Abstract
The mechanisms that underpin the production of small molecules and cytokines that lead to inflammation or programmed cell death are intricately intertwined. So much so that some of the proteins that contribute to the transcriptional up regulation of cytokines can switch their role in the right circumstances to generate cell death-inducing complexes. This entwinement is reflected in the fact that inflammation helps an organism fight pathogens and that therefore pathogens are under an evolutionary pressure to interfere with this process. Cell death is therefore a defensive measure that may serve to deny pathogens a host cell, expose pathogens to the immune system and also provide additional inflammatory information to the host. Clearly such a system must be tightly regulated and ubiquitylation is a post-translational protein modification that is at the heart of this regulation. In this review, we discuss the regulatory ubiquitin events that dictate the formation and activation of death-inducing complexes containing RIPK1/FADD/caspase-8, and examine how these events collectively determine cell fate.
Facts
Ubiquitin is a small molecule that can be post translationally attached to proteins in different orientations.
E3 Ligases attach ubiquitin to substrates, DUBs deubiquitylate substrates and can counter the activity of E3 Ligases.
The DISC, Complex 2 and the Ripoptosome are complexes that all contain the core components RIPK1, FADD and caspase-8.
caspase-8 is activated by oligomerisation and can induce apoptosis and inhibit necroptosis.
MLKL is the key necroptotic effector and can be activated by RIPK3 phosphorylation.
Open Questions
How many ubiquitin molecules exist within a single ubiquitin chain and how is ubiquitin chain length determined?
What is the stoichiometry of RIPK1/FADD/caspase-8 containing complexes?
Outside of ubiquitin, how many other post-translational events regulate the formation and activation of FADD/caspase-8 containing complexes?
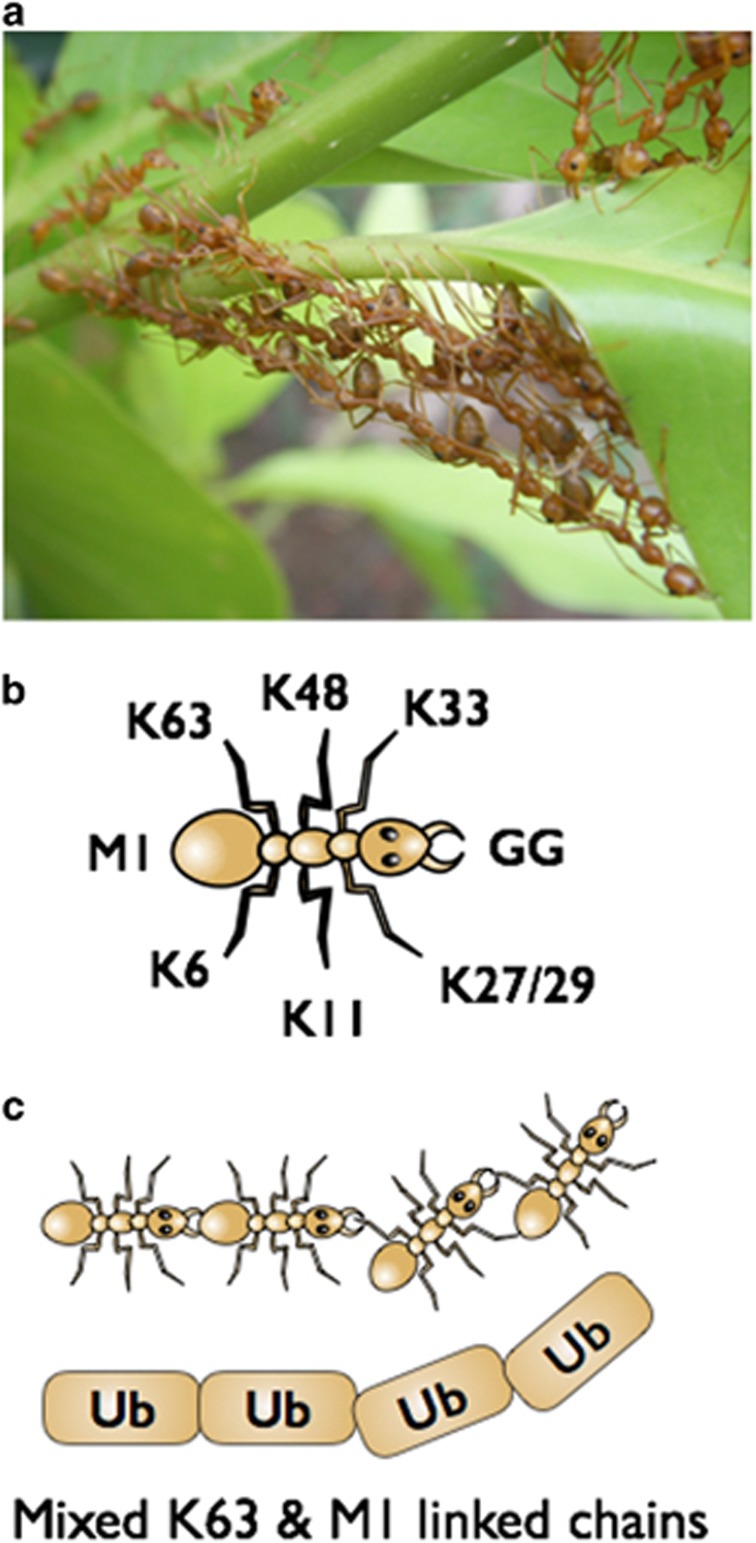
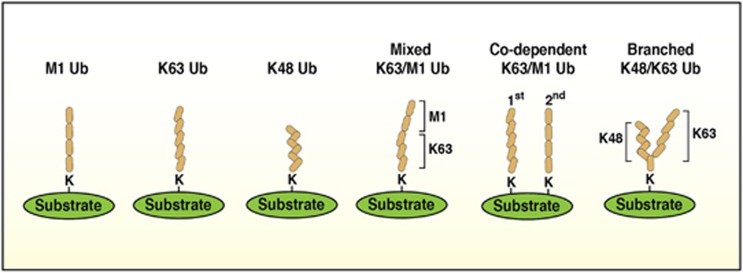
Accomplishing higher-level tasks by multiplexing many smaller individual units is a strategy used in many biological systems. Such complex self-assemblages are seen in the insect world, where social insects like weaver or army ants use their bodies to form chains or large scaffolds and structures to benefit their colony1 (Figure 1a). The small 7-kDa ubiquitin protein, like these ants, is a hard working multi-tasker that can perform a wide variety of tasks by linking up with other ubiquitin molecules in different combinations. Ubiquitin can either be linked via the free ε-amine in lysine residues or the α-amine in the N-terminal methionine of substrate proteins by ubiquitin E3 ligases to the c-terminal glycine of ubiquitin in an isopeptide bond. Single ubiquitin molecules can also be attached to target proteins and this is termed mono- or multi mono-ubiquitylation. However because ubiquitin contains seven lysine residues and the free N-terminal methionine, ubiquitin molecules can be linked together to form a poly-ubiquitin chain. The configuration of the ubiquitin chain is dependent on whether a particular lysine or the N-terminal methionine within the ubiquitin molecule is utilised. These chains can contain single linkage types (e.g.: K63 only) or alternatively contain multiple linkage types where ubiquitin molecules are attached in different orientations within the same chain (Figure 2). The different linkages that form are M1-, K6-, K11-, K27-, K29-, K33-, K48- and K63-linked ubiquitin (Figures 1b and c). In an elegant study by Emmerich et al.2 M1-linked poly-ubiquitin chains were shown to be heavily dependent on the presence of K63-linked chains presenting as either hybrid chains (M1 & K63 in the same chain) or separate chains linked to the same protein (Figure 2). This dependency of one linkage type for another is interesting when ubiquitin-binding domain proteins are considered, as different linkages existing in close proximity to one another may facilitate the recruitment of distinct protein complexes to mediate their activation.
