ABSTRACT
The oral microbiota associated with the initiation and progression of dental caries has yet to be fully characterized. The Human Oral Microbe Identification Using Next-Generation Sequencing (HOMINGS) approach was used to analyze the microbiomes of site-specific supragingival dental plaques from children with different caries status. Fifty-five children (2 to 7 years of age) were assessed at baseline and at 12 months and grouped as caries free (CF), caries active with enamel lesions (CAE), and caries active with dentin carious lesions (CA). Plaque samples from caries-free tooth surfaces (PF) and from enamel carious lesions (PE) and dentin carious lesions (PD) were collected. 16S community profiles were obtained by HOMINGS, and 408 bacterial species and 84 genus probes were assigned. Plaque bacterial communities showed temporal stability, as there was no significant difference in beta diversity values between the baseline and 12-month samples. Irrespective of collection time points, the microbiomes of healthy tooth surfaces differed substantially from those found during caries activity. All pairwise comparisons of beta diversity values between groups were significantly different (P < 0.05), except for comparisons between the CA-PF, CAE-PE, and CA-PE groups. Streptococcus genus probe 4 and Neisseria genus probe 2 were the most frequently detected taxa across the plaque groups, followed by Streptococcus sanguinis, which was highly abundant in CF-PF. Well-known acidogenic/aciduric species such as Streptococcus mutans, Scardovia wiggsiae, Parascardovia denticolens, and Lactobacillus salivarius were found almost exclusively in CA-PD. The microbiomes of supragingival dental plaque differ substantially among tooth surfaces and children of different caries activities. In support of the ecological nature of caries etiology, a steady transition in community species composition was observed with disease progression.
KEYWORDS: bacteria, caries, children, microbiome, supragingival, biofilms, dental plaque
INTRODUCTION
Oral bacteria that colonize the teeth form dental plaque, a biofilm community that exists in equilibrium with host defenses and is generally compatible with the integrity of the tooth tissues (1). The development of carious lesions on tooth tissues involves a dynamic biological process wherein acids produced by bacterial fermentation of dietary carbohydrates effect the demineralization of dental tissues. Repeated acidification results in the emergence of acid-producing and highly acid-tolerant organisms, a selective process that upsets pH homeostasis and shifts the demineralization-remineralization balance toward the loss of tooth minerals (2–5). Oral environmental perturbations associated with changes in the availability of dietary carbohydrates and pH can influence microbial metabolic activity, which in turn can modify the environment and induce microbial selection to create a more pathogenic microbiome (6). Thus, changes in both the compositions and biochemical activities of oral biofilms are essential etiologic determinants of dental caries.
Advances in DNA sequencing and bioinformatics have facilitated the disclosure of associations of numerous oral bacterial taxa with dental health or with caries activity (7–13). The use of next-generation sequencing (NGS) technology has revealed the complexity of microbiomes at unprecedented levels and is providing a foundation to understand how hundreds of bacterial species coinhabit and functionally interact to maintain homeostasis, to discourage the establishment of pathogens, and, when conditions are favorable, to cause disease (14, 15). However, most studies correlating the composition of the oral microbiome with caries activity and etiology have examined either saliva or dental plaque samples pooled from multiple tooth surfaces (16–18), which diminishes their clinical relevance considering that carious lesions occur at specific tooth sites (13). Moreover, there is now clear evidence that the different oral habitats that exist in the human mouth (tissues and location) are colonized by distinct microbial communities (9, 13, 19). To date, only a few studies have examined the microbial profiles of site-specific supragingival plaque (10, 20). For a better understanding of the caries process and the functions of specific organisms in the transition from dental health to the different stages of caries, it is imperative that the plaque sampling methods be site specific.
Despite growing efforts to define the composition and activities of the oral microbiome in health and disease (7, 10, 21), the current understanding of the basis for the intraindividual and interindividual differences in microbial profiles is limited. One of the major challenges facing oral heath researchers today is distinguishing which of the potential host-microbial interactions are critical for the maintenance of dental health. Defining the composition of the oral microbiome is the first logical step in achieving this goal by providing essential information for future metagenomic and metabolomic studies correlating microbial community composition and metabolism with health and caries status. It is even more critical to evaluate these processes in young children because of the high caries prevalence in this population (22). In fact, untreated caries in deciduous teeth affects over 600 million children worldwide and represents a major biological, social, and financial burden on individuals and health care systems (23). Accordingly, the main purposes of this study were to define the microbiomes of site-specific supragingival dental plaques of young children with different caries status and to determine whether the microbial profiles change or remain stable over a 12-month period. New insights into the stability or variability of the oral microbiome and into the physiological or ecological relevance of microbial communities will be needed to evaluate existing and future outcomes of caries interventions.
RESULTS
The mean age of the children (n = 55) enrolled in this study was 4.3 years at baseline. With regard to their genders and races, 53% of the children were male, 47% were female, 86% were Caucasian, 7% were African-American, and 7% were of other races. At baseline, 67% of the children had primary dentition, 33% had mixed dentition, 47% were caries free (CF), 27% were caries active with enamel lesions (CAE), and 26% were caries active with dentin carious lesions (CA). At 12 months, 58% of the children had primary dentition, 42% had mixed dentition, 38% were CF, 36% were CAE, and 26% were CA. Most of the children (n = 50; 90%) had the same caries status after 12 months, and only 5 children (10%) changed from CF at baseline to CAE at 12 months.
