ABSTRACT
Bordetella bronchiseptica is pervasive in swine populations and plays multiple roles in respiratory disease. Additionally, B. bronchiseptica is capable of establishing long-term or chronic infections in swine. Bacterial biofilms are increasingly recognized as important contributors to chronic bacterial infections. Recently the polysaccharide locus bpsABCD has been demonstrated to serve a critical role in the development of mature biofilms formed by the sequenced laboratory strain of B. bronchiseptica. We hypothesized that swine isolates would also have the ability to form mature biofilms and the bpsABCD locus would serve a key role in this process. A mutant containing an in-frame deletion of the bpsABCD structural genes was constructed in a wild-type swine isolate and found to be negative for poly-N-acetylglucosamine (PNAG)-like material by immunoblot assay. Further, the bpsABCD locus was found to be required for the development and maintenance of the three-dimensional structures under continuous-flow conditions. To investigate the contribution of the bpsABCD locus to the pathogenesis of B. bronchiseptica in swine, the KM22Δbps mutant was compared to the wild-type swine isolate for the ability to colonize and cause disease in pigs. The bpsABCD locus was found to not be required for persistence in the upper respiratory tract of swine. Additionally, the bpsABCD locus did not affect the development of anti-Bordetella humoral immunity, did not contribute to disease severity, and did not mediate protection from complement-mediated killing. However, the bpsABCD locus was found to enhance survival in the lower respiratory tract of swine.
KEYWORDS: Bordetella, biofilms
INTRODUCTION
Bordetella bronchiseptica is a Gram-negative bacterium closely related to Bordetella pertussis and Bordetella parapertussis with a broad host range that naturally infects a wide variety of wild, domestic, and companion animals. In swine, B. bronchiseptica is widespread and is an important contributor to respiratory disease. In young pigs, it is a primary cause of bronchopneumonia, and in older pigs, it contributes to secondary pneumonia (1–3). It is the primary etiologic agent of nonprogressive atrophic rhinitis, a mild to moderately severe reversible condition, and it promotes colonization by toxigenic strains of Pasteurella multocida, which leads to severe progressive atrophic rhinitis (4, 5). Numerous studies have demonstrated that coinfection with B. bronchiseptica increases colonization and exacerbates the severity of disease caused by both viral and bacterial pathogens, including swine influenza virus (SIV), porcine reproductive and respiratory syndrome virus (PRRSV), porcine respiratory coronavirus (PRCV), Haemophilus parasuis, Pasteurella multocida, and Streptococcus suis (6–13).
A universal trait of all B. bronchiseptica infections is that they result in long-term to lifelong carriage (14–19). This holds true despite the use of vaccines, as B. bronchiseptica is frequently isolated from the nasal cavities of vaccinated animals, suggesting that vaccines fail to protect animals from colonization (20, 21). More importantly, vaccinated animals then serve as asymptomatic carriers that continue to shed and transmit B. bronchiseptica to cohorts (22–27). A major barrier impeding the development of improved vaccines and intervention strategies is the gap in our understanding of the mechanisms and the identity and function of gene products contributing to chronic asymptomatic carriage of B. bronchiseptica in the respiratory tract.
Bacterial biofilms are increasingly recognized as important contributors to chronic or persistent infections (28–31). Biofilms are defined as an adherent community of microorganisms encased within a complex matrix that protects the encased community from a variety of environmental stresses, such as shear flow forces, antimicrobial compounds, and host immune and clearance mechanisms (28–35). Recent studies have demonstrated that both B. pertussis and B. bronchiseptica are capable of forming biofilms on abiotic surfaces (36–41) and in the mouse respiratory tract (37, 42–44).
Bordetella species produce an exopolysaccharide, known as the Bordetella polysaccharide (Bps), which is encoded by the bpsABCD operon (39). Previous studies have demonstrated that Bps is required for Bordetella biofilm formation and persistent colonization of the mouse respiratory tract (42, 43). Thus, while Bps has emerged as a critical factor contributing to the pathogenesis of B. bronchiseptica and B. pertussis in mouse models, no data exist regarding the role of Bps in the pathogenesis of B. bronchiseptica in swine. In this report, we begin investigating factors contributing to biofilm formation and how they impact Bordetella pathogenesis in swine by constructing an in-frame deletion of the bpsABCD genes in B. bronchiseptica strain KM22, a virulent swine isolate, and comparing this mutant to KM22 for its ability to form mature biofilms, colonize, and cause disease in swine.
RESULTS
Biofilm development is a conserved phenotype among B. bronchiseptica strains isolated from multiple animal species.
The laboratory reference strain of B. bronchiseptica, RB50, was originally isolated from a rabbit and has previously been demonstrated to form biofilms (38–40). We hypothesized that if biofilms served a beneficial role in the infectious and ecological success for this species, then this phenotype would be conserved and manifested by strains isolated from animals other than rabbits. To experimentally address this hypothesis, we examined several B. bronchiseptica strains isolated from a variety of animal species and investigated the ability of these strains to form biofilms. Biofilm formation was quantitated by standard microtiter crystal violet (CV) assays (38, 40). RB54, a B. bronchiseptica Bvg− phase-locked derivative of RB50 that has been previously demonstrated to be defective in biofilm formation, was used as a negative control (40) (Fig. 1). All of the B. bronchiseptica strains isolated from animal hosts tested formed robust biofilms (Fig. 1). Further, several B. bronchiseptica strains (M584/99, KM22, OSU054, and SO3287) isolated from seal, pig, turkey, and sea otter hosts exhibited a statistically significant greater capacity to form biofilms than RB50 (Fig. 1). These data strongly support our hypothesis that the ability to form a biofilm is a beneficial and conserved phenotype among B. bronchiseptica isolates and accentuate the need to investigate factors contributing to biofilm development in virulent bacterial isolates. For the rest of the study, we focused on the swine isolate KM22.
FIG 1.
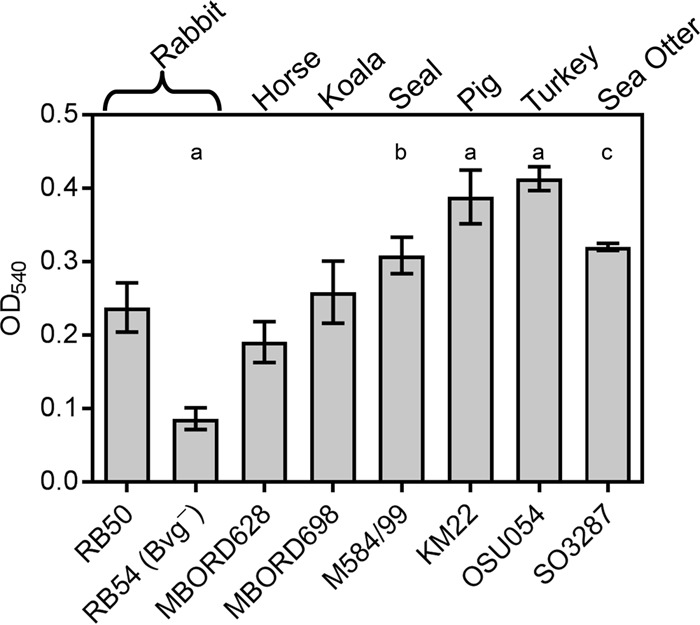
Biofilm-forming capacity of B. bronchiseptica isolates. Strains tested are shown along the x axis, and the respective mammalian host from which each isolate was obtained is provided at the top. The indicated strains were grown statically for 72 h. Biofilm formation was quantified by standard microtiter assays and measuring the absorbance at 540 nm, plotted along the y axis. Bars represent the average of absorbances obtained from 3 independent plates representing biological replicates, each containing triplicate replicates. Error bars represent ± standard deviations (SD). Statistical differences: P = 0.0001 for RB54, KM22, and OSU054 compared to RB50 (a); P = 0.0081 for M584/99 compared to RB50 (b); P = 0.0019 for SO3287 compared to RB50 (c). A P value of <0.05 was considered significant.
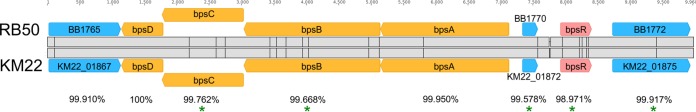
The KM22 bpsABCD locus is highly conserved and similar to RB50 bpsABCD and is required for the production of a polysaccharide antigenically similar to Staphylococcus aureus PNAG.
To initially characterize the KM22 bpsABCD locus, the nucleotide sequence of the region harboring the bpsABCD locus for strain KM22 was compared to the nucleotide sequence of the corresponding region in the laboratory reference strain RB50. The nucleotide region is highly conserved between the strains, with an overall sequence identity of 99.789% (Fig. 2). Focusing on specific genes, the percent identity for bpsA was observed to be 99.95% with 1 single nucleotide polymorphism (SNP), bpsB was 99.668% identical with 7 SNPs, bpsC was 99.762% identical with 3 SNPs, bpsD was 100% identical, and bpsR was 98.971% identical with 5 SNPs (Fig. 2). These SNPs are predicted to result in the following amino acid replacements: W6R and P486L in BpsB, A95T in BpsC, and L8P and D110G in BpsR (Fig. 2). Two nucleotide insertions were identified within the intergenic region between KM22_01872 and bpsR (Fig. 2).