Figure 1.
Weaver ant chains parallel ubiquitin chains. (a) A photograph of the chain-like structures formed by the weaver ant (genus Oecophylla). Weaver ants form head to tail chains in order to reach adjacent leaves during the building of their nests. The leaves are then pulled closer together and larval secretions are used to make a seal. (b) A Schematic of an ant used as to represent the lysine residues (K6, K11, K27, K29, K33, K48 and K63), the N-terminal methionine (M1) and the C-terminal diGly (GG) residues within ubiquitin. (c) A Schematic using ants (top) and ubiquitin (Ub) (bottom) to represent a mixed K63/M1 ubiquitin chain
Figure 2.
Ubiquitin linkages. Schematic depicting a few possible scenarios for ubiquitin chain orientations and combinations. Ubiquitin chains are thought to occur in single, mixed or branched forms. Single chains occur when only one ubiquitin linkage is represented in the chain, whereas mixed linkages have more than one linkage type represented. Branched chains on the other hand occur when two chain types are represented but when these chain types occur in distinct chains. Alternatively, co-dependency may occur if the conjugation of one chain type (the 1st chain type) is dependent on another (the 2nd chain type)
Ubiquitin researchers are beginning to understand and decipher the code that enables cells to interpret different chain configurations. New tools that allow researchers to investigate the occurrence of different ubiquitin linkages are aiding in this endeavour. For example, Tandem Ubiquitin Binding Entities (TUBEs), that bind to and also stabilise ubiquitin chains, allow for relatively simple purification of ubiquitylated proteins that can then be analysed by western blot or mass spectrometry. Linkage-specific TUBEs can also be used to specifically purify K63-, K48- and M1-linked ubiquitylated proteins. Linkage-specific ubiquitin antibodies can also be used similarly to TUBEs and some have the additional benefit that they can be used on denatured proteins. Furthermore, the use of linkage-specific deubiquitinases (DUBs) can implicate the types of ubiquitin linkages present in a sample and can help reveal mixed and branched ubiquitin chains. Ubiquitin Chain Restriction Analysis (UbiCREST) kits that contain all relevant DUBs are available and allow researchers to characterise their chain of interest with minimal effort.3 Targeted Mass Spectrometry is without question the gold standard to prove ubiquitin modification and has been used to identify residues targeted with ubiquitin, some of which we discuss below.
The beauty of ubiquitin is that it can be combined in an almost limitless number of potential combinations and these can be dynamically regulated. It is therefore highly suited to orchestrate complex, temporally regulated, signalling events and many different signalling platforms utilise it. One that stands out in the literature due to the intense research in the area, and the early recognition of the important roles ubiquitin plays in this pathway, is TNFR1 signalling. Upon binding of TNF, TNFR1 generates both a membrane bound complex (complex 1) and a cytoplasmic complex (complex 2) and it appears, although no definitive data has been provided, that the cytoplasmic complex evolves from the membrane bound form. This sequence of events was proposed based on the fact that the cytoplasmic complex can be detected after the formation of the membrane bound complex and that both the membrane bound and cytoplasmic complex contain the molecule RIPK1.4 Although this data suggests a chronological sequence it is noteworthy that certain treatments can generate a cytoplasmic complex, containing RIPK1, Fas-associated protein with death domain (FADD) and caspase-8 without the assistance of TNF-TNFR1 signalling. The complex that forms independently of cell surface receptor engagement has been dubbed the Ripoptosome, but in our opinion is likely to be very similar in composition to the cytoplasmic TNFR1 complex 2.5 Complex 2 and the Ripoptosome are both capable of activating apoptosis and in the presence of the necroptosis-inducing molecule RIPK3, these complexes also have the potential to drive necroptosis. This review focuses on the different FADD/Caspase-8 containing complexes and what is currently known about the ubiquitylation of their components and how these ubiquitylation events dictate signalling responses.
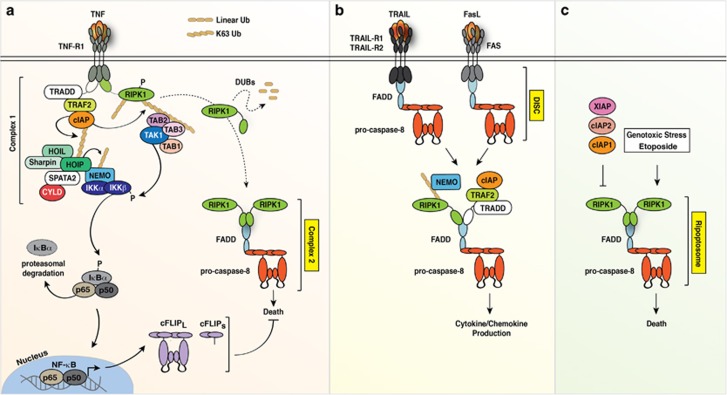
Complex Complexes – the Death-Inducing Signalling Complex, Complex 2 and the Ripoptosome
Complex 2 and the Ripoptosome are not the only complexes containing caspase-8, FADD and RIPK1. In fact, the death-inducing signalling complex (DISC) was the first complex reported to contain these key players. This complex forms downstream of the death receptors FAS/CD95,6 TRAIL-R1 (DR4) and TRAIL-R2 (DR5).7 The complex is generated as a result of homotypic interactions between the death domain (DD) present in the cytoplasmic tail of the receptor and the molecule FADD, followed by death effector domain (DED) mediated recruitment, oligomerisation and activation of caspase-8 or 10 (Figure 3). The DISC also contains the caspase-8 inhibitor cellular FLICE inhibitory protein (cFLIP), which exerts critical functions that determine cellular fate. Importantly, and distinctly to the Ripoptosome and Complex 2 the DISC is tethered to the cytoplasmic side of the plasma membrane by the receptors (Figure 3). Notably, secondary signalling complexes that do not contain the death receptor can also be observed.8 The cytoplasmic TNFR1 complex 2, forms downstream of the DD containing receptor TNFR1, however differently to TRAIL-R1/R2 (DR4/5) and FAS/CD95 it does not complex around the DDs of TNFR1 but rather forms via the DDs of TRADD and/or RIPK1. The Ripoptosome on the other hand forms irrespective of ligand-receptor engagement at the cell surface and instead forms spontaneously upon cIAP depletion.9, 10 Because chemotherapeutic agents such as Etoposide can trigger IAP depletion these drugs can also promote the formation of the Ripoptosome (Figure 3). Although this ~2 MDa complex contains the core components caspase-8, FADD and RIPK1, in certain cell types and on particular stimuli this complex can also recruit cFLIP and RIPK3 (ref. 11) and also potentially the regulatory proteins NEMO (IKKγ), TAK1 and ATM.12 RIPK3 is a necroptosis-inducing RHIM-containing protein that can be activated by other RHIM-containing proteins such as RIPK1, Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF), and DNA-dependent activator of interferon regulatory factors (DAI).13 Outside of these three well defined complexes, other reports have demonstrated that a complex between FADD, caspase-8, RIPK1 and RIPK3 forms on TLR ligation to regulate inflammasome activation and IL-1β secretion.14, 15, 16 Regardless of the entry point the formation and activation of this complex is highly regulated to avoid aberrant activation.
