Accurate DNA replication is essential for genome maintenance. Two recent reports have uncovered new molecular mechanisms controlling the termination phase of DNA replication in higher eukaryotes and established crucial roles for the CRL2LRR1 ubiquitin ligase and the p97 segregase in replisome unloading from chromatin.
Eukaryotic DNA replication can be divided into three distinct steps. During licensing, pre-replication complexes (pre-RCs) assemble at DNA replication origins in the G1 phase of the cell cycle. This is followed by replication initiation at the G1-S phase transition, when CDKs (cyclin-dependent kinases) and DDKs (DBF4-dependent kinases) promote the recruitment of the GINS complex and CDC45 to assemble an active CMG (Cdc45-MCM-GINS) helicase that initiates bidirectional DNA synthesis.1 When DNA synthesis is completed, the CMG helicase is disassembled and unloaded from chromatin during replication termination.
A multitude of studies have demonstrated that the early phases of DNA replication are regulated by ubiquitylation. For instance, the E3 ubiquitin ligase complexes cullin-RING ligase-1 (CRL1) and cullin-RING ligase-4 (CRL4) prevent re-replication and the occurrence of genome instability by targeting pre-RC components for proteasomal degradation (reviewed in Truong et al.2). Cullin-RING ligases (CRLs) constitute a protein family of >200 modular E3s and are composed of eight distinct subfamilies containing different cullins, namely CUL1, CUL2, CUL3, CUL4A, CUL4B, CUL5, CUL7 and CUL9.3 Cullins work as molecular scaffolds assembling the different complex subunits, that is, a RING-finger protein (RBX1 or RBX2), which interacts with the ubiquitin-conjugating enzyme, an adaptor protein and one of many substrate-receptor subunits. The activity of CRLs is primarily controlled at the level of substrate recruitment. The direct recognition of the target protein by the substrate-receptor subunit and its recruitment to the core CRL platform are in fact regulated in response to specific stimuli. Moreover, all CRLs are activated through the covalent attachment of the ubiquitin-like protein Nedd8 to the cullin subunit.
It has been recently shown that also the final phase of DNA replication, that is, replication termination, is controlled by CRL-mediated ubiquitylation.4, 5 In budding yeast, the CRL1DIA2 ubiquitin ligase mediates the K48-linked ubiquitylation of the CMG subunit MCM7 at the end of DNA replication. This finding prompted Sonneville et al.6 and Dewar et al.7 to identify the CRL involved in DNA replication termination in higher eukaryotes. To that goal, they used different strategies now described in two recently published reports.
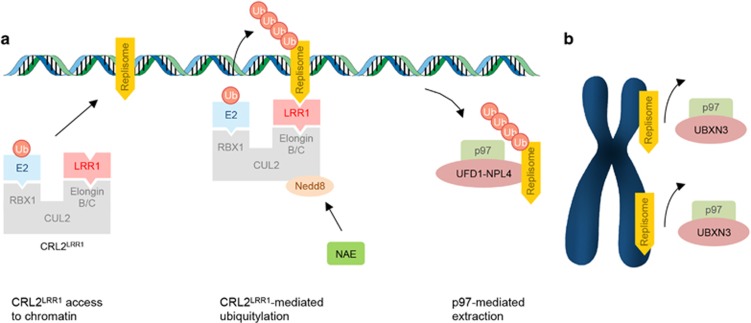
Dewar et al.7 employed a proteomic screen in Xenopus egg extracts aimed at identifying proteins associated with terminated CMG complex. A key factor of their approach was the utilization of MLN4924, a small-molecule inhibitor that blocks CRL activity by preventing its neddylation. The rationale behind this choice was that inhibiting the activity of the CRL responsible for MCM7 ubiquitylation would trap the CRL on chromatin. Using this strategy, they found that CRL2LRR1, a CRL complex in which CUL2 bridges RBX1 to the adaptor elongin B/C and the substrate-receptor LRR1, is enriched on chromatin when replisome unloading is inhibited. They also determined that during DNA replication termination, CRL2LRR1 mediates MCM7 ubiquitylation, triggering replisome unloading from chromatin.
