Abstract
Background
The Passiflora genus comprises hundreds of wild and cultivated species of passion fruit used for food, industrial, ornamental and medicinal purposes. Efforts to develop genomic tools for genetic analysis of P. edulis, the most important commercial Passiflora species, are still incipient. In spite of many recognized applications of microsatellite markers in genetics and breeding, their availability for passion fruit research remains restricted. Microsatellite markers in P. edulis are usually limited in number, show reduced polymorphism, and are mostly based on compound or imperfect repeats. Furthermore, they are confined to only a few Passiflora species. We describe the use of NGS technology to partially assemble the P. edulis genome in order to develop hundreds of new microsatellite markers.
Results
A total of 14.11 Gbp of Illumina paired-end sequence reads were analyzed to detect simple sequence repeat sites in the sour passion fruit genome. A sample of 1300 contigs containing perfect repeat microsatellite sequences was selected for PCR primer development. Panels of di- and tri-nucleotide repeat markers were then tested in P. edulis germplasm accessions for validation. DNA polymorphism was detected in 74% of the markers (PIC = 0.16 to 0.77; number of alleles/locus = 2 to 7). A core panel of highly polymorphic markers (PIC = 0.46 to 0.77) was used to cross-amplify PCR products in 79 species of Passiflora (including P. edulis), belonging to four subgenera (Astrophea, Decaloba, Distephana and Passiflora). Approximately 71% of the marker/species combinations resulted in positive amplicons in all species tested. DNA polymorphism was detected in germplasm accessions of six closely related Passiflora species (P. edulis, P. alata, P. maliformis, P. nitida, P. quadrangularis and P. setacea) and the data used for accession discrimination and species assignment.
Conclusion
A database of P. edulis DNA sequences obtained by NGS technology was examined to identify microsatellite repeats in the sour passion fruit genome. Markers were submitted to evaluation using accessions of cultivated and wild Passiflora species. The new microsatellite markers detected high levels of DNA polymorphism in sour passion fruit and can potentially be used in genetic analysis of P. edulis and other Passiflora species.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-017-3881-5) contains supplementary material, which is available to authorized users.
Keywords: De novo genome assembly, Microsatellite, Cross-species transferability
Background
Passiflora is a highly diverse genus with approximately 520 species distributed in tropical regions of America, Asia and Africa [1]. Despite taxonomical uncertainties, approximately 96% of the Passiflora species are found in South and Central America [2]. Major centers of diversity include regions of Brazil and Colombia [3, 4], both countries with hundreds of species catalogued. However, just a few Passiflora species are used in agriculture, mostly for production of fruits, which are consumed in natura or as juice. Passion fruit species are also used as ornamentals, in the food industry and for medicinal purposes.
Sour passion fruit (P. edulis) is by far the most important commercial Passiflora species worldwide. It is an allogamous species, displaying a well documented variability of shapes and colors of fruits, flowers and plants. Genetic diversity in P. edulis has been assessed by morphological descriptors [5–7] and agronomic traits [8–10]. Detection of DNA polymorphism in P. edulis has been pursued with different types of molecular markers, such as Inter Simple Sequence Repeat (ISRR) [11], Random Amplified Polymorphic DNA (RAPD) [12–14], Amplified Fragment Length Polymorphism (AFLP) [15, 16] and microsatellites [17–19]. High levels of genetic variability have been recorded in morphological and agronomic evaluations of sour passion fruit, as well as in most marker systems. However, the use of microsatellite markers in genetic analysis of P. edulis underscores low DNA polymorphism [16–18] in an otherwise highly diverse species.
Advantages of microsatellite markers over other technologies include high reproducibility, co-dominance, high polymorphic information content (PIC) and multi-allelism [20–22]. Less than 200 microsatellite markers have been developed for P. edulis [17, 19, 23] and only a small fraction of these markers have been validated and used in genetic studies [16, 23–25]. The few polymorphic P. edulis microsatellite markers are based on compound or imperfect motifs, which are hard to interpret on routine genotyping assays due to allele binning difficulties [26, 27]. This could be a constraint to some applications, especially for population genetic studies [28, 29]. Perfect microsatellite markers (i.e. repeat of the same nucleotide motif without interruption or variation) would be more suitable, but they are only a small fraction (~10%) of the total number of P. edulis markers [17, 19, 23]. Also, the use of microsatellite markers in Passiflora has been limited to a few species, such as P. edulis [17, 19, 23], P. alata [30, 31], P. cincinnata [18, 19], P. setacea [19], and P. contracta [32]. This is only a tiny fraction (~1%) of the known Passiflora species. Similar constraints to microsatellite marker availability and use are also observed in other Passiflora species. Therefore, although there is a wide number of applications of microsatellite markers in genetics and breeding, their development and availability for passion fruit research is still restricted.
Microsatellite detection and isolation has been most often based on enrichment of genomic libraries by selective hybridization [33] or by primer extension [34]. Another approach is to identify microsatellite repeats in DNA databases such as EST sequences [35]. The development of microsatellite markers in Passiflora has been based on the construction of genomic libraries enriched for simple sequence repeats [17–19, 23, 30–32]. This is an effective but time and labor consuming technique that can lead to microsatellite discovery and marker development. However, new approaches such as next-generation sequencing (NGS) can provide a large number of high quality genome sequences that can be obtained faster and at reduced costs, facilitating the detection of thousands of microsatellite sites in the genome of a target species [36–39].
In the present study we used NGS to sequence the P. edulis genome. We then screened contig sequences obtained by partial de novo assembly to detect perfect microsatellite sites. This data was used to develop and validate microsatellite markers using P. edulis accessions of the germplasm bank. Markers were then evaluated for quality and polymorphism in P. edulis and five closely related Passiflora species and, also, for cross-species transferability to 78 Passiflora species belonging to four subgenera (Astrophea, Decaloba, Distephana and Passiflora), recently collected in Brazil.
Methods
DNA extraction and genome sequencing – Fresh young leaves of the accession Passiflora edulis CPGA1, a sour yellow rind commercial cultivar of passion fruit, were used for DNA extraction with the standard CTAB protocol [40]. The construction of the genomic DNA fragment library and massive parallel paired-end sequencing by synthesis using an Illumina GAII sequencer followed the Illumina protocol.
De novo genome assembly – The presence of non-nuclear and/or exogenous DNA sequences on the passion fruit DNA database was verified by BLASTing it against a database of chloroplast, mitochondrial and potential contaminant DNA (fungi, bacteria and virus). Extraneous sequences were removed from the analysis. The short-read correction tool of SOAPdenovo (Release 1.05), used to correct Illumina GA reads for large plant and animal genomes [41], was applied to FASTQ formatted files containing DNA sequencing reads. The CLC trimmer function (default limit = 0.05) (CLC Genomics Workbench 4.1 software, CLC Bio, Aarhus, Denmark) was then used to eliminate Illumina sequencing adapters and low quality reads. ErrorCorrection routines and KmerFreq were run with default parameters (seed length = 17, quality cutoff = 5). Final FASTQ files were submitted to de novo assembly routines using a bubble size of 50 bp on the CLC Genomics Workbench (Assembly Length Fraction = 0.5; Similarity = 0.8), followed by a scaffolding procedure by MipScaffolder [42]. Mismatch, deletion and insertion cost parameters were set to 2, 3 and 3, respectively. The k-mer size on CLC Bio assembler was set to 25 bp and the coverage cutoff to 10X. During assembly, the default word length parameter was adjusted to 25, using k-mer (de Bruijn graph k-mer) overlap information in order to assure unambiguous paths of resulting contigs. The fraction of short insert size contigs >160 bp was considered in the analysis. Overlaps between sequences were depicted by de Bruijn graph structures [43].
Identification of microsatellite sites and marker development – The partial de novo sequence assembly results were submitted to simple sequence repeat loci identification using PHOBOS [44]. The location and number of di-, tri-, and tetra-nucleotide SSRs in the draft de novo genome assembly were listed and quantified. Sequence repeats located in putative coding regions were identified with the gene model version TAIR 9 using P. edulis contigs blasted against Arabidopsis thaliana transcripts (AtGDB171). An ab initio prediction of coding regions was also performed using geneid [45] [http://genome.crg.es/software/geneid/]. Both analyses were considered for the selection of microsatellites located in structural and coding regions. Only microsatellite sites located in genomic regions with minimum 15X coverage were considered for marker development. A database of simple sequence repeats with four or more di-nucleotide repeats and three or more tri- and tetra-nucleotide repeats was created. Microsatellite loci showing a simple motif exactly repeated in tandem (“perfect microsatellite”) were listed and those with compound (more than one motif) or imperfect repeats were set aside. Perfect microsatellites with minimum 3× motif repeat and located on contigs with minimum 2.5 Kb length and 20X average coverage, as an attempt to maximize loci independence and marker quality, composed the group of selected markers. Finally, PCR primer pairs for 816 perfect microsatellite loci were developed with Primer3Plus [46].
Plant materials and microsatellite marker descriptive statistics – Ten accessions of sour passion fruit (P. edulis Sims), maintained by the Passion Fruit Germplasm Bank, Embrapa Cerrados, Planaltina, DF, were used to evaluate if the new set of markers is suitable for genetic analysis of passion fruit. Passport data of the passion fruit accessions used in the present study is described on Table 1 (rows 1 to 10). These accessions represent a diverse group of cultivars and local varieties collected in different regions of Brazil. The only exception is accession “Gulupa” from Colombia. This accession, however, is believed to have been originally collected in Brazil and later introduced in Colombia [47, 48] and was, therefore, also used in the analysis. These ten P. edulis accessions were genotyped with a random sample of 60 di- and tri-nucleotide microsatellites. Marker polymorphism, number of alleles, heterozigosity, PIC values and other statistics were estimated by CERVUS [49].
Table 1.