Figure 3.
Complex 2, the DISC and the Ripoptosome. (a) Schematic depicting the activation of RIPK1/FADD/caspase-8 containing complexes. Complex 2 forms downstream of complex 1 upon ligation of TNF with its receptor TNFR1. TNF induces timerisation of TNFR1 and this stimulates recruitment the downstream signalling effectors, culminating in the activation of NF-κB. Key ubiquitylation and phosphorylation events are important to limit the association of RIPK1 with FADD/caspase-8 in order to prevent cell death. (b) The DISC forms upon ligation of the ligands, TRAIL or FasL to their cognate receptors. Ligand binding induces receptor trimerisation and the recruitment of FADD and caspase-8. In type I cells caspase-8 activation can induce apoptosis via direct cleavage of downstream caspases. Through an incompletely defined mechanism a secondary signalling complex can also form downstream of the DISC comprising signalling proteins, such as RIPK1 and the cIAPs, which stimulates the upregulation of cytokines and chemokines. (c) The Ripoptosome, comprising the core components RIPK1/FADD/caspase-8, does not require ligand/receptor engagement and is a cytoplasmic complex that is kept in check by the IAP proteins. The Ripoptosome can be induced upon genotoxic stresses such as Etoposide or removal/inhibition of the IAP proteins
Weaving a Stronger Signal – Ubiquitin-Mediated Regulation of RIPK1
Weaver ants work together to create intricate chains that allow them to ratchet together leaves that are beyond the reach of a single ant. These chains then allow other ants to bind together the leaf seams using larval secretions to make a strong enclosed nest. Similarly to the chains that assist the weaver ant to build its nest, ubiquitin chains on RIPK1 allow it to perform a variety of functions that culminate in both survival and death signals. RIP kinases are studied intensively due to their roles in both caspase-dependent and – independent cell death and their ability to function downstream of signalling receptors. Confusingly for researchers new to the field, in most signalling cascades these kinases play a structural/scaffolding role in their respective signalling complexes and their kinase activity takes a back seat. Thus while RIPK1 knockout mice are perinatal lethal with a massive systemic inflammatory syndrome,17, 18, 19, 20 RIPK1 kinase dead mice are viable and healthy.21, 22 Indeed even where RIPK2 kinase inhibitors indubitably block NOD signalling it appears that they do so by preventing a conformational change necessary for it to perform its structural role.23
The regulation of RIPK1 by ubiquitylation is well described downstream of TNFR1. Ligation of TNF with TNFR1 stimulates the recruitment of TRADD and RIPK1 to the cytoplasmic DD of TNFR1. TRADD facilitates the recruitment of TRAF2, allowing the recruitment of the cIAP proteins and subsequently the recruitment of LUBAC (HOIP, HOIL and Sharpin) to complex 1. These events, in addition to the K63-linked non-degradative ubiquitylation of RIPK1 within complex 1, stimulate the activation of the downstream signalling effectors in certain cell types.24, 25, 26 Deregulation of complex 1 by disrupting the ubiquitylation events causes the dismantling of complex 1 and stimulates the formation and activation of complex 2 (Figure 3). The primary E3 ligases responsible for this ubiquitylation in most cell types are the cellular Inhibitor of APoptosis proteins, cIAP1 and cIAP2.27, 28, 29, 30 However, although TNF-induced ubiquitylation of RIPK1 is almost completely absent in cIap1−/−cIap2−/− double knockout cells immediately following TNF stimulation there are still some high molecular weight bands that can be visualised via western blot. This may indicate that all the E3 ligases that regulate RIPK1 directly are still not defined, or that additional ubiquitin-like linkages such as SUMO or NEDD8 are attached to RIPK1 following TNF stimulation. Other E3 ligases may play an indirect role in regulating RIPK1 ubiquitylation, such as RNF11 that has been shown to play a role in allowing A20 to deubiquitylate RIPK1 and terminate this signalling.31 IAP driven ubiquitylation is critically important to prevent the cytotoxic activities of RIPK1, potentially by delaying or limiting the recruitment of RIPK1 to a FADD/caspase-8 containing complex. This is demonstrated by the fact that mutation of a critical lysine in RIPK1, K377, to arginine decreased TNF-induced ubiquitylation of RIPK1 and increased the susceptibility of RIPK1 K377R expressing cells to apoptosis.24, 32 The significance of IAP regulation of RIPK1-mediated cytotoxicity is highlighted by the fact that the early embryonic lethality observed in cIap1−/−cIap2−/− and cIap1−/−Xiap−/− mice is prevented by genetic deletion of RIPK1.30 Further evidence is provided from studies using chemotherapeutic agents termed Smac-mimetics (SM), which mimic the endogenous IAP antagonist Smac/DIABLO and inhibit the action of IAP proteins. These compounds act by inducing cIAP dimerisation which in turn promotes their auto-ubiquitylation and proteasomal degradation33, 34 and by relieving XIAP mediated suppression of caspases. TNF-induced ubiquitylation of RIPK1 is significantly reduced if cells are pre-treated with SM compounds suggesting that IAPs mediate the majority of RIPK1 ubiquitylation.28, 29, 35, 36 Finally mutation of cIAP E3 ligase activity also prevents RIPK1 ubiquitylation upon TNF stimulation highlighting a central role for the cIAPs in ubiquitylation of RIPK1.29, 37 cIAP1-mediated ubiquitylation of RIPK1 and other complex 1 components facilitates the recruitment of the linear ubiquitin assembly complex (LUBAC),29 which generates M1-linked ubiquitin chains. This set of ubiquitin events at the TNFR1 complex 1 are relatively well described, and the role for these chains in promoting NF-κB and MAPK activation which in turn leads to cFLIP up regulation are well understood. Without wishing to detract from all these studies it is worth noting that a proteome-wide approach to examine ubiquitin modifications following TNFR1 signalling did not detect ubiquitylated RIPK1 although it was able to identify many other well characterised ubiquitin substrates (including TRAF2, cIAP1 and NEMO).38 This is despite the fact that the authors were able to readily identify