Abstract
Twist1, a basic helix–loop–helix transcription factor is implicated as a key mediator of epithelial–mesenchymal transition (EMT) and metastatic dissemination in p53-deficient cancer cells. On the other hand, checkpoint kinase 2 (Chk2), a major cell cycle regulatory protein provides a barrier to tumorigenesis due to DNA damage response by preserving genomic stability of the cells. Here we demonstrate that Chk2 induction proficiently abrogates invasion, cell scattering and invadopodia formation ability of p53-mutated invasive cells by suppressing Twist1, indicating Chk2 confers vital role in metastasis prevention. In addition, ectopic Chk2, as well as its (Chk2) induction by natural podophyllotoxin analog, 4′-demethyl-deoxypodophyllotoxin glucoside (4DPG), strongly restrain Twist1 activity along with other mesenchymal markers, for example, ZEB-1, vimentin and Snail1, whereas the epithelial markers such as E-cadherin and TIMP-1 expression augmented robustly. However, downregulation of endogenous Chk2 by siRNA as well as Chk2 selective inhibitor PV1019 implies that 4DPG-mediated inhibition of Twist1 is Chk2-dependent. Further, mechanistic studies unveil that Chk2 negatively regulates Twist1 promoter activity and it (Chk2) interacts steadily with Snail1 protein to curb EMT. Strikingly, Chk2 overexpression triggers premature senescence in these cells with distinctive increase in senescence-associated β-galactosidase (SA-β-gal) activity, G2/M cell cycle arrest and induction of senescence-specific marker p21waf1/Cip1. Importantly, stable knockdown of Twist1 by shRNA markedly augments p21 expression, its nuclear accumulation, senescence-associated heterochromatin foci (SAHF) and amplifies the number of SA-β-gal-positive cells. Moreover, our in vivo studies also validate that 4DPG treatment significantly abrogates tumor growth as well as metastatic lung nodules formation by elevating the level of phospho-Chk2, Chk2 and suppressing Twist1 activity in mouse mammary carcinoma model. In a nutshell, this report conceives a novel strategy of Twist1 suppression through Chk2 induction, which prevents metastatic dissemination and promotes premature senescence in p53-defective invasive cancer cells.
Epithelial–mesenchymal transition (EMT), a key biological process is manifested during development by which epithelial cells acquire mesenchymal, fibroblast-like phenotype and predominantly exhibit reduced intercellular adhesion.1, 2 In cancer, EMT plays a critical role in imparting more aggressive behavior to the cells for metastatic spread of the disease.1 Twist1, a basic helix–loop–helix transcription factor is overexpressed during the process in various carcinomas, sarcomas, neuroblastomas, gliomas and melanomas.3, 4 At the molecular level, Twist1 facilitates MDM2-mediated degradation of p53, renders the cancer cells more aggressive and resistant to therapies. Hence, high levels of Twist1 expression is a predominant feature in p53-defective cancer cells.5, 6 Twist1 downregulates epithelial markers such as E-cadherin, TIMP1 and augments the mesenchymal markers namely, Vimentin, ZEB-1, MMP-2 and N-cadherin driving the cells toward malignancy.3, 4 Twist1 has been demonstrated to suppress p21 at the promoter level; thus preventing the cells from undergoing senescence and activates EMT program.7 Hence, regulating Twist1 activity to prevent aberrant invasive signaling and to turn on senescence machinery could divulge new therapeutic strategies.
Chk2 is a key tumor suppressor of the DNA damage checkpoint pathway, which is activated in response to exogenous insults including ionizing radiation and chemotherapeutics.8 DNA double-strand breaks (DSBs) stimulate ataxia–telangiectasia-mutated (ATM) protein kinase that subsequently phosphorylates Chk2 at Thr68 and activates it.9 However, hyperactive Chk2, in turn, directly phosphorylates and regulates the downstream functions of p53, CDC25 and BRCA1 implying checkpoint activation and G2/M cell cycle arrest.10 Emerging evidence demonstrates that Chk2 kinase significantly contributes in inducing both replicative and premature cellular senescence.11, 12 High level of virally transduced Chk2 in human lung cancer A549 cells leads to transcriptional induction of p21, activation of p53, G2/M cell cycle arrest and senescence.13 However, Chk2-dependent senescence and transcriptional induction deeply corroborate in p53-defective SK-BR-3 (breast carcinoma) and HaCaT (immortalized keratinocyte) cells, suggesting a p53-independent role of Chk2 in p21 induction and senescence.13 Albeit, the functions of Chk2 in DNA damage response and cell cycle regulation is well documented, how it regulates excessive oncogenic signaling leading to invasion and metastasis and favors cellular senescence as a protective mechanism, is yet to be studied.
This report for the first time describes a functional role of Chk2 induction by ectopic overexpression as well as by treatment with (4′-demethyl-deoxypodophyllotoxin glucoside (4DPG) attenuating Twist1-mediated invasion and metastasis in p53-defective cancer cells from diverse tissue origin. Furthermore, this Chk2-mediated rescuing of p21 to promote premature senescence as a tumor suppressive safeguard mechanism in these cells warrants a novel strategy for pharmacological intervention.
Results
Chk2 overexpression abrogates motility/invasion of cancer cells and regulates the EMT-related markers
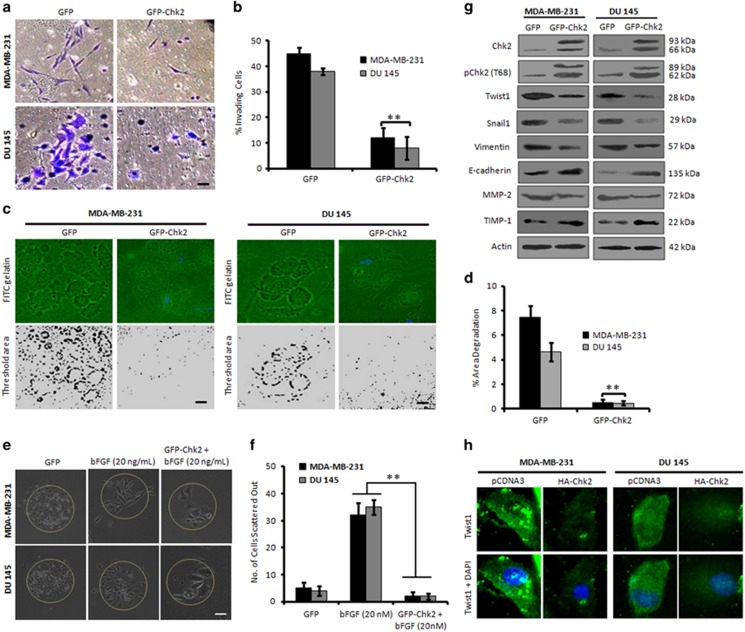
Mutations in p53 tumor suppressor gene directly contributes to EMT by regulating the expression of some metastasis-related genes.14 We examined the effect of Chk2 overexpression on the invasion of three p53-defective aggressive cancer cell lines namely, MDA-MB-231, DU 145 and PANC-1. Following transient transfection of the cells with GFP and GFP-Chk2, the matrigel invasion assay results revealed that Chk2 attenuates invasion of these cells 48–72 h post transfection (Figures 1a, b and Supplementary Figure S1a).We also performed the same experiment on p53 null human prostate cancer PC-3 cells. Our results demonstrated that Chk2 expression strongly impeded the invasion of aggressive PC-3 cells (Supplementary Figure S1b). Invadopodia formation is an important platform to study the EMT process,15 and therefore we examined the effects of GFP-Chk2 on the invadopodia formation ability of MDA-MB-231 and DU 145 cells. The results implied sufficient degradation of FITC-gelatin matrix in GFP-transfected cells, the degradation ability was significantly reduced in cells transfected with GFP-Chk2 (Figures 1c and d). Furthermore, GFP-transfected cells scattered out of the colonies upon stimulation with bFGF; whereas, GFP-Chk2 convincingly halted the bFGF-induced cell migration (Figures 1e, f and Supplementary Figure S1c).
Figure 1.