Germplasm accessions of passion fruit (Passiflora spp.) collected in different regions of Brazil and genotyped with the new microsatellite markers
| Species | Subgenus | Origin | |
|---|---|---|---|
| 1 | Passiflora edulis Sims | Passiflora | Selection Embrapa CPGA1, Distrito Federal |
| 2 | Passiflora edulis Sims | Passiflora | Selection Embrapa CPMSC1, Paraná |
| 3 | Passiflora edulis Sims | Passiflora | Selection Maguary, Minas Gerais |
| 4 | Passiflora edulis Sims | Passiflora | Cafuringa, Distrito Federal |
| 5 | Passiflora edulis Sims | Passiflora | Niquelândia, Goiás |
| 6 | Passiflora edulis Sims | Passiflora | Oliveira, Minas Gerais |
| 7 | Passiflora edulis Sims | Passiflora | Búzios, Rio de Janeiro |
| 8 | Passiflora edulis Sims | Passiflora | Criciúma, Santa Catarina |
| 9 | Passiflora edulis Sims | Passiflora | Jundiaí, São Paulo |
| 10 | Passiflora edulis Sims | Passiflora | Gulupa, Colombia (originally from Brazil) |
| 11 | Passiflora edulis Sims | Passiflora | BRS Maracujá Jaboticaba, Distrito Federal |
| 12 | Passiflora actinia Hook. | Passiflora | Curitiba, Paraná |
| 13 | Passiflora acuminata DC. | Passiflora | Manaus, Amazonas |
| 14 | Passiflora alata Curtis | Passiflora | Monte Verde, Minas Gerais |
| 15 | Passiflora alata Curtis | Passiflora | Selection Embrapa, Distrito Federal |
| 16 | Passiflora alata Curtis | Passiflora | Selection Embrapa, Distrito Federal |
| 17 | Passiflora alata Curtis | Passiflora | Selection Embrapa, Distrito Federal |
| 18 | Passiflora alata Curtis | Passiflora | Trancoso, Bahia |
| 19 | Passiflora ambigua Hemsl. | Passiflora | Confresa, Mato Grosso |
| 20 | Passiflora amethystina Mikan | Passiflora | Monte Verde, Minas Gerais |
| 21 | Passiflora araujoi Sacco | Distephana | Santarém, Pará |
| 22 | Passiflora auriculata Kunth | Decaloba | Manaus, Amazonas |
| 23 | Passiflora bahiensis Klotzsch | Passiflora | Lençóis, Bahia |
| 24 | Passiflora biflora Lam. | Decaloba | Novo Airão, Amazonas |
| 25 | Passiflora boticarioana Cervi | Passiflora | Conceição do Mato Dentro, Minas Gerais |
| 26 | Passiflora caerulea L. | Passiflora | Bento Gonçalves, Rio Grande do Sul |
| 27 | Passiflora capsularis L. | Decaloba | Planaltina, Distrito Federal |
| 28 | Passiflora cerasina Annonay & Feuillet | Passiflora | Presidente Figueiredo, Amazonas |
| 29 | Passiflora cerradense Sacco | Astrophea | Planaltina, Distrito Federal |
| 30 | Passiflora cervii Milward-de-Azevedo | Decaloba | Caeté, Minas Gerais |
| 31 | Passiflora chlorina L. K. Escobar | Astrophea | Caeté, Minas Gerais |
| 32 | Passiflora cincinnata Mast. | Passiflora | Rio Pardo de Minas, Minas Gerais |
| 33 | Passiflora coccinea Aubl. | Passiflora | Pontes e Lacerda, Mato Grosso |
| 34 | Passiflora decaisneana G. Nicholson | Passiflora | Planaltina, Distrito Federal |
| 35 | Passiflora edmundoi Sacco | Passiflora | Rio Pardo, Minas Gerais |
| 36 | Passiflora eichleriana Mast. | Passiflora | Criciúma, Santa Catarina |
| 37 | Passiflora elegans Mast. | Passiflora | Patos de Minas, Minas Gerais |
| 38 | Passiflora ferruginea Mast. | Decaloba | Rio Branco, Acre |
| 39 | Passiflora foetida L. | Passiflora | Belém, Pará |
| 40 | Passiflora galbana Mast. | Passiflora | Ponte Nova, Minas Gerais |
| 41 | Passiflora gardneri Mast. | Passiflora | Silvania, Goiás |
| 42 | Passiflora gibertii Brown | Passiflora | Poconé, Mato Grosso |
| 43 | Passiflora glandulosa Cav. | Passiflora | Igarapé-açú, Pará |
| 44 | Passiflora haematostigma Mart. ex Mast. | Astrophea | Natividade, Tocantins |
| 45 | Passiflora hatschbachii Cervi | Passiflora | Jaíba, Minas Gerais |
| 46 | Passiflora hypoglauca Harms | Passiflora | Ouro Preto, Minas Gerais |
| 47 | Passiflora incarnata L. | Passiflora | Centroflora, Botucatu, São Paulo |
| 48 | Passiflora jilekii Wawra | Passiflora | Manhuaçu, Minas Gerais |
| 49 | Passiflora junqueirae Imig & Cervi | Passiflora | Caparaó, Minas Gerais |
| 50 | Passiflora kermesina Link & Otto | Passiflora | São José do Laranjal, Minas Gerais |
| 51 | Passiflora laurifolia L. | Passiflora | Picos, Piauí |
| 52 | Passiflora ligularis Juss. | Passiflora | Commercial Orchard |
| 53 | Passiflora loefgrenii Vitta | Passiflora | Criciúma, Santa Catarina |
| 54 | Passiflora luetzelburgii Harms | Passiflora | Rio Pardo de Minas, Minas Gerais |
| 55 | Passiflora malacophylla Spruce ex Mast. | Passiflora | Rio das Ostras, Rio de Janeiro |
| 56 | Passiflora maliformis L. | Passiflora | Selection Embrapa, Distrito Federal |
| 57 | Passiflora maliformis L. | Passiflora | Boa Vista, Roraima |
| 58 | Passiflora maliformis L. | Passiflora | Guajará Mirim, Rondônia |
| 59 | Passiflora mendoncaei Harms | Passiflora | Monte Verde, Minas Gerais |
| 60 | Passiflora micropetala Mast. | Decaloba | Iranduba, Amazonas |
| 61 | Passiflora miersii Mast. in Mart. | Passiflora | Monte Verde, Minas Gerais |
| 62 | Passiflora misera Kunth | Decaloba | Trancoso, Bahia |
| 63 | Passiflora morifolia Mast. in Mart. | Decaloba | Lavras, Minas Gerais |
| 64 | Passiflora mucronata Lam. | Passiflora | Campos dos Goytacazes, Rio de Janeiro |
| 65 | Passiflora nitida Kunth | Passiflora | Presidente Figueiredo, Amazonas |
| 66 | Passiflora nitida Kunth | Passiflora | Planaltina, Distrito Federal |
| 67 | Passiflora nitida Kunth | Passiflora | Marabá, Pará |
| 68 | Passiflora odontophylla Harms ex Glaz. | Passiflora | Caeté, Minas Gerais |
| 69 | Passiflora organensis Gardn. | Decaloba | Serra dos Órgãos, Rio de Janeiro |
| 70 | Passiflora pedata L. | Passiflora | Manaus, Amazonas |
| 71 | Passiflora picturata Ker | Passiflora | Álter do Chão, Pará |
| 72 | Passiflora pohlii Mast. in Mart. | Decaloba | Planaltina, Distrito Federal |
| 73 | Passiflora porophylla Vell. | Decaloba | Caeté, Minas Gerais |
| 74 | Passiflora quadrangularis L. | Passiflora | Silvania, Goiás |
| 75 | Passiflora quadrangularis L. | Passiflora | Commercial Orchard |
| 76 | Passiflora quadrifaria Vanderpl. | Distephana | Manaus, Amazonas |
| 77 | Passiflora quadriglandulosa Rodschied | Distephana | Porto Velho, Rondônia |
| 78 | Passiflora racemosa Brot. | Passiflora | Búzios, Rio de Janeiro |
| 79 | Passiflora recurva Mast. in Mart. | Passiflora | Rio Pardo de Minas, Minas Gerais |
| 80 | Passiflora rhamnifolia Mast. | Astrophea | Caeté, Minas Gerais |
| 81 | Passiflora riparia Mart. | Passiflora | Confresa, Mato Grosso |
| 82 | Passiflora rubra L. | Decaloba | Monte Verde, Minas Gerais |
| 83 | Passiflora saxicola Gontsch. | Decaloba | Porto Seguro, Bahia |
| 84 | Passiflora sclerophylla Harms | Astrophea | Manaus, Amazonas |
| 85 | Passiflora setacea DC. | Passiflora | Tapiramutá, Bahia |
| 86 | Passiflora setacea DC. | Passiflora | Planaltina, Distrito Federal |
| 87 | Passiflora setacea DC. | Passiflora | Manhuaçu, Minas Gerais |
| 88 | Passiflora setacea DC. | Passiflora | Janaúba, Minas Gerais |
| 89 | Passiflora sidaefolia M. Roemer | Passiflora | Caparaó, Minas Gerais |
| 90 | Passiflora speciosa Gardn. | Passiflora | Manhuaçu, Minas Gerais |
| 91 | Passiflora suberosa L. | Decaloba | Macapá, Amapá |
| 92 | Passiflora subrotunda Mast. in Mart. | Passiflora | Natal, Rio Grande do Norte |
| 93 | Passiflora tenuifila Killip | Passiflora | Patos de Minas, Minas Gerais |
| 94 | Passiflora tholozanii Sacco | Distephana | Girau, Rondônia |
| 95 | Passiflora tricuspis Mast. in Mart. | Decaloba | Planaltina, Distrito Federal |
| 96 | Passiflora triloba Ruiz & Pav. ex DC. | Passiflora | Cruzeiro do Sul, Acre |
| 97 | Passiflora trintae Sacco | Passiflora | Rio Pardo, Minas Gerais |
| 98 | Passiflora variolata Poepp. & Endl. | Distephana | Manaus, Amazonas |
| 99 | Passiflora vespertilio L. | Decaloba | Manaus, Amazonas |
| 100 | Passiflora villosa Vell. | Passiflora | Alto Paraíso, Goiás |
| 101 | Passiflora vitifolia Kunth | Passiflora | Poconé, Mato Grosso |
Cross-species transferability of P. edulis microsatellite markers
In order to test the potential cross-species transferability of novel P. edulis microsatellite markers to other Passiflora species, we genotyped 90 accessions belonging to 78 Passiflora species native to Brazil (Table 1, rows 12 to 101), maintained by the Passion Fruit Germplasm Bank, Embrapa Cerrados, Planaltina, DF. These passion fruit species belong to four subgenera (Astrophea, Decaloba, Distephana and Passiflora). These accessions were genotyped with 18 polymorphic markers out of a sample of 60 markers initially selected for testing. Successful PCR amplifications were recorded as presence or absence of amplicons if the allele sizes were detected in the approximate expected range.