FIG 2.
Sequence alignment of the bps locus in RB50 and KM22. Genes constituting the bpsABCD operon (orange arrows) are labeled, the regulatory gene bpsR (red arrow) is labeled, and genes not associated with the bps locus (blue arrows) are labeled. Percent pairwise identity is shown below each pair of genes. The vertical lines in the gray alignment box indicate the location of nucleotide polymorphisms. Asterisks indicate nucleotide differences present in genes predicted to result in amino acid differences.
It has previously been demonstrated that the bpsABCD locus is required for the production of the Bps polysaccharide that is antigenically and biochemically similar to poly-β-1,6-N-acetylglucosamine (PNAG/PGA) family of polysaccharides by the B. bronchiseptica laboratory reference strain RB50 (39). To determine whether the bpsABCD operon is required for the production of Bps by KM22, cell surface extracts were obtained for B. bronchiseptica strains RB50, KM22, and KM22Δbps, harboring an in-frame deletion of the bpsABCD genes, and used in an immunoblot assay with antisera raised against deacetylated PNAG from S. aureus (39, 45). Both RB50 and KM22 produced a polysaccharide antigenically similar to that of S. aureus PNAG (Fig. 3). In contrast, KM22Δbps exhibited a severe loss in the production of the antibody cross-reactive material (Fig. 3). These data demonstrate that the nucleotide regions containing the bpsABCD loci for KM22 and RB50 are highly conserved and over 99% identical. Further, similar to what is seen with RB50, the bpsABCD locus is required for the production of PNAG by KM22.
FIG 3.
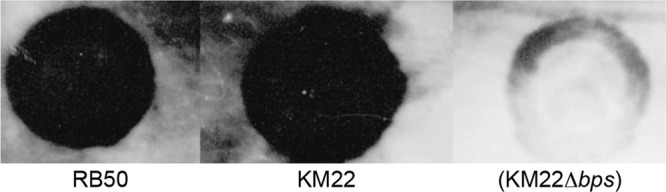
Detection of crude exopolysaccharides by immunoblot analysis. Boiled EDTA surface extracts from the indicated B. bronchiseptica strains were treated with pronase and spotted onto a nitrocellulose membrane followed by detection using an S. aureus anti-dPNAG antibody as described in Materials and Methods.
The bpsABCD locus is required for the maintenance of the 3D structure of B. bronchiseptica KM22 biofilms.
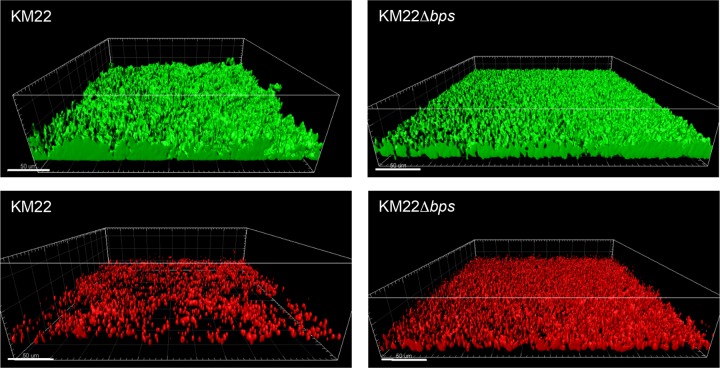
To investigate the role of the bpsABCD locus in the maintenance and architecture of biofilms formed by B. bronchiseptica KM22, structural features of biofilms formed by KM22 or KM22Δbps under continuous-flow conditions were evaluated using a flow cell assay. KM22 formed robust biofilms consisting of large tower-shaped structures (Fig. 4). In contrast, KM22Δbps formed a thin uniform matlike layer of cells on the surface that lacked the large tower-shaped structures observed in KM22 biofilms (Fig. 4). When the biofilms formed by KM22 or KM22Δbps were quantitatively analyzed, no statistical difference in the biovolume or the biomass was observed (Table 1). However, KM22 biofilms exhibited statistically greater maximum thickness (P = 0.0219) and a statistically greater roughness coefficient (P = 0.0145) than did KM22Δbps biofilms (Table 1). Next, we evaluated differences in the amount of dead bacterial cells present in the biofilms formed by KM22 or KM22Δbps. A larger amount of dead cells were observed in the KM22Δbps biofilms than in the KM22 biofilms, and this observation was supported by quantitative measurements in which KM22Δbps biofilms exhibited statistically greater biovolume (P = 0.0145) and biomass (P = 0.0145) than did KM22 biofilms (Table 1). Together, these data demonstrate that the bpsABCD locus is required for the maintenance of the 3-dimensional (3D) structure of B. bronchiseptica KM22 biofilms.
FIG 4.
Flow cell biofilm formation of KM22 and KM22Δbps. B. bronchiseptica strain KM22 or KM22Δbps was inoculated directly into continuous-flow chambers. Bacteria were allowed to attach for 90 min; the flow of SS/Sm medium was initiated at a rate of 0.3 ml/min. Biofilms were allowed to grow for 90 h at 37°C. Mature biofilms were stained with 1 μl/ml Syto 9 (green; indicates live bacterial cells; top images) and 1 μl/ml propidium iodide (red; indicates dead bacterial cells; bottom images). Three independent experiments were performed. Representative volumetric 3D image reconstructions are shown.
TABLE 1.
Quantitative analysis of biofilms formed by B. bronchiseptica KM22 or KM22Δbps grown under continuous-flow conditions
| Parameter (unit) | KM22a | KM22Δbpsa | P value |
|---|---|---|---|
| Biovolume (μm3) | 6.06 × 105 (2.21 × 104) | 5.04 × 105 (6.04 × 103) | 0.0628 |
| Biomass (μm3/μm2) | 12.72 (0.46) | 10.58 (0.13) | 0.0628 |
| Max thickness (μm) | 37.71 (2.11) | 24.05 (0.49) | 0.0219 |
| Roughness coefficient | 4.32 (0.29) | 2.19 (0.08) | 0.0145 |
| Biovolume (dead) (μm3) | 26.07 (1.68) | 56.44 (3.85) | 0.0145 |
| Biomass (dead) (μm3/μm2) | 5.47 × 10−4 (3.52 × 10−5) | 1.18 × 10−3 (8.09 × 10−5) | 0.0145 |
Average values of parameters obtained from three independent experiments. Standard errors are provided in parentheses.
The KM22 bpsABCD locus enhances survival in the lower respiratory tract of swine but is not required for persistence in the upper respiratory tract or disease severity.