Chk2 expression negatively regulates invasion of p53-deficient cancer cells. (a) MDA-MB-231 and DU 145 cells were transiently transfected with GFP and GFP-Chk2 plasmid construct for 48 h and checked for the invasion of cells through matrigel invasion assay. Invading cells were observed and photographed under an inverted microscope at × 20 magnification. Scale bar: 20 μm. (b) Bar graph showing results from quantification of invading cells (n=3, error bars indicate±S.D.).**P<0.01. (c) Cells were transfected with GFP and GFP-Chk2 construct and assessed for their invadopodia formation and matrix degradation ability over FITC-gelatin matrix coverslips. Blue parts show the nucleus stained with DAPI mounting media. Images were taken at × 20 magnification, and the threshold area of degradation was determined with the help of Image J software. Scale bar: 20 μm. (d) Bar graphs showing the percent area of degradation quantified through Image J analysis (n=3, error bars indicate±S.D.).**P<0.01. (e) MDA-MB-231 and DU 145 cells were allowed to form the distinct colonies for 5 days, stimulated with bFGF (20 ng/ml) and then transfected with either GFP or GFP-Chk2 for 48 h. Cells were then observed for their scattering out of the colonies and photographed under × 20 magnification. Scale bar: 20 μm. (f) A total of five random colonies from each field were analyzed and counted manually under the microscope for the number of scattered cells and was adjusted to the total cells in particular colony. Bar graphs showing quantification of the average number of cells scattered out of the colonies (n=3, error bars indicate±S.D.).**P<0.01. (g) Cells were transfected with GFP or GFP-Chk2 for 48 h; whole-cell lysates were prepared, and an equal amount of protein (20 μg) was subjected to western blot analysis for the expression of mesenchymal markers like; Twist1, Snail1, vimentin, MMP-2 and epithelial markers such as E-cadherin and TIMP-1 (n=3). (h) Immunofluorescence staining of Twist1 in MDA-MB-231 and DU 145 cells after transiently transfecting the cells with pCDNA3 and HA-Chk2 for 48 h (original magnification × 20)
Since Chk2 overexpressed cells displayed retarded invasion ability, we sought to examine the major mesenchymal markers of EMT and invasion in these cells. Recombinant fused GFP-Chk2 expression was adequate along with a phospho-T68 signal indicating an optimum threshold activation of the recombinant protein.16 Further, the immunoblot results indicated a steady-state downregulation of Twist1 with concomitant decrease in Snail1, MMP-2, vimentin protein levels and upregulation of epithelial markers such as E-cadherin and TIMP-1 in GFP-Chk2-transfected cells compared to the GFP (Figure 1g and Supplementary Figure S1d). In addition, the immunofluorescence analysis illustrated a marked reduction of Twist1 in Chk2 overexpressing cells compared to the vector-transfected cells (Figure 1h). These results collectively establish Chk2 induction abrogates EMT in p53-defective invasive cancer cells.
Chk2 activation by 4DPG negatively regulates Twist1-mediated invasion in cancer cells
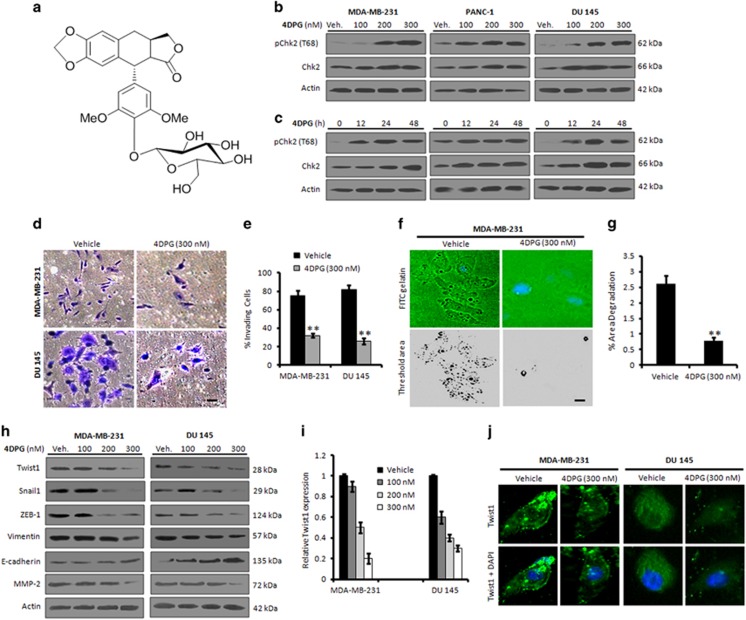
4DPG, a novel natural podophyllotoxin congener, has recently been reported by our group for its promising anticancer activities in diverse cancer cell lines (Figure 2a).17 The cytotoxicity results indicated that 4DPG effectively inhibited the proliferation of MDA-MB-231, PANC-1 and DU 145 cells with IC50 values of 0.256±0.062, 0.272±0.049 and 0.281±0.035 μM, respectively, whereas the IC50 value of 4DPG in normal human breast epithelial (fR2) cells was found to be 24.5±5.125, signifying the selectivity of the molecule toward cancer cells(Supplementary Figure S2 and Supplementary Table S1). To initiate a detailed investigation of the effects of 4DPG on Chk2 activity, we performed concentration-dependent western blot analysis. Although the results unveiled a robust upregulation of pChk2 (T68) as well as total Chk2 at the 200 and 300 nM doses of 4DPG, a substantial p-Chk2 induction was visible even at the initial concentration (100 nM) for all the three cell lines tested (Figure 2b). Further, time-dependent experimental data demonstrated a shoot-up in the expression of pChk2 and total Chk2 even at the 12 h, which gradually augmented to the 48 h (Figure 2c). To further confirm, whether the compound exclusively upregulates Chk2 expression, we conducted western blot analysis for Chk1. The results showed unaltered expression of Chk1 in these cells even at the higher concentration (Supplementary Figures S3a and b). Furthermore, our matrigel invasion assay results in MDA-MB-231 and DU 145 cells unveiled a sufficient reduction in the number of invading cells in 4DPG (300 nM)-treated wells compared to the untreated control (Figures 2d and e). We also observed a remarkable reduction in the area of degradation over the FITC-gelatin matrix, indicating a significant loss of invasive potential in 4DPG (300 nM)-treated cells (Figures 2f and g). 4DPG treatment also abrogated the growth factor-induced scattering of highly invasive mouse mammary carcinoma (4T1) cells (Supplementary Figure S4). Rationally, we investigated whether 4DPG could modulate the expression of mesenchymal/epithelial markers to suppress the invasion of p53-defective aggressive cells; as expected, the immunoblot data revealed a significant downregulation of Twist1, Snail1, ZEB-1, vimentin and MMP-2 expression with concomitant increase in epithelial markers E-cadherin and TIMP-1 in presence of 4DPG (Figures 2h and i). The immunocytochemistry data also demonstrated a significant decrease in Twist1 level in cells treated with 4DPG (300 nM) for 48 h (Figure 2j). Taken together, these results imply that 4DPG suppresses invasion of p53-deficient metastatic cancer cells via activation of Chk2.
Figure 2.
Chk2 induction by 4DPG suppresses EMT and invasion in cells with low p53 level. (a) Chemical structure of 4DPG. (b and c) Dose-dependent and time-dependent expression patterns of Chk2 after 4DPG treatment. MDA-MB-231, PANC-1 and DU 145 cells were exposed to increasing concentrations of 4DPG (100, 200 and 300 nM) for 48 h and 300 nM of 4DPG for 0, 12, 24 and 48 h. Whole-cell lysates were prepared and checked for the expression of phospho-Chk2 (T68) and total-Chk2 through western blot analysis. (d) MDA-MB-231 and DU 145 cells are exposed to 4DPG (300 nM) along with vehicle (DMSO) for 48 h and checked for their ability to invade through the matrigel with the help of matrigel invasion (Boyden chamber) assay system. Images were captured under an inverted microscope at × 20 magnification. Scale bar: 20 μm. (e) Bar graphs showing the percent invading cells (n=3, error bars indicate±S.D.). **P<0.01. (f) MDA-MB-231 cells were cultured over FITC-gelatin matrix coverslips and treated with 4DPG (300 nM) for 48 h. Cells were subsequently washed, stained with DAPI mounting media and photographed under Floid Cell Imaging Station at × 20 magnification. Blue region shows the nucleus stained with DAPI mounting media. The threshold area of degradation was determined with the help of Image J software. Scale bar: 20 μm. (g) Bar graphs showing the percent area of degradation quantified through Image J analysis (n=3, error bars indicate±S.D.). **P<0.01. (h) MDA-MB-231 and DU 145 cells were treated with indicated concentrations of 4DPG along with vehicle for 48 h; whole-cell lysates were prepared and subjected to western blot analysis for the expression of Twist1, Snail1, ZEB-1, vimentin, MMP-2 and E-cadherin. (i) Bar graphs showing the results of densitometric analysis of bands obtained from the western blot analysis (n=3, error bars indicate±S.D.). (j) Immunocytochemistry of Twist1 in MDA-MB-231 and DU 145 cells after 48 h exposure of the cells to 4DPG (300 nM) (original magnification × 20)
Inhibition of Chk2 augments Twist1-mediated EMT and invasion
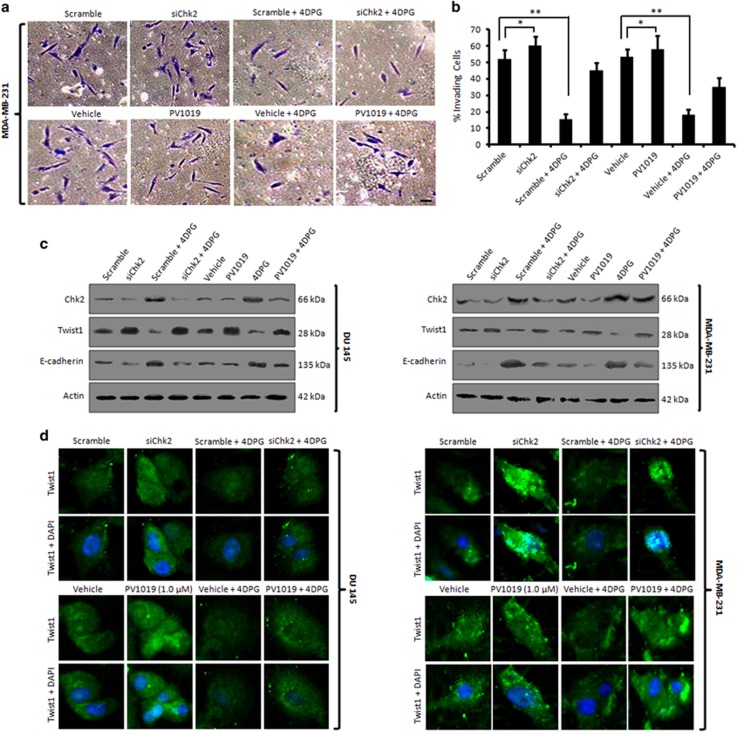
Next, we were curious to examine whether silencing of endogenous Chk2 could revert the invasiveness of cancer cells. In order to do this, siRNA-mediated silencing of endogenous Chk2 was carried out in MDA-MB-231cells following subjected to matrigel invasion assay. Indeed, the results depicted a significant increase in cells invading through the matrigel in siChk2-transfected cells compared to the scramble transfected cells, indicating Chk2 directly/indirectly modulated the penetration potential of MDA-MB-231 cells through matrigel. In addition, siChk2 plus 4DPG treatment could not mimic the effect of alone 4DPG treatment. Moreover, in the presence of PV1019 (1 μM), a potent, selective inhibitor of Chk2, significant invasion through the membrane was achieved even with 4DPG treatment as well as siChk2 plus 4DPG treatment (Figures 3a and b) elucidating activated Chk2 modulates the invasion of p53-defective tumor cells. Similar results of Chk2 silencing were also obtained from the experiment conducted in p53 null PC-3 cells (Supplementary Figures S5a and b). To further validate, western blot experiments were carried out with the lysates from above set in both DU 145 and MDA-MB-231 cells revealed a striking upregulation of Twist1 in the lanes (siChk2, siChk2 plus 4DPG and PV1019 treatments) with a simultaneous decrease in E-cadherin expression in the corresponding lanes. On the other hand, 4DPG treatment significantly amplified the Chk2 levels in the scramble plus 4DPG and 4DPG treatment cells, respectively (Figure 3c). The immunocytochemistry data also demonstrated a robust increase in Twist1 expression when Chk2 was partially knocked down through siRNA transfection as well as PV1019 treatment compared to scramble/vehicle treatment (Figure 3d). To examine the cellular morphology, we used TGF-β as an inducer of EMT and noted that PANC-1 cells treated with TGF-β (10 ng/ml) for 24 h exhibited prominent mesenchymal phenotypes. Conversely, Chk2 overexpression with HA-Chk2 or treatment with 4DPG (300 nM) reverted this mesenchymal morphology in these cells compared to vector or untreated control (Supplementary Figure S6). All these results clearly demonstrate that 4DPG-mediated Twist1 inhibition was Chk2-dependent.