RIPK1 ubiquitylation by western blot. This may be indicative of the fact that there are many different RIPK1 variants that make detecting individual targeted peptides by global Mass Spectrometry approaches difficult, as others have successfully detected ubiquitin on RIPK1 by combining two-dimensional separation with multiple reaction monitoring mass spectrometry.39 This technical detail aside it is well established that RIPK1 gets rapidly ubiquitylated at the membrane bound complex following TNF signalling in a predominantly cIAP1-dependent manner (Figure 3). However, less is known about the ubiquitylation of RIPK1 on its ‘journey’ to the cytoplasmic complex and how this might limit or promote the killing potential of RIPK1. Other post translation modifications such as phosphorylation have been reported to limit the cytotoxic activities of RIPK1 following TNF40 indicating that this transition from complex 1 to complex 2 is likely to be a highly regulated process. Given that loss of the IAPs sensitises cells to TNF-induced death and results in unmodified RIPK1 it is plausible that ubiquitin acts to restrain RIPK1 and limits its association with FADD/caspase-8. This may occur indirectly through transcriptional up regulation of cFLIP, or it may promote proteasomal turnover of RIPK1. Consistent with proteasomal degradation, RIPK1 becomes K48 and K11 ubiquitylated upon TNF signalling.37, 41 Alternatively the presence of non-degradative ubiquitin chains might preclude ubiquitylated RIPK1 from generating cytoplasmic RIPK1 complexes. Consistent with this idea Cylindromatosis (CYLD), a K63- and M1-linked deubiquitylating enzyme, and A20, a DUB capable of interacting with M1-linked chains that appears to, counter-intuitively, limit M1-linked ubiquitin chain cleavage, seem to regulate such complex formation and cell death.42, 43, 44, 45 Loss of CYLD reduces TNF SM induced formation of the cytoplasmic RIPK1 complex while loss of A20 increases complex 2 formation.46 The fact that SMs reduce RIPK1 modification in complex 1 coupled with the fact that DUBs help generate complex 2 is suggestive that RIPK1 in complex 2, the DISC or the Ripoptosome is likely to be minimally ubiquitylated. However, Geserick et al.8 showed that RIPK1 in the DISC was quite strongly modified, even in the presence of IAP depletion using SMs, and Cullen and colleagues47 also observed prominent RIPK1 modification without SM treatment. Likewise in a Ripoptosome generated by genotoxic stress or SM treatment, RIPK1 bound to caspase-8 (although largely unmodified) still retained higher molecular weight species, and RIPK1 in the cytoplasmic complex-II is clearly modified when bound to caspase-8.4, 9, 10
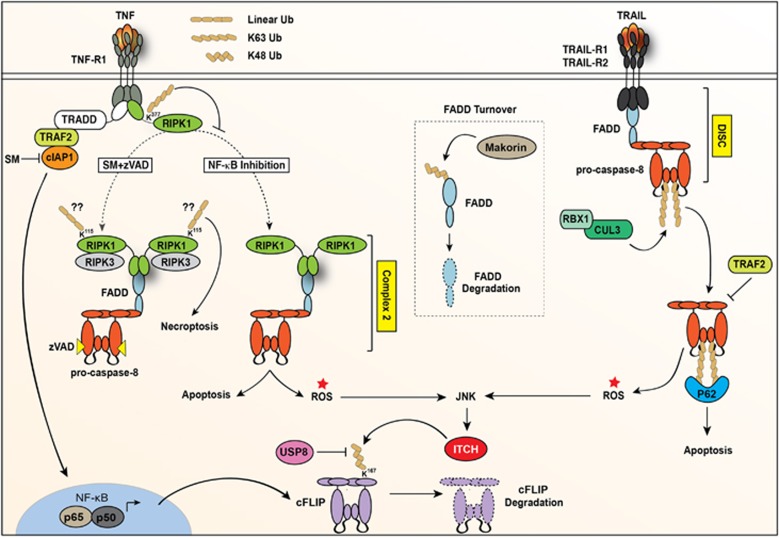
Ubiquitin modification of RIPK1 has also been explored in the context of necroptosis. In situations where caspase inhibitors block the activity of caspase-8 (and caspase-8/cFLIP heterodimers), the cytoplasmic complex (complex 2, DISC or Ripoptosome) matures into a RIPK3 and MLKL containing complex. Immunoprecipitation of this necroptosis-inducing complex revealed that RIPK1 is modified with K63- and M1-linked ubiquitin.48 The observation that linear ubiquitin is present strongly implies the presence of LUBAC, a heterotrimeric complex that forms through interaction of HOIP, HOIL-1 and Sharpin. Consistent with this, De Almagro et al. demonstrated that knockdown of HOIL-1 and HOIP reduced the linear ubiquitylation of RIPK1 in complex 2. The ubiquitylation site on RIPK1 under these conditions was later found to be K115 (Figure 4), and mutation of this site to arginine decreased both linear and K63-linked ubiquitylation, suggesting either mixed or branched chains form at that particular site.48 An alternative explanation could be that one linkage type could be required for the other linkage to be conjugated (Figure 2), but this would require additional study. Expression of RIPK1 K115R in cells reduced the killing potential of RIPK1 under necroptotic conditions and importantly this mutation did not effect RIPK1 ubiquitylation at the TNFR1 signalling complex or alter the activation of NF-κB and JNK following TNF.48 Thus, ubiquitylation at this site appears to specifically determine the signalling capabilities of RIPK1 in complex 2. It also indicates that in contrast to ubiquitylation at K377 this particular ubiquitylation event at K115 actually promotes the killing activity of RIPK1. Interestingly the kinase activity of RIPK1 is required for this ubiquitylation event because a kinase dead version of RIPK1 was unable to be ubiquitylated. However, ubiquitylation at this site does not dictate the kinase activity of RIPK1, because mutation of this residue does not alter the ability of RIPK1 to auto-phosphorylate. One might imagine a situation whereby under conditions that induce the killing potential of RIPK1, the kinase activity of RIPK1 is activated and this stimulates the ubiquitylation of RIPK1 at K115. It remains to be confirmed whether this ubiquitylation is dictated by phosphorylation of RIPK1 itself, which may induce a conformational change that increases lysine availability, or whether phosphorylation of an interacting partner is required, potentially an E3-ubiquitin ligase that requires phosphorylation for activation. It also remains to be seen whether these ubiquitylation events at K115 are the same events that occur on RIPK1 in the Ripoptosome and the DISC, as one could also argue that the modification seen on RIPK1 when bound to FADD/caspase-8 is simply left over ubiquitin chains from DUB cleavage events that have taken place upstream of formation of these complexes. In line with this argument, the prediction that LUBAC might also associate with complex 2 might simply represent that linear ubiquitin events have taken place in complex 1 and have evolved to be part of complex 2, and not that LUBAC directly associates with complex 2. Nevertheless, it has been demonstrated that LUBAC is recruited to the DISC upon TRAIL stimulation and functions to linearly ubiquitinate RIPK1 and caspase-8.49 Together these data suggest that the formation of complex 2 can be influenced by ubiquitin events, but that multiple factors determine whether there is sufficient complex 2 activity to induce cell death and even the nature of that death.
