME1 protein levels in HCT116, DLD1 KRAS-mutant CRC cells (Supplementary Figure S1B). ME1 is a critical ME responsible for converting malate to pyruvate, which supplies fuels of tricarboxylic acid flux, and generates NADPH. Emerging studies suggested that KRAS-driven cancers exhibited aberrant metabolism pathways to promote tumor growth.28, 29 Thus upregulated ME1 might contribute to the oncogenic metabolism pathways in KRAS-mutant CRCs.
Human KRAS-mutant CRC cells are hypersensitive to ME1 suppression
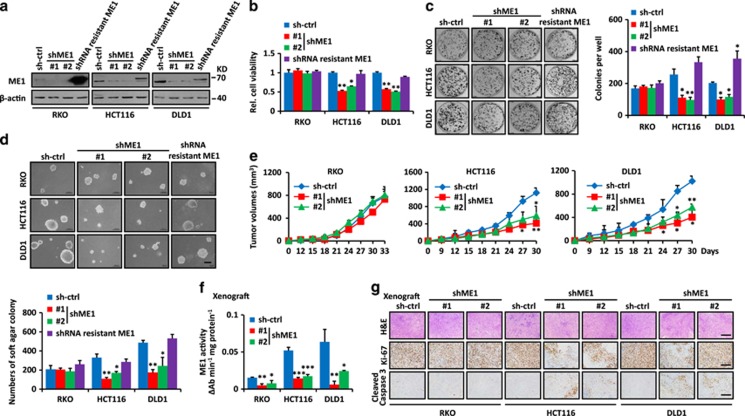
To explore the function of ME1, we applied two distinct shRNAs to stably knockdown ME1 expression and rescued ME1 expression by expressing shRNA-resistant ME1 in CRC cells (Figure 5a). Silencing of ME1 selectively inhibited both anchorage-dependent and -independent growth only in KRAS-mutant HCT116, DLD1 CRC cells, while it showed no significant effect in KRAS WT CRC cells (RKO, SW48 and HT29) (Figures 5b–d; Supplementary Figure S10). ME1 suppression decreased the cellular NADPH, triglyceride and fatty acids levels only in KRAS-mutant HCT116, DLD1 CRC cells (Supplementary Figure S11). Restoring ME1 expression abolished the inhibition (Figures 5b–d; Supplementary Figures S10 and S11). Following subcutaneous inoculation into the flanks of nude mice, only KRAS-mutant HCT116, DLD1 CRC cells stably knockdowning ME1 showed significantly retarded tumor growth (Figure 5e; Supplementary Figure S10). Consistent with their different rates of growth, these tumors showed decreased ME1 activity (Figure 5f), low levels of Ki-67 and high levels of caspase-3 staining (Figure 5g). These data reveal that human KRAS-mutant CRC cells are hypersensitive to ME1 suppression.
Figure 5.
ME1 is required for cell growth and tumorigenicity of human KRAS-mutant CRC cells. (a) Inhibition of ME1 was confirmed by immunoblot. ME1 protein levels were rescued by overexpressing shME1-resistant ME1. β-actin was used as the loading control. (b–d) ME1 suppression inhibits cell growth (b), focus formation (c) and anchorage-independent soft agar growth (d) in KRAS-mutant CRC cells. Restoring ME1 expression rescued the defects in HCT116 and DLD1 cells. Scale bar, 200 μm. Colonies of focus formation and anchorage-independent soft agar were counted and showed in bar graphs. (e) ME1 inhibition attenuated tumor growth (n=5). (f) ME1 activity in xenografts from (e). (g) ME1 inhibition attenuated cell proliferation and increased apoptosis, as indicated by Ki-67 and cleaved caspase-3 staining in xenografts from e. Scale bar, 200 μm. Data are shown as the means±S.D. *P<0.05, **P<0.01, ***P<0.001
miR-30a overexpression and ME1 suppression attenuates AOM-/DSS-induced colorectal tumorigenesis
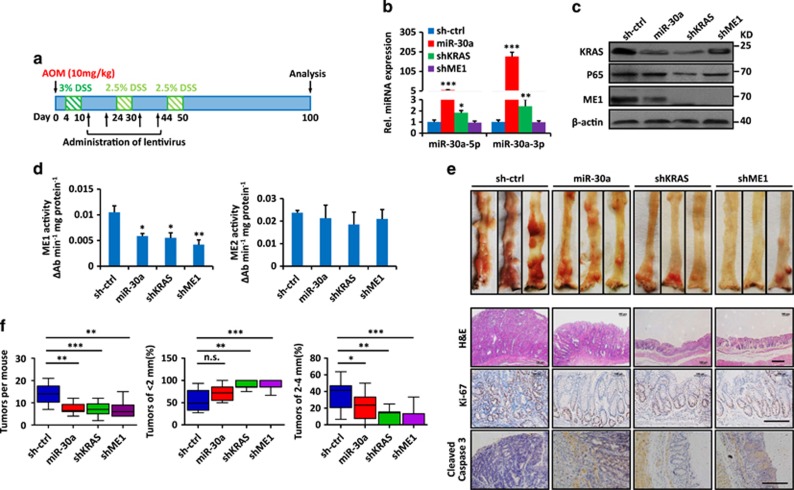
As miR-30a overexpression and ME1 knockdown selectively impaired growth and tumorigenicity of KRAS-mutant CRC cells, AOM-/DSS-induced CRC mouse model, which is a widely used animal model for CRC induction and harbors ~33% frequency of KRAS mutations,30 was used to evaluate their effects on colorectal tumorigenesis (Figure 6a). Female mice were injected intraperitoneally with AOM, followed by three rounds of DSS treatment and alternating four times of indicated lentivirus intrarectal instillation. Mice were killed 100 days after AOM injections