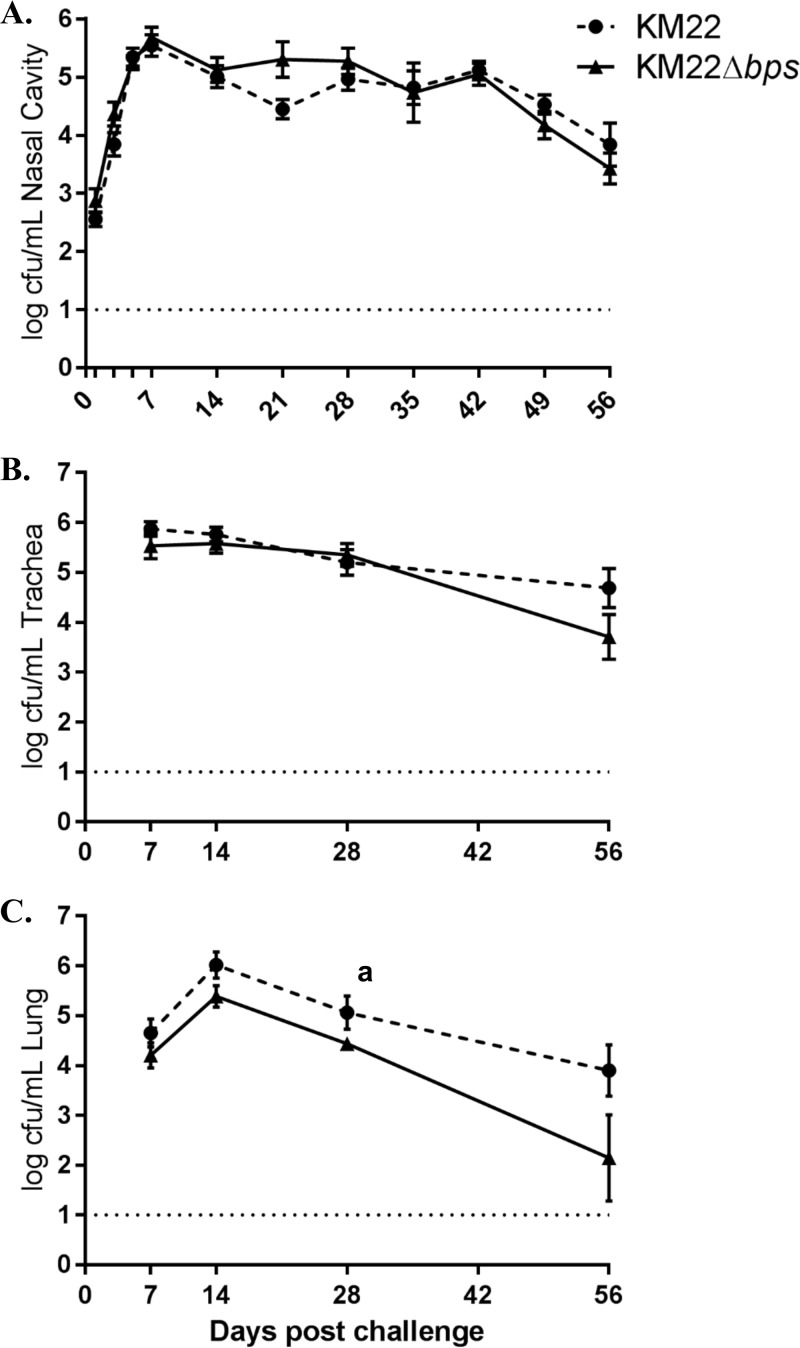
To investigate the contribution of the bpsABCD locus to colonization and clinical disease in swine, groups of 1-week-old piglets were intranasally inoculated with KM22, KM22Δbps, or phosphate-buffered saline (PBS). Colonization of the nasal cavity was evaluated on days 1, 3, 5, and 7 and weekly thereafter until day 56 postinfection (Fig. 5A). No B. bronchiseptica CFU were recovered from any site in the respiratory tracts of piglets mock inoculated with PBS. No significant difference in the number of CFU recovered from the nasal cavity of KM22Δbps-infected pigs compared to KM22-infected pigs was found for any of the time points examined (Fig. 5A). Colonization of sites in the lower respiratory tract was assessed at 7, 14, 28, 42, and 56 days postinfection. No significant difference in the number of CFU recovered from the tracheas of KM22Δbps-infected pigs compared to those of the KM22-infected pigs was found for any of the days examined (Fig. 5B). Overall, significantly lower CFU levels were recovered from the lungs of KM22Δbps-infected pigs than from those infected with the wild-type KM22 for all days examined (P = 0.0075) (Fig. 5C).
FIG 5.
Colonization of the swine respiratory tract by wild-type B. bronchiseptica strain KM22 or KM22Δbps. Groups of 16 pigs were inoculated intranasally with KM22 (circles) or KM22Δbps (triangles). Bacterial load in the nasal cavity (A) was quantified 1, 3, 5, 7, 14, 21, 28, 35, 42, 49, and 56 days postinoculation. Bacterial load in the trachea (B) and the lungs (C) was quantified 7, 14, 28, and 56 days postinoculation. The x axis indicates days postinoculation, and the y axis indicates the mean CFU expressed as the mean log10 ± the standard error (error bars). The dashed line indicates the limit of detection (log10 = 1). Statistical difference for results in panel C: P = 0.0075 for KM22 compared to KM22Δbps for all days examined (a). A P value of <0.05 was considered significant.
Clinical signs were noted only in piglets from groups inoculated with KM22 or the KM22Δbps mutant and consisted of sneezing and coughing. Lung pathology was assessed at 7, 14, 28, 42, and 56 days postinfection by determining the mean percentage of the lung affected by pneumonia. Overall, no pneumonia was observed in PBS-infected piglets, whereas a similar degree of pneumonia was observed in KM22Δbps- and KM22-infected pigs. The pneumonia exhibited was mild, typical of B. bronchiseptica pneumonia, and consisted of areas of red, observed 7 and 14 days postinfection, to tan, seen 42 and 56 days postinfection, consolidation with well-demarcated borders and a cranial ventral distribution (Table 2). Combined, these data demonstrate that the bpsABCD locus enhances survival in the lung or lower respiratory tract of swine; however, it is not required for persistence in the upper respiratory tract or disease severity.
TABLE 2.
Mean percentages of the lungs affected by pneumonia in pigs inoculated with B. bronchiseptica KM22 or KM22Δbps
| Day of necropsy | Group | Mean % (range) affected by pneumonia |
|---|---|---|
| 7 | PBS control | 0 (0) |
| KM22 | 2.025 (0–5.1) | |
| KM22Δbps | 1.363 (0–4) | |
| 14 | PBS control | 0 (0) |
| KM22 | 1 (0–2) | |
| KM22Δbps | 1.5 (0–4) | |
| 28 | PBS control | 0 (0) |
| KM22 | 0.25 (0–1) | |
| KM22Δbps | 0.95 (0–3) | |
| 56 | PBS control | 0 (0) |
| KM22 | 0.875 (0–3.5) | |
| KM22Δbps | 0.375 (01.5) |
The KM22 bpsABCD locus did not the affect the development of anti-Bordetella humoral immunity in swine.
It has been demonstrated that PNAG is expressed during infections and elicits an antibody response (46, 47). Furthermore, both B. bronchiseptica and B. pertussis have been shown to express Bps within the nasal cavities of mice during infections, and sera from B. pertussis-infected individuals are reactive against Bps by immunoblotting, demonstrating that Bps is expressed during infections (42, 43). We therefore tested if the bpsABCD locus contributed to differences in serum anti-Bordetella antibody titers, which could have contributed to the low CFU levels recovered from the lungs of KM22Δbps-infected pigs compared to those infected with the wild-type KM22. To investigate the effect of the bpsABCD locus on the development of anti-Bordetella humoral immunity in swine, serum levels of anti-Bordetella IgG were quantified in pigs infected with KM22 or KM22Δbps, or mock infected with PBS, by enzyme-linked immunosorbent assay (ELISA) using heat-killed KM22 or KM22Δbps whole cells as antigen. The data represent samples collected from pigs at day 56 postchallenge, with four pigs per group. When heat-killed KM22 whole cells were used as antigen, similar serum IgG levels were detected in KM22- and KM22Δbps-infected pigs; however, higher serum anti-Bordetella IgG levels were detected in KM22- and KM22Δbps-infected pigs than in PBS-inoculated pigs (Fig. 6A). Similarly, when heat-killed KM22Δbps whole cells were used as antigen, similar serum IgG levels were detected in KM22- and KM22Δbps-infected pigs and higher serum anti-Bordetella IgG levels were detected in KM22 and KM22Δbps-infected pigs than in PBS-inoculated pigs (Fig. 6B). These data demonstrate that the bpsABCD locus did not affect the development of anti-Bordetella humoral immunity in swine.
FIG 6.
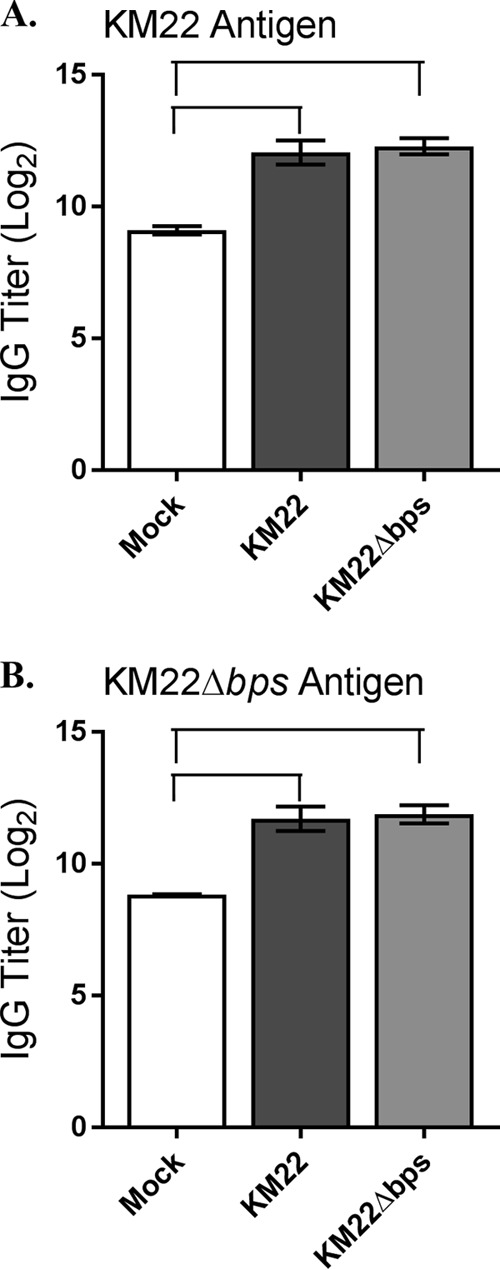
Serum anti-Bordetella IgG titers. Serum was collected 56 days postchallenge from pigs intranasally inoculated with KM22 of KM22Δbps or mock challenged with PBS. Titers of anti-Bordetella antibodies were measured by ELISA using B. bronchiseptica KM22 (A) or KM22Δbps (B) as the antigen. The x axis indicates the treatment group, and the y axis indicates the log2 mean relative titer ± the standard error (error bars). A one-way ANOVA with Tukey's posttest was used for statistical analysis, and P values of <0.05 are indicated with connecting bars.