Figure 4.
Ubiquitylation of RIPK1/FADD/caspase-8 containing complexes. Schematic depicting the ubiquitylation events that take place to regulate the activation of FADD/caspase-8 containing complexes. In situations where the IAPs are removed by the addition of SM compounds and cells are stimulated with TNF, RIPK1 forms a prominent complex with FADD and caspase-8. Under these conditions RIPK1 is modified with ubiquitin at lysine (K) 115 and this modification aids in its ability to induce apoptosis. “??” indicate that these chains may not be exclusively linear chains and may be a mixture of K63 and linear chains. Upon stimulation with TRAIL, FADD and caspase-8 interact with the cytoplasmic tail of the receptor to form the DISC. Caspase-8 is subsequently ubiquitylated by the cullin E3 ligase CUL3, and upon dissociation of caspase-8 from the plasma membrane complex p62 is recruited to these ubiquitin chains to aid in full activation of caspase-8 and apoptosis. This activation of caspase-8 is reported to be negatively regulated by the E3-ubiquitin ligase TRAF2. Both TNF-induced and TRAIL-induced activation of caspase-8 can drive the activation of ROS, which stimulates the activation of JNK. JNK can subsequently drive phosphorylation and activation of the E3-ubiqutin ligase ITCH that can then regulate the levels of cFLIP by ubiquitylation at K167. Ubiquitylation of cFLIP can be negatively regulated by the de-ubiquitinase USP8 that acts to reverse these events. Similarly, the regulation of FADD occurs outside of caspase-8 containing complexes and is mediated by the E3-ubiquitin ligase Makorin, which acts to regulate the steady state levels of FADD
Stronger Together – Ubiquitylation of Caspase-8 Dimers
Caspases require an upstream signal to trigger their conversion from an inactive to an active conformation. Caspase-8 is an apoptotic protease and like all initiator caspases, relies on adaptor-mediated dimerisation of monomeric caspase-8.50 Dimerisation at the FADD DD, leads to further oligomers of dimers and leads to autoproteolysis of the linker, separating the large and small subunits of the catalytic domain. Dimerisation is the main signal leading to caspase-8 activation, and kinetic activation studies have shown that linker proteolysis enhances the equilibrium for caspase-8 dimerisation.50, 51 Consistent with the idea that dimerisation is the main signal for caspase-8 activation, caspase-8 cleavage mutants were reported to efficiently promote apoptosis irrespective of processing.52 Another study using caspase-8 knockout mice reconstituted with a BAC transgenic caspase-8 cleavage mutant were somewhat resistant to FasL induced killing, at least over short term experiments, however, it was notable that the mice developed normally and did not appear to develop the lymphoproliferative disease observed in FAS (lpr) and FasL (gld) mutant mice.53 On the other hand studies using an in vitro DISC reconstitution method, consisting of either the FAS or TRAIL-R1 intracellular domain linked to beads, FADD and caspase-8, showed that a non-cleavable mutant displayed only intra-DISC activity (could cleave another caspase-8 molecule or cFLIP) but was unable to cleave a fluorescent substrate.54, 55 Taken together it appears that while linker proteolysis is not required for caspase-8 activation and for intra-dimer cleavage, it nevertheless helps the dimeric active form of caspase-8 persist.
Germane to the theme of this review, it has been suggested that caspase-8 dimerisation requires a ubiquitin-dependent event. This ubiquitylation was shown to be mediated by the CUL3-RBX E3 ligase complex (Figure 4). Silencing of CUL3 decreased TRAIL stimulated ubiquitylation of caspase-8, limited recruitment of caspase-8 to the TRAIL-R1 DISC and reduced caspase-8 activity and TRAIL induced cell death.56 CUL3 was found associated with the DISC and the ubiquitylation site on caspase-8 was identified as lysine 461. Ubiquitylation of this residue helped recruit the ubiquitin-binding protein p62/sequestsome-1, which recruited active caspase dimers into aggresomes to cleave substrate proteins. Modification of caspase-8 has also been detected upon TLR ligation under conditions where the IAPs are removed.15 This stimulation induces Ripoptosome formation but unexpectedly the modification of caspase-8 is somewhat dependent on RIPK3, because Ripk3−/− BMDMs have reduced caspase-8 modification upon LPS treatment in combination with SMs.15 Given that RIPK3 is required for full caspase-8 activity under these conditions it is plausible that this modification contributes to caspase-8 activation.