Figure 3.
Effect of decreased Chk2 expression on EMT and invasion. (a) MDA-MB-231 cells were either transfected with scramble, siChk2 or treated with vehicle, 4DPG (300 nM), PV1019 (1.0 μM) alone each and combination of scramble plus 4DPG, siChk2 plus 4DPG and PV1019 plus 4DPG for 48 h. Cells were then checked for their ability to invade through the matrigel with the help of Boyden chamber invasion assay system. Images were captured under an inverted microscope at × 20 magnification. Scale bar: 20 μm. (b) Bar graphs showing the quantification of percent invading cells (n=3, error bars indicate±S.D.). *P<0.05, **P<0.01. (c) DU 145 and MDA-MB-231 cells were either transfected with or treated with above conditions (same as in invasion assay) for 48 h; whole-cell lysates were employed for immunoblotting of Chk2, Twist1 and E-cadherin. (d) DU 145 and MDA-MB-231 cells were seeded in eight-well chamber slides and treated with the same conditions as given in the matrigel invasion assay above and subjected to immunocytochemistry of Twist1 (original magnification × 20)
Chk2 induction promotes premature senescence in p53-defective cancer cells
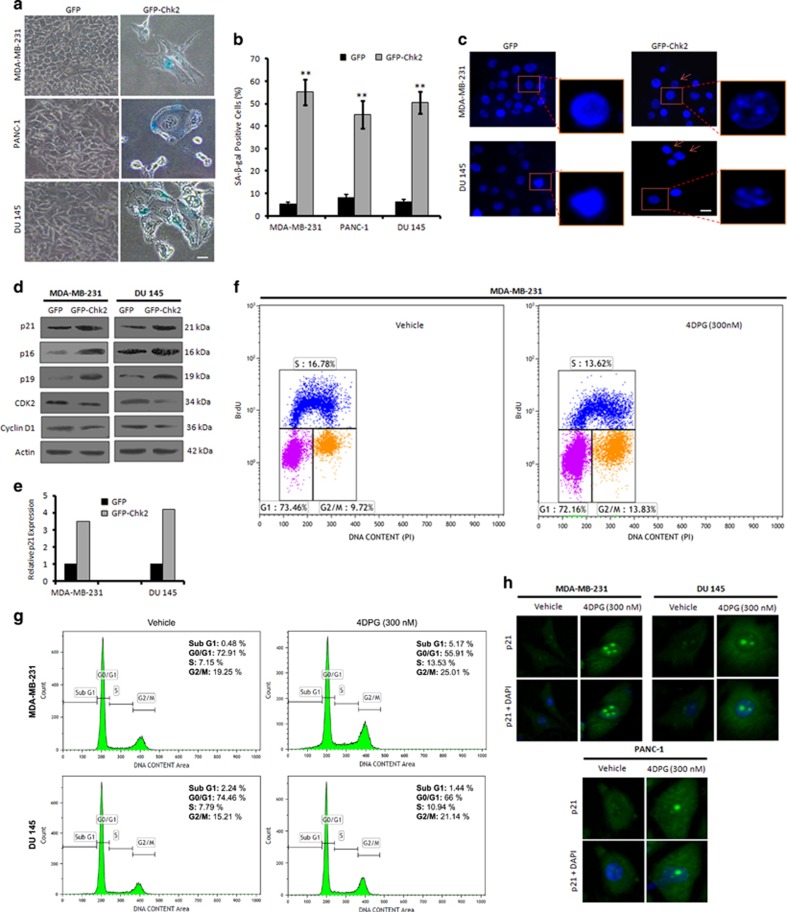
To examine whether the anti-invasive effect of GFP-Chk2 leads to activation of senescence in these aggressive cells, SA-β-gal staining was performed. We observed a tremendous increase in the percent of SA-β-gal-positive cells with flattened shape and bluish nuclear stains in GFP-Chk2-transfected MDA-MB-231, PANC-1 and DU 145 cells (not in GFP transfection), indicating signatures of cellular senescence (Figures 4a and b). To counterproof, we investigated the presence of SAHF, a newly characterized hallmark of senescence,18 in these cells. Beaded nuclei were clearly visible in GFP-Chk2-transfected cells depicting the typical hetero-chromatinized regions within the nucleus, whereas GFP-transfected cells were devoid of such appearances (Figure 4c). Importantly, senescent cells exhibit alterations in gene expressions including, cyclin-dependent kinase (CDK) inhibitors p21Waf1/Cip1, p19Arf and p16INK4a.13, 19 Intriguingly, GFP-Chk2 expression showed elevated expression of senescence-specific marker p21 with concomitant induction of p16, p19 as well as diminished expression of CDK2 and cyclin D1 within 72 h of transfection (Figures 4d and e). Although premature senescence is associated with inhibition of cell proliferation per se20we noted that GFP-Chk2 significantly hindered the colony-forming ability of all the three cancer cell lines (Supplementary Figure S7). To determine the effect of 4DPG, a potential agonist of Chk2, on cell cycle, we performed bromo-deoxyuridine (BrdU) incorporation assay in MDA-MB-231 cells. Interestingly, we found an increase in G2/M population of the BrdU-positive cells (13.83%) in 4DPG-treated MDA-MB-231 cells compared to 9.72% in vehicle (Figure 4f). Our cell cycle analysis through PI staining also depicted increase in G2/M DNA content (25.01% in MDA-MB-231 and 21.14% in DU 145) in 4DPG-treated cells compared to the vehicle treatment (19.25% in MDA-MB-231 and 15.21% in DU 145) (Figure 4g), explicating Chk2-dependent checkpoint response. Similarly, to examine the effects of 4DPG treatment on senescence induction, the SA-β-gal staining results clearly illustrated that 300 nM of 4DPG triggered senescence phenotype in MDA-MB-231, PANC-1 and DU 145 cells (Supplementary Figures S8a and b). Furthermore, analysis of downstream effectors of Chk2 function demonstrated a marked increase in p21 expression accompanied with p19 and deceleration in CDK2 level in a dose-dependent manner (Supplementary Figure S8c). Our immunocytochemistry results also depicted that 4DPG (300 nM) was able to induce nuclear accumulation of p21 in MDA-MB-231, PANC-1 and DU 145 cells (Figure 4h). It is reported that Chk2 can activate senescence by inducing p21 in p53 intact cells.13 Supporting, our results revealed that GFP-Chk2-transfected MCF7 cells undergone premature senescence forcibly compared to their respective GFP control cells. However, in TGF-β-treated wells, EMT features become predominated (Supplementary Figures S9a and b).Together, these results show that Chk2 activation mimics the induction of checkpoint response to DNA damage, activating the p53-independent p21pathway to promote senescence.