The KM22 bpsABCD locus does not mediate protection from complement-mediated killing.
It has previously been demonstrated that the bpsABCD locus confers resistance to complement-mediated clearance of B. pertussis in mice as well as provides protection from in vitro complement-mediated killing assays (48). Thus, we hypothesized that by providing protection from in vitro complement-mediated killing, the bpsABCD locus promotes the survival of KM22 in the swine lungs. To test this hypothesis, sera collected on day 56 postinfection from pigs infected with KM22 or KM22Δbps or mock infected with PBS were used in an antibody-complement killing assay along with B. bronchiseptica target strains. Sera from KM22-, KM22Δbps-, or mock-infected pigs induced similar levels of antibody-complement killing of wild-type KM22 and the KM22Δbps mutant (Fig. 7). Taken together, these results show that the bpsABCD locus does not mediate protection from complement-mediated killing of B. bronchiseptica KM22.
FIG 7.
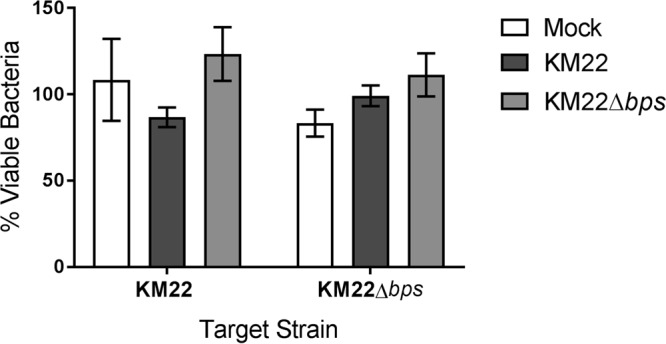
Bacterial killing by immune serum-complement-mediated lysis. Sera collected on day 56 postchallenge from pigs inoculated with KM22, KM22Δbps, or PBS (Mock) were incubated with the indicated B. bronchiseptica target strain (x axis) in the presence of guinea pig serum (complement source) as described in Materials and Methods. After incubation, bacteria were enumerated to determine the percent viable bacteria (y axis) expressed as the means ± standard errors (error bars). A one-way ANOVA with Tukey's posttest was used for statistical analysis, and a P value of 0.05 or less was considered significant.
DISCUSSION
Regardless of vaccination status, B. bronchiseptica infections result in long-term persistent colonization of the host (22–27). Bacterial biofilms are considered substantial contributors to chronic or persistent infections, given that living within a biofilm community protects bacteria from a variety of environmental stresses, such as antimicrobials, as well as host immune and clearance mechanisms (28–35). Several studies have demonstrated that both B. pertussis and B. bronchiseptica are capable of forming biofilms on abiotic surfaces (36–41) and in the mouse respiratory tract (37, 42–44, 49). To examine if other strains of B. bronchiseptica isolated from a diversity of host species could form biofilms in a manner similar to that seen with the laboratory reference strain, in vitro microtiter plate assays were used to quantify biofilm formation produced by a variety of B. bronchiseptica isolates. While all of the B. bronchiseptica isolates examined formed robust biofilms, several isolates exhibited a greater capacity to form biofilms than the reference strain. These data suggest that the ability to form a biofilm is a beneficial and conserved phenotype among B. bronchiseptica isolates.
The bpsABCD locus is required for the production of the Bps polysaccharide by the B. bronchiseptica reference strain RB50 (39). To begin investigating the role of Bps in the pathogenesis of B. bronchiseptica in swine, the nucleotide region harboring bpsABCD from KM22 was compared to that of RB50 and observed to be highly conserved, with an overall sequence identity of 99.789%. An in-frame deletion of the bpsABCD genes was subsequently constructed in the KM22 swine isolate and tested for the production of the Bps polysaccharide. Using an immunoblot assay, we found that deletion of the bpsABCD locus in KM22 resulted in a severe loss in the production of the cross-reactive material, indicating that similar to RB50, the bpsABCD locus is required for the production of this polysaccharide by KM22. Next, the functional role that the bpsABCD locus serves in the architectural and structural features of biofilms formed by KM22 was investigated under continuous-flow conditions. Biofilms formed by wild-type KM22 exhibited statistically greater thickness and roughness than did KM22Δbps biofilms. These structural features are typically quantified when evaluating bacterial biofilms, as they are indicative of towers or pillars and other three-dimensional architectural structures. In addition to more-complex and more-mature three-dimensional structures, biofilms formed by wild-type KM22 contained significantly fewer dead bacterial cells than KM22Δbps biofilms. Combined, these data demonstrate that the bpsABCD locus is required for the development and maintenance of the three-dimensional structure of B. bronchiseptica KM22 biofilms.
To investigate the contribution of the bpsABCD locus to the pathogenesis of B. bronchiseptica in swine, we then compared the KM22Δbps mutant to the wild-type swine isolate for the ability to colonize and cause disease in pigs following intranasal inoculation. Based on previous studies, we hypothesized that the bpsABCD locus would be required for persistent colonization of both the upper and lower respiratory tracts of swine (42, 43). However, colonization of the KM22Δbps mutant in the nasal cavity was similar to that of KM22. These data demonstrate that the bpsABCD locus is not required for persistence in the upper respiratory tract of swine. This finding was unexpected, given that the bpsABCD locus for KM22 and RB50 is highly conserved and a previous study demonstrated that the bpsABCD locus is required for the persistent or long-term colonization of mouse nasal cavities by the B. bronchiseptica reference strain RB50 (43). Additionally, the bpsABCD locus is required for persistent colonization of mouse nasal cavities by B. pertussis (42). Variations in nasal colonization by bps mutants between mice and pigs are likely dependent on either host-specific differences or the requirements of strain-specific gene products. Due to immune and/or competitive-exclusion forces, the factors contributing to KM22 persistently colonizing the upper respiratory tract of swine may be quite different from the factors contributing to B. bronchiseptica and B. pertussis persistently colonizing mice. For example, the microbial communities present in the upper respiratory tract of swine differ from those present in mice and often include bacterial species such as Actinobacillus, Haemophilus, Pasteurella, Mycoplasma, Streptococcus, and Staphylococcus (50–52), all of which are known to form biofilms and many of which are known to secrete biofilm-enhancing factors (53–59). Field isolates of Actinobacillus pleuropneumoniae are known to produce a polysaccharide similar to S. aureus PNAG (60). The possibility that the presence of these bacterial species enhanced and/or aided the KM22Δbps mutant to persistently colonize the nasal cavity of swine cannot be excluded. Focusing on KM22-specific factors other than Bps, the assembly and annotation of the KM22 genome sequence were recently obtained, and after coding sequences in KM22 and RB50 were directly compared, 169 KM22-specific coding sequences were identified (61, 62). KM22-specific genes that could potentially serve a role in colonizing the upper respiratory tract of swine and/or biofilm formation include tracheal colonization factor (tcfA), serum resistance protein (brkA), seven transcriptional regulators, and putative adhesins such as KM22_00109, a pilus assembly protein (KM22_04913), and a fimbrial subunit gene (KM22_03370) (62). Both tcfA and brkA are predicted to be expressed as full-length functional proteins in KM22 and are annotated as pseudogenes in RB50 (62).
Similar to the colonization levels for the RB50Δbps mutant strain in mice (43), the colonization levels of the KM22Δbps mutant in the trachea were similar to those of KM22 for all days examined. When evaluating colonization of the lower respiratory tract, the KM22Δbps mutant was recovered from the lungs at significantly lower levels than the wild type for all days examined. The decreased bacterial burden of the KM22Δbps mutant observed early in the infectious process is different from that reported for the corresponding RB50Δbps mutant in mice, where RB50 and the RB50Δbps mutant were recovered from the lungs of mice at similar levels at 15 days postchallenge, followed by a decreased bacterial burden of the RB50Δbps mutant at 38 days postchallenge. The decreased bacterial burden of the KM22Δbps mutant also differs from lung colonization reported for a B. pertussis Δbps mutant in mice, in a study where a B. pertussis Δbps mutant was recovered at lower levels than wild-type B. pertussis at early time points, days 1 to 6 postchallenge; however, by day 7, similar bacterial loads of wild-type B. pertussis and the B. pertussis Δbps mutant were recovered from the lungs of mice (42).
Clinical signs and the degree of pneumonia observed in KM22Δbps-infected pigs were indistinguishable from those of KM22-infected pigs. Additionally, similar serum anti-Bordetella IgG levels were detected in KM22- and KM22Δbps-infected pigs. Combined, these data demonstrate that the bpsABCD locus does not affect the development of anti-Bordetella humoral immunity and does not contribute to B. bronchiseptica-associated pathology and disease severity in swine. However, the bpsABCD locus is required for survival rates equivalent to wild-type levels in the lung or lower respiratory tract of swine.