For most Passiflora species, only one accession was represented in the Germplasm Bank. However, for those species with two to five accessions available, cross-amplification and marker polymorphism could be computed (Table 1). Allele frequencies observed in 27 accessions of six species (P. edulis, P. alata, P. maliformis, P. nitida, P. quadrangularis and P. setacea) plus accession 11 (Table 1, BRS Maracujá Jaboticaba) were used to estimate pairwise genetic distances using the Band coefficient [50]. BRS Maracujá Jaboticaba is an autogamous variety of sour passion fruit of unknown phylogeny which produces small fruits of purple rind. Genetic similarities detected by microsatellite markers was explored by Principal Coordinate Analysis (PCoA) using NTSYSpc v.2.10 [51]. An analysis of population structure and ancestry of these 28 accessions based on Bayesian statistics, without prior assignment to species, was also performed using Structure v.2.3.4 [52, 53]. Batch runs with correlated and independent allele frequencies among inferred clusters were tested with population parameters set to admixture model (burn-in 250,000; run-length 500,000). In order to identify the number of clusters in the sample of Passiflora accessions, the values of ln P(D) were obtained for tests of K ranging from 1 to10 using 20 independent runs for each K (length of burnin period: 50,000; number of MCMC reps after burnin: 50,000). The most probable value of K for each test was detected by delta K [54]. Passiflora accessions were allocated to a cluster if Q values were greater or equal to 0.70, or otherwise considered as intermediate or admixed. DNA extraction and quantification of all passion fruit accessions followed the procedures described above.
Microsatellite marker PCR assays - Multiplex panels for simultaneous evaluation of microsatellite markers were designed using Multiplex Manager [55]. PCR assays were carried in a final volume of 5 μL containing 5 ηg of genomic DNA, 1X QIAGEN Multiplex PCR Kit Master Mix (QIAGEN), 0.5X Q-Solution (QIAGEN), and 0.2 μM of each primer. Reactions were performed on a Veriti™ Thermal Cycler (Applied Biosystems, USA) using the following amplification program: 95 °C for 15 min; 35 cycles at 94 °C for 30 s, 55, 57 or 60 °C for 90 s, and 72 °C for 60 s; followed by a final extension step at 60 °C for 60 min. We added 9 μL of Hi-Di™ Formamide (Applied Biosystems, USA) and a ROX-labeled internal size standard to 1 μL of the PCR product and denatured at 94 °C for 5 min. Denatured products were injected on an ABI3730 (Applied Biosystems, USA) automated sequencer. Allele size calling and genotyping were carried out with GeneMapper® v4.1 (Applied Biosystems, USA). Automated allelic binning was performed with Tandem [56]. Fisher’s exact test was used to test the association between the level of marker polymorphism and the repeat size (di- or tri-nucleotide) using the MedCalc Statistical Software v.12.7.7 [http://www.medcalc.org; 2013].
Results
Partial sequencing and de novo assembly of the Passiflora genome for microsatellite site detection.
Sequence assembly was based on 225,293,527 short read DNA sequences (average length = 62.65 bp), representing 14.1 Gbp (Table 2), which corresponds to ~4.5× coverage of the passion fruit genome, assuming a genome size of 3126 Mbp [57]. A total of 234,239 contig segments showing variation in size from 166 to 45,662 bp, average size of 707 bp and covering 165,702,691 bp, were examined for the presence of microsatellite sites. The genome sequences of the P. edulis genome have been deposited in GenBank under the BioProject ID SUB2376276.
Table 2.
Summary of Illumina paired-end read sequence data, de novo assembly and detection of microsatellite repeats in the Passiflora edulis genome
| Sequence information | Total # | Size variation (bp) | Average length (bp) | Total (bp) |
| Illumina paired-end reads | 225,293,527 | 52–76 | 62.65 | 14,113,860,125 |
| Contigs | 234,239 | 166–45,662 | 707 | 165,702,691 |
| Microsatellite sequences | Total # | >5 repeats | ||
| Compound and/or imperfect microsatellites | 1,544,549 | - | ||
| Perfect di-nucleotides | 360,162 | 13,391 | ||
| Perfect tri-nucleotides | 60,669 | 1436 | ||
| Perfect tetra-nucleotides | 7463 | 186 | ||
| Total | 1,972,843 | - |
A batch of 1,972,843 microsatellite sites matched the criteria set for simple sequence repeat discovery in the assembled contig segments (Table 2). Perfect microsatellite included 360,162 di-nucleotide repeats with the number of repeats ranging from 3 to 20 (13,391 > 5 repeats). Perfect tri-nucleotide repeats included 60,669 sites ranging from 3 to 14 repeats (1436 > 5 repeats). Perfect tetra-nucleotide repeats included 7463 sites ranging from 3 to 13 repeats (186 > 5 repeats).
Sequence analysis of P. edulis contigs allowed 37,761 gene annotations and identified 5947 sequence repeats located in putative coding regions, of which 2990 hits were non redundant. An ab initio prediction of coding regions resulted in the compilation of 101,361 hits in exon regions of the 47,706 scaffolds evaluated.
Using a minimum 15X average coverage as a cut off, a total of 1300 perfect microsatellite sites were selected in functional and structural genomic regions of sour passion fruit. In this sample of microsatellite sites, tri-nucleotide repeats were the most abundant class (534 sites), followed by tetra-nucleotide (475) and di-nucleotide (294) (Fig. 1a). The most frequent types of microsatellite sequences observed on each class were AT/TA, GAA/TTC and AAAT/ATTT (Fig. 1b). The most frequent di-nucleotide repeat motif (AT) was also the most abundant one, comprising (5.3%) of the perfect microsatellite region detected on contigs with at least 15X coverage. On the other hand, tri- and tetra-nucleotide repeat motifs had a more balanced distribution among different classes.
Fig. 1.
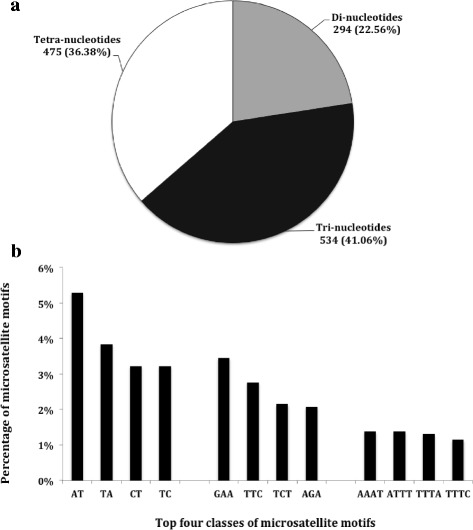
a Distribution of di-, tri-, and tetra-nucleotide perfect microsatellites on contigs with a minimum 15X average coverage; b Distribution of most frequent repeat motifs on contigs with a minimum 15X average coverage
The list of 1300 microsatellite sites was further examined for PCR primer development (Additional file 1). Primer pairs flanking the DNA repeats could be developed for 816 microsatellite sites, which were suitable for design within each contig, showing no adjacent simple sequence repeat loci and attending the minimal specified requirements which have been previously described. The new microsatellite markers were given the “BrPe” prefix. The list includes 149 di-, 329 tri- and 338 tetra-nucleotide markers. Approximately 56% of the markers are located in functional regions of the P. edulis genome (60 di-, 263 tri- and 139 tetra-nucleotide markers) and the remaining in structural regions.
A random sample of 60 markers (50 di- and 10 tri-nucleotide repeats) was labeled with fluorescent dyes and combined for simultaneous amplification in duos or trios in order to test their genotyping efficiency and marker polymorphism on passion fruit accessions. We tested 25 panels, usually containing two markers each, for simultaneous allele amplification. A total of 52 markers could readily amplify PCR products in all 25 duo panels without any adjustment in PCR amplification conditions (Fig. 2). Five markers worked better in solo amplifications (BrPe0014, BrPe0021, BrPe0033, BrPe0042, BrPe0043). PCR amplicons were not obtained for only three markers (5%) (BrPe0004, BrPe0005, BrPe0048), although further attempts to adjust PCR were not pursued. This represents a very high rate of PCR amplification success for microsatellite markers.
Fig. 2.
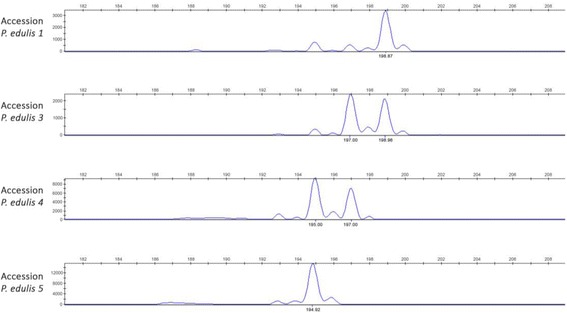
Electropherograms of marker BrPe0006 showing amplification patterns and DNA polymorphism between four accessions of P. edulis (accessions1, 3, 4, 5) (Y axis = pfu; X axis = allele size)
Descriptive statistics of microsatellite markers
Among the 57 markers which produced amplicons, 42 (~74%) were polymorphic when tested on a sample of ten P. edulis germplasm accessions, providing the detection of 137 alleles (Table 3). Fifteen markers were not polymorphic (nine di-nucleotide and six tri-nucleotide repeat markers) (Additional file 1). The number of observed alleles for all polymorphic microsatellite markers ranged from 2 to 7, with an average value of 3.26 alleles per locus (Table 3). Marker expected heterozigosity (He) values ranged from 0.19 to 0.84, with an average of 0.55. Observed heterozigosity (Ho) values ranged from 0.00 to 1.00, with an average of 0.35. Polymorphism Information Content (PIC) values ranged from 0.16 to 0.77, with an average of 0.45 (Table 3).
Table 3.