Figure 4.
Chk2 induction promotes premature senescence in p53-mutated invasive cancer cells. (a) MDA-MB-231, PANC-1 and DU 145 cells were transfected with GFP and GFP-Chk2 construct; 72 h of post transfection, cells were incubated with X-gal staining solution for 48 h and then checked for the SA-β-gal activity of the cells. Images were captured under bright-field microscope at × 20 magnification. Scale bar: 20 μm. (b) Bar graph indicates quantification of SA-β-gal-positive cells (n=3, error bars indicate±S.D.). **P<0.01. (c) MDA-MB-231 and DU 145 cells after transfection with GFP and GFP-Chk2 construct for 72 h, stained with DAPI containing mounting media and observed for the SAHF under Floid Cell Imaging Station at × 20 magnification. Scale bar: 20 μm. Enlarged scale-up images to properly show the formation of heterochromatin foci. (d) Seventy-two hours of post transfection with GFP and GFP-Chk2, whole-cell lysates were prepared and subjected to western blot analysis of the various senescence specific markers like; p21, p16, p19, CDK2 and cyclin D1. (e) Bar graph showing relative p21 expression from densitometric analysis of the obtained bands from the western blotting experiment. (f) MDA-MB-231 cells were treated with 4DPG (300 nM) for 48–72 h and then pulse labeled with BrdU (30 μM) for 1 h. Cells were then processed for BrdU flow cytometric analysis. (g) Cells were treated with 4DPG (300 nM) along with vehicle for 72 h and checked for cell cycle analysis (PI staining) through flow cytometry. (h) MDA-MB-231, PANC-1 and DU 145 cells were treated with 4DPG (300 nM) along with vehicle and employed for the immunocytochemical analysis of p21 localization (original magnification × 20)
Chk2 activation negatively regulates Twist1 promoter activity and it (Chk2) interacts with Snail1
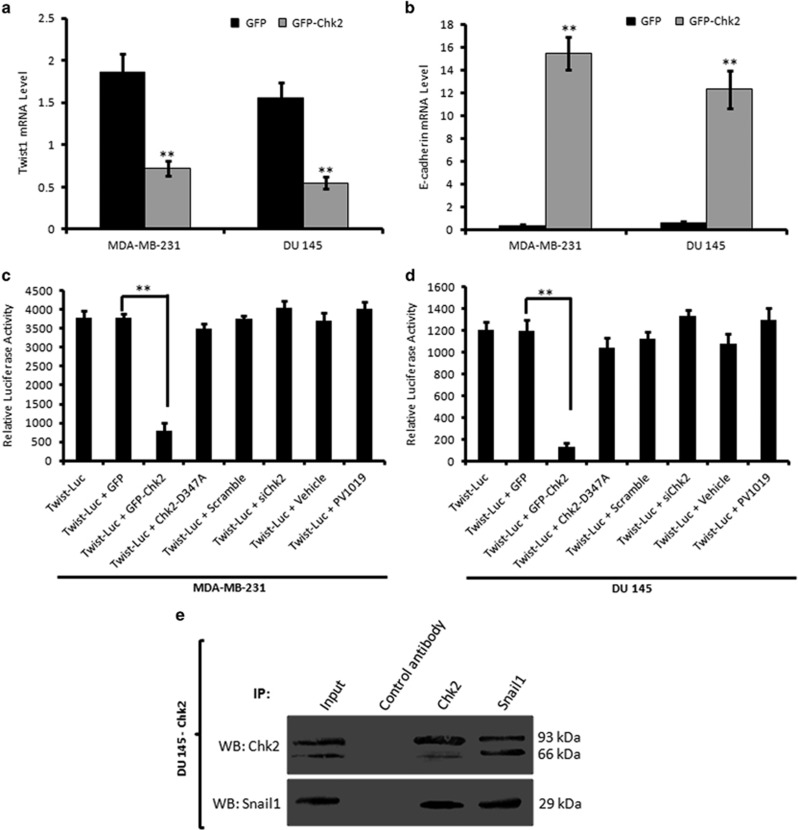
So far, our results clearly pointed out that Chk2 induction was consistently modulating the major EMT regulator Twist1; therefore, we hypothesized that Chk2 could regulate Twist1 promoter activity and blocked the formation of Twist1 mRNA. As expected, the quantification real-time reverse transcription-PCR results implied a sharp attenuation in Twist1 mRNA level in cells transfected with GFP-Chk2 (2.5-fold in MDA-MB-231 and 2.6 fold in DU 145) (Figure 5a), whereas the E-cadherin mRNA level augmented markedly (15-fold in MDA-MB-231 and 12.5-fold in DU 145) in GFP-Chk2-transfected cells (Figure 5b). Similarly, 4DPG (300 nM) treatment resulted a significant reduction in mRNA level of Twist1 (14-fold in MDA-MB-231 and 6-fold in DU 145) in 4DPG-treated cells compared to untreated control. Conversely, the mRNA level of E-cadherin increased in 4DPG-treated tumor cells (5.2-fold in MDA-MB-231 and 4-fold in DU 145) (Supplementary Figures S10a and b). To further determine whether Chk2 regulates Twist1 transcription, we analyzed Twist1 promoter activity through a Dual-Glo Luciferase Assay System (Promega). Notably, the results demonstrated that the relative luciferase activity of cells co-transfected with Twist-Luc and GFP-Chk2 reduced significantly (6.2-fold in MDA-MB-231 and 8.5-fold in DU 145) compared to the cells co-transfected with Twist-Luc and GFP, indicating a negative regulation of Twist1 promoter activity by Chk2 (Figures 5c and d). However, co-transfection of a kinase-dead Chk2 (hChk2-D347A) along with Twist-Luc could not suppress the Twist1 promoter activity in these cells. Conversely, the Twist-Luc activity augmented in siChk2-transfected as well as PV1019-treated cells (Figures 5c and d). Furthermore, when subjected to 4DPG (300 nM), the luciferase activity of Twist1 reduced convincingly (3-fold in MDA-MB-231 and 2.3-fold in DU 145) compared to the vehicle (Supplementary Figures S11a and b). For comprehensive understanding of the molecular mechanism by which Chk2 negatively regulated Twist1 transcriptional activity and protein expression, we were in hunch of upstream regulators that activate Twist1-mediated EMT and invasion. Snail1, a zinc-finger E-box-binding protein initiates EMT programme by repressing epithelial genes such as E-cadherin by binding to its promoter.1 We were curious to investigate whether any physical interaction exist in between Chk2 and Snail1 to regulate EMT in p53-defective cancer cells. Interestingly, our coimmunoprecipitation analysis demonstrated that Snail1 antibody coimmunoprecipitated Chk2 in GFP-Chk2 stable clone of DU 145 cells; similarly Chk2 antibody, but not the control antibody coimmunoprecipitated Snail1 in these cells (Figure 5e). All these results collectively suggest that Chk2 regulates Twist1 at the transcriptional level by suppressing its (Twist1) promoter activity and Chk2 interacts with Snail1 protein in p53-deficient cancer cells.
Figure 5.