and miR-30a overexpression was confirmed (Figure 6b). Mice administrated lentivirus containing overexpressing miR-30a, knockdown of KRAS or ME1 showed decreased ME1, KRAS protein levels and ME1 activity rather than ME2 (Figures 6c and d). Also mice treated with lentivirus containing overexpressing miR-30a, knockdown of KRAS or ME1 developed significantly decreased numbers and size of tumors throughout the middle and distal colon (Figures 6e and f). Such treatment showed decreased proliferation and increased apoptosis as indicated by the decreased Ki-67 and increased cleavage caspase-3 staining (Figure 6e). These results suggested that miR-30a overexpression and ME1 suppression attenuate AOM-/DSS-induced colorectal tumorigenesis.
Figure 6.
miR-30a overexpression and ME1 suppression attenuate AOM-/DSS-induced colorectal tumorigenesis in mice. (a) Treatment schedule with lentivirus containing miR-30a expression vector during AOM-/DSS-induced colorectal tumorigenesis in mice. Indicated lentivirus was intrarectally administrated into mice (n=10 per group) weekly for four times. Mice were killed 100 days after AOM injection. (b) Relative miR-30a-5p/3p levels were detected in colorectal tissues by RT-qPCR from (a). (c) Protein levels of KRAS, P65 and ME1 were detected in colorectal tissues by immunoblot. β-actin was used as the loading control. (d) ME1 and ME2 activity of colorectal tissues from (a). (e) Representative images of mouse colorectal tumors were showed. Immunohistochemistry of mouse colorectal tumors of miR-30a overexpression, KRAS and ME1 suppression showed decreased Ki-67 and increased cleaved caspase-3 staining compared with the control group. (f) The numbers of tumors in the mouse colorectum were counted and percentages of tumors of <2 mm or 2–4 mm in diameter were quantified. Data are shown as the means ± S.D. *P<0.05, **P<0.01, ***P<0.001
Discussion
Oncogenic KRAS mutations contribute to colorectal tumorigenesis. Here we found that miR-30a selectively impaired growth of KRAS-mutant CRC cells in vitro and in vivo. miR-30a has been shown to have the tumor suppressor function in colorectal cancer HCT116, Lovo, SW480 and SW620 cells.31, 32, 33 Consistent with our results, these cell lines are KRAS-mutant CRC cells. miR-30a was reported to be regulated by p53 and inhibit autophagy by directly targeting Beclin1.34, 35 To our knowledge, oncogenic KRAS activation promotes autophagy to recycle the nutrients for tumorigenesis.36, 37 Thus miR-30a overexpression might selectively block the nutrients supplying in KRAS-mutant tumors. In the present study, we found that miR-30a directly inhibit ME1 and decrease its activity. ME1 suppression inhibits NADPH production, lipogenesis and fatty acid uptake in KRAS-mutant CRC cells. Also miR-30a directly inhibits KRAS. A previous study reported that miR-126 selectively inhibited the viability of KRAS-mutant CRC cells, but not the KRAS WT CRC cells through repression of multiple genes essential for the survival of KRAS-mutant cells.38