A previous study from our laboratory demonstrated that the bpsABCD locus conferred resistance to complement-mediated clearance of B. pertussis in vivo as well as provided protection from in vitro complement-mediated killing assays (48). The same study went on to demonstrate that ectopic expression of bpsABCD genes in Escherichia coli provided resistance to complement-mediated killing and complement deposition (48). Given that significantly fewer CFU were recovered from the lungs of pigs infected with the KM22Δbps over all the time points examined, we hypothesized that the bpsABCD locus could provide protection from in vitro complement-mediated killing of B. bronchiseptica KM22 and therefore confer resistance to complement-mediated clearance from the lower respiratory tract. However, sera from KM22-, KM22Δbps-, or mock-infected pigs exhibited similar levels of antibody-complement killing of both the wild-type KM22 and the KM22Δbps mutant, demonstrating that the bpsABCD locus does not mediate protection from complement-mediated killing of B. bronchiseptica KM22. As previously mentioned, KM22 harbors a brkA gene that is predicted to be expressed as a full-length functional protein, whereas the brkA gene in RB50 is annotated as a pseudogene (62). For B. pertussis, BrkA has been shown to confer resistance to antibody-mediated complement lysis (63, 64). Similar to RB50, KM22 harbors the genes encoding lipopolysaccharide (LPS) and an O-antigen (62). The products of some of these genes, for example, PagP, have been shown to confer resistance to antibody-mediated complement lysis (65). It is possible that one of these factors and/or other polysaccharides produced from KM22 and the KM22Δbps mutant helped to confer protection from complement-mediated killing such that an increased sensitivity to antibody-mediated complement lysis could be ascertained using the in vitro assays performed.
A common outcome of B. bronchiseptica infections is long-term carriage. The data presented in this report reveal strain-specific differences in Bordetella respiratory tract colonization from different animal species. These findings highlight the importance of studying bacterial strains isolated from different animal species and the use of different animal models for the development of improved vaccines and intervention strategies to mitigate chronic carriage of B. bronchiseptica within the respiratory tract. Further, given that PNAG is a conserved surface/capsular polysaccharide expressed by a large number of bacterial, fungal, and eukaryotic pathogens, it has become a target for vaccine development (46, 47, 66). As previously mentioned, the upper respiratory tract of swine is typically colonized by bacterial species that are known to form biofilms such as Actinobacillus, Haemophilus, Pasteurella, Mycoplasma, Streptococcus, and Staphylococcus (50–52) and produce a polysaccharide similar to S. aureus PNAG (Fig. 2) (60). Vaccination strategies targeting PNAG could significantly benefit the swine industry by reducing the burden of respiratory infections without relying on the use of antimicrobials.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. bronchiseptica strain KM22 is a virulent swine isolate (61, 62), and strain KM22Δbps is a bpsABCD in-frame deletion mutant of KM22. B. bronchiseptica strains RB50 and RB54 (a Bvg− phase-locked derivative of RB50) have been previously described (67). B. bronchiseptica strain MBORD628 was isolated from a horse (68), strain M584/99 was isolated from a seal, strain OSU054 was isolated from a turkey (68), and strain SO3287 was isolated from a sea otter. All B. bronchiseptica strains were maintained on Bordet-Gengou (BG) agar (Difco, Sparks, MD) supplemented with 10% sheep's blood. Liquid-cultured bacteria were grown at 37°C to mid-log phase in Stainer-Scholte (SS) medium. E. coli One Shot TOP10 (Invitrogen, Carlsbad, CA) was used for all cloning steps, and E. coli SM10λpir was used to mobilize plasmids into B. bronchiseptica KM22. When appropriate, antibiotics were included at the following concentrations: carbenicillin, 100 μg/ml; streptomycin, 40 μg/ml.
Genome analysis.
Genomic sequences were obtained from GenBank for B. bronchiseptica RB50 (NC_002927) and KM22 (JNHR00000000), and nucleotide sequence comparisons were performed using Geneious Pro 8.1.5 (Biomatters Inc., Auckland, New Zealand) and the NCBI BLAST program (http://blast.ncbi.nlm.nih.gov/). The KM22 locus tags for the genes studied are as follows: bpsA, KM22_01871; bpsB, KM22_01870; bpsC, KM22_01869; bpsD, KM22_01868; and bpsR, KM22_01874.
Microtiter plate assay of Bordetella biofilm formation.
Static biofilm formation was evaluated by the microtiter plate assay as described previously (40). Briefly, Bordetella strains were grown in a total volume of 100 μl of SS medium at 37°C in 96-well polyvinyl chloride (PVC) microtiter plates. The plates were inoculated at an optical density at 600 nm (OD600) of 0.05 for the respective B. bronchiseptica strains. The microtiter plates were sealed with cellophane tape and incubated under stationary conditions. The growth medium was exchanged with fresh SS broth every 24 h. At 72 h, the nonattached and loosely adherent bacteria were removed by discarding the medium from the wells, which were then washed thoroughly with water. Cells that remained adhered to the wells were stained with a 0.1% solution of crystal violet (CV) and were incubated at room temperature for 30 min. The washing process was repeated, and the CV staining the cells was solubilized with 200 μl of 95% ethanol. Biofilm formation was quantitated by measuring the OD540 for each well by transferring 125 μl of the solubilized CV stain to a fresh polystyrene microtiter dish. Statistical significance was determined using a one-way analysis of variance (ANOVA) with Dunnett's posttest. P values of <0.01 were considered significant.
Deletion of bpsABCD in B. bronchiseptica KM22.
Strain KM22Δbps, containing an in-frame deletion of the bpsABCD genes, was constructed as follows. The 1,175 bp downstream and the last 6 codons of the bpsD gene were PCR amplified using bpsD-for and bpsD-rev (Table 3). The resulting PCR product was purified, ligated into the pCR-BluntII-TOPO vector (Invitrogen, Carlsbad, CA) to create pTN33, and transformed into Top10 cells. The insert of pTN33, containing the downstream region of bpsD, was cloned into pUC19 (New England BioLabs, Ipswich, MA) via EcoRI and XbaI sites to obtain pTN40. The 1,554 bp upstream and the first 20 codons of the bpsA gene were PCR amplified using primers bpsA-for and bpsA-rev (Table 3). The resulting PCR product was purified, ligated into the pCR-BluntII-TOPO vector to create pTN34, and transformed into Top10 cells. The insert, containing the upstream region of bpsA, was cloned into pTN40 via XbaI and HindIII sites to obtain pTN44. The DNA fragment containing both the upstream and downstream regions of bpsA and bpsD from pTN44 was PCR amplified using primers bpsAD-for and bpsAD-rev (Table 3). The resulting PCR product was purified, ligated into the pCR-BluntII-TOPO vector to create pTN47, and transformed into Top10 cells. The insert, containing both the upstream and downstream regions of bpsA and bpsD, was cloned into pSS4245 (69) via NotI and BamHI sites to obtain pTN49. pTN49 was introduced into E. coli SM10λpir and transconjugated into KM22 as previously described (69). Cointegrants containing pTN49 were positively selected on BG-streptomycin-carbenicillin–50 mM MgSO4 plates and incubated for 4 days at 37°C. The resulting cointegrants were then streaked onto BG plates without added MgSO4 (Bvg+ phase conditions) and incubated for 2 days at 37°C, which resulted in colonies lacking pTN49 and containing strains either of the wild type or with bpsABCD genes deleted. The absence of pTN49 was confirmed by growth on BG-streptomycin plates and lack of growth on BG-carbenicillin plates. To further confirm the wild-type or bps deletion genotype, colonies were screened by PCR using screening primers bpsADck-for and bpsADck-rev (Table 3) to detect either the wild-type locus (8,820 bp) or the bpsABCD deletion (2,914 bp). A single colony selected for subsequent use, designated KM22Δbps, was confirmed to harbor the bpsABCD deletion (2,914 bp), and the resulting PCR amplicon encompassing regions upstream and downstream of bpsA and bpsD was confirmed by DNA sequence analysis.
TABLE 3.
Primers used in this study
| Primer name or gene target | Sequence (5′ to 3′) |
|---|---|
| bpsD-for | GAATTCGGCCGTCCGCTTATCCGAGAGGTAATC |
| bpsD-rev | TCTAGACGCGCCTTGACCCTGTGAGCGCTCAG |
| bpsA-for | TCTAGAAGTCCGCCGCAGGCCACGCACGG |
| bpsA-rev | AAGCTTGTTCGGACCGGCACCCTATAACCACAT |
| bpsAD-for | GCGGCCGCGGCCGTCCGCTTATCCGAGAGGTAATC |
| bpsAD-rev | GGATCCAAGCTTGTTCGGACCGGCACCCTATAACCACAT |
| bpsADck-for | GCCGGGTCGGCCAATGGGTGTTCTACC |
| bpsADck-rev | TTCCCCTTGAGTTGACGACGGTATCACTGC |
Immunoblot analysis.