Chk2 negatively regulates transcriptional activity of Twist1 and it (Chk2) binds to Snail1. (a and b) MDA-MB-231 and DU 145 cells were transiently transfected with GFP and GFP-Chk2 construct for 48 h and checked for the mRNA level of Twist1 and E-cadherin through real-time PCR analysis. β-actin gene was used as an internal control for normalization (n=3, error bars indicate±S.D.). **P<0.01. (c and d) MDA-MB-231 and DU 145 cells were transfected with Twist-Luc promoter and co-transfected with Twist-Luc plus GFP, Twist-Luc plus GFP-Chk2, Twist-Luc plus Chk2-D347A, Twist-Luc plus scramble siRNA, Twist-Luc plus siChk2 and/or treated with Twist-Luc plus vehicle as well as Twist-Luc plus PV1019 for 48 h. Luciferase activity was measured with the help of a Dual-Glo Luciferase assay system (Promega). Normalization was done with luciferase activity of PGL3 vector (n=3, error bars indicate±S.D.). **P<0.01. (e) Activated Chk2 binds to endogenous Snail1 in p53-deficient cancer cells. Whole-cell extracts of Chk2-DU 145 stable cells were prepared and subjected to immunoprecipitation with Chk2 antibody, Snail1 antibody and control antibody. The immunoprecipitates and inputs were subjected to western blot analysis for Chk2 and Snail1 expression
Stable knockdown of Twist1 promotes premature senescence through induction of p21
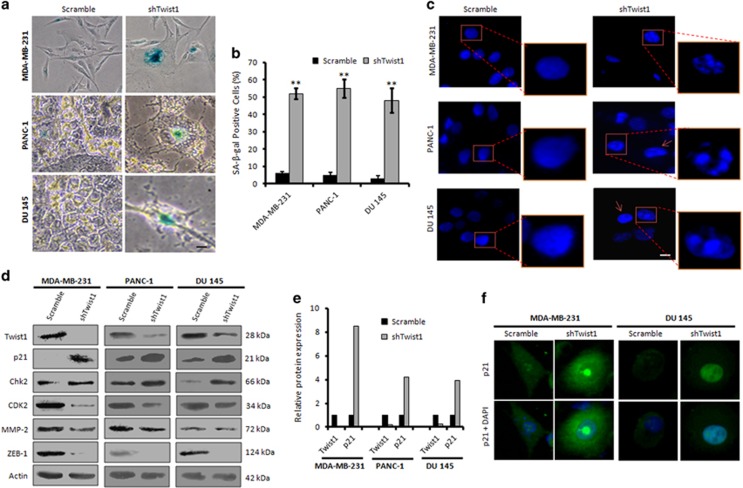
Twist1 has been previously documented to halt premature senescence by suppressing p21 at the transcriptional level by directly binding to its promoter in a p53-independent manner.21 To examine, whether stable knockdown of Twist1 can avert EMT through inducing senescence by rescuing p21, we prepared stable Twist1 knockdown clones of MDA-MB-231, DU 145 and PANC-1 cells employing Twist1 shRNA. Our SA-β-gal staining of the shTwist1 clones implicated a marked boost in SA-β-gal-positive staining (46% in MDA-MB-231, 55% in PANC-1 and 48% in DU 145) compared to the scramble-transfected clones (Figures 6a and b). We also observed an increase in SAHF in the shTwist1 stable clones of MDA-MB-231, PANC-1 and DU 145 compared to the scramble (Figure 6c). Our immunoblotting and immunocytochemistry experiments further validated a significant upregulation of p21 (8.5-fold in MDA-MB-231, 4.2-fold in PANC-1 and 3.9-fold in DU 145) in shTwist1 stable clones with concomitant reduction of Twist1, CDK2, ZEB-1 and MMP-2 expressions along with the increase in Chk2 expression (Figures 6d and e). DDR stimulates ATM protein kinase, which in turn phosphorylates Chk2 (T68) for its activation.9 The same ATM kinase also stabilizes Snail1 by phosphorylating it on S100, and thus instigating in tumor invasion and metastasis.22 Hence, there might be some orchestration of feedback loop interplaying through downstream effectors like Twist1 affecting Chk2 activity through this axis. Our immunocytochemistry results also confirmed a steady nuclear accumulation of p21 in shTwist1 stable clones in comparison to the scramble in these cells (Figure 6f and Supplementary Figure S12). Collectively, these results provide evidences that suppression of Twist1 activity can promote premature senescence in p53-mutated cancer cells by inducing the expression and nuclear accumulation of p21.
Figure 6.
Stable knockdown of Twist1 induces premature senescence in invasive cells. (a) Stable clones of MDA-MB-231, PANC-1 and DU 145 expressing scramble and shTwist1 were employed for the X-gal staining to detect the SA-β-gal activity of the cells. Images were taken under bright-field microscope at × 20 magnification. Scale bar: 20 μm. (b) Bar graphs showing quantification of the SA-β-gal-positive cells (n=3, error bars indicate±S.D.). **P<0.01. (c) Stable clones of shTwist1 were subjected to DAPI staining of the nucleus for the detection of SAHF. Fluorescent micrographs were captured under Floid Cell Imaging Station at × 20 magnification. Scale bar: 20 μm. Scale-up images were taken to properly visualize the heterochromatin foci formation. (d) Whole-cell lysates were prepared from the stable clones of MDA-MB-231, PANC-1 and DU 145 cells bearing scramble and shTwist1 and western blot analysis of the samples was carried out for the expression of Twist1, p21, Chk2 and CDK2. (e) Bar graphs showing relative protein expression of Twist1 and p21 determined by densitometric analysis of the bands obtained from immunoblotting. (f) Immunocytochemistry of p21 was carried out with the stable clones of shTwist1 (original magnification × 20)
In vivo tumor growth and metastasis study
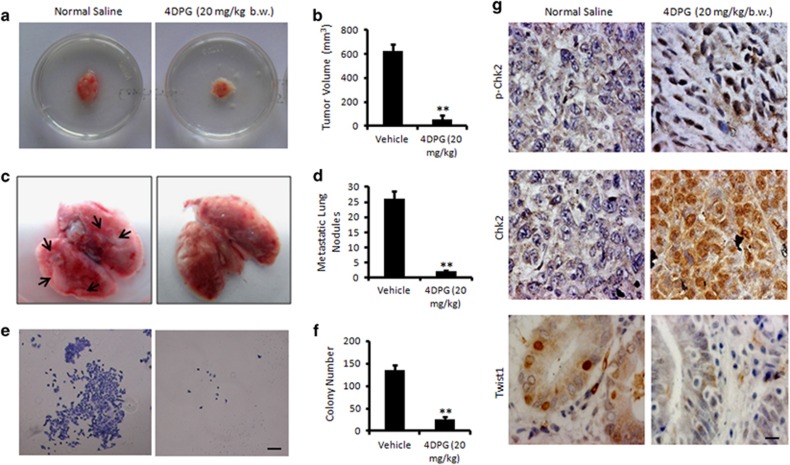
To evaluate the effect of cellular Chk2 inducer 4DPG on tumor growth and metastases in vivo, we employed 4T1 mouse mammary carcinoma model. The primary tumors showed 86% inhibition in tumor volume in the 4DPG-treated group compared to the normal saline (Figures 7a and b). Interestingly, a series of metastatic lung nodules (yellowish-white patches) were observed spreading over the lungs in control animals, whereas lungs dissected from the animals treated with 4DPG were mostly devoid of metastatic lesions indicating abrogation of lung metastases from the mammary pads (Figures 7c and d). Isolation of the 4T1 cells from the lung suspension further depicted a large number of colonies in the flask containing the lung suspension from saline-treated animals, whereas, a few colonies were identified in the 4DPG-treated group, confirming the loss of metastatic ability of 4T1 cells from the primary tumor following exposure to 4DPG (Figures 7e and f). The immunohistochemistry results revealed a marked increase in pChk2 and Chk2 along with diminish Twist1 expression in tumors dissected from 4DPG-treated group compared to the vehicle treatment (Figure 7g). Together, these data strongly imply that 4DPG is a potential inhibitor of tumor growth and metastases.
Figure 7.
In vivo antitumor and antimetastatic activity of 4DPG. (a) The effect of 4DPG (20 mg/kg/b.w.) on tumor growth in mouse mammary carcinoma (4T1) model. (b) Tumor volume was measured with the help of a Vernier Caliper (n=2, error bars indicate±S.D.).**P<0.01. (c) Lungs of each animal (control and treated) were dissected carefully, and the number of lung metastatic nodules were quantified through manual counting. (d) Bar graph represents the quantification of number of lung metastatic nodules in both saline and 4DPG-treated animals (n=2, error bars indicate±S.D.). **P<0.01. (e) Selection of 4T1 colonies from the lung suspension of saline and 4DPG-treated animals. Colonies were observed and photographed under an inverted microscope at × 4 magnification. Scale bar: 20 μm. (f) Bar graphs showing the quantification of colony number (n=2, error bars indicate±S.D.). **P<0.01. (g) Immunohistochemistry of the tumor tissue samples obtained from the saline and 4DPG-treated animals for the expression of pChk2 (T68), Chk2 and Twist1 (original magnification × 40). Scale bar: 20 μm
Discussion
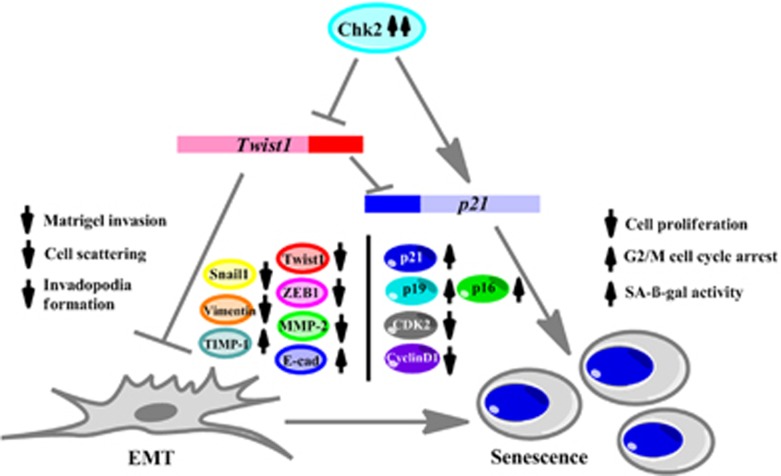
In this report, we conceive Chk2 as a major target to curb EMT in p53-defective aggressive cancer cells by its (Chk2) ectopic overexpression or exposure to natural product 4DPG. In a broader sense, the active Chk2 not only halt/reduce EMT but more precisely turn on senescence cascade in these cancer cells. Therefore, novelty of this study does not limit the circumference of the therapeutic usage of 4DPG through opening up new avenues of treatment regime in metastatic cancer cells. Interestingly, this study underscores that Chk2 activation is an essential prerequisite for the abrogation of invasion in cancer cells by Chk2-mediated transcriptional repression of Twist1 and stable knockdown of Twist1 promotes senescence in these p53-defective cells (Figure 8).
Figure 8.