Identification of synthetic lethal interactions with KRAS mutations is an alternative approach to conquer KRAS-mutant driven cancers. Recently, genome-wide RNA interference screens identified a list of candidate genes including the mitotic kinase PLK1,9 transcription factor GATA2,39 anti-apoptotic BH3 family member BCL-XL8 and SUMO E2 ligase UBC9.40 These genes span diverse cellular progression including cell mitosis, cell apoptosis, transcription and protein modification. We employed a high-content miRNA library-based screens to identify candidate miRNAs and found that miR-30a selectively attenuates KRAS-mutant-driven colorectal tumorigenesis. Interestingly, UBC9 is a direct target of miR-30a in human adipocytes.41
ME1 is upregulated in various cancers including CRCs and associated with poor overall survival.26, 42 Furthermore, we revealed that expression of ME1 is elevated in KRAS-mutant CRCs compared with WT CRCs. KRAS mutation reprograms glutamine metabolism flux in pancreatic cancer and ME1 is one of critical genes involved in glutamine metabolism.28 An association between KRAS mutations and ME1 was reported in non-small-cell lung cancers.43 Our research revealed the positive association between KRAS mutations and ME1 in CRCs, and evaluated the therapeutic potential by animal experiments.
In summary, our data identified critical roles of miR-30a and ME1 in the development of KRAS-mutant CRCs. Manipulating the miR-30a and ME1 expression levels might have therapeutic potentials for this kind of disease.
Materials and Methods
Human CRC specimens
Human CRC tissues were obtained from patients at Union Hospital in Wuhan, China. Clinical characteristics of human CRC samples are provided in Supplementary Table 2. Informed consent was obtained at the Union Hospital in Wuhan, China. The diagnosis of CRC was confirmed in each case by histological review and none of them received chemotherapy prior to resection.
Constructs
Human Pri-miR-30a, including ~400 bp of stem-loop structures, was PCR-amplified from HEK-293 genomic DNA and cloned into the lentiviral vector pHAGE-CMV-MCS-PGK-GFP. For shME1-resistant ME1 expression vector, first the human ME1 coding sequence was amplified from HEK-293 cDNA and cloned into the lentiviral vector pHAGE-CMV-MCS-PGK-puro. Then mutation in the shRNA-binding sites was generated by overlap extension PCR. ME1 and KRAS shRNAs were purchased from GENECHEM (Shanghai, China). The non-specific sequence (5′-TTCTCCGAACGTGTCACGT-3′) was used as control. The targeted sequences and primers for construction are provided in Supplementary Table 3.
Cell culture and transfection
Human HCT116, DLD1, SW48, HT29 and RKO CRC cells, and HEK-293 cells were cultured under standard conditions as described.4, 8, 9, 10, 11, 15 Cell lines stably expressing shRNAs were maintained in Dulbecco-modified eagle’s minimum essential medium with 1–2 μg/ml puromycin. For transfection, cells were seeded in six-well plates 24 h before and grown to 70–80% confluence for transfection. Plasmids (3–5 μg) in 50 μl Opti-MEM (GIBCO, Life Technologies, Grand Island, NY, USA) were mixed with 5 μl Lipofectamine (Invitrogen, Carlsbad, CA, USA) in 50 μl Opti-MEM, allowing complexes to form for at least 30 min at room temperature, and then added to the cells. Stable cell lines expressing the indicated miRs, cDNAs or shRNAs were generated by lentiviral transduction in the presence of 8 μg/ml polybrene followed by selection with 1–2 μg/ml puromycin for at least 10 days. Stable cell lines were detected for the expression of miRs, mRNA or shRNA by RT-qPCR or immunoblot.