Descriptive statistics of novel Passiflora edulis microsatellite markers
| # | Marker | Primer Sequences 5′-3′ | RepeatMotif | Ta | AlleleN | Allele Size Range (bp) |
He | Ho | PIC |
|---|---|---|---|---|---|---|---|---|---|
| 1 | BrPe0032 | F:TTGCACAATGACCAATGTTGT R:CTGAGCACCTTGTCAAAATACA |
(AT)13 | 60 °C | 7 | 137–157 | 0.84 | 1.00 | 0.77 |
| 2 | BrPe0028 | F:CAAAAGGAACAGGGAAGA R:GAAAGAGAGAAAGACAGAGA |
(TA)6 | 55 °C | 5 | 90–110 | 0.80 | 0.50 | 0.72 |
| 3 | BrPe0024 | F:CCCTACCTTTCTCTGCTT R:CATCTCCTCTATCTCCTTC |
(TC)7 | 55 °C | 4 | 221–231 | 0.76 | 0.70 | 0.68 |
| 4 | BrPe0031 | F:AGGTCGGTGGGTGTGTTTAG R:CATTCAACTCCCCAAAAGGT |
(TA)9 | 60 °C | 5 | 134–150 | 0.77 | 0.67 | 0.67 |
| 5 | BrPe0014 | F:AATATGGCTGGGGAAAAC R:TTCCTGTCTTTGGACCTT |
(AG)7 | 57 °C | 5 | 215–227 | 0.75 | 0.50 | 0.67 |
| 6 | BrPe0033 | F:GCCATGAGAGACTTGGGAGA R:CGGTTGCCAAAAAGAAGAGA |
(AT)8 | 60 °C | 5 | 237–249 | 0.72 | 0.30 | 0.65 |
| 7 | BrPe0038 | F:TTTCAACTTTTCGTGTGTGC R:TGTTGTTGCTTGGAAGGATG |
(AT)6 | 60 °C | 5 | 154–176 | 0.73 | 0.60 | 0.64 |
| 8 | BrPe0042 | F:CATGCATTCATTTGTTTTTCTTG R:GATGCTGGGAAAAAGAGTGC |
(AT)8 | 60 °C | 6 | 142–160 | 0.71 | 0.80 | 0.63 |
| 9 | BrPe0003 | F:CTTTCTCTCCCTATACCC R:CCCTCCATAATCACATAAC |
(TC)11 | 55 °C | 5 | 277–291 | 0.70 | 0.40 | 0.62 |
| 10 | BrPe0043 | F:TCATACATGGATGTCAAATCGATAC R:GCGGACCAAGAAAATTCAAA |
(AT)8 | 60 °C | 4 | 199–207 | 0.71 | 0.50 | 0.60 |
| 11 | BrPe0006 | F:AAGGAAAAGAACAGCCTCA R:CGCTCTCAAATCAGTCAAA |
(TC)10 | 55 °C | 4 | 193–199 | 0.68 | 0.40 | 0.59 |
| 12 | BrPe0002 | F:AAAGCCCAGATGAAGTGAA R:GGCTCCAATCAGAAGTGT |
(AG)12 | 55 °C | 3 | 177–185 | 0.69 | 0.56 | 0.58 |
| 13 | BrPe0021 | F:ACTTCCTCATCATTCG R:GCTATGCCTCTTTTTG |
(TA)7 | 55 °C | 3 | 158–164 | 0.67 | 0.25 | 0.56 |
| 14 | BrPe0036 | F:TCGGACCTTAAAACCGAGAA R:CAGCACCAAAATTTGACGAG |
(TC)6 | 60 °C | 4 | 197–203 | 0.65 | 0.10 | 0.54 |
| 15 | BrPe0023 | F:AGATACCACACCCAATAG R:TTGGAGTTGTTGGGGA |
(CT)7 | 55 °C | 6 | 118–132 | 0.57 | 0.40 | 0.52 |
| 16 | BrPe3011 | F:CCGGTCTTCCTGATTGACTC R:CCTCTCTCACCTGGAACTGC |
(TTC)4 | 60 °C | 3 | 157–163 | 0.62 | 0.30 | 0.50 |
| 17 | BrPe0037 | F:TGATAATGCAGCGAAAGAGC R:TCACACTCCATTTGCTCTGC |
(TG)6 | 60 °C | 3 | 227–231 | 0.61 | 0.20 | 0.49 |
| 18 | BrPe0010 | F:GAAGAAAAAAGGGCTTG R:GTTAGGGTTTGGAGGA |
(TC)9 | 55 °C | 3 | 200–204 | 0.60 | 0.40 | 0.48 |
| 19 | BrPe0001 | F:GTTGAGAGGATTGTGTTTG R:ATGGTAGAGGAGGAGAGA |
(CT)14 | 55 °C | 3 | 143–157 | 0.56 | 0.14 | 0.46 |
| 20 | BrPe0012 | F:AGAGAGAGAGAGAGAG R:ACATCATACTCCTCATCC |
(AG)8 | 55 °C | 3 | 214–218 | 0.58 | 0.13 | 0.45 |
| 21 | BrPe0008 | F:TTTTCAGCCTCCACTCTT R:TACACCACCAACACTCAC |
(AG)9 | 55 °C | 3 | 264–274 | 0.57 | 1.00 | 0.44 |
| 22 | BrPe0025 | F:CAAGGAACCAGAACAAGAAGAA R:GAAGAACAAGCCAGCCCA |
(GA)6 | 55 °C | 3 | 114–126 | 0.57 | 0.11 | 0.44 |
| 23 | BrPe0039 | F:GCTGCTCCACTGTGAATGTC R:AACCTAGCCCCGTCACAGTA |
(AT)6 | 60 | 3 | 193–203 | 0.57 | 0.10 | 0.44 |
| 24 | BrPe0050 | F:TCAAGGGTATCTTTGGTGCTG R:AGCTTCAGCGAGACAAAACC |
(TG)7 | 60 °C | 3 | 197–205 | 0.56 | 0.20 | 0.44 |
| 25 | BrPe0013 | F:GATCGAGGTGAGGTACTG R:GGTTTGGCTTTAATGGAGG |
(AG)8 | 55 °C | 2 | 169–171 | 0.53 | 0.00 | 0.38 |
| 26 | BrPe0020 | F:TAAAGCATCAGGTCAG R:TAGATAGATTTGACGGG |
(GT)7 | 55 °C | 2 | 295–297 | 0.53 | 0.00 | 0.38 |
| 27 | BrPe0034 | F:CCTGTGGTGAAAATGGAACC R:GAGCCCTGGACTGACACATT |
(CT)15 | 60 °C | 2 | 217–227 | 0.56 | 1.00 | 0.38 |
| 28 | BrPe0049 | F:GGGAATCAAAACCATGCAGT R:CTCCCAGCTTCCACTCACTC |
(TA)9 | 60 °C | 2 | 189–191 | 0.53 | 0.11 | 0.38 |
| 29 | BrPe3012 | F:CGCCCTTTCTGAAGATAATCC R:GCAATGCTAAGAAGGCCAAG |
(TCT)4 | 60 °C | 2 | 181–183 | 0.53 | 0.11 | 0.38 |
| 30 | BrPe0018 | F:TCCTTCCTTCTCCTCC R:ACACTTGTCTCTCATCT |
(CT)7 | 55 °C | 3 | 135–149 | 0.43 | 0.30 | 0.37 |
| 31 | BrPe0022 | F:GGCATAGAAGTGGAAGGG R:GGAAGGGAAGTGAAGGGA |
(AG)7 | 55 °C | 2 | 98–104 | 0.51 | 0.20 | 0.37 |
| 32 | BrPe0047 | F:TGGGCCATTTCTTTTCTCTC R:GAATCCTGCATGAGTTGAGGA |
(CT)9 | 60 °C | 2 | 186–192 | 0.48 | 0.30 | 0.35 |
| 33 | BrPe3014 | F:CGGAAGCGTGCTCATAAAGT R:AAGCCTGTGAGGTTGATTCG |
(AGA)5 | 60 °C | 2 | 218–220 | 0.48 | 0.30 | 0.35 |
| 34 | BrPe0007 | F:AAAGCCCAGATGAAGTGAA R:GGCTCCAATCAGAAGTGT |
(AG)9 | 55 °C | 2 | 177–179 | 0.40 | 0.50 | 0.31 |
| 35 | BrPe0027 | F:TCCAATCTTCTCAACC R:CAAACTAGTAAACCCC |
(TA)6 | 46 °C | 3 | 97–101 | 0.35 | 0.20 | 0.30 |
| 36 | BrPe3027 | F:CCAAAATGCCCAAAATGTCT R:GTCCGTGAGGAGATGTCGAT |
(GGT)4 | 60 °C | 3 | 178–202 | 0.35 | 0.40 | 0.30 |
| 37 | BrPe0019 | F:AAAGAGAAGGATGGATG R:AAAAAGGACGAGGAAGA |
(TC)7 | 55 °C | 2 | 210–214 | 0.36 | 0.14 | 0.28 |
| 38 | BrPe0044 | F:GGACGCTAAGAGACCCATTG R:TAAAAGCCCCACTTGCAATC |
(TA)6 | 60 °C | 2 | 217–219 | 0.33 | 0.38 | 0.26 |
| 39 | BrPe0016 | F:TGGTTGGTGGGTCTTGT R:CTCTTTCCTCTCTCTCTCTCT |
(AG)7 | 55 °C | 2 | 277–279 | 0.21 | 0.22 | 0.18 |
| 40 | BrPe0045 | F:CGCTTCCACTTTACCAGCTC R:GACCAACAACAGGCACAATG |
(GT)8 | 60 °C | 2 | 183–185 | 0.21 | 0.22 | 0.18 |
| 41 | BrPe0011 | F:GTTCTACTCCCTCATT R:CTTCTTAACATCCCCA |
(CT)8 | 53 °C | 2 | 74–80 | 0.19 | 0.20 | 0.16 |
| 42 | BrPe0017 | F:TTGTCTCTCGGTTCTCT R:CAAACACAAAACCCCC |
(AG)7 | 55 °C | 2 | 86–90 | 0.19 | 0.00 | 0.16 |
| AVERAGE | 3.26 | 0.55 | 0.35 | 0.45 |
F: forward primer; R: reverse primer; Ta: annealing temperature; Ho: observed heterozygosity; He: expected heterozygosity; PIC: polymorphic information content
We checked whether the size ranges for the polymorphic loci included their expected product size on P. edulis. Expected product sizes for each microsatellite marker are based on sequence information generated by the de novo assembly process. The proportion of markers that generated amplicons within 5% of their expected sizes was 100% (42 out of 42). Approximately 55% of the polymorphic markers generated amplicons with product size exactly as expected (23 out of 42).
Out of 50 di-nucleotide markers tested for DNA polymorphism, 17 were located on structural genomic regions and 33 on putative functional sites of the P. edulis genome (Additional file 1). We found no significant association (Fisher’s exact test p-value = 0.64) between the level of marker polymorphism and repeat size (di- or tri-nucleotide).
Microsatellite marker cross-amplification in Passiflora species.