Schematic representation of the proposed mechanism of action of Chk2 in metastasis prevention and induction of senescence. Chk2 induction inhibits EMT through suppression of matrigel invasion, cell scattering and invadopodia-formation ability of invasive p53-defective cancer cells. At the molecular level, Chk2 inhibits Twist1 transcriptional activity, thereby, negatively regulating pro-EMT markers such as Snail1, ZEB-1, vimentin, MMP-2 and favoring anti-EMT markers such as E-cadherin and TIMP-1. This role of Chk2 further activates premature senescence with a decrease in cell proliferation, G2/M cell cycle arrest and increase in SA-β-gal activity in these cells coupled with induction of senescence-specific marker p21 expression along with p16, p19 and downregulation of CDK2 and cyclin D1 expression
In response to oncogenic stimuli, nuclear factor-kB (NF-kB), a transcription factor positively regulates transcription of major mesenchymal genes including Snail1 and Twist1 to further aggravate the disease.23, 24 NF-kB (p65) is crucial for TNF-α-induced expression of Twist1 through a canonical signaling.24 However, a study by Solanas et al.25 demonstrates that overexpression of Snail1 can upregulate transcriptional activity of NF-kB in SW-480 and MiaPaca-2 cells, providing evidences that there might be existing some feedback mechanism behind this regulation. Keeping in mind, the above crosstalk between Snail1 and NF-kB in activating Twist1, we hypothesized that Chk2 can interact with Snail1 to affect Twist1 transcriptional activity via NF-kB. Indeed, our coimmunoprecipitation analysis implies that in stably expressing Chk2-DU 145 cells, Chk2 binds to Snail1 through protein–protein interaction. Here, for the first time, we report a novel interaction between Chk2 and Snail1 in the regulation of Twist1-mediated EMT and invasiveness in p53-defective cancer cells.
Senescence could be triggered by diverse stress signals, including DNA damage responses, irrespective of the p53 status of the cell.13 Supporting the evidence, GFP-Chk2 overexpression/4DPG treatment of three different p53-defective aggressive cancer cells consistently augmented robust nuclear accumulation of p21, increasing p16, p19 and decelerated CDK2 levels to promote senescence. A recent study by Galanos et al.26 demonstrates that in p53 mutant human cancers, sustained p21 accumulation inhibit ubiqutin ligases CRL4–CDT2 leading to deregulated origin licensing, genomic instability and aggressiveness. Studies also demonstrate that the oncogenic function of p21 is attributed to unconventional cytoplasmic localization of p21 that inactivates pro-apoptotic proteins.27 However, the present study focuses on the activation of Chk2 repressing the transcriptional activity of Twist1 through its binding to Snail1, thus preventing invasion. In addition, Chk2 also induces premature senescence in these cells by neutralizing the inhibitory effect of Twist1 on p21 expression. Moreover, stably expressing Chk2-DU 145 cells showed sufficient senescent phenotypes with increase in SA-β-gal activity and p21 expression indicating, Chk2 activation promotes and maintains senescence in these aggressive cells (Supplementary Figures S13a–c).
Senescence is a complex biological process that hinders incipient cancer cells from acquiring the malignant transformation.12 Since p53 is a whistleblower to determine the fate of the victimized cell whether to opt for apoptosis or senescence; recent evidence suggest that senescence gets priority to rescue from such catastrophic condition in the p53-defective background.12 In those consequences, Chk2 would be an appropriate target to restore cellular senescence, since chemotherapies are most likely to be efficacious in those tumors that senesce compared to those that do not.28, 29, 30 4DPG, an aryltetralin-type lignan was first isolated from Podophyllum emodi31 and showed remarkable potency (15-fold more potent than its natural congener 4′-demethyl-deoxypodophyllotoxin).17 In the present study, we report 4DPG is a potent inducer of cellular Chk2, not Chk1 and targeting Chk2-mediated anti-tumor and anti-metastatic approach would be promising leap for the development of next-generation phytomedicine.
Materials and methods
Cell culture
All the cell lines used in this study were procured from American Type Culture Collection (ATCC, Manassas, VA, USA). The p53 status of the cell lines (i.e., MDA-MB-231: p53 R280K, DU 145: p53 P223L, PANC-1: p53 R273H, PC-3: p53 null, 4T1: p53 null, MCF7: p53 intact) were confirmed from p53 database of the TP53 website. MDA-MB-231 cells were cultured in Leibovitz’s L-15 medium, DU 145, MCF7 cells in RPMI-1640 medium and PANC-1 cells in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, USA) and 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO, USA) in a humidified CO2 incubator (New Brunswick Galaxy 170 R) with 5% CO2. Mouse mammary carcinoma 4T1 cells (a gift from Dr Avinash Bajaj, Regional Center for Biotechnology, New Delhi, India) were cultured in the RPMI-1640 medium.
Matrigel invasion assay
The cell invasion through matrigel was carried out with the BD Biocoat Tumor Invasion Assay System (BD Biosciences, Bedford, MA, USA) according to the manufacturer’s instruction and described previously.32
In situ fluorescent gelatin degradation assay
The matrix gelatin degradation assay was performed to assess the formation of invadopodia or podosomes. Following transfection of the MDA-MB-231 and DU 145 cells with GFP and GFP-Chk2 plasmid or 4DPG treatment was analyzed by fluorescence microscope according to the method previously described.32
Cell scattering assay and clonogenic assay
The cell scattering and colony formation were determined according to the earlier described experimental protocol.32 For cell scattering, bFGF (20 ng/ml) was employed to stimulate cell motility. A total of five random colonies from each field were analyzed and counted manually under the microscope for the number of scattered cells and was adjusted to the total cells in particular colony.
Transient transfection
MDA-MB-231, DU 145, PANC-1 and PC-3 cells were harvested and transfected with GFP and GFP-Chk2, pCDNA3 and HA-Chk2 plasmid construct (generously gifted by Dr. Domenico Delia, Fondazione IRCCS Istituto Nazionale Tumori, Italy); hChk2-D347A (kindly gifted by Ann C. Tuma, Jann Sarkaria’s Laboratory, Mayo Clinic, Rochester, MN, USA); using Neon Transfection System (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction.
siRNA knockdown experiments
MISSION esiRNAs for human CHEK2(EHU158481), were procured from Sigma-Aldrich (St. Louis, MO, USA) and transfection experiments were performed using oligofectamine (Life Technologies, Carlsberg, CA, USA) according to the manufacturer’s instructions.
Cell viability assay
The cell viability was assessed in the presence of 4DPG by standard MTT dye uptake method according to the procedure previously described.33
SA-β-gal staining and SAHF detection
The procedure was followed as described previously.34Briefly, cells (0.4 × 106) were seeded in six-well plates and transfected with GFP, GFP-Chk2 or treated with vehicle and 4DPG (300 nM) for 72 h. Cells were accordingly washed twice with PBS, fixed with 4% paraformaldehyde for 5 min, rinsed twice with PBS and incubated with freshly prepared staining solution (40 mM citric acid/Na phosphate buffer, 5 mM K4[Fe(CN)6] 3H2O, 5 mM K3[Fe(CN)6], 150 mM sodium chloride, 2 mM magnesium chloride and 1 mg/ml X-gal in distilled water, pH 6.0) for 48 h at 37 °C. Successively, stained cells were thoroughly washed and air dried in dark. Cells were then observed under bright-field microscope (NIKON, Nikon Corporation, Chiyoda-ku, Tokyo, Japan) for the SA-β-gal-positive cells and images were captured at × 20 magnification.
The senescence-associated heterochromatin foci (SAHF) detection method was carried out as per the previously described method.18, 35 On the previous day, about 20 000 MDA-MB-231, DU 145 cells were seeded in eight-well chamber slide following transfection with GFP and GFP-Chk2 or 4DPG treatment (next day). After 10 days, the cells were washed with PBS and fixed with 4 % paraformaldehyde (w/v) at room temperature for 10 min. After discarding the paraformaldehyde, cells were washed with PBS, stained and mounted with UltraCruz mounting media containing DAPI (Invitrogen). Fluorescence images were captured with Floid cell imaging station (Invitrogen) using × 20 objective.
BrdU cell proliferation assay
Briefly, MDA-MB 231 cells were seeded at 0.3 × 106 cells per well on 35-mm Petri dishes and incubated for overnight. Next day, the medium was changed and cells were treated with vehicle and 4DPG (300 nM) for 72 h. The cells were pulse labeled with 30 μM BrdU (Sigma) for 1 h in the dark prior to harvesting. Cells were then trypsinized, centrifuged at 2000 r.p.m. for 3 min, washed twice in ice-cold PBS containing 1% BSA and fixed with ice-cold 70% ethanol (v/v) for 2 h at 1 °C. Cells were then washed twice with PBS and resuspended in denaturing buffer (2 N HCl/0.5%Triton X-100) to expose labeled DNA for an additional 30 min at room temperature. After denaturation, cells were pelleted and washed with PBS/1% BSA containing 0.1% Triton X-100. Cells were then treated with 0.1 M sodium borate buffer (pH 8.5) for 2 min at room temperature to neutralize the acid and washed out. The pellet was washed twice with PBS/1% BSA containing 0.1% Triton X-100 and the cells were stained with fluorescein isothiocyanate (FITC)-conjugated anti-BrdU antibody (Thermo Fisher Scientific, Boston, MA, USA) in 0.5% Tween 20/1% BSA/PBS for 1 h at 37 °C in dark. Cells were washed twice with PBS/1% BSA and resuspended in PBS containing RNAse A (200 μg/ml) and propidium iodide (PI) (50 μg/ml) in dark prior to flow cytometry. Cells were acquired with Gallios 10-color flow cytometer (Beckman Coulter Inc., Fullerton, CA, USA) and data were analyzed using Kaluza analysis software (Beckman Coulter, version 1.3).