Interestingly caspase-10 was not ubiquitylated in a similar CUL3/RBX-dependent manner, even though the target lysine is conserved in both caspases.56 It is also notable that in the elegant in vitro DISC reconstitution experiments, already discussed, that the minimal requirements for caspase-8 oligomerisation and activation are a DD, FADD and caspase-8.54, 55 Typical of the multi-functional nature of ubiquitin, ubiquitylation has also been proposed to shut down the activity of caspase-8 and limit its death-inducing capabilities. This ubiquitylation is proposed to be driven by TRAF2 in a RING-mediated event that stimulates the K48-linked ubiquitylation and proteasomal degradation of active caspase-8 subunits57 (Figure 4). Other authors have been unable to demonstrate that the TRAF2 RING is able to interact with E2s or ubiquitylate substrates arguing against the possibility that the RING of TRAF2 ubiquitylates caspase-8.58 However failure to find activity does not prove its absence and in other experiments Traf2 knockout cells reconstituted with a TRAF2 ΔRING mutant activated NF-κB in response to TNF normally but were unable to protect cells from TNF-induced death, strongly indicating a protective role for the TRAF2 RING.59
Ubiquitin – Destroyer of Proteins – Ubiquitylation of cFLIP Induces Apoptosis
cFLIP is an NF-κB-inducible anti-apoptotic protein that directly limits the full activation of caspase-8. cFLIP serves as a checkpoint to determine the signalling outputs of complex 2. cFlip comes in a variety of flavours attributed to the multiple cFlip mRNA splice isoforms that give rise to either a long 55 kDa cFLIP (cFLIPL), a short 26 kDa cFLIP (cFLIPS) or a smaller 23 kDa cFLIP (cFLIPR). cFlip clearly arose from a genetic duplication event and is structurally highly similar to caspase-8, the main point of difference being that its pseudo-caspase domain lacks the critical catalytic residue required for full caspase activation. Loss of cFLIP causes embryonic lethality at embryonic day E10.5 and conditional deletion in several tissues results in cell death and lethality.60, 61 In addition to the transcriptional regulation already described, cFLIP can also be regulated by ubiquitin-mediated degradation. It has been proposed that the HECT E3-ubiquitin ligase ITCH, can be phosphorylated and activated by TNF-TNFR1 activated JNK, and that it in turn then K48 ubiquitylates and degrades cFLIP62, 63, 64 (Figure 4). Interestingly cFLIP is protected from degradation by RIPK1 and TRAF2 because in the absence of these two proteins cFLIP becomes rapidly degraded and this degradation is blocked by proteosomal inhibitors.59, 65 It has been shown that cFLIP can be modified on Lysine 167, and this ubiquitylation is controlled through a phosphorylation event that occurs on the neighbouring threonine residue at position 166 in response to ROS up regulation but the relevance to cFLIP stability following Death Receptor signalling was not explored.62 Conversely, the deubiquitylating enzyme USP8 has been shown to interact with cFLIP, reduce steady state ubiquitylation of cFLIP, and thereby increase steady state levels of cFLIP (Figure 4). As expected, USP8 depletion promoted FasL-, TRAIL- and TNF-induced apoptosis, but whether over expression of USP8 could help stabilise cFLIP post death ligand signalling was not explored.66
Turned Over by Ubiquitin – Post-Translational Regulation of FADD
FADD is an essential core component of the DISC, the Ripoptosome and complex 2. The role of FADD is simple, but nonetheless important, because it functions to bridge together the killers, caspase-8 or -10, with the activators, RIPK1 or TRADD. This role of FADD in these complexes is attributed to its unique domain architecture as FADD contains both a DD and a DED. Surprisingly the information regarding the regulation of FADD by post-translational modifications is limited, and to date only one group has demonstrated the regulation of FADD by ubiquitylation.67 Nevertheless, Lee et al. uncovered a ubiquitylation event that regulates the stability of FADD and consequently the ability of FADD to mediate apoptosis. This ubiquitylation regulates the steady state levels of FADD and is driven by the E3-ubiqutin ligase Makorin RING Finger Protein 1 (MKRN1; Figure 4). Silencing of MKRN1 elevated the protein levels of FADD, reduced the degradation of FADD upon cyclohexamide (CHX) treatment, reduced ubiquitylation of FADD and increased the susceptibility of cells to TNF- TRAIL- and FAS-induced apoptosis. This data demonstrates that FADD is constitutively turned over by MKRN1, providing a regulatory mechanism to limit inappropriate apoptosis driven by FADD spontaneously associating with caspase-8 or 10. FADD is turned over by the proteasome as the degradation of FADD induced by CHX treatment is blocked by treatment with MG132, suggesting that MKRN1 induces K48-linked ubiquitylation of FADD. This research pinpoints MKRN1 as a potential therapeutic target, which could be beneficial in combination with treatments that activate extrinsic apoptosis such as TRAIL. However, it will be important to also consider the impact of globally reregulating cellular FADD levels and the consequence this may have on cellular fate if MKRN1 was targeted.
Concluding Remarks
Post-translational modification of proteins with ubiquitin opens a pandora’s box of potential outcomes and it will take significant experimental effort to fully understand the combinatorial complexity of ubiquitin events in signalling complexes. The emergence of new tools to study ubiquitylation, such as TUBEs, linkage-specific antibodies and the UbiCREST system provides the hope that we will one day be able to appreciate all of the intricacies. Nevertheless, it is very likely that there are some simple rules underlying these complex processes. If we take inspiration from the weaver ant, chains begin with a single ant reaching out to a distant leaf, a second ant climbs aboard and ant by ant the living chain grows until their goal is reached. Interestingly, the higher the numbers of worker ants present in a chain the higher the probability to join that chain. Could ubiquitin chain length be regulated in a similarly simple manner? In the case of ants the chain length is determined by reaching the leaf goal but it seems unlikely that ubiquitin chains bridge between different molecules and more likely that they function as assembly platforms. If the goal of ubiquitin chains is to bring two proteins such as the IKKs and TAK1 together one can imagine a scenario where TAB2/TAB3/TAK1 and LUBAC bind to K63-linked chains, LUBAC then extends the K63 chains with linear ubiquitin chains until NEMO binds. If a K63 DUB is associated with the TAB/TAK complex and a linear specific DUB is associated with NEMO then one can imagine how chain length could be precisely regulated. Thus recruitment of DUBs together with a chain protecting entity (e.g., TAB, NEMO or A20) could serve to cap chains.
Cell death platforms have been of focus in recent years due to their obvious therapeutic potential for cancer treatment and their involvement in various auto-inflammatory diseases. Much is understood about the upstream events that lead to their formation, however, there are gaps in our understanding of the protein composition and the post-translational modifications. This is highlighted by the fact that the Ripoptosome is a ~2 MDa complex which indicates that it cannot simply be made up of a single unit of RIPK1 (~75 KDa), FADD (~23 KDa) and a caspase-8 dimer (~110 kDa). Because there is no data on the stoichiometry of these known components it is impossible to know whether all components have been described? Nor, as should already be apparent from this review, is it clear what post-translational modifications are essential to regulate the activity of this complex. It remains possible that there are other ubiquitin-like modifications such as SUMO and NEDD8 that contribute to the downstream signalling responses, and certainly IAPs that regulate these death complexes have been shown to be able to neddylate substrates.68
Ants account for ~20% of the animal biomass on the planet, found on almost every landmass,69 and as the famed myrmecologist EO Wilson noted, ‘if ants and other insects disappeared the environment would collapse’, partly because of their role the recycling of organic matter. The ubiquitin system in cells accounts for just over 1% of total cellular protein.70 Although it has been most recognised for its role in tagging other proteins for destruction with K48-linked ubiquitin chains, or waste disposal, it is worth considering that approximately only 20% of cellular ubiquitin is accounted for in any type of chain.70 Like an ant, ubiquitin is undoubtedly a seemingly tireless multi-tasking worker. Others have used an economy analogy to describe the ubiquitin system which we have found inspiring.70 Picturing it as an ant functioning in a colony has likewise stimulated us to think about how ubiquitin might work in the context of a cell, and we hope that this analogy is similarly thought provoking to the reader.
Acknowledgments
Rebecca Feltham and John Silke are supported by an National Health and Medical Research Project grant (1081272) and fellowship (1107149), respectively.
Footnotes
Edited by M Gyrd-Hansen
The authors declare no conflict of interest.