Markers were ranked by PIC values and used to evaluate their cross amplification in 79 Passiflora species (including P. edulis). The average PIC value for the 18 selected markers was 0.60, varying from 0.46 to 0.77 (Table 3, markers 1–16, 18, 19). A survey on the potential cross-amplification of these microsatellite markers in a collection of Passiflora species showed that 72% of the marker/species combinations resulted in positive amplifications (Table 4), with cross-amplification values ranging from 33% to 94%. Such a large proportion of marker transferability was not anticipated. Three markers produced PCR products in all 79 Passiflora species (BrPe0032, BrPe0038, BrPe3011). BrPe0032 had the highest PIC and number of alleles in the tested sample of P. edulis accessions. Primers BrPe0001, BrPe0034 and BrPe0042 also worked in most of the species tested, with the exception of P. porophylla (BrPe0001), Passiflora triloba and P. vitifolia (BrPe0034), and P. capsularis and P. gibertii (BrPe0042). Interestingly, at least 14 markers (BrPe0032, BrPe0038, BrPe3011, BrPe0001, BrPe0034, BrPe0042, BrPe0036, BrPe0006, BrPe0010, BrPe0028, BrPe0031, BrPe0021, BrPe0003, BrPe0033) could cross amplify PCR products in 17 species (P. cerasina, P. coccinea, P. decaisneana, P. quadrangularis, P. riparia, P. variolata, P. mendoncaei, P. nitida, P. racemosa, P. recurva, P. ligularis, P. maliformis, P. odontophylla, P. pedata, P. tenuifila, P. alata, P. setacea). Fifty percent of the markers produced amplicons in all but two of the species tested, P. pohlii (Decaloba) and P. sclerophylla (Astrophea).
Table 4.
Cross-species transferability of 18 P. edulis microsatellite markers to 78 Passiflora species
| Species | BrPe0043 | BrPe0014 | BrPe0002 | BrPe0024 | BrPe0033 | BrPe0003 | BrPe0021 | BrPe0031 | BrPe0028 | BrPe0006 | BrPe0010 | BrPe0036 | BrPe0034 | BrPe0042 | BrPe0001 | BrPe0032 | BrPe0038 | BrPe3011 | % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Passiflora pohlii | − | − | − | − | − | − | − | − | − | − | − | − | + | + | + | + | + | + | 33% |
| Passiflora sclerophylla | − | − | − | − | + | − | − | − | + | − | − | − | + | + | + | + | + | + | 44% |
| Passiflora suberosa | − | − | − | − | − | − | − | − | + | + | + | − | + | + | + | + | + | + | 50% |
| Passiflora eichleriana | − | − | − | − | + | − | + | − | − | + | + | − | + | + | + | + | + | + | 56% |
| Passiflora hatschbachii | − | − | − | − | + | + | − | + | − | − | − | + | + | + | + | + | + | + | 56% |
| Passiflora picturata | − | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | 56% |
| Passiflora porophylla | − | + | − | − | − | − | − | + | + | + | − | + | + | + | − | + | + | + | 56% |
| Passiflora saxicola | − | − | − | − | − | − | − | + | + | − | + | + | + | + | + | + | + | + | 56% |
| Passiflora tricuspis | − | − | − | − | − | + | − | − | + | + | − | + | + | + | + | + | + | + | 56% |
| Passiflora araujoi | − | − | − | − | − | − | + | + | + | + | − | + | + | + | + | + | + | + | 61% |
| Passiflora auriculata | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | 61% |
| Passiflora edmundoi | − | − | − | − | − | + | − | − | + | + | + | + | + | + | + | + | + | + | 61% |
| Passiflora ferruginea | − | − | − | + | − | − | + | + | + | − | − | + | + | + | + | + | + | + | 61% |
| Passiflora haematostigma | − | − | − | + | + | − | − | − | + | + | − | + | + | + | + | + | + | + | 61% |
| Passiflora loefgrenii | − | − | − | + | − | − | + | − | − | + | + | + | + | + | + | + | + | + | 61% |
| Passiflora micropetala | − | − | − | + | − | − | − | + | + | − | + | + | + | + | + | + | + | + | 61% |
| Passiflora misera | − | − | − | + | − | − | + | − | + | + | + | − | + | + | + | + | + | + | 61% |
| Passiflora morifolia | − | − | − | − | − | + | + | − | − | + | + | + | + | + | + | + | + | + | 61% |
| Passiflora rubra | − | + | − | + | − | − | − | − | + | + | − | + | + | + | + | + | + | + | 61% |
| Passiflora vespertilio | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | 61% |
| Passiflora acuminata | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | 67% |
| Passiflora caerulea | − | − | + | − | − | − | + | − | + | + | + | + | + | + | + | + | + | + | 67% |
| Passiflora capsularis | + | − | − | + | − | − | + | + | − | + | + | + | + | − | + | + | + | + | 67% |
| Passiflora cervii | − | − | − | + | − | − | − | + | + | + | + | + | + | + | + | + | + | + | 67% |
| Passiflora jilekii | − | − | − | + | − | − | + | + | + | + | + | − | + | + | + | + | + | + | 67% |
| Passiflora rhamnifolia | − | − | − | − | + | − | − | + | + | + | + | + | + | + | + | + | + | + | 67% |
| Passiflora tholozanii | − | − | − | − | − | + | − | + | + | + | + | + | + | + | + | + | + | + | 67% |
| Passiflora triloba | − | + | − | − | + | + | + | − | + | + | + | − | − | + | + | + | + | + | 67% |
| Passiflora vitifolia | + | − | − | − | + | − | − | + | + | + | + | + | − | + | + | + | + | + | 67% |
| Passiflora ambigua | + | − | − | + | − | − | + | − | + | + | + | + | + | + | + | + | + | + | 72% |
| Passiflora biflora | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | 72% |
| Passiflora boticarioana | − | − | + | + | + | + | − | − | − | + | + | + | + | + | + | + | + | + | 72% |
| Passiflora cerradense | − | − | − | + | + | + | − | − | + | + | + | + | + | + | + | + | + | + | 72% |
| Passiflora chlorina | + | − | − | + | + | + | − | − | − | + | + | + | + | + | + | + | + | + | 72% |
| Passiflora cincinnata | − | − | − | + | + | + | − | − | + | + | + | + | + | + | + | + | + | + | 72% |
| Passiflora elegans | − | − | + | − | + | + | − | + | − | + | + | + | + | + | + | + | + | + | 72% |
| Passiflora foetida | − | − | − | − | + | + | + | + | − | + | + | + | + | + | + | + | + | + | 72% |
| Passiflora gardneri | − | − | + | + | + | + | + | − | − | − | + | + | + | + | + | + | + | + | 72% |
| Passiflora gibertii | − | − | − | + | + | − | + | + | + | + | + | + | + | − | + | + | + | + | 72% |
| Passiflora incarnata | + | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | 72% |
| Passiflora junqueirae | − | − | + | − | + | − | + | + | + | − | + | + | + | + | + | + | + | + | 72% |
| Passiflora kermesina | − | + | + | − | − | + | − | + | + | − | + | + | + | + | + | + | + | + | 72% |
| Passiflora organensis | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | 72% |
| Passiflora quadrifarial | − | − | − | − | + | + | − | + | + | + | + | + | + | + | + | + | + | + | 72% |
| Passiflora trintae | − | − | − | + | − | + | + | − | + | + | + | + | + | + | + | + | + | + | 72% |
| Passiflora villosa | − | − | + | − | + | + | − | + | − | + | + | + | + | + | + | + | + | + | 72% |
| Passiflora actinia | − | − | + | − | + | + | + | − | + | + | + | + | + | + | + | + | + | + | 78% |
| Passiflora amethystina | − | − | + | + | + | − | + | − | + | + | + | + | + | + | + | + | + | + | 78% |
| Passiflora cerasina | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 78% |
| Passiflora coccinea | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 78% |
| Passiflora decaisneana | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 78% |
| Passiflora galbana | − | − | + | + | − | + | + | + | + | − | + | + | + | + | + | + | + | + | 78% |
| Passiflora hypoglauca | − | − | + | − | + | + | − | + | + | + | + | + | + | + | + | + | + | + | 78% |
| Passiflora malacophylla | − | − | + | + | + | + | + | − | − | + | + | + | + | + | + | + | + | + | 78% |
| Passiflora miersii | − | − | + | − | + | + | + | + | + | − | + | + | + | + | + | + | + | + | 78% |
| Passiflora mucronata | − | − | − | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | 78% |
| Passiflora quadrangularis | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 78% |
| Passiflora quadriglandulosa | + | − | − | − | + | + | + | + | + | + | + | − | + | + | + | + | + | + | 78% |
| Passiflora riparia | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 78% |
| Passiflora sidaefolia | − | − | − | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | 78% |
| Passiflora subrotunda | − | − | − | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | 78% |
| Passiflora variolata | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 78% |
| Passiflora bahiensis | + | − | − | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | 83% |
| Passiflora luetzelburgii | − | − | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | 83% |
| Passiflora mendoncaei | − | − | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 83% |
| Passiflora nitida | − | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 83% |
| Passiflora racemosa | − | − | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 83% |
| Passiflora recurva | − | − | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 83% |
| Passiflora speciosa | + | + | − | − | + | + | + | − | + | + | + | + | + | + | + | + | + | + | 83% |
| Passiflora glandulosa | + | + | − | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | 89% |
| Passiflora laurifolia | + | − | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | 89% |
| Passiflora ligularis | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 89% |
| Passiflora maliformis | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 89% |
| Passiflora odontophylla | − | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 89% |
| Passiflora pedata | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 89% |
| Passiflora tenuifila | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 89% |
| Passiflora alata | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 94% |
| Passiflora setacea | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 94% |
| Passiflora edulis | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 100% |
The new microsatellite markers uncovered genetic diversity in P. edulis (Fig. 2, Table 3) and also in other related species (Fig. 3). PCoA analysis based on marker polymorphism assessed by 18 markers on 28 accessions belonging to six species (P. edulis, P. alata, P. maliformis, P. nitida, P. quadrangularis and P. setacea) allowed their separation in four main clusters. The variation captured by eigenvalue of the first three axis was high (axis 1 = 32.08%, axis 2 = 14.20% and axis 3 = 11.10%). Interestingly, the P. edulis accessions formed two clusters (Fig. 3a) and could be easily separated from the accessions of the other Passiflora species. The only exception was BRS Maracujá Jaboticaba (Table 1, accession 11), which did not cluster with the two P. edulis groups and seems to be closely associated to a cluster formed by P. setacea accessions. The fourth cluster included accessions of P. nitida, P. quadrangularis, P. alata and P. maliformis. Although the accessions of these four species could be discriminated with this set of microsatellite markers, they were all included in the same cluster. An analysis of population structure and ancestry of these 28 accessions with no prior assignment of species also inferred the existence of four main clusters, estimated by plotting values of K vs Delta K, for K varying from 1 to 10 (Fig. 3b). Again the accessions of P. edulis were allocated to two clusters, P. setacea accessions were separated in a third group, while accessions of P. nitida, P. quadrangularis, P. alata and P. maliformis formed a fourth group. All accessions were allocated to one of the four clusters with Q value ≥70, with the exception of BRS Maracujá Jaboticaba (accession e11), which showed an admixed or intermediate profile.