Flow cytometric cell cycle analysis
MDA-MB-231 and DU 145 cells were seeded at 0.3 × 106 cells per well on 35-mm dishes and incubated for overnight at 37 °C and 5% CO2. Next day, the cells were treated with vehicle and 4DPG (300 nM) for 72 h. After the completion of the incubation period, adherent cells were trypsinized, centrifuged at 2000 rpm for 3 min, washed twice with ice-cold PBS and fixed with ice-cold 70% ethanol (v/v) for 1 h at 4 °C. Cells were washed with PBS and incubated with 50 μg/ml of PI and 200 μg/ml of RNase A in PBS at room temperature for 30 min in dark. Acquisition was performed on Gallios 10-color flow cytometer (Beckman Coulter Inc.) and data were analyzed using Kaluza analysis software (Beckman Coulter, version 1.3).
Western blotting
Western blot analysis was carried out as per the indicated condition (figure legends) as previously described.33 The list of antibodies and their working dilutions are described in Supplementary Table S2.
Immunofluorescent staining
MDA-MB-231, PANC-1 and DU 145 cells were seeded in eight-well chamber slide at a density of 0.5 × 105 cells per well. These cells were further treated as per the indicated condition (figure legends). Successively, immunocytochemical studies were carried out following the published protocol.36 The antibodies used and their working dilutions were provided in Supplementary Table S2. Images were captured under Floid Cell Imaging Station (Invitrogen) at × 20 magnification.
RNA extraction and quantitative real-time PCR
Total RNA was isolated from cells using TRIzol reagent (Invitrogen, Life Technologies) according to the manufacturer’s protocol. A total of 5 μg of RNA was treated with DNase (Ambion TURBO DNA-free, Life Technologies). First-strand cDNA synthesis was carried out using the ImProm-II Reverse Transcription System (Promega, Madison, WI, USA) according to the manufacturer’s instructions, with an anchored oligo-dT12 primer (FirstChoice RLM-RACE Kit, Ambion, Life Technologies) and 1 μg of DNase-treated RNA as a template. Gene-specific primers used for quantitative real-time RT-PCR (qPCR) study was designed using LightCycler Probe Design Software 2.0 (Hoffmann-La Roche, Basel, Switzerland). Primer details are mentioned in Supplementary Table S3. Primers were verified using conventional endpoint PCR for single-band amplification.
The real-time quantitative PCR assay was carried out using the LightCycler 96 Real-Time PCR System (Hoffmann-La Roche). Each PCR reaction (10 μl) contained 1 × LightCycler 480 SYBR Green I Master (Hoffmann-La Roche), 1 μM primers (Integrated DNA Technologies, Coralville, IA, USA) and cDNA (appropriately diluted). Thermal cycling conditions for the qPCR were as follows: pre-incubation at 95 °C for 10 min, followed by 50 cycles of three-step amplification (95 °C for 10 s, 55 °C for 15 s and 72 °C for 15 s). The PCR was followed by a dissociation curve analysis (heating to 95 °C for 10 s at normal ramping, cooling to 65 °C for 60 s at normal ramping followed by slow heating to 97 °C for 1 s at a reduced ramping rate of 0.2 °C/s) to ensure qPCR reaction specificity. Each assay was carried out in triplicate, and a non-template negative control was included. β-actin gene was chosen as a house-keeping, internal control for normalization. The threshold cycle (Ct) of the amplification curve was used for the calculations. The relative expression level was analyzed using the 2−ΔΔCtmethod,37 where -ΔΔCt= (Ct, target−Ct, actin)time x−(Ct, target−Ct, actin)time 0.
Preparation of stable cell lines
Stable Twist1 short-hairpin RNA or scramble transfected clones were prepared by transfecting the cells with MISSION shRNA for Twist1 (CCGGAGTCCGCAGTCTTACGAGGAGCTCGAGCTCCTCGTAAGACTGCGGACTTTTTTG) (Sigma) through electroporation by Neon Transfection system (Invitrogen). Clones were selected by periodic treatment of puromycin to the cells for 15 days, then pooled and tested for the Twist1 expression through confocal microscopy as well as western blot analysis.
Stable GFP and GFP–Chk2-transfected clones were prepared by transfecting DU 145 cells with Lipofectamine 2000 (Invitrogen) as per the manufacturer’s instruction. Clones were selected by periodic treatment of geneticin (G418) (Sigma-Aldrich) to the cells for 15 days. Clones were then pooled and tested for the Chk2 ectopic expression through confocal microscopy and western blotting.
Luciferase reporter assay
We employed Twist1 promoter containing genes for luciferase activity (Twist-Luc) of +1 to −824 bp in a PGL3-Luc vector generously gifted by Dr. Mien-Chie Hung (MD Anderson Cancer Center, The University of Texas, USA). The promoter region contains a TATAA box at nt −32 to −28 bp, relative to the transcription start site. The proximal promoter region contains two putative STAT3-binding elements and the critical NF-kB (p65) DNA-binding elements are located at the 120-bp region of the promoter.24, 38 MDA-MB-231 and DU 145 cells were transfected with Twist-Luc and incubated overnight in 96-well tissue culture plates at 10 × 103 cells per well. Cells were then either co-transfected with either GFP, GFP-Chk2, Chk2-D347A, scramble, siChk2 and treated with vehicle, PV1019 and/or 4DPG for 48 h. The luciferase activity was measured with the help of a Dual-Glo Luciferase assay system (Promega) according to the manufacturer’s protocol. Normalization was done with the luciferase activity of PGL3 vector in each condition.
Coimmunoprecipitation analysis
The procedure was followed as previously described with minor modifications.39 Briefly, stable clones of GFP and GFP-Chk2-transfected DU 145 cells (2 × 106each) were cultured in 90 mm Petri dishes in a humidified CO2 incubator for 24 h. Cells were then harvested with cell scrapper, washed with PBS and resuspended in lysis buffer (containing 1 × PBS (pH 7.5), 1% Triton X-100, 0.1% SDS, 5 mM EDTA, 1 mM PMSF, 1% NP-40, 100 μM Na3VO4and 1 × protease inhibitor cocktail from Roche, Inc. (Roche, Basel, Switzerland)), centrifuged at 14 000 r.p.m. for 10 min to remove cell debris, and precleared by adding 50 μg of protein G PLUS-Agarose beads. Precleared lysates were subjected to immunoprecipitation with 5 μg of antibody conjugated to 50 μl of protein G PLUS-Agarose beads. The immunoprecipitates were washed with lysis buffer and employed for western blot analysis.
Experimental animals
All animals used in this study were bred and maintained at the central animal facility of Indian Institute of Integrative Medicine, Jammu, India. Animals were maintained at 20–25 °C in a 12 h light–dark cycle, routinely monitored for their diet and water consumption and proper sanitations were maintained to avoid any risk of possible pathogenic contamination. Animal studies were performed in accordance with the experimental guidelines that were approved by the Animal Ethics Committee of the institute ‘CPCSEA’ (IAEC No. 51/02/15). During the animal experiments, special handling and care were adopted in a humane way, so that no extra pains/injuries were imparted to the animals. To minimize the mortality of animals during experimentation, only a limited number of animals were employed to yield the statistically significant results.
Mouse model for tumor growth and metastasis
The experiment was performed according to the pre-standardized protocol with minor modifications.40 To evaluate the in vivo anti-tumor and anti-metastatic efficacy of 4DPG, healthy female Balb/c mice (25–30 g) were taken. Animals were randomized into two groups, and six animals were taken per group. For the tumor cells implantation, mouse mammary carcinoma 4T1 cells (1 × 106 per 200 μl) diluted in serum-free RPMI medium were injected subcutaneously into the mammary pad of each mouse around the second right mammary gland. A week after tumor cell implantation, when the palpable mammary tumors develop, mice were injected intraperitoneally with either vehicle (normal saline) or 4DPG (20 mg/kg/b.w.) in each alternative day for 2 weeks. Tumor sizes were measured in each alternate day after tumor cell injection, and the body weight was recorded once in a week. Mice were killed on the 15th day after treatment initiation, and tumors were dissected out carefully from the mammary pad area. Lungs were removed out, and lung metastatic nodules were photographed as well as quantified under a dissecting microscope. Lungs were then minced, treated with collagenase/DNase I solution for 2 h at 37 °C and passed through the 70 μm cell strainer (BD) to obtain the single-cell suspension. The suspension was then diluted three to four times, resuspended in selection medium containing 6-thioguanine and further incubated for 15 days to isolate the 4T1 colonies. Colonies were then washed with PBS, fixed with methanol and stained with 0.2 % crystal violet solution and quantified under an inverted microscope.