Crude exopolysaccharide extracts were prepared using a previously described method for purification of PNAG in Staphylococcus species (39, 70). Approximately 5 × 109 cells of different strains grown overnight at 37°C in broth culture were harvested by centrifugation, resuspended in 100 μl of 0.5 M EDTA, and boiled for 5 min at 100°C. Cells were removed by centrifugation, and the supernatant was treated with 1 mg proteinase K/ml for 60 min at 60°C. At the end of the incubation period, samples were heated to 85°C for 15 min to inactivate the protease followed by phenol chloroform extraction. A 5-μl volume of the extract was spotted on a nitrocellulose membrane and allowed to dry overnight. The membrane was blocked with 5% nonfat milk and probed with a 1:5,000 dilution of a goat antibody raised against S. aureus deacetylated PNAG (dPNAG) conjugated to diphtheria toxoid (45). A secondary mouse anti-goat IgG antibody conjugated to horseradish peroxidase (Pierce) was used at a concentration of 1:20,000 for detection in conjunction with the Amersham ECL (enhanced chemiluminescence) Western blotting detection kit (GE Healthcare Bio-Sciences, Pittsburgh, PA).
CSLM and image analysis.
Overnight cultures of B. bronchiseptica KM22 and KM22Δbps were grown in SS medium with 40 μg/ml streptomycin (SS/Sm). The overnight cultures were diluted to an OD of approximately 0.250 in fresh SS/Sm and were used to inoculate ibidi μ-Slide I0.4 Luer sterile single-use tissue culture treated (ibiTreat) flow cells with a channel height of 400 μm (ibidi USA, Madison, WI). The flow cells were incubated for 90 min at room temperature to allow initial bacterial adherence. Following this incubation, the flow of SS/Sm medium was initiated at a rate of 0.3 ml/min. Biofilms were allowed to grow for 90 h at 37°C. After growth, mature biofilms were stained with 1 μl/ml Syto 9 and 1 μl/ml propidium iodide using the FilmTracer Live/Dead biofilm viability kit (Invitrogen). Imaging was performed using a Nikon A1R+ confocal laser scanning microscope (CSLM) with a 20× objective. Images were acquired at 1,024 × 1,024 pixels using a Z-step of 0.95 μm. Syto 9 staining was detected using an excitation wavelength of 487.4 nm and an emission wavelength of 525 nm. Propidium iodide staining was detected using an excitation wavelength of 562 nm and an emission wavelength of 595 nm. Three independent experiments were performed. The image acquisition software used was Nikon NIS-Elements AR 4.40, and postimage analysis was performed using Imaris software (Bitplane, Concord, MA). Means of quantitative parameters were analyzed for significance using an unpaired two-tailed Student t test. A 5% level of significance (P < 0.05) was considered significant.
Experimental infection in swine.
B. bronchiseptica strains KM22 and KM22Δbps were cultured on BG agar supplemented with 10% sheep's blood at 37°C for 40 h. Suspensions of these cultures were prepared in PBS to contain approximately 2 × 109 CFU/ml, and 106 CFU diluted in PBS was used for inoculation of pigs. Serial dilutions of inoculum for each strain were plated on BG agar plates to determine CFU per milliliter and to confirm the expected colony morphology and hemolytic phenotype. Cultured dilutions of inocula of strains KM22 and TN29 (Δbps mutant) contained 5.6 × 105 and 4.3 × 105 CFU/ml, respectively. All colonies from all inocula displayed the expected colony morphology and hemolytic characteristics. Each experimental group of pigs was housed in a separate isolation room in biosafety level 2 (BSL2) containment facilities, and the swine were cared for in compliance with the Institutional Animal Care and Use Committee of the National Animal Disease Center. B. bronchiseptica was not isolated from any nasal swabs collected from all piglets prior to the start of the experiment. Forty naturally farrowed early-weaned piglets were divided into 2 experimental groups of 16 pigs each and 1 control group of 8. Pigs were inoculated intranasally at 1 week of age with 1 ml (0.5 ml/nostril) of a bacterial suspension containing KM22 or KM22Δbps or with 1 ml of sterile PBS. Bacterial colonization of the nasal cavity was quantified by nasal swabs on days 1, 3, and 5 and by nasal washes at all other sample days postinoculation. On days 7, 14, 28, and 56 postinoculation, 4 pigs from each experimental group (KM22 or KM22Δbps) and 2 pigs from the mock-infected (PBS) group were euthanized for sample collection to evaluate the bacterial burden in the respiratory tract, immune responses, and lesion severity. The trachea was then severed just below the larynx, and the trachea and lung were removed.
Determination of colonization.
Nasal swabs were placed into tubes containing 500 μl PBS and vortexed. Nasal washes were performed by instilling 5 ml of PBS into the nasal cavity through one nostril and collecting the effluent into a beaker. Tracheal washes were performed by placing a segment of trachea approximately 8 cm in length in a 15-ml centrifuge tube with 5 ml of PBS and shaking vigorously. Lung lavage was performed by filling the lungs with 50 ml of sterile PBS, gently massaging, and aspirating; approximately 25 ml of the PBS was recovered. Serial 10-fold dilutions were made from nasal swab, nasal and tracheal wash, and lung lavage fluids, and the number of CFU of B. bronchiseptica per milliliter was determined by plating 100 μl of the dilutions on duplicate selective blood agar plates containing 20 μg/ml penicillin, 10 μg/ml amphotericin B, 10 μg/ml streptomycin, and 10 μg/ml spectinomycin. The limit of detection was 10 CFU/ml. B. bronchiseptica was identified on the basis of colony morphology. Colonies from plates representing each respiratory tract site (nasal cavity, trachea, and lung) were randomly screened by PCR to confirm that recovered colonies were the same as the challenge strain for all B. bronchiseptica-inoculated groups using the screening primers and PCR conditions listed above. Statistical analyses of the nasal colonization data were performed using a mixed linear model (SAS 9.3 for Windows XP; SAS Institute Inc., Cary, NC, USA) for repeated measures and a heterogeneous autoregressive covariance structure to best account for unequal study day intervals. Linear combinations of the least-squares mean estimates for log10 CFU were used in a priori contrasts after testing for either a significant (P < 0.05) effect of the bacterial challenge strains or a significant strain-and-time interaction. Comparisons were made between challenge groups for each isolate at each time point using a 5% level of significance (P < 0.05) to assess statistical differences. Endpoint data for tracheal and lung bacterial loads were analyzed using analysis of variance and a general linear model for unbalanced data that included treatment group and study day, with CFU as the dependent variable. A 5% level of significance (P < 0.05) was used to assess statistical differences.
Pathological evaluation of the lung.
At necropsy, an estimate of gross lung involvement was assigned based on the percentage of each lung lobe affected and the percentage of total lung volume that each lobe represented. The percentage of total lung volume of each lobe was estimated to be 10% for the left cranial, 10% for the left middle, 25% for the left caudal, 10% for the right cranial, 10% for the right middle, 25% for the right caudal, and 10% for the intermediate lung lobes. Means were analyzed for significance using a two-way ANOVA with a Tukey's multiple-comparison test to compare differences among groups. A 5% level of significance (P < 0.05) was considered significant.
Antibody response.
Serum was collected from infected and control pigs on day 56 postchallenge. Titers of anti-Bordetella antibodies were measured by ELISA as previously described (14, 15). Briefly, plates were coated with heat-killed B. bronchiseptica KM22 or KM22Δbps (grown to an OD600 of 0.6). Sera were serially diluted, and B. bronchiseptica-specific antibody was detected using secondary antibodies specific for swine immunoglobulin G. Means were analyzed for significance using ANOVA with Tukey's multiple-comparison test to compare differences among groups. A 5% level of significance (P < 0.05) was considered significant.
Immune serum-complement-mediated lysis.
Sera collected on day 56 postinfection from KM22-, KM22Δbps-, or PBS-treated pigs were heat inactivated by incubation at 56°C for 30 min. Each B. bronchiseptica target strain was prepared in the same manner as the bacterial suspensions used for inoculation of the pigs. Suspensions of these cultures were prepared in PBS and diluted to 2 × 106 CFU/ml. Each antibody-complement killing assay was run in triplicate and set up as follows: 10 μl of swine serum, 5 μl of guinea pig serum (complement source), and 100 μl of 2 × 106 CFU/ml of the target strain were added to a single well of a 96-well round-bottom plate. SS broth containing 0.06% bovine serum albumin was added to bring the final volume to 0.3 ml per well. Wells containing infected swine serum only, guinea pig serum only, or bacteria only were included as controls. Plates were then incubated at 37°C for 4 h with gentle agitation. Serial dilutions in PBS were then plated on blood agar plates to determine CFU counts, which were then used to calculate the ratio of bacteria in treatment wells to the bacteria in control wells and reported as the percent viable bacteria. Means were analyzed for significance using ANOVA with Tukey's multiple-comparison test to compare differences among groups. A 5% level of significance (P < 0.05) was considered significant.
ACKNOWLEDGMENTS
We thank Steven Kellner for his excellent technical assistance.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Research in the laboratory of A.R.H. is supported by a Merit Award (I01 BX002711) from the Department of Veteran Affairs and NIH grant AI083211 (Project 3). Work in the laboratory of R.D. was supported by the following grants: R01AI125560, 1R21AI123805-01, and contract no. HHSN272201200005C.