miRNA library screen
miRNA expression library containing 1255 miRNA-expressing vectors was described previously.22 For miRNA library screening, HCT116 and RKO cells were seeded in triplicate at a density of 103 cells per well in 96-well plates 24 h before transfection. Then cells were transfected with 200 ng of individual miRNA expression vectors or empty control vector. Cell viability was measured by MTT assay according to the manufacturer’s instructions (Promega, Madison, WI, USA) 96 h after transfection compared with parallel wells that transfected with control vector.
RT-qPCR
Total RNA was isolated from cells by TRizol Reagent (Invitrogen). Then 1 μg RNA was reversely transcribed into cDNA by RevertAid First-strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). Primers for protein-coding genes were oligo(dT)18. Primers for non-coding miRNAs were: miR-30a-5p, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTTCCA-3′ miR-30a-3p, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCTGCA-3′. RT-qPCR was performed by SYBR Green Supermix (Bio-Rad, Hercules, CA, USA). Primers of RT-qPCR are listed in Supplementary Table 3. GAPDH, β-actin and U6 RNA were used as an internal control for protein-coding genes and miRNAs, respectively.
Luciferase reporter assays
The DNA fragments containing the potential miR-30a-binding region in KRAS 3′UTR and ME1 3′UTR were amplified by PCR, and were cloned into a pGL3-promoter vector (Promega). Regions of miR-30a promoter were also amplified and cloned into pGL3-promoter vector. Mutations in the miR-30a seed-matching sequences and potential P65-binding sites were generated by overlap extension PCR. Primers for the constructs and mutations are listed in Supplementary Table S3. For luciferase reporter assay of KRAS and ME1 3′UTR, cells overexpressing or deleted miR-30a were plated 24 h before transfection in 24-well plates and transiently transfected with 500 ng of the reporter plasmid, and 10 ng Renilla reporter. For luciferase reporter assay of miR-30a promoter, cells were plated 24 h before transfection in 24-well plates, and transiently transfected with 500 ng of the reporter plasmid and 10 ng Renilla reporter in cells stably expressing control-shRNA, P65 shRNA-#1 or #2, respectively. Then the luciferase activity was measured by the Dual Luciferase Reporter assay (Promega). Transfection efficiency was normalized on the basis of the Renilla luciferase activity.
Chromatin immunoprecipitation
ChIP was previously described.44 Briefly, intracellular protein–DNA complexes from HEK-293 cells transiently transfected with Flag-tagged P65, HCT116 or DLD1 cells were cross-linked in situ by 1% of formaldehyde. Total lysates were then sonicated, and subjected to chromatin-conjugated immunoprecipitation using specific antibodies including mouse anti-Flag monoclonal antibody (F3165, Sigma-Aldrich, St. Louis, MO, USA) and rabbit anti-P65 polyclonal antibody (A2547, Abclonal, Woburn, MA, USA). After reversal of cross-links, precipitated DNA was purified and analyzed by DNA gel electrophoresis or qPCR with specific primers listed in Supplementary Table 2.
Isolation of RNA-induced silencing complex-associated RNA
RNA immunoprecipitation was performed according to the methods previously described.45, 46 Briefly, 107 RKO, HCT116 or DLD1 cells stablely overexpressing miR-30a or control vector were UV-irradiated and lysed with RIP buffer supplemented with RNase Inhibitor (Thermo Scientific) and then treated with DNase I (Thermo Scientific). The separated supernatant was incubated with 15 μg rabbit monoantibody against AGO-2 (D2C9, Cell Signaling Technology, Beverly, MA, USA) or 15 μg normal rabbit IgG (Thermo Scientific) for 4 h and then was incubated with protein G agarose beads (Thermo Scientific). One-fifth of AGO-2-RNA immunoprecipitation was analyzed by immunoblot. The other precipitation was digested by proteinase K (Thermo Scientific) and the precipitated RNA was isolated by TRizol Reagent (Invitrogen), and the expression levels of miR-30a-5p/3p, ME1 and KRAS were analyzed by RT-qPCR.