References
- Reid CR, Lutz MJ, Powell S, Kao AB, Couzin ID, Garnier S. Army ants dynamically adjust living bridges in response to a cost-benefit trade-off. Proc Natl Acad Sci USA 2015; 112: 15113–15118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerich CH, Ordureau A, Strickson S, Arthur JS, Pedrioli PG, Komander D et al. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc Natl Acad Sci USA 2013; 110: 15247–15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospenthal MK, Mevissen TE, Komander D. Deubiquitinase-based analysis of ubiquitin chain architecture using Ubiquitin Chain Restriction (UbiCRest). Nat Protoc 2015; 10: 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 2003; 114: 181–190. [DOI] [PubMed] [Google Scholar]
- Murphy JM, Silke J. Ars Moriendi; the art of dying well - new insights into the molecular pathways of necroptotic cell death. EMBO Rep 2014; 15: 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH et al. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J 1995; 14: 5579–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprick MR, Weigand MA, Rieser E, Rauch CT, Juo P, Blenis J et al. FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity 2000; 12: 599–609. [DOI] [PubMed] [Google Scholar]
- Geserick P, Hupe M, Moulin M, Wong WW, Feoktistova M, Kellert B et al. Cellular IAPs inhibit a cryptic CD95-induced cell death by limiting RIP1 kinase recruitment. J Cell Biol 2009; 187: 1037–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F et al. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell 2011; 43: 432–448. [DOI] [PubMed] [Google Scholar]
- Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M et al. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell 2011; 43: 449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C et al. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 2011; 471: 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Xia F, Hermance N, Mabb A, Simonson S, Morrissey S et al. A cytosolic ATM/NEMO/RIP1 complex recruits TAK1 to mediate the NF-kappaB and p38 mitogen-activated protein kinase (MAPK)/MAPK-activated protein 2 responses to DNA damage. Mol Cell Biol 2011; 31: 2774–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Berghe T, Hassannia B, Vandenabeele P. An outline of necrosome triggers. Cell Mol Life Sci 2016; 73: 2137–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriwaki K, Bertin J, Gough PJ, Chan FK. A RIPK3-caspase 8 complex mediates atypical pro-IL-1beta processing. J Immunol 2015; 194: 1938–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vince JE, Wong WW, Gentle I, Lawlor KE, Allam R, O'Reilly L et al. Inhibitor of apoptosis proteins limit RIP3 kinase-dependent interleukin-1 activation. Immunity 2012; 36: 215–227. [DOI] [PubMed] [Google Scholar]
- Zinngrebe J, Rieser E, Taraborrelli L, Peltzer N, Hartwig T, Ren H et al. —LUBAC deficiency perturbs TLR3 signaling to cause immunodeficiency and autoinflammation. J Exp Med 2016; 213: 2671–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WJ, Daley-Bauer LP, Thapa RJ, Mandal P, Berger SB, Huang C et al. RIP1 suppresses innate immune necrotic as well as apoptotic cell death during mammalian parturition. Proc Natl Acad Sci USA 2014; 111: 7753–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard JA, O'Donnell JA, Evans JM, Lalaoui N, Poh AR, Rogers T et al. RIPK1 regulates RIPK3-MLKL-driven systemic inflammation and emergency hematopoiesis. Cell 2014; 157: 1175–1188. [DOI] [PubMed] [Google Scholar]
- Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity 1998; 8: 297–303. [DOI] [PubMed] [Google Scholar]
- Dillon CP, Weinlich R, Rodriguez DA, Cripps JG, Quarato G, Gurung P et al. RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell 2014; 157: 1189–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K, Dugger DL, Wickliffe KE, Kapoor N, de Almagro MC, Vucic D et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science 2014; 343: 1357–1360. [DOI] [PubMed] [Google Scholar]
- Berger SB, Kasparcova V, Hoffman S, Swift B, Dare L, Schaeffer M et al. Cutting Edge: RIP1 kinase activity is dispensable for normal development but is a key regulator of inflammation in SHARPIN-deficient mice. J Immunol 2014; 192: 5476–5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachbur U, Stafford CA, Bankovacki A, Zhan Y, Lindqvist LM, Fiil BK et al. A RIPK2 inhibitor delays NOD signalling events yet prevents inflammatory cytokine production. Nat Commun 2015; 6: 6442. [DOI] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell 2006; 22: 245–257. [DOI] [PubMed] [Google Scholar]
- Wong WW, Gentle IE, Nachbur U, Anderton H, Vaux DL, Silke J. RIPK1 is not essential for TNFR1-induced activation of NF-kappaB. Cell Death Differ 2010; 17: 482–487. [DOI] [PubMed] [Google Scholar]
- Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected]. Nat Cell Biol 2006; 8: 398–406. [DOI] [PubMed] [Google Scholar]
- Mahoney DJ, Cheung HH, Mrad RL, Plenchette S, Simard C, Enwere E et al. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc Natl Acad Sci USA 2008; 105: 11778–11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varfolomeev E, Goncharov T, Fedorova AV, Dynek JN, Zobel K, Deshayes K et al. c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation. J Biol Chem 2008; 283: 24295–24299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell 2009; 36: 831–844. [DOI] [PubMed] [Google Scholar]
- Moulin M, Anderton H, Voss AK, Thomas T, Wong WW, Bankovacki A et al. IAPs limit activation of RIP kinases by TNF receptor 1 during development. EMBO J 2012; 31: 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shembade N, Parvatiyar K, Harhaj NS, Harhaj EW. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-kappaB signalling. EMBO J 2009; 28: 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell MA, Legarda-Addison D, Skountzos P, Yeh WC, Ting AT. Ubiquitination of RIP1 regulates an NF-kappaB-independent cell-death switch in TNF signaling. Curr Biol 2007; 17: 418–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltham R, Bettjeman B, Budhidarmo R, Mace PD, Shirley S, Condon SM et al. Smac mimetics activate the E3 ligase activity of cIAP1 protein by promoting RING domain dimerization. J Biol Chem 2011; 286: 17015–17028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber EC, Schoeffler AJ, Lingel A, Elliott JM, Fedorova AV, Giannetti AM et al. Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science 2011; 334: 376–380. [DOI] [PubMed] [Google Scholar]
- Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell 2007; 131: 682–693. [DOI] [PubMed] [Google Scholar]
- Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell 2008; 30: 689–700. [DOI] [PubMed] [Google Scholar]
- Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J 2010; 29: 4198–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner SA, Satpathy S, Beli P, Choudhary C. SPATA2 links CYLD to the TNF-alpha receptor signaling complex and modulates the receptor signaling outcomes. EMBO J 2016; 35: 1868–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 2011; 471: 591–596. [DOI] [PubMed] [Google Scholar]
- Dondelinger Y, Jouan-Lanhouet S, Divert T, Theatre E, Bertin J, Gough PJ et al. NF-κB-independent role of IKKalpha/IKKbeta in preventing RIPK1 kinase-dependent apoptotic and necroptotic cell death during TNF signaling. Mol Cell 2015; 60: 63–76. [DOI] [PubMed] [Google Scholar]
- Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 2008; 134: 668–678. [DOI] [PubMed] [Google Scholar]
- Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell 2008; 133: 693–703. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 2003; 424: 797–801. [DOI] [PubMed] [Google Scholar]
- Moquin DM, McQuade T, Chan FK. CYLD deubiquitinates RIP1 in the TNFalpha-induced necrosome to facilitate kinase activation and programmed necrosis. PLoS ONE 2013; 8: e76841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draber P, Kupka S, Reichert M, Draberova H, Lafont E, de Miguel D et al. LUBAC-recruited CYLD and A20 regulate gene activation and cell death by exerting opposing effects on linear ubiquitin in signaling complexes. Cell Rep 2015; 13: 2258–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Yamaguchi N. The seventh zinc finger motif of A20 is required for the suppression of TNF-alpha-induced apoptosis. FEBS Lett 2015; 589: 1369–1375. [DOI] [PubMed] [Google Scholar]
- Cullen SP, Henry CM, Kearney CJ, Logue SE, Feoktistova M, Tynan GA et al. Fas/CD95-induced chemokines can serve as "find-me" signals for apoptotic cells. Mol Cell 2013; 49: 1034–1048. [DOI] [PubMed] [Google Scholar]
- de Almagro MC, Goncharov T, Newton K, Vucic D. Cellular IAP proteins and LUBAC differentially regulate necrosome-associated RIP1 ubiquitination. Cell Death Dis 2015; 6: e1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafont E, Kantari-Mimoun C, Draber P, De Miguel D, Hartwig T, Reichert M et al. The linear ubiquitin chain assembly complex regulates TRAIL-induced gene activation and cell death. EMBO J 2017; 36: 1147–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM et al. A unified model for apical caspase activation. Mol Cell 2003; 11: 529–541. [DOI] [PubMed] [Google Scholar]
- Pop C, Fitzgerald P, Green DR, Salvesen GS. Role of proteolysis in caspase-8 activation and stabilization. Biochemistry 2007; 46: 4398–4407. [DOI] [PubMed] [Google Scholar]
- Fiandalo MV, Schwarze SR, Kyprianou N. Proteasomal regulation of caspase-8 in cancer cell apoptosis. Apoptosis 2013; 18: 766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang TB, Oh GS, Scandella E, Bolinger B, Ludewig B, Kovalenko A et al. Mutation of a self-processing site in caspase-8 compromises its apoptotic but not its nonapoptotic functions in bacterial artificial chromosome-transgenic mice. J Immunol 2008; 181: 2522–2532. [DOI] [PubMed] [Google Scholar]
- Hughes MA, Harper N, Butterworth M, Cain K, Cohen GM, MacFarlane M. Reconstitution of the death-inducing signaling complex reveals a substrate switch that determines CD95-mediated death or survival. Mol Cell 2009; 35: 265–279. [DOI] [PubMed] [Google Scholar]
- Hughes MA, Powley IR, Jukes-Jones R, Horn S, Feoktistova M, Fairall L et al. Co-operative and hierarchical binding of c-FLIP and caspase-8: a unified model defines how c-FLIP isoforms differentially control cell fate. Mol Cell 2016; 61: 834–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 2009; 137: 721–735. [DOI] [PubMed] [Google Scholar]
- Gonzalvez F, Lawrence D, Yang B, Yee S, Pitti R, Marsters S et al. TRAF2 Sets a threshold for extrinsic apoptosis by tagging caspase-8 with a ubiquitin shutoff timer. Mol Cell 2012; 48: 888–899. [DOI] [PubMed] [Google Scholar]
- Yin Q, Lamothe B, Darnay BG, Wu H. Structural basis for the lack of E2 interaction in the RING domain of TRAF2. Biochemistry 2009; 48: 10558–10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vince JE, Pantaki D, Feltham R, Mace PD, Cordier SM, Schmukle AC et al. TRAF2 must bind to cIAPs for TNF to efficiently activate NF-{kappa}B and to prevent TNF-induced apoptosis. J Biol Chem 2009; 284: 35906–35915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayotova-Dimitrova D, Feoktistova M, Ploesser M, Kellert B, Hupe M, Horn S et al. cFLIP regulates skin homeostasis and protects against TNF-induced keratinocyte apoptosis. Cell Rep 2013; 5: 397–408. [DOI] [PubMed] [Google Scholar]
- Piao X, Komazawa-Sakon S, Nishina T, Koike M, Piao JH, Ehlken H et al. c-FLIP maintains tissue homeostasis by preventing apoptosis and programmed necrosis. Sci Signal 2012; 5: ra93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie-Grantham RP, Matsuzawa S, Reed JC. Novel phosphorylation and ubiquitination sites regulate reactive oxygen species-dependent degradation of anti-apoptotic c-FLIP protein. J Biol Chem 2013; 288: 12777–12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan K, Pathak C. FADD regulates NF-kappaB activation and promotes ubiquitination of cFLIPL to induce apoptosis. Sci Rep 2016; 6: 22787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Kamata H, Solinas G, Luo JL, Maeda S, Venuprasad K et al. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell 2006; 124: 601–613. [DOI] [PubMed] [Google Scholar]
- Gentle IE, Wong WW, Evans JM, Bankovacki A, Cook WD, Khan NR et al. In TNF-stimulated cells, RIPK1 promotes cell survival by stabilizing TRAF2 and cIAP1, which limits induction of non-canonical NF-kappaB and activation of caspase-8. J Biol Chem 2011; 286: 13282–13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong M, Lee EW, Seong D, Seo J, Kim JH, Grootjans S et al. USP8 suppresses death receptor-mediated apoptosis by enhancing FLIPL stability. Oncogene 2016; 36: 458–470. [DOI] [PubMed] [Google Scholar]
- Lee EW, Kim JH, Ahn YH, Seo J, Ko A, Jeong M et al. Ubiquitination and degradation of the FADD adaptor protein regulate death receptor-mediated apoptosis and necroptosis. Nat Commun 2012; 3: 978. [DOI] [PubMed] [Google Scholar]
- Broemer M, Tenev T, Rigbolt KT, Hempel S, Blagoev B, Silke J et al. Systematic in vivo RNAi analysis identifies IAPs as NEDD8-E3 ligases. Mol Cell 2010; 40: 810–822. [DOI] [PubMed] [Google Scholar]
- Schultz TR. In search of ant ancestors. Proc Natl Acad Sci USA 2000; 97: 14028–14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague MJ, Heride C, Urbe S. The demographics of the ubiquitin system. Trends Cell Biol 2015; 25: 417–426. [DOI] [PubMed] [Google Scholar]