The Labib and Gambus laboratories came to similar conclusions using a different strategy. They employed an elegant cytological assay in Caenorhabditis elegans embryos in which the unloading of GFP-tagged CMG components from chromatin was followed by time-lapse microscopy after RNAi-mediated depletion of CRL subunits. The authors found that depletion of CRL2LRR1 subunits resulted in the persistence of the CMG helicase on prophase chromatin. The genetic experiments carried out in C. elegans were complemented by reconstitution experiments in vitro using Xenopus egg extracts. The authors showed that CRL2LRR1 associates with the replisome during DNA replication termination promoting the ubiquitylation of MCM7.6
An additional mechanism involved in replisome unloading was uncovered in both studies. In their proteomic screen, the Räschle and Walter laboratories recovered four adaptors (UFD1, NPL4, UBXN7 and ASPC1) of the p97 segregase, which is known to extract polyubiquitylated proteins from chromatin,8 and proposed that during replication termination, p97 promotes the unloading of the replisome ubiquitylated by CRL2LRR1 (Figure 1a). In agreement with this model, the Gambus and Labib groups showed that late in S phase a p97-dependent mechanism is required to remove the replisome from chromatin.
Figure 1.
Replisome unloading is controlled by two pathways. (a) During DNA replication termination, the CRL2LRR1 ubiquitin ligase and the p97 segregase trigger replisome unloading from chromatin. (b) An additional backup mechanism that depends on the p97 adaptor UBXN3 drives replisome unloading from mitotic chromatin. See text for details. For the sake of clarity, the different components of the replisome are not shown
Interestingly, the Gambus and Labib groups observed that although the expression of LRR1 was silenced by RNAi, the CMG helicase was still released from mitotic chromatin after prophase. The authors found that combined depletion of LRR1 and UBXN3, an additional p97 adaptor, resulted in persistence of several CMG components on mitotic chromatin, suggesting that an additional mechanism, mediated by UBXN3, is responsible for replisome disassembly in mitosis (Figure 1b). Notably, whereas inhibition of either the mitotic or the S phase CMG disassembly pathway has marginal effect on worm viability, combined inhibition of the two pathways, obtained by silencing the expression of both LRR1 and UBXN3, drastically reduced cell viability. These results imply a backup function of the mitotic pathway, and highlight the relevance of replisome disassembly for cellular and organismal survival.
These two seminal studies raise several questions related to the control of DNA replication by ubiquitylation. For instance, how is LRR1 regulated? What is the mechanism of substrate recognition by CRL2LRR1? Additional work is required to map the interaction between LRR1 and MCM7, and assess whether post-translational modifications, cofactors, or alternative binding partners contribute to replisome unloading by controlling the binding between the leucine-rich repeats of LRR1 and a specific domain of MCM7. Alternatively, it is possible that modifications of MCM7 are not critical for its interaction with LRR1, rather, it is the temporally and spatially controlled access to chromatin of CRL2LRR1 that represents the main regulatory factor.
Inactivation of LRR1 in early C. elegans embryos is known to induce a DNA damage response that is mediated by ATL-1, the C. elegans functional homologue of ATR in vertebrates.9, 10 However, the type of DNA damage induced by LRR1 inactivation remains to be determined. Moreover, if the only function of LRR1 is replisome unloading, these findings suggest that cells can use the mitotic backup pathway to prevent cell death at the cost of increased genome instability. In this regard, the identification of the molecular mechanisms controlling the mitotic pathway might reveal novel synthetic lethality strategies for cancer therapies. Indeed, many cancer types bear deletion of FAF1, the human homologue of UBXN3. In these cancers, small-molecule inhibitors of CRL2LRR1 would lead to cell death due to failure of replisome unloading. Further studies will reveal whether replisome disassembly is an exploitable weakness of cancer cells.
Acknowledgments
VD’A is supported by the Medical Research Council MC_PC_12007. DG is supported by the KWF grant HUBR 2014-6806.
Footnotes
The authors declare no conflict of interest.
References
- Diffley JF. Philos Trans R Soc Lond B Biol Sci 2011; 366: 3545–3553. [DOI] [PMC free article] [PubMed]
- Truong LN et al. J Mol Cell Biol 2011; 3: 13–22. [DOI] [PMC free article] [PubMed]
- Petroski MD et al. Nat Rev Mol Cell Biol 2005; 6: 9–20. [DOI] [PubMed]
- Maric M et al. Science 2014; 346: 1253596. [DOI] [PMC free article] [PubMed]
- Moreno SP et al. Science 2014; 346: 477–481. [DOI] [PubMed]
- Sonneville R et al. Nat Cell Biol 2017; 19: 468–479. [DOI] [PMC free article] [PubMed]
- Dewar JM et al. Genes Dev 2017; 31: 275–290. [DOI] [PMC free article] [PubMed]
- Ramadan K et al. Chromosoma 2017; 126: 17–32. [DOI] [PubMed]
- Starostina NG et al. Dev Cell 2010; 19: 753–764. [DOI] [PMC free article] [PubMed]
- Merlet J et al. Development 2010; 137: 3857–3866. [DOI] [PMC free article] [PubMed]