Bioinformatic prediction of miR-30a targets
Computational prediction of miRNA targets was performed in online databases miRDB (http://mirdb.org/miRDB, Version 5.0) and TargetScan (http://www.targetscan.org/vert_71/, Release 7.1). The GenBank accession numbers used for KRAS, ME1 and miR-30a were NM_004985, NM_002395 and NR_029504, respectively.
Cell viability, foci, soft agar and sub-G1 DNA sorting
These were performed as previously described.22, 44, 47 Briefly, for cell viability assays, 103 indicated cells were seeded in triplicate in 96-well plates and grown for 5 days. Cell viability was measured using MTT assay according to the manufacturer’s instructions (Promega). For colony formation assays, 103 cells were seeded in triplicate in six-well plates and grown for 2 weeks. Colonies were counted and photographed after methanol fixation and methylene blue staining. For soft agar colony assays, 4000 cells were plated in triplicate in soft agar (0.35% low-melting point agarose on top of 0.7% bottom agarose) in six-well plates and fed with Dulbecco-modified eagle’s minimum essential medium. Colonies were counted and photographed after 2 weeks. Cell cycle analyses were performed on propidium iodide-stained nuclei using a MoFlo XDP-Flow Cytometer (FACS, Beckman Coulter, Brea, CA, USA). Percent of sub-G1 DNA content was analyzed by single-histogram statistics.
Wound-healing assay
About 5 × 105 indicated cells were plated in six-well plates and allowed to grow overnight to 90% confluence. Then a 200-μl pipette tip was used to scrap the cell culture slowly and vertically. The plates were washed three times with pre-warmed PBS before the fresh medium was added. The wounds were photographed every 24 h.
Cell migration and invasion assay
Migration assay was performed using a Transwell Assay Chamber (PET track-etched membrane; 24-well inserts, pore size, 8 μm, Corning Incorporated, Corning, NY, USA). For the invasion assays, the cells were seeded into Matrigel-coated Invasion Chambers, which were pre-coated with 10 mg/ml growth factor-reduced BD Matrigel matrix (BD Biosciences, Franklin Lakes, NJ, USA). Briefly, indicated cells were incubated in a medium containing 1% FBS for 24 h. Then 2 × 105 indicated cells suspended in 200 μl medium containing 1% FBS were seeded in the top chambers. About 500 μl fresh medium supplemented with 10% FBS was added to the bottom chambers. After incubation for 24 h at 37 °C, non-migrating cells on the top side of the chambers were removed by cotton swab. Cells migrating to the bottom side were fixed and stained with methylene blue. Then the numbers of migrated or invaded cells were counted and photographed.
Subcutaneous xenograft
All animal studies were approved by the Animal Care Committee of Wuhan University. Four-week-old female BALB/c nude mice were purchased from SLAC Laboratory Animal (Changsha, China) and maintained in microisolator cages. Tumorigenicity assays and tumor volume measurements were performed as previously described.44 In brief, 107 cells were suspended in 100 μl serum-free DMEM and injected subcutaneously in the flanks of animals (n=5 per group). Tumor growth was monitored every 3 days for a total period of ~30 days. Tumor volumes were calculated by the equation V (mm3)=a × b × c/2, where a is the length, b is the width and c is the height. Tumors were harvested for DNA, RNA and protein analyses.
Administration of lentivirus to AOM-/DSS-induced CRC mouse model
Viral production and infection were performed following the principles previously described.48 Briefly, lentivirus was produced using calcium–phosphate-mediated transfection of a second-generation packaging system in HEK-293 cells. Lentivirus containing supernatant was harvested at both 48 and 72 h after transfection and was filtered using 0.45 μm filter. Acquired lentivirus was mixed with PEG solution (5% PEG8000 and 0.5 M NaCl) overnight and concentrated by centrifugation at 7000 g for 30 min in 4 °C. Virus titers were determined by HIV-1 p24 ELISA (Perkin Elmer, Waltham, MA, USA) and the virus was concentrated to 108 titers.