Immunohistochemistry
For immunohistochemical detection, paraffin-embedded tissue sections of the tumors (4DPG treated and untreated) were prepared. After dewaxination and hydration, tissue sections were subjected to endogenous peroxide blocking with 3% hydrogen peroxide in peroxidase blocking solution for 25 min. Antigen retrieval was done with citrate buffer (1 ×) for 25 min and then blocked for 1 h with 1% BSA. Sections were then incubated with primary antibodies for phospho-Chk2, total Chk2 and Twist1 (dilution 1:150), overnight at 4 °C, then extensively washed with PBS and further incubated with HRP-conjugated secondary antibody (anti-rabbit) for 1 h at room temperature. Sections were exposed to 3,3′-diaminobenzidine (DAB) substrate, counterstained with Gill’s hematoxylin no. 1 and finally dehydrated and mounted. Microscopic observation was carried out with the help of Zeiss fluorescence microscope Axioimager.Z2 (Carl Zeiss Light Microscopy, Jena, Germany).
Statistical analysis
Data were expressed as the mean±S.D. of three independent experiments performed and analyzed by Student’s t-test. IC50 values were determined with the help of GraphPad Prism software Version 5.0 (GraphPad Software, La Jolla, CA, USA) by taking the log of inhibitor versus response. A two-sided value of *P<0.05 was considered significant in all cases.
Acknowledgments
The work was supported by the institutional internal grant MLP-6002 with institutional publication number IIIM/2013/2017. We thank our Director Dr RA Vishwakarma for encouraging us to accomplish this work. The authors are grateful to Council of Scientific and Industrial Research (CSIR) and Department of Biotechnology, Govt. of India, for providing fellowship to the research scholars.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by J Bartek
The authors declare no conflict of interest.
Supplementary Material
References
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2012; 15: 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue L, Bellacosa A. Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3-kinase/AKT pathways. Oncogene 2005; 24: 7443–7454. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004; 117: 927–939. [DOI] [PubMed] [Google Scholar]
- Kwok WK, Ling M-T, Lee T-W, Lau TCM, Zhou C, Zhang X et al. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res 2005; 65: 5153–5162. [DOI] [PubMed] [Google Scholar]
- Piccinin S, Tonin E, Sessa S, Demontis S, Rossi S, Pecciarini L et al. A ‘‘Twist box’’ code of p53 inactivation: twist box:p53 interaction promotes p53 degradation. Cancer Cell 2012; 22: 404–415. [DOI] [PubMed] [Google Scholar]
- Qin Q, Xu Y, He T, Qin C, Xu J. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res 2012; 22: 90–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit MA, Peeper DS. Deregulating EMT and senescence: double impact by a single twist. Cancer Cell 2008; 14: 5–7. [DOI] [PubMed] [Google Scholar]
- Bartek J, Falck J, Lukas J. CHK2 kinase-a busy messenger. Nat Rev Mol Cell Biol 2001; 2: 877–886. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 2007; 26: 7741–7748. [DOI] [PubMed] [Google Scholar]
- Falck J, Mailand N, Syljuåsen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 2001; 410: 842–847. [DOI] [PubMed] [Google Scholar]
- Gire V, Roux P, Wynford-Thomas D, Brondello J-M, Dulic V. DNA damage checkpoint kinase Chk2 triggers replicative senescence. EMBO J 2004; 23: 2554–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 2007; 8: 729–740. [DOI] [PubMed] [Google Scholar]
- Aliouat-Denis Cc-M, Dendouga N, Van den Wyngaert I, Goehlmann H, Steller U, van de Weyer I et al. p53-independent regulation of p21Waf1/Cip1 expression and senescence by Chk2. Mol Cancer Res 2005; 3: 627–634. [DOI] [PubMed] [Google Scholar]
- Sun Y, Wicha M, Leopold WR. Regulation of metastasis-related gene expression by p53: a potential clinical implication. Mol Carcinog 1999; 24: 25–28. [PubMed] [Google Scholar]
- Martin KH, Hayes KE, Walk EL, Ammer AG, Markwell SM, Weed SA. Quantitative measurement of invadopodia-mediated extracellular matrix proteolysis in single and multicellular contexts. J Vis Exp 2012; 66: e4119–e4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JK, Lovly CM, Piwnica-Worms HRegulation of the Chk2 protein kinase by oligomerization-mediated cis and trans-phosphorylation. Mol Cancer Res 2003; 1: 598–609. [PubMed]
- Zilla MK, Nayak D, Amin H, Nalli Y, Rah B, Chakraborty S et al. 4'-Demethyl-deoxypodophyllotoxin glucoside isolated from Podophyllum hexandrum exhibits potential anticancer activities by altering Chk-2 signaling pathway in MCF-7 breast cancer cells. Chem Biol Interact 2014; 224: 100–107. [DOI] [PubMed] [Google Scholar]
- Aird KM, Zhang R. Detection of senescence-associated heterochromatin foci (SAHF). Methods Mol Biol 2013; 965: 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest 2004; 114: 1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan LC, Chiang WF, Liang CH, Tsai YT, Wong TY, Chen KC et al. α-Catulin knockdown induces senescence in cancer cells. Oncogene 2011; 30: 2610–2621. [DOI] [PubMed] [Google Scholar]
- Ansieau Sp, Bastid J, Doreau A, Morel A-P, Bouchet BP, Thomas CM et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell 2008; 14: 79–89. [DOI] [PubMed] [Google Scholar]
- Sun M, Guo X, Qian X, Wang H, Yang C, Brinkman KL et al. Activation of the ATM-Snail pathway promotes breast cancer metastasis. J Mol Cell Biol 2012; 4: 304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y-d, Zhou BP. TNF-α/NF-kB/Snail pathway in cancer cell migration and invasion. Brit J Cancer 2010; 102: 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-W, Xia W, Huo L, Lim S-O, Wu Y, Hsu JL et al. Epithelial-mesenchymal transition induced by TNF-α requires NF-kB-mediated transcriptional upregulation of Twist1. Cancer Res 2012; 72: 1290–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanas G, Porta-de-la-Riva M, Agusti C, Casagolda D, Sanchez-Aguilera F, Larriba MJ et al. E-cadherin controls β-catenin and NF-kB transcriptional activity in mesenchymal gene expression. J Cell Sci 2008; 121: 2224–2234. [DOI] [PubMed] [Google Scholar]
- Galanos P, Vougas K, Walter D, Polyzos A, Maya-Mendoza A, Haagensen EJ et al. Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing. Nat Cell Biol 2016; 18: 777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roninson IB. Oncogenic functions of tumour suppressor p21 Waf1/Cip1/Sdi1: association with cell senescence and tumour-promoting activities of stromal fibroblasts. Cancer Lett 2002; 179: 1–14. [DOI] [PubMed] [Google Scholar]
- Roberson RS, Kussick SJ, Vallieres E, Chen S-YJ, Wu DY. Escape from therapy-induced accelerated cellular senescence in p53-null lung cancer cells and in human lung cancers. Cancer Res 2005; 65: 2795–2803. [DOI] [PubMed] [Google Scholar]
- Schmitt CA, Fridman JS, Yang M, Lee S, Baranov E, Hoffman RM et al. A senescence program controlled by p53 and p16 INK4a contributes to the outcome of cancer therapy. Cell 2002; 109: 335–346. [DOI] [PubMed] [Google Scholar]
- te Poele RH, Okorokov AL, Jardine L, Cummings J, Joel SP. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res 2002; 62: 1876–1883. [PubMed] [Google Scholar]
- Iida N, Inatomi Y, Murata H, Murata J, Lang FA, Tanaka T et al. New phenylpropanoid glycosides from Juniperus communis var. depressa. Chem Pharm Bull 2010; 58: 742–746. [DOI] [PubMed] [Google Scholar]
- Amin H, Nayak D, Chakraborty S, Kumar A, Yousuf K, Sharma PR et al. Par-4 dependent modulation of cellular β-catenin by medicinal plant natural product derivative 3-azido Withaferin A. Mol Carcinog 2016; 55: 864–881. [DOI] [PubMed] [Google Scholar]
- Rah B, Amin H, Yousuf K, Khan S, Jamwal G, Mukherjee D et al. A novel MMP-2 inhibitor 3-azidowithaferin A (3-azidoWA) abrogates cancer cell invasion and angiogenesis by modulating extracellular Par-4. PLoS ONE 2012; 7: e44039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-β-gal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc 2009; 4: 1798–1806. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, ur Rasool R, Kumar S, Nayak D, Rah B, Katoch A et al. Cristacarpin promotes ER stress-mediated ROS generation leading to premature senescence by activation of p21waf-1. AGE 2016; 38: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rah B, Ru Rasool, Nayak D, Yousuf SK, Mukherjee D, Kumar LD et al. PAWR-mediated suppression of BCL2 promotes switching of 3-azido withaferin A (3-AWA)-induced autophagy to apoptosis in prostate cancer cells. Autophagy 2015; 11: 314–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lo H-W, Hsu S-C, Xia W, Cao X, Shih J-Y, Wei Y et al. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res 2007; 67: 9066–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami A, Qiu S, Dexheimer TS, Ranganathan P, Burikhanov R, Pommier Y et al. Par-4 binds to topoisomerase 1 and attenuates its DNA relaxation activity. Cancer Res 2008; 68: 6190–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasool Ru, Rah B, Amin H, Nayak D, Chakraborty S, Rawoof A et al. Dual modulation of Ras-Mnk and PI3K-AKT-mTOR pathways: a novel c-FLIP inhibitory mechanism of 3-AWA mediated translational attenuation through dephosphorylation of eIF4E. Sci Rep 2016; 6: 18800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.