Fig. 3.
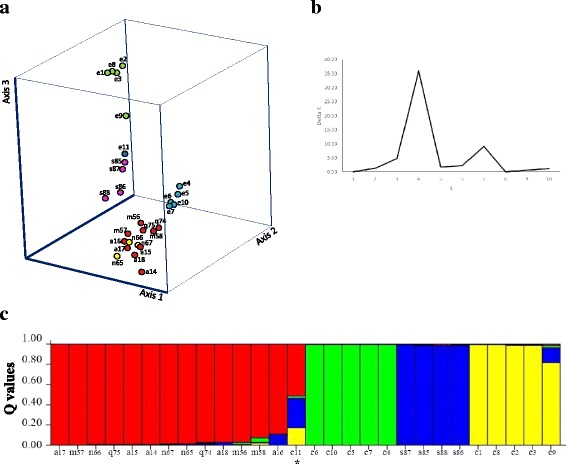
a Principal coordinates analysis of 28 accessions of Passiflora belonging to six closely related passion fruit species based on microsatellite polymorphism. Samples are identified according to accession number on Table 1, preceded by species initial: P. edulis (samples e1 to e11); P. alata (samples a14 to a18); P. maliformis (samples m56 to m58); P. nitida (samples n65 to n67); P. quadrangularis (samples q74 and q75) and P. setacea (samples s85 to s88); b Plot of K vs Delta K values to define the most probable number of clusters in the analysis of population structure and ancestry of 28 Passiflora accessions without prior assignment to species; c Passiflora accessions were allocated to clusters based on Q values (Q > 0.70) for K = 4. Admixed or intermediate samples identified with an asterisk
Discussion
Most microsatellite markers of P. edulis and other Passiflora species developed so far were obtained by sequencing of genomic libraries enriched with simple sequence repeat regions [17–19, 23, 30–32]. There are only ~200 microsatellite di-nucleotide markers available for P. edulis [17, 19, 23]. Here we describe the efficient use of NGS to obtain a large amount of sequence data and applied bioinformatics tools to develop a novel sample of 816 microsatellite markers for this species. The lack of a significant set of polymorphic microsatellite markers for P. edulis and the majority of the Passiflora species was one of the main justifications of the present study. Microsatellite marker technology is used routinely in many genetic and breeding applications in different organisms, but it has had very limited use in passion fruit research. Other marker technologies, such as Single Nucleotide Polymorphism (SNP), have recently become accessible to several plant species and should soon be also available for sour passion fruit.
It has been observed that most microsatellite markers developed for P. edulis usually detect low polymorphism, estimated as varying from 15% [19] to 24.7% [23]. These results have been interpreted as evidence that genetic diversity in P. edulis is low [18, 19], contrasting with the high morphological [7] and agronomic diversity [8, 10] observed in this species. In order to verify how polymorphic is the new set of microsatellite markers, we tested a random sample of 60 new markers on ten accessions of P. edulis collected in different regions of Brazil and estimated genetic parameters such as Ho, PIC and number of alleles. Approximately 74% of the di- and tri-nucleotide markers with amplicon products were polymorphic, and PIC, Ho and allele number were high. PIC values for 80.9% (38/47) of the di-nucleotides markers ranged from 0.26 to 0.77, and for 40% (4/10) of the tri-nucleotides markers from 0.30 to 0.50. Using DNA fingerprinting based on only two markers (BrPe0028 and BrPe0032), one could discriminate all P. edulis accessions used in the present study. These estimates are similar to values found for other allogamous species where NGS technology was used for microsatellite development, such as the forage Brachiaria ruziziensis [39] or radish Raphanus sativus [58]. Therefore, we did not find evidence of low microsatellite polymorphism in P. edulis as assessed by the new set of microsatellite markers. Quite contrary, the majority of the markers tested were highly polymorphic. It is possible that the low polymorphism in P. edulis assessed by previous studies with microsatellite markers was actually caused by hidden genetic relatedness of passion fruit samples used in the screening, or simply because the markers tested were located in more conserved regions of the sour passion fruit genome.
Perfect microsatellite markers represent only a small fraction (~10%) of the total number of P. edulis markers available so far. The vast majority are compound or imperfect motif markers, which are hard to interpret on routine genotyping assays due to allele binning difficulties [26, 27]. Also, most of the studies with P. edulis microsatellite markers were based on allelic discrimination in agarose gels [59, 60] or polyacrylamide gels [16, 17, 19, 61, 62], what added more challenge to the analysis of compound and imperfect microsatellite markers. This could be a constraint to some applications, especially for population genetic studies [28]. All new markers are based on repeat of the same nucleotide motif without interruption or variation, what should facilitate genetic analysis.
We tested the new set of P. edulis microsatellite markers on other 78 Passiflora species. The percentage of cross-species transferability to other species of the subgenus Passiflora was high (75.4%), similar to Distephana (71.11%). However, it decreased to species belonging to Astrophea (63.33) and Decaloba (59.72%). Oliveira et al. [24] obtained similar results for cross-species transferability of P. edulis microsatellite markers to subgenera Passiflora (>73%) and Decaloba (54%). It is interesting to notice that P. edulis PCR products were obtained for at least 50% of the tested markers in all 90 accessions of other Passiflora species, with the exception of P. pohlii (Decaloba) and P. sclerophylla (Astrophea). This is an indication that a substantial proportion of the new P. edulis microsatellite markers can potentially be used in genetic studies of a great range of Passiflora species.
A combined analysis of 28 germplasm accessions of six Passiflora species (P. edulis, P. alata, P. maliformis, P. nitida, P. quadrangularis and P. setacea) using the new microsatellite markers demonstrated their efficiency to uncover genetic diversity in passion fruit. P. edulis accessions formed two clusters that could be easily separated from the accessions of the other Passiflora species. These P. edulis accessions were obtained in different regions of Brazil but there was no correlation between genetic clustering and geographic origin (data not shown). One of the clusters, however, is comprised of sour passion fruit accessions (Table 1, accessions 1–3 and 8) that have been widely used commercially (ex. accessions Maguary, CPGA1 and CPMSC1) and possibly derived from population of common ancestry [23]. Accession 8 (Criciúma, Santa Catarina) was originally collected in area close to Passiflora orchards, and its fruits might have been derived from cross-pollination with commercial cultivars. Although classified as P. edulis, the accession BRS Maracujá Jaboticaba (Table 1, accession 11) did not cluster with accessions of the two P. edulis groups. BRS Maracujá Jaboticaba seems to be closely associated to a third cluster formed by P. setacea accessions (Fig. 3a), although the estimated probability of inclusion in this group was not high (Q value = 0.52) (Fig. 3c). Recent analysis of the BRS Maracujá Jaboticaba mating system indicates that this accession is preferentially autogamous, while most P. edulis accessions are allogamous [63], what could explain its genetic distance to other sour passion fruit accessions. Further analysis on the role of different mating systems and mating plasticity in P. edulis genetic diversity should be pursued.
The fourth cluster included accessions of P. nitida, P. quadrangularis, P. alata and P. maliformis. Molecular phylogeny analysis of Passiflora species using nrITS, trnL-trnF and rps4 polymorphism grouped P. alata, P. quadrangularis, P. maliformis, P. setacea and P. edulis [64]. Plastid DNA analysis also found that P. alata, P. nitida, P. edulis and P. maliformis are closely related [65]. Paiva et al. [60] using microsatellites markers of Oliveira [23] and Pádua et al. [31] identified molecular similarity among P. edulis and P. setacea. Passiflora phylogeny is indeed very complex, with more than 520 species distributed in several continents. Microsatellite markers might help to understand genetic relationships within species and among accessions of closely related species.
Anthropic pressure at the centers of diversity is contributing to genetic erosion of many plant species, including Passiflora [66–68]. Intensive in situ conservation of native flora as well as efforts to collect wild species, landraces and local varieties for ex situ conservation are necessary for current and future use of passion fruit. Short term seed viability remains an important constraint to conservation [69, 70] and most collections rely on vegetative propagation for storage. It is a challenge to keep large numbers of passion fruit accessions by vegetative propagation of germplasm collections with usually restricted human and economic resources. Since vegetative propagation is the main form of conservation, each accession of passion fruit is usually comprised of one or a few plants per species or variety, imposing limits to ex situ genetic diversity storage. Routine activities of germplasm conservation and breeding demand the application of genome technology, including microsatellite markers, in conservation and use of passion fruit genetic resources.
Conclusion
NGS technology was used to obtain a large amount of sequence data, which was applied to the development of hundreds of microsatellite markers for P. edulis. The new markers detected high levels of DNA polymorphism in P. edulis and could be used to assess genetic diversity in sour passion fruit accessions and in closely related species. The levels of cross-species transferability varied from 33% to 89% after testing 78 Passiflora species belonging to four subgenera (Passiflora, Distephana, Astrophea and Decaloba), indicating that a great number of P. edulis microsatellite markers could be potentially used in genetic analysis of other Passiflora species. This new set of microsatellite markers has many applications to germplasm conservation, breeding programs and genetic studies of passion fruit.
Acknowledgements
We would like to thank CAPES for a partial scholarship to SA. This research was also supported by CNPq, Rede Passitec, Embrapa Cerrados and Embrapa Genetic Resources and Biotechnology.
Funding
The design of the study, collection, analysis, and interpretation of data and manuscript writing were sponsored by Embrapa Macroprograma 2 (Grant # 02.12.02.006.00.03) and Rede Passitec II (CNPq 404,847/2012–9). A partial Ph.D. scholarship was provided by CAPES to SA.
Availability of data and materials
Information about contigs, repeat motifs, primer sequences, melting temperatures and expected product sizes for 816 di-, tri- and tetra-nucleotide microsatellite markers is available on Additional file 1.