AOM-/DSS-induced CRC model was performed following the principles previously described.49 Eight-week-old female C57BL/6 mice were injected intraperitoneally with 10 mg/kg AOM (Sigma-Aldrich). Five days later, mice were given drinking water containing 3% DSS (MP Biomedicals, Santa Ana, CA, USA) for 6 days followed by 2 weeks regular drinking water. Then mice were fed two rounds with 2.5% DSS water for 1 week and killed on day 100.
For lentivirus administration, it was performed as described previously.50 Briefly, mice were anesthetized and given an intrarectal enema of 100 μl 50% ethanol. Three hours after the enema, 100 μl lentivirus solution containing miR-30a (108 titers) or control vector was instilled intrarectally from the anus of the mice. The mice were inverted for 30 s after administration of intrarectal products to prevent leakage. Forty mice were randomly divided into four groups and administrated lentivirus-expressing miR-30a, KRAS shRNAs, ME1 shRNAs or control vector, respectively. Lentivirus administration was performed once a week for four times.
Metabolic and protein analyses
The levels of NADPH, triglyceride and fatty acids in cultured cells were determined using a NADP+/NADPH quantification Kit, Triglyceride Assay Kit and Fatty Acids Assay Kit (all from BioVision, Milpitas, CA, USA) respectively, following the manufacturer’s instructions. ME1 and ME2 activity was determined using cytosolic extracts and lysate of mitochondria as described, respectively.26
For immunoblot, cell pellets and tissues were lysed with SDS-PAGE lysis buffer, boiled and resolved with SDS-PAGE electrophoresis. Proteins were then transferred to nitrocellulose membranes, which were blocked with 5% non-fat milk and probed with corresponding antibodies including mouse anti-KRAS monoclonal antibody (sc-30, 1:200, Santa Cruz Biotechnology, Dallas, TX, USA), mouse anti-ME1 monoclonal antibody (sc-100569, 1:1000, Santa Cruz Biotechnology), rabbit anti-P65 polyclonal antibody (A2547, 1:1000, Abclonal), mouse anti-Flag monoclonal antibody (F3165, 1:10 000, Sigma-Aldrich) and mouse anti-β-actin monoclonal antibody (sc-47778, 1:2000, Santa Cruz Biotechnology). The blots were then incubated with a 1:1000–1:5000 dilution of HRP-conjugated IgG (Santa Cruz Biotechnology), washed and subjected to chemiluminescent detection as previously described.44
H&E and immunohistochemistry
Sections from colorectum of the mice and xenografts were fixed with formalin for 4 h, embedded in paraffin, sectioned and stained with hematoxylin and eosin according to the standard protocol. Ki-67 (Sc-15402, 1:500, Santa Cruz Biotechnology) staining was performed according to the manufacturer’s protocol. For apoptosis detection, caspase-3 (ab4051, 1:1000, Abcam, Cambridge, UK) staining was performed according to the manufacturer’s instruction. After staining, the photographs were taken by optical microscopy.
TCGA, CCLE and GSE data analyses
Data sets of CRC were downloaded from the TCGA data portal (http://tcga-data.nci.nih.gov). ME1 expression was assessed in human CRC tissues and adjacent non-cancer tissues from the TCGA mRNA data set. ME1 and ME2 expression levels of various cancer cell lines were obtained from the CCLE (http://www.broadinstitute.org/ccle/home). For miR-30a expression analysis, we found a published GSE data set (GSE18392) from NCBI GEO Data Sets and analyzed miR-30a-5p/3p expression in CRC tissues and adjacent non-tumor tissues.
Statistics
For data analysis, the SPSS statistical package for Windows (SPSS 16, SPSS Incorporated, Chicago, IL, USA), the GraphPad Prism Software (version 5.01, GraphPad Software, San Diego, CA, USA) and Microsoft Excel (Excel in Microsoft Office 2013 for Windows, Microsoft Corporation, Redmond, WA, USA) were used. Data are expressed as the mean±S.D. or S.E.M. Other statistical analysis was performed using the two-tailed Student’s t-test, Mann–Whitney U-test or Log-rank test, and P<0.05 was considered statistically significant.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81472549 and 31521091) and the 973 Program of China (2016YFC1302300 and 2014CB910600).