We thank Gerald B. Pier for a generous gift of the dPNAG antibody.
REFERENCES
- 1.Duncan JR, Ramsey RK, Switzer WP. 1966. Pathology of experimental Bordetella bronchiseptica infection in swine: pneumonia. Am J Vet Res 27:467–472. [PubMed] [Google Scholar]
- 2.Duncan JR, Ross RF, Switzer WP, Ramsey FK. 1966. Pathology of experimental Bordetella bronchiseptica infection in swine: atrophic rhinitis. Am J Vet Res 27:457–466. [PubMed] [Google Scholar]
- 3.Palzer A, Ritzmann M, Wolf G, Heinritzi K. 2008. Associations between pathogens in healthy pigs and pigs with pneumonia. Vet Rec 162:267–271. doi: 10.1136/vr.162.9.267. [DOI] [PubMed] [Google Scholar]
- 4.Chanter N, Magyar T, Rutter JM. 1989. Interactions between Bordetella bronchiseptica and toxigenic Pasteurella multocida in atrophic rhinitis of pigs. Res Vet Sci 47:48–53. [PubMed] [Google Scholar]
- 5.Magyar T, Lax AJ. 2002. Atrophic rhinitis, p 169–197. In Brogden KA, Guthmiller J (ed), Polymicrobial diseases. ASM Press, Washington, DC. [PubMed] [Google Scholar]
- 6.Brockmeier SL. 2004. Prior infection with Bordetella bronchiseptica increases nasal colonization by Haemophilus parasuis in swine. Vet Microbiol 99:75–78. doi: 10.1016/j.vetmic.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Brockmeier SL, Loving CL, Nicholson TL, Palmer MV. 2008. Coinfection of pigs with porcine respiratory coronavirus and Bordetella bronchiseptica. Vet Microbiol 128:36–47. doi: 10.1016/j.vetmic.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockmeier SL, Palmer MV, Bolin SR. 2000. Effects of intranasal inoculation of porcine reproductive and respiratory syndrome virus, Bordetella bronchiseptica, or a combination of both organisms in pigs. Am J Vet Res 61:892–899. doi: 10.2460/ajvr.2000.61.892. [DOI] [PubMed] [Google Scholar]
- 9.Brockmeier SL, Palmer MV, Bolin SR, Rimler RB. 2001. Effects of intranasal inoculation with Bordetella bronchiseptica, porcine reproductive and respiratory syndrome virus, or a combination of both organisms on subsequent infection with Pasteurella multocida in pigs. Am J Vet Res 62:521–525. doi: 10.2460/ajvr.2001.62.521. [DOI] [PubMed] [Google Scholar]
- 10.Brockmeier SL, Register KB. 2007. Expression of the dermonecrotic toxin by Bordetella bronchiseptica is not necessary for predisposing to infection with toxigenic Pasteurella multocida. Vet Microbiol 125:284–289. doi: 10.1016/j.vetmic.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 11.Loving CL, Brockmeier SL, Vincent AL, Palmer MV, Sacco RE, Nicholson TL. 2010. Influenza virus coinfection with Bordetella bronchiseptica enhances bacterial colonization and host responses exacerbating pulmonary lesions. Microb Pathog 49:237–245. doi: 10.1016/j.micpath.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Vecht U, Wisselink HJ, van Dijk JE, Smith HE. 1992. Virulence of Streptococcus suis type 2 strains in newborn germfree pigs depends on phenotype. Infect Immun 60:550–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vecht U, Arends JP, van der Molen EJ, van Leengoed LA. 1989. Differences in virulence between two strains of Streptococcus suis type II after experimentally induced infection of newborn germ-free pigs. Am J Vet Res 50:1037–1043. [PubMed] [Google Scholar]
- 14.Nicholson TL, Brockmeier SL, Loving CL. 2009. Contribution of Bordetella bronchiseptica filamentous hemagglutinin and pertactin to respiratory disease in swine. Infect Immun 77:2136–2146. doi: 10.1128/IAI.01379-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholson TL, Brockmeier SL, Loving CL, Register KB, Kehrli ME Jr, Shore SM. 2014. The Bordetella bronchiseptica type III secretion system is required for persistence and disease severity but not transmission in swine. Infect Immun 82:1092–1103. doi: 10.1128/IAI.01115-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholson TL, Brockmeier SL, Loving CL, Register KB, Kehrli ME Jr, Stibitz SE, Shore SM. 2012. Phenotypic modulation of the virulent Bvg phase is not required for pathogenesis and transmission of Bordetella bronchiseptica in swine. Infect Immun 80:1025–1036. doi: 10.1128/IAI.06016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodnow RA. 1980. Biology of Bordetella bronchiseptica. Microbiol Rev 44:722–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akerley BJ, Cotter PA, Miller JF. 1995. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80:611–620. doi: 10.1016/0092-8674(95)90515-4. [DOI] [PubMed] [Google Scholar]
- 19.Mattoo S, Cherry JD. 2005. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev 18:326–382. doi: 10.1128/CMR.18.2.326-382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin EK, Jung R, Hahn TW. 2007. Polymorphism of pertactin gene repeat regions in Bordetella bronchiseptica isolates from pigs. J Vet Med Sci 69:771–774. doi: 10.1292/jvms.69.771. [DOI] [PubMed] [Google Scholar]
- 21.Shin EK, Seo YS, Han JH, Hahn TW. 2007. Diversity of swine Bordetella bronchiseptica isolates evaluated by RAPD analysis and PFGE. J Vet Sci 8:65–73. doi: 10.4142/jvs.2007.8.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellis JA. 2015. How well do vaccines for Bordetella bronchiseptica work in dogs? A critical review of the literature 1977-2014. Vet J 204:5–16. doi: 10.1016/j.tvjl.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Z, Wang C, Xue Y, Tang X, Wu B, Cheng X, He Q, Chen H. 2011. The occurrence of Bordetella bronchiseptica in pigs with clinical respiratory disease. Vet J 188:337–340. doi: 10.1016/j.tvjl.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 24.Bemis DA, Carmichael LE, Appel MJ. 1977. Naturally occurring respiratory disease in a kennel caused by Bordetella bronchiseptica. Cornell Vet 67:282–293. [PubMed] [Google Scholar]
- 25.Bemis DA. 1992. Bordetella and Mycoplasma respiratory infections in dogs and cats. Vet Clin North Am Small Anim Pract 22:1173–1186. doi: 10.1016/S0195-5616(92)50308-4. [DOI] [PubMed] [Google Scholar]
- 26.Coutts AJ, Dawson S, Binns S, Hart CA, Gaskell CJ, Gaskell RM. 1996. Studies on natural transmission of Bordetella bronchiseptica in cats. Vet Microbiol 48:19–27. doi: 10.1016/0378-1135(95)00128-X. [DOI] [PubMed] [Google Scholar]
- 27.Schulz BS, Kurz S, Weber K, Balzer HJ, Hartmann K. 2014. Detection of respiratory viruses and Bordetella bronchiseptica in dogs with acute respiratory tract infections. Vet J 201:365–369. doi: 10.1016/j.tvjl.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 30.Brook I. 2016. Microbiology of chronic rhinosinusitis. Eur J Clin Microbiol Infect Dis 35:1059–1068. doi: 10.1007/s10096-016-2640-x. [DOI] [PubMed] [Google Scholar]
- 31.Nazzari E, Torretta S, Pignataro L, Marchisio P, Esposito S. 2015. Role of biofilm in children with recurrent upper respiratory tract infections. Eur J Clin Microbiol Infect Dis 34:421–429. doi: 10.1007/s10096-014-2261-1. [DOI] [PubMed] [Google Scholar]
- 32.Prat C, Lacoma A. 2016. Bacteria in the respiratory tract—how to treat? Or do not treat? Int J Infect Dis 51:113–122. doi: 10.1016/j.ijid.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 34.Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, Otto M. 2004. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem 279:54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 35.Anderson GG, O'Toole GA. 2008. Innate and induced resistance mechanisms of bacterial biofilms. Curr Top Microbiol Immunol 322:85–105. [DOI] [PubMed] [Google Scholar]
- 36.Hoffman C, Eby J, Gray M, Heath Damron F, Melvin J, Cotter P, Hewlett E. 2017. Bordetella adenylate cyclase toxin interacts with filamentous haemagglutinin to inhibit biofilm formation in vitro. Mol Microbiol 103:214–228. doi: 10.1111/mmi.13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conover MS, Mishra M, Deora R. 2011. Extracellular DNA is essential for maintaining Bordetella biofilm integrity on abiotic surfaces and in the upper respiratory tract of mice. PLoS One 6:e16861. doi: 10.1371/journal.pone.0016861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irie Y, Mattoo S, Yuk MH. 2004. The Bvg virulence control system regulates biofilm formation in Bordetella bronchiseptica. J Bacteriol 186:5692–5698. doi: 10.1128/JB.186.17.5692-5698.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parise G, Mishra M, Itoh Y, Romeo T, Deora R. 2007. Role of a putative polysaccharide locus in Bordetella biofilm development. J Bacteriol 189:750–760. doi: 10.1128/JB.00953-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mishra M, Parise G, Jackson KD, Wozniak DJ, Deora R. 2005. The BvgAS signal transduction system regulates biofilm development in Bordetella. J Bacteriol 187:1474–1484. doi: 10.1128/JB.187.4.1474-1484.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serra D, Bosch A, Russo DM, Rodriguez ME, Zorreguieta A, Schmitt J, Naumann D, Yantorno O. 2007. Continuous nondestructive monitoring of Bordetella pertussis biofilms by Fourier transform infrared spectroscopy and other corroborative techniques. Anal Bioanal Chem 387:1759–1767. doi: 10.1007/s00216-006-1079-9. [DOI] [PubMed] [Google Scholar]