Abbreviations
- AFLP
Amplified Fragment Length Polymorphism
- BLAST
Basic Local Alignment Search Tool
- CTAB
Cetyl Trimethylammonium Bromide
- DNA
Deoxyribonucleic Acid
- He
Expected Heterozigosity
- Ho
Observed Heterozigosity
- ISSR
Inter Simple Sequence Repeat
- MCMC
Markov Chain Monte Carlo
- NGS
Next-Generation Sequencing
- nrITS
Nuclear Ribosomal Internal Transcribed Spacer
- PCoA
Principal Coordinate Analysis
- PCR
Polymerase Chain Reaction
- PIC
Polymorphic Information Content
- RAPD
Random Amplified Polymorphic DNA
- SNP
Single Nucleotide Polymorphism
- SOAP
Short Oligonucleotide Alignment Program
Additional file
Contig sequence information, repeat motifs, primer sequences, melting temperatures and expected product sizes of 816 di-, tri- and tetra-nucleotide microsatellite markers developed for the sour passion fruit (Passiflora edulis L.). (XLSX 244 kb)
Authors’ contributions
SA worked on DNA extraction, primer design, multiplex panel development, marker genotyping, statistical analyses and drafted the manuscript. AMM prepared genomic libraries, worked on genome assembly and detection of microsatellite sequences. AMC helped with the discussion. NTJ helped to identify and collect all the accessions. FGF supervised the study and helped with the discussion. MEF conceived the study, supervised the laboratory work, performed statistical analyses and drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study has been conducted in accordance with the Brazilian legislation (Law 13.123–5/20/2015) and the “Convention on the Trade in Endangered Species of Wild Fauna and Flora”. All plant accessions used in the study are deposited at and maintained by the “Flor da Paixão” Passion Fruit Germplasm Bank, Embrapa Cerrados (CPAC), BR 020 Km 18, Planaltina, DF, 73,310–970, Brazil. The Passion Fruit Germplasm Bank was established by resolution n° 041/2011/SECEX/CGEN, Process n° 02000.001723/2010–31, Genetic Heritage Management Council (CGEN). The authorizations for conducting scientific research and bio-prospection were provided by the Brazilian Institute of the Environment and Renewable Natural Resources (Special Authorization n° 002/2008, Process N° 02001.008153/2009–67) and CGEN (Special Authorization n° 001- B/2013, Process n° 02000.001704/2009–71). Voucher specimens of the plant accessions used in the present work have been deposited at the Embrapa Genetic Resources and Biotechnology Herbarium, Brasilia, DF, Brazil. The experimental research described here comply with institutional, national, and international guidelines. Seeds and cuttings can be available by request.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-017-3881-5) contains supplementary material, which is available to authorized users.
Contributor Information
Susan Araya, Email: susan_araya@hotmail.com.
Alexandre M Martins, Email: mm.alexandre@gmail.com.
Nilton T V Junqueira, Email: nilton.junqueira@embrapa.br.
Ana Maria Costa, Email: ana-maria.costa@embrapa.br.
Fábio G Faleiro, Email: fabio.faleiro@embrapa.br.
Márcio E Ferreira, Email: marcio.ferreira@embrapa.br, Email: marcio.ferreira@ars.usda.gov.
References
- 1.MacDougal J, Feuillet C. Systematics. In: Ulmer T, MacDougal J, editors. Passiflora: passionflowers of the world. Portland, OR: Timber Press; 2004. pp. 27–31. [Google Scholar]
- 2.Cerqueira-Silva CB, Jesus ON, Santos ESL, Corrêa RX, Souza AP. Genetic breeding and diversity of the genus Passiflora: progress and perspectives in molecular and genetic studies. Int J Mol Sci. 2014;15:14122–14152. doi: 10.3390/ijms150814122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferreira FR, Oliveira JC. Germoplasma de Passiflora no Brasil. In: São José AR, editor. A cultura do maracujá no Brasil. Jaboticabal: FUNEP; 1991. pp. 187–200. [Google Scholar]
- 4.Ocampo J, D’Eeckenbrugge G, Jarvis A. Distribution of the genus Passiflora L. diversity in Colombia and its potential as an indicator for biodiversity management on the coffee growing zone. Diversity. 2010;2:1158–1180. doi: 10.3390/d2111158. [DOI] [Google Scholar]
- 5.Plotze R d O, Falvo M, Pádua JG, Bernacci LC, MLC V, GCX O, et al. Leaf shape analysis using the multiscale Minkowski fractal dimension, a new morphometric method: a study with Passiflora (Passifloraceae) Can J Bot. 2005;83:287–301. doi: 10.1139/b05-002. [DOI] [Google Scholar]
- 6.Viana AJC, Souza MM, Araújo IS, Corrêa RX, Ahnert D. Genetic diversity in Passiflora species determined by morphological and molecular characteristics. Biol Plant. 2010;54:535–538. doi: 10.1007/s10535-010-0094-z. [DOI] [Google Scholar]
- 7.Crochemore ML, Molinari HB, Stenzel NMC. Caracterização agromorfológica do maracujazeiro (Passiflora spp.) Rev Bras Frutic. 2003;25:5–10. doi: 10.1590/S0100-29452003000100004. [DOI] [Google Scholar]
- 8.Cerqueira-Silva CBM, Moreira CN, Figueira AR, Corrêa RX, Oliveira AC. Detection of a resistance gradient to passion fruit woodiness virus and selection of “ yellow ” passion fruit plants under field conditions. Genet Mol Res. 2008;7:1209–1216. doi: 10.4238/vol7-4gmr484. [DOI] [PubMed] [Google Scholar]
- 9.Abreu S d PM, Peixoto JR, Vilela NT, De Figueiredo MA. Características agronômicas de seis genótipos de maracujazeiro-azedo cultivados no Distrito Federal. Rev Bras Frutic. 2009;31:920–924. doi: 10.1590/S0100-29452009000300042. [DOI] [Google Scholar]
- 10.Meletti LMM, Soares-Scott MD, Bernacci LC. Caracterização fenotípica de três seleções de maracujazeiro-roxo (Passiflora edulis Sims) Rev Bras Frutic. 2005;27:268–272. doi: 10.1590/S0100-29452005000200020. [DOI] [Google Scholar]
- 11.Santos LF, Oliveira EJ, Santos Silva A, Carvalho FM, Costa JL, Pádua JG. ISSR markers as a tool for the assessment of genetic diversity in Passiflora. Biochem Genet. 2011;49:540–554. doi: 10.1007/s10528-011-9429-5. [DOI] [PubMed] [Google Scholar]
- 12.Fajardo D, Angel F, Grum M, Tohme J, Lobo M, Roca WM. Genetic variation analysis of the genus Passiflora L. using RAPD markers. Euphytica. 1998;101:341–347. doi: 10.1023/A:1018395229851. [DOI] [Google Scholar]
- 13.Bellon G, Faleiro FG, Junqueira KP, Junqueira NTV, Santos EC, Braga MF, et al. Variabilidade genética de acessos silvestres e comerciais de Passiflora edulis Sims. com base em marcadores RAPD. Rev Bras Frutic. 2007;29:124–127. doi: 10.1590/S0100-29452007000100027. [DOI] [Google Scholar]
- 14.Crochemore ML, Molinari HBC, Vieira LGE. Genetic diversity in passion fruit (Passiflora spp.) evaluated by RAPD markers. Braz Arch Biol Technol. 2003;46:521–527. doi: 10.1590/S1516-89132003000400005. [DOI] [Google Scholar]
- 15.Segura S, D’Eeckenbrugge G, Bohorquez A, Ollitrault P, Tohme J. An AFLP diversity study of the genus Passiflora focusing on subgenus Tacsonia. Genet Resour Crop Evol. 2002;0:1–0. [Google Scholar]
- 16.Ortiz DC, Bohórquez A, Duque MC, Tohme J, Cuéllar D, Mosquera VT. Evaluating purple passion fruit (Passiflora edulis Sims F. edulis) genetic variability in individuals from commercial plantations in Colombia. Genet Resour Crop Evol. 2012;59:1089–1099. doi: 10.1007/s10722-011-9745-y. [DOI] [Google Scholar]
- 17.Oliveira EJ, Pádua JG, Zucchi MI, Camargo LE a, MHP F, MLC V. Development and characterization of microsatellite markers from the yellow passion fruit (Passiflora edulis f. flavicarpa) Mol Ecol Notes. 2005;5:331–333. doi: 10.1111/j.1471-8286.2005.00917.x. [DOI] [Google Scholar]
- 18.Cerqueira-Silva CB, Santos ES. L, Souza AM, Mori GM, Oliveira EJ, Corrêa RX, et al. development and characterization of microsatellite markers for the wild south American Passiflora cincinnata (Passifloraceae) Am J Bot. 2012;99:e170–e172. doi: 10.3732/ajb.1100477. [DOI] [PubMed] [Google Scholar]
- 19.Cerqueira-Silva CB, Santos ESL, Vieira JGP, Mori GM, Jesus ON, Corrêa RX, et al. New microsatellite markers for wild and commercial species of Passiflora (Passifloraceae) and cross-amplification. Appl Plant Sci. 2014;2:1300061. doi: 10.3732/apps.1300061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powell W, Morgante M, Andre C, Hanafey M, Vogel S, Tingey S, et al. The comparision of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed. 1996;13:391–393. [Google Scholar]
- 21.Brondani RPV, Brondani C, Tarchini R, Grattapaglia D. Development, characterization and mapping of microsatellite markers in Eucalyptus grandis and E. urophylla. TAG Theor Appl Genet. 1998;97:816–827. doi: 10.1007/s001220050961. [DOI] [Google Scholar]
- 22.Litt M, Luty JA. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Hum Genet. 1989;44:397–401. [PMC free article] [PubMed] [Google Scholar]
- 23.Oliveira EJ. Desenvolvimento e uso de marcadores microssatélites para construção e integração de mapas genéticos de maracujá-amarelo (Passiflora edulis Sims f . flavicarpa Deg.). Universidade de São Paulo; 2006.