Author contributions
HS and CX performed all experiments. HS and KC performed bioinformatics and statistical analyses. YL helped to perform the experiments of administration of lentivirus and other animal experiments. JZ provided human CRC specimens and pathology identification. RD and XZ provided reagents. HS and YL designed the study and wrote the manuscript.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by S Fulda
The authors declare no conflict of interest.
Supplementary Material
References
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol 2008; 9: 517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen activated protein kinase cascade for the treatment of cancer. Oncogene 2007; 26: 3291–3310. [DOI] [PubMed] [Google Scholar]
- Ahmed D, Eide PW, Eilertsen IA, Danielsen SA, Eknæs M, Hektoen M et al. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013; 2: e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford H. Cancer: the Ras renaissance. Nature 2015; 520: 278–280. [DOI] [PubMed] [Google Scholar]
- Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov 2014; 13: 828–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG Jr. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer 2005; 5: 689–698. [DOI] [PubMed] [Google Scholar]
- Corcoran RB, Cheng KA, Hata AN, Faber AC, Ebi H, Coffee EM et al. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell 2013; 23: 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 2009; 137: 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl C, Frohling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 2009; 137: 821–834. [DOI] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009; 462: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Polytarchou C, Hommes DW, Palumbo T, Hatziapostolou M, Koutsioumpa M, Koukos G et al. MicroRNA214 is associated with progression of ulcerative colitis, and inhibition reduces development of colitis and colitis-associated cancer in mice. Gastroenterology 2015; 149: 981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Chen Y, Cheng L, Prochownik EV, Li Y. microRNA-206 impairs c-Myc-driven cancer in a synthetic lethal manner by directly inhibiting MAP3K13. Oncotarget 2016; 7: 16409–16419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crippa E, Folini M, Pennati M, Zaffaroni N, Pierotti MA, Gariboldi M. miR-342 overexpression results in a synthetic lethal phenotype in BRCA1-mutant 1937 breast cancer cells. Oncotarget 2016; 7: 18594–18604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkowski R, Du L, Zhao Z, McMillan E, Kosti A, Yang CR et al. Genetic mutation of p53 and suppression of the miR-17 approximately 92 cluster are synthetic lethal in non-small cell lung cancer due to upregulation of vitamin D Signaling. Cancer Res 2015; 75: 666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent OA, Chivukula RR, Mullendore M, Wentzel EA, Feldmann G, Lee KH et al. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev 2010; 24: 2754–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca A, Valvo C, Fabrizi E, di Martino S, Biffoni M, Runci D et al. Analysis of the combined action of miR-143 and miR-145 on oncogenic pathways in colorectal cancer cells reveals a coordinate program of gene repression. Oncogene 2013; 32: 4806–4813. [DOI] [PubMed] [Google Scholar]
- Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell 2014; 25: 469–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer 2011; 11: 849–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Li W, Shen H, Zhang J, Zhu Y, Li Y. microRNA-129-5p, a c-Myc negative target, affects hepatocellular carcinoma progression by blocking the Warburg effect. J Mol Cell Biol 2016; 8: 400–410. [DOI] [PubMed] [Google Scholar]
- Sarver AL, French AJ, Borralho PM, Thayanithy V, Oberg AL, Silverstein KA et al. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer 2009; 9: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL. Ras oncogenes in human cancer: a review. Cancer Res 1989; 49: 4682–4689. [PubMed] [Google Scholar]
- Daily K, Patel VR, Rigor P, Xie X, Baldi P. MotifMap: integrative genome-wide maps of regulatory motif sites for model species. BMC Bioinformatics 2011; 12: 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Du W, Mancuso A, Wellen KE, Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature 2013; 493: 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puyol M, Martin A, Dubus P, Mulero F, Pizcueta P, Khan G et al. A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell 2010; 18: 63–73. [DOI] [PubMed] [Google Scholar]
- Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 2013; 496: 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr EM, Gaude E, Turrell FK, Frezza C, Martins CP. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature 2016; 531: 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Le Leu RK, Young GP. Detection of K-ras mutations in azoxymethane-induced aberrant crypt foci in mice using LNA-mediated real-time PCR clamping and mutant-specific probes. Mutat Res 2009; 677: 27–32. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Tang Q, Qin D, Yu L, Huang R, Lv G et al. Role of microRNA 30a targeting insulin receptor substrate 2 in colorectal tumorigenesis. Mol Cell Biol 2015; 35: 988–1000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wei W, Yang Y, Cai J, Cui K, Li RX, Wang H et al. MiR-30a-5p suppresses tumor metastasis of human colorectal cancer by targeting ITGB3. Cell Physiol Biochem 2016; 39: 1165–1176. [DOI] [PubMed] [Google Scholar]
- Liu M, Huang F, Zhang D, Ju J, Wu XB, Wang Y et al. Heterochromatin protein HP1gamma promotes colorectal cancer progression and is regulated by miR-30a. Cancer Res 2015; 75: 4593–4604. [DOI] [PubMed] [Google Scholar]
- Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J. Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin Cancer Res 2006; 12: 2014–2024. [DOI] [PubMed] [Google Scholar]
- Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X et al. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy 2009; 5: 816–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 2011; 25: 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgendy M, Sheridan C, Brumatti G, Martin SJ. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell 2011; 42: 23–35. [DOI] [PubMed] [Google Scholar]
- Hara T, Jones MF, Subramanian M, Li XL, Ou O, Zhu Y et al. Selective targeting of KRAS-mutant cells by miR-126 through repression of multiple genes essential for the survival of KRAS-mutant cells. Oncotarget 2014; 5: 7635–7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Hancock DC, Molina-Arcas M, Steckel M, East P, Diefenbacher M et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell 2012; 149: 642–655. [DOI] [PubMed] [Google Scholar]
- Yu B, Swatkoski S, Holly A, Lee LC, Giroux V, Lee CS et al. Oncogenesis driven by the Ras/Raf pathway requires the SUMO E2 ligase Ubc9. Proc Natl Acad Sci USA 2015; 112: E1724–E1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh EH, Chen Y, Bader DA, Hamilton MP, He B, York B et al. Mitochondrial activity in human white adipocytes is regulated by the ubiquitin carrier protein 9/microRNA-30a axis. J Biol Chem 2016; 291: 24747–24755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen D, Liu D, Tang J, Dong L, Liu Y, Tao Z et al. Malic enzyme 1 induces epithelial-mesenchymal transition and indicates poor prognosis in hepatocellular carcinoma. Tumour Biol 2015; 36: 6211–6221. [DOI] [PubMed] [Google Scholar]
- Chakrabarti G. Mutant KRAS associated malic enzyme 1 expression is a predictive marker for radiation therapy response in non-small cell lung cancer. Radiat Oncol 2015; 10: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Sun D, Li W, Shen H, Zhu Y, Li C et al. A c-Myc-MicroRNA functional feedback loop affects hepatocarcinogenesis. Hepatology 2013; 57: 2378–2389. [DOI] [PubMed] [Google Scholar]
- Wang H, Sun T, Hu J, Zhang R, Rao Y, Wang S et al. miR-33a promotes glioma-initiating cell self-renewal via PKA and NOTCH pathways. J Clin Invest 2014; 124: 4489–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009; 460: 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Li W, Tian Y, Xu P, Wang H, Zhang J et al. Upregulation of miR-362-3p modulates proliferation and anchorage-independent growth by directly targeting Tob2 in hepatocellular carcinoma. J Cell Biochem 2015; 116: 1563–1573. [DOI] [PubMed] [Google Scholar]
- Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med 2008; 14: 1271–1277. [DOI] [PubMed] [Google Scholar]
- Man SM, Zhu Q, Zhu L, Liu Z, Karki R, Malik A et al. Critical role for the DNA Sensor AIM2 in stem cell proliferation and cancer. Cell 2015; 162: 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H, Kimura T, Haga K, Kasahara N, Anton P, McGowan I. Effective in vivo and ex vivo gene transfer to intestinal mucosa by VSV-G-pseudotyped lentiviral vectors. BMC Gastroenterol 2010; 10: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.