- 42.Conover MS, Sloan GP, Love CF, Sukumar N, Deora R. 2010. The Bps polysaccharide of Bordetella pertussis promotes colonization and biofilm formation in the nose by functioning as an adhesin. Mol Microbiol 77:1439–1455. doi: 10.1111/j.1365-2958.2010.07297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sloan GP, Love CF, Sukumar N, Mishra M, Deora R. 2007. The Bordetella Bps polysaccharide is critical for biofilm development in the mouse respiratory tract. J Bacteriol 189:8270–8276. doi: 10.1128/JB.00785-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cattelan N, Dubey P, Arnal L, Yantorno OM, Deora R. 2016. Bordetella biofilms: a lifestyle leading to persistent infections. Pathog Dis 74:ftv108. doi: 10.1093/femspd/ftv108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maira-Litran T, Kropec A, Goldmann DA, Pier GB. 2005. Comparative opsonic and protective activities of Staphylococcus aureus conjugate vaccines containing native or deacetylated staphylococcal poly-N-acetyl-beta-(1-6)-glucosamine. Infect Immun 73:6752–6762. doi: 10.1128/IAI.73.10.6752-6762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenney D, Pouliot KL, Wang Y, Murthy V, Ulrich M, Doring G, Lee JC, Goldmann DA, Pier GB. 1999. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science 284:1523–1527. doi: 10.1126/science.284.5419.1523. [DOI] [PubMed] [Google Scholar]
- 47.Kelly-Quintos C, Kropec A, Briggs S, Ordonez CL, Goldmann DA, Pier GB. 2005. The role of epitope specificity in the human opsonic antibody response to the staphylococcal surface polysaccharide poly N-acetyl glucosamine. J Infect Dis 192:2012–2019. doi: 10.1086/497604. [DOI] [PubMed] [Google Scholar]
- 48.Ganguly T, Johnson JB, Kock ND, Parks GD, Deora R. 2014. The Bordetella pertussis Bps polysaccharide enhances lung colonization by conferring protection from complement-mediated killing. Cell Microbiol 16:1105–1118. doi: 10.1111/cmi.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serra DO, Conover MS, Arnal L, Sloan GP, Rodriguez ME, Yantorno OM, Deora R. 2011. FHA-mediated cell-substrate and cell-cell adhesions are critical for Bordetella pertussis biofilm formation on abiotic surfaces and in the mouse nose and the trachea. PLoS One 6:e28811. doi: 10.1371/journal.pone.0028811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lowe BA, Marsh TL, Isaacs-Cosgrove N, Kirkwood RN, Kiupel M, Mulks MH. 2012. Defining the “core microbiome” of the microbial communities in the tonsils of healthy pigs. BMC Microbiol 12:20. doi: 10.1186/1471-2180-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kernaghan S, Bujold AR, MacInnes JI. 2012. The microbiome of the soft palate of swine. Anim Health Res Rev 13:110–120. doi: 10.1017/S1466252312000102. [DOI] [PubMed] [Google Scholar]
- 52.Correa-Fiz F, Fraile L, Aragon V. 2016. Piglet nasal microbiota at weaning may influence the development of Glasser's disease during the rearing period. BMC Genomics 17:404. doi: 10.1186/s12864-016-2700-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paharik AE, Horswill AR. 2016. The staphylococcal biofilm: adhesins, regulation, and host response. Microbiol Spectr 4(2). doi: 10.1128/microbiolspec.VMBF-0022-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brady LJ, Maddocks SE, Larson MR, Forsgren N, Persson K, Deivanayagam CC, Jenkinson HF. 2010. The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol Microbiol 77:276–286. doi: 10.1111/j.1365-2958.2010.07212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao G, Tang H, Zhang S, Ren H, Dai J, Lai L, Lu C, Yao H, Fan H, Wu Z. 2017. Streptococcus suis small RNA rss04 contributes to the induction of meningitis by regulating capsule synthesis and by inducing biofilm formation in a mouse infection model. Vet Microbiol 199:111–119. doi: 10.1016/j.vetmic.2016.12.034. [DOI] [PubMed] [Google Scholar]
- 56.Simmons WL, Dybvig K. 2015. Catalase enhances growth and biofilm production of Mycoplasma pneumoniae. Curr Microbiol 71:190–194. doi: 10.1007/s00284-015-0822-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loera-Muro A, Jacques M, Avelar-Gonzalez FJ, Labrie J, Tremblay YD, Oropeza-Navarro R, Guerrero-Barrera AL. 2016. Auxotrophic Actinobacillus pleurpneumoniae grows in multispecies biofilms without the need for nicotinamide-adenine dinucleotide (NAD) supplementation. BMC Microbiol 16:128. doi: 10.1186/s12866-016-0742-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kubera A, Thamchaipenet A, Shoham M. 2017. Biofilm inhibitors targeting the outer membrane protein A of Pasteurella multocida in swine. Biofouling 33:14–23. doi: 10.1080/08927014.2016.1259415. [DOI] [PubMed] [Google Scholar]
- 59.Bello-Orti B, Deslandes V, Tremblay YD, Labrie J, Howell KJ, Tucker AW, Maskell DJ, Aragon V, Jacques M. 2014. Biofilm formation by virulent and non-virulent strains of Haemophilus parasuis. Vet Res 45:104. doi: 10.1186/s13567-014-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Izano EA, Sadovskaya I, Vinogradov E, Mulks MH, Velliyagounder K, Ragunath C, Kher WB, Ramasubbu N, Jabbouri S, Perry MB, Kaplan JB. 2007. Poly-N-acetylglucosamine mediates biofilm formation and antibiotic resistance in Actinobacillus pleuropneumoniae. Microb Pathog 43:1–9. doi: 10.1016/j.micpath.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nicholson TL, Shore SM, Bayles DO, Register KB, Kingsley RA. 2014. Draft genome sequence of the Bordetella bronchiseptica swine isolate KM22. Genome Announc 2(4):e00670-14. doi: 10.1128/genomeA.00670-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nicholson TL, Shore SM, Register KB, Bayles DO, Kingsley RA, Brunelle BW. 2016. Comparative genomic analysis of the swine pathogen Bordetella bronchiseptica strain KM22. Vet Microbiol 182:87–94. doi: 10.1016/j.vetmic.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barnes MG, Weiss AA. 2001. BrkA protein of Bordetella pertussis inhibits the classical pathway of complement after C1 deposition. Infect Immun 69:3067–3072. doi: 10.1128/IAI.69.5.3067-3072.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fernandez RC, Weiss AA. 1994. Cloning and sequencing of a Bordetella pertussis serum resistance locus. Infect Immun 62:4727–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pilione MR, Pishko EJ, Preston A, Maskell DJ, Harvill ET. 2004. pagP is required for resistance to antibody-mediated complement lysis during Bordetella bronchiseptica respiratory infection. Infect Immun 72:2837–2842. doi: 10.1128/IAI.72.5.2837-2842.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skurnik D, Cywes-Bentley C, Pier GB. 2016. The exceptionally broad-based potential of active and passive vaccination targeting the conserved microbial surface polysaccharide PNAG. Expert Rev Vaccines 15:1041–1053. doi: 10.1586/14760584.2016.1159135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cotter PA, Miller JF. 1994. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect Immun 62:3381–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Register KB, Ivanov YV, Jacobs N, Meyer JA, Goodfield LL, Muse SJ, Smallridge WE, Brinkac L, Kim M, Sanka R, Harvill ET, Losada L. 2015. Draft genome sequences of 53 genetically distinct isolates of Bordetella bronchiseptica representing 11 terrestrial and aquatic hosts. Genome Announc 3(2):e00152-15. doi: 10.1128/genomeA.00152-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buboltz AM, Nicholson TL, Weyrich LS, Harvill ET. 2009. Role of the type III secretion system in a hypervirulent lineage of Bordetella bronchiseptica. Infect Immun 77:3969–3977. doi: 10.1128/IAI.01362-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kropec A, Maira-Litran T, Jefferson KK, Grout M, Cramton SE, Gotz F, Goldmann DA, Pier GB. 2005. Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect Immun 73:6868–6876. doi: 10.1128/IAI.73.10.6868-6876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]