- 24.Oliveira GAF, Pádua JG, Costa JL, Jesus ON, Carvalho FM, Oliveira EJ. Cross-species amplification of microsatellite loci developed for Passiflora edulis Sims. In related Passiflora species. Braz Arch Biol Technol. 2013;56:785–792. doi: 10.1590/S1516-89132013000500009. [DOI] [Google Scholar]
- 25.Reis RV, Oliveira EJ, Viana AP, Pereira TNS, Pereira MG, Silva MG de M. Diversidade genética em seleção recorrente de maracujazeiro amarelo detectada por marcadores microssatélites. Pesq Agrop Brasileira. 2011;46:51–57. doi: 10.1590/S0100-204X2011000100007. [DOI] [Google Scholar]
- 26.Lim KG, Kwoh CK, Hsu LY, Wirawan A. Review of tandem repeat search tools: a systematic approach to evaluating algorithmic performance. Brief Bioinform. 2013;14:67–81. doi: 10.1093/bib/bbs023. [DOI] [PubMed] [Google Scholar]
- 27.Domaniç NO, Preparata FP. A novel approach to the detection of genomic approximate tandem repeats in the levenshtein metric. J Comput Biol. 2007;14:873–891. doi: 10.1089/cmb.2007.0018. [DOI] [PubMed] [Google Scholar]
- 28.Ma ZQ, Röder M, Sorrells ME. Frequencies and sequence characteristics of di-, tri-, and tetra-nucleotide microsatellites in wheat. Genome. 1996;39:123–130. doi: 10.1139/g96-017. [DOI] [PubMed] [Google Scholar]
- 29.Goldstein DB, Clark AG. Microsatellite variation in north American populations of Drosophila melanogaster. Nucleic Acids Res. 1995;23:3882–3886. doi: 10.1093/nar/23.19.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penha HA, Pereira GDS, Zucchi MI, Diniz AL, Vieira MLC. Development of microsatellite markers in sweet passion fruit, and identification of length and conformation polymorphisms within repeat sequences. Plant Breed. 2013;132:731–735. doi: 10.1111/pbr.12083. [DOI] [Google Scholar]
- 31.Padua JG, Oliveira EJ, Zucchi MI, Oliveira GCX, Camargo LEA, Vieira MLC. Isolation and characterization of microsatellite markers from the sweet passion fruit (Passiflora alata Curtis: Passifloraceae) Mol Ecol Notes. 2005;5:863–865. doi: 10.1111/j.1471-8286.2005.01090.x. [DOI] [Google Scholar]
- 32.Cazé ALR, Kriedt RA, Beheregaray LB, Bonatto SL, Freitas LB. Isolation and characterization of microsatellite markers for Passiflora contracta. Int J Molec Sci. 2012;13:11343–11348. doi: 10.3390/ijms130911343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karagyozov L, Kalcheva ID, Chapman VM. Construction of random small-insert genomic libraries highly enriched for simple sequence repeats. Nucleic Acids Res. 1993;21:3911–3912. doi: 10.1093/nar/21.16.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paetkau D. Microsatellites obtained using strand extension: an enrichment protocol. BioTechniques. 1999;26:690–697. doi: 10.2144/99264st05. [DOI] [PubMed] [Google Scholar]
- 35.Morgante M, Olivieri AM. PCR-amplified microsatellites as markers in plant genetics. Plant J. 1993;3:175–182. doi: 10.1111/j.1365-313X.1993.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 36.Abdelkrim J, Robertson BC, Stanton J-A, Gemmell N. Fast, cost-effective development of species-specific microsatellite markers by genomic sequencing. BioTechniques. 2009;46:185–192. doi: 10.2144/000113084. [DOI] [PubMed] [Google Scholar]
- 37.Castoe TA, Poole AW. Gu W, Jason de Koning AP, Daza JM, smith EN, et al. rapid identification of thousands of copperhead snake (Agkistrodon contortrix) microsatellite loci from modest amounts of 454 shotgun genome sequence. Mol Ecol Resour. 2010;10:341–347. doi: 10.1111/j.1755-0998.2009.02750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Csencsics D, Brodbeck S, Holderegger R. Cost-effective, species-specific microsatellite development for the endangered dwarf bulrush (Typha minima) using next-generation sequencing technology. J Hered. 2010;101:789–793. doi: 10.1093/jhered/esq069. [DOI] [PubMed] [Google Scholar]
- 39.Silva PI, Martins AM, Gouvea EG, Pessoa-Filho M, Ferreira ME. Development and validation of microsatellite markers for Brachiaria ruziziensis obtained by partial genome assembly of Illumina single-end reads. BMC Genomics. 2013;14:17. doi: 10.1186/1471-2164-14-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doyle J, Doyle JL. Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 41.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 42.Salmela L, Mäkinen V, Välimäki N, Ylinen J, Ukkonen E. Fast scaffolding with small independent mixed integer programs. Bioinformatics. 2011;27:3259–3265. doi: 10.1093/bioinformatics/btr562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayer C. Phobos 3.3.11. 2006-2010.
- 45.Guigó R, Knudsen S, Drake N, Smith T. Prediction of gene structure. J Mol Biol. 1992;226:141–157. doi: 10.1016/0022-2836(92)90130-C. [DOI] [PubMed] [Google Scholar]
- 46.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM. Primer3Plus, an enchanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fonseca-Trujillo N, Márquez-Cardona MP, Moreno-Osorio JH, Terán-Pérez W, Schuler-García I. Caracterización molecular de materiales cultivados de gulupa (Passiflora edulis f . edulis) Univ Sci. 2009;14:135–140. doi: 10.11144/javeriana.SC14-2-3.cmdm. [DOI] [Google Scholar]
- 48.Rendón JS, Ocampo J, Urrea R. Estudio sobre polinización y biología floral en Passiflora edulis F. edulis Sims, como base para el premejoramiento genético. Acta Agronóm. 2013;62:232–241. [Google Scholar]
- 49.Marshall TC, Slate J, Kruuk LEB, Pemberton JM. Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- 50.Lynch M. The similarity index and DNA fingerprinting. Mol Biol Evol. 1990;7:478–484. doi: 10.1093/oxfordjournals.molbev.a040620. [DOI] [PubMed] [Google Scholar]
- 51.Rohlf F. Numerical taxonomy and multivariate analysis system (NTSYS-pc) New York: Departament of Ecology and Evolution; 1990. [Google Scholar]
- 52.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pritchard JK, Wen X, Falush D. Documentation for structure software: version 2.3. Chicago, IL: University of Chicago; 2010. [Google Scholar]
- 54.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 55.Holleley CE, Geerts PG. Multiplex manager 1.0: a cross-platform computer program that plans and optimizes multiplex PCR. BioTechniques. 2009;46:511–517. doi: 10.2144/000113156. [DOI] [PubMed] [Google Scholar]
- 56.Matschiner M, Salzburger W. TANDEM: integrating automated allele binning into genetics and genomics workflows. Bioinformatics. 2009;25:1982–1983. doi: 10.1093/bioinformatics/btp303. [DOI] [PubMed] [Google Scholar]
- 57.Souza MM, Palomino G, Pereira TNS, Pereira MG, Viana AP. Flow cytometric analysis of genome size variation in some Passiflora species. Hereditas. 2004;141:31–38. doi: 10.1111/j.1601-5223.2004.01739.x. [DOI] [PubMed] [Google Scholar]
- 58.Zhai L, Xu L, Wang Y, Cheng H, Chen Y, Gong Y, et al. Novel and useful genic-SSR markers from de novo transcriptome sequencing of radish (Raphanus sativus L.) Mol Breed. 2014;33:611–624. doi: 10.1007/s11032-013-9978-x. [DOI] [Google Scholar]
- 59.Castro J. Conservação dos recursos genéticos de Passiflora e seleção de descritores mínimos para caracterização de maracujazeiro: Universidade Federal do Recôncavo da Bahia; 2012.
- 60.Paiva C, Pio Viana A, Azevedo Santos E, de Oliveira Freitas JC, Oliveira Silva RN, de Oliveira EJ. Genetic variability assessment in the genus Passiflora by SSR marker. Chilean J Agric Res. 2014;74:355–360. doi: 10.4067/S0718-58392014000300015. [DOI] [Google Scholar]
- 61.Oliveira EJ, Vieira MLC, Garcia AAF, Munhoz CF, Margarido GRA, Consoli L, et al. An integrated molecular map of yellow passion fruit based on simultaneous maximum-likelihood estimation of linkage and linkage phases. J Am Soc Hortic Sci. 2008;133:35–41. [Google Scholar]
- 62.Cerqueira-Silva CBM, Jesus ON, Oliveira EJ, Santos ESL, Souza AP. Characterization and selection of passion fruit (yellow and purple) accessions based on molecular markers and disease reactions for use in breeding programs. Euphytica. 2015;202:345–359. doi: 10.1007/s10681-014-1235-9. [DOI] [Google Scholar]
- 63.Araya S. Desenvolvimento, validação e aplicação de marcadores microssatélite em estudos genéticos de Passifloras: Universidade de Brasília; 2016.
- 64.Muschner VC, Lorenz AP, Cervi AC, Bonatto SL, Souza-Chies TT, Salzano FM, et al. A first molecular phylogenetic analysis of Passiflora (Passifloraceae) Am J Bot. 2003;90:1229–1238. doi: 10.3732/ajb.90.8.1229. [DOI] [PubMed] [Google Scholar]
- 65.Hansen AK, Gilbert LE, Simpson BB, Downie SR, Cervi AC, Jansen RK. Phylogenetic relationships and chromosome number evolution in Passiflora. Syst Bot. 2006;31:138–150. doi: 10.1600/036364406775971769. [DOI] [Google Scholar]
- 66.Ferreira F. Recursos genéticos de Passiflora. In: Faleiro FG, Junqueira NTV, Braga MF, editors. Maracujá: germoplasma e melhoramento genético. Planaltina, DF: Embrapa Cerrados; 2005. pp. 41–52. [Google Scholar]
- 67.Myers N, Mittermeier RA, Mittermeier CG, Da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 68.Rodrigues RR, Lima RAF, Gandolfi S, Nave AG. On the restoration of high diversity forests: 30 years of experience in the Brazilian Atlantic Forest. Biol Conserv. 2009;142:1242–1251. doi: 10.1016/j.biocon.2008.12.008. [DOI] [Google Scholar]
- 69.Khurana E, Singh JS. Ecology of seed and seedling growth for conservation and restoration of tropical dry forest : a review. Environ Conserv. 2001;28:39–52. doi: 10.1017/S0376892901000042. [DOI] [Google Scholar]
- 70.Dobson AP, Bradshaw AD, Baker AJM. Hopes for the future: restoration ecology and conservation biology. Scand J Stat. 1997;277:515–522. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Information about contigs, repeat motifs, primer sequences, melting temperatures and expected product sizes for 816 di-, tri- and tetra-nucleotide microsatellite markers is available on Additional file 1.