Illumina sequencing produced an average of 66,918 reads/sample (total of 186 samples). After low-quality reads were removed, the Human Oral Microbe Identification Using Next-Generation Sequencing (HOMINGS) approach produced a total of 12,014,038 operational taxonomic units (OTUs). Numbers of counts/sample ranged from 444 to 199,356 (mean, 64,591.6). Two samples with low OTU counts (444 and 20,650) were removed, and all remaining samples were then normalized to a count of 34,621. This resulted in a total of 492 taxonomic assignments: 408 bacterial species and 84 genus probes across all samples.
To profile bacterial communities, the samples were grouped as plaques from caries-free tooth surfaces of CF children (CF-PF), CAE-PF, plaques from enamel carious lesions of CAE children (CAE-PE), CAE-PE, CA-PF, CA-PE, and plaques from CA children with dentin carious lesions (CA-PD). Permutational multivariate analysis of variance (PERMANOVA) showed no significant differences in the beta diversity values of plaque communities when the groups were compared at baseline and at 12 months (see Table S1 in the supplemental material). Consequently, sequencing data from the two time points for each sample group were combined for subsequent analyses; e.g., CF-PF data at baseline were combined with CF-PF data at 12 months.
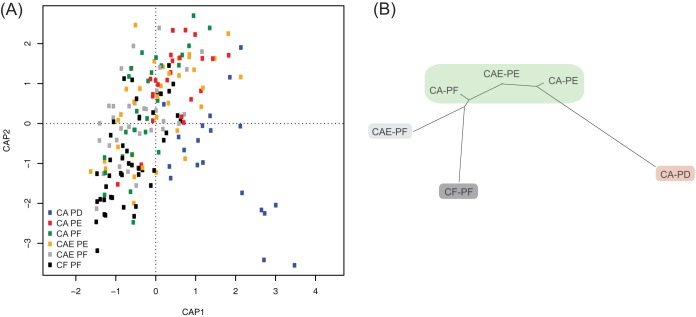
Distance-based redundancy analysis (db-RDA) showed that the bacterial communities of CA-PD samples were the most dissimilar from those of the other groups, with these samples occurring mostly and almost exclusively in the bottom right quadrant of Fig. 1A. CF-PF and CA-PE samples were also well differentiated from each other, with CF-PF samples occurring mostly in the bottom left quadrant and CA-PE samples occurring mostly in the upper right quadrant of Fig. 1A. A neighbor-joining dendrogram was constructed by using the variance components obtained from the pairwise PERMANOVAs (Table 1 and Fig. 1B). CF-PF communities showed the most separation from CA-PD communities. The CA-PF, CAE-PE, and CA-PE groups were connected by relatively short branch lengths and were not significantly differentiated from one another (Table 1 and Fig. 1B). All remaining comparisons among the different groups (plaque communities) were significant.
FIG 1.
(A) Constrained analysis of principal coordinates (CAP) of plaque bacterial communities using distance-based redundancy analysis (db-RDA). (B) Neighbor-joining phylogeny based on pairwise PERMANOVA components for beta diversity. Branch lengths represent the degrees to which bacterial communities are differentiated. Groups shaded in green showed no significant differences in beta diversity values. CF, caries-free children; CAE, caries-active children with enamel carious lesions; CA, caries-active children with dentin carious lesions; PF, supragingival plaque from caries-free tooth surfaces; PE, plaque from active, enamel carious lesions; PD, plaque from active, dentin carious lesions.
TABLE 1.
Pairwise PERMANOVAs for significant differences in beta diversity measures (Bray-Curtis) among the different groups of site-specific plaque samplesa
| Group |
P value or pseudo-F value |
|||||
|---|---|---|---|---|---|---|
| CF-PF | CAE-PF | CAE-PE | CA-PF | CA-PE | CA-PD | |
| CF-PF | 0.0011* | 0.0060* | 0.0183* | 0.0003* | 0.0003* | |
| CAE-PF | 3.225 | 0.0103* | 0.0205* | 0.0003* | 0.0003* | |
| CAE-PE | 2.668 | 2.373 | 0.0877 | 0.1926 | 0.0003* | |
| CA-PF | 2.156 | 1.975 | 1.580 | 0.0942 | 0.0003 | |
| CA-PE | 4.575 | 3.382 | 1.275 | 1.489 | 0.0003* | |
| CA-PD | 7.795 | 6.640 | 4.145 | 5.358 | 3.678 | |
The top right values are P values corrected by using the FDR. The bottom left values are pseudo-F values (variance components). *, significant P values; CF, caries-free children; CAE, caries-active children with enamel carious lesions; CA, caries-active children with dentin carious lesions; PF, supragingival plaque from caries-free tooth surfaces; PE, plaque from active, enamel carious lesions; PD, plaque from active, dentin carious lesions. Tests were run using 10,000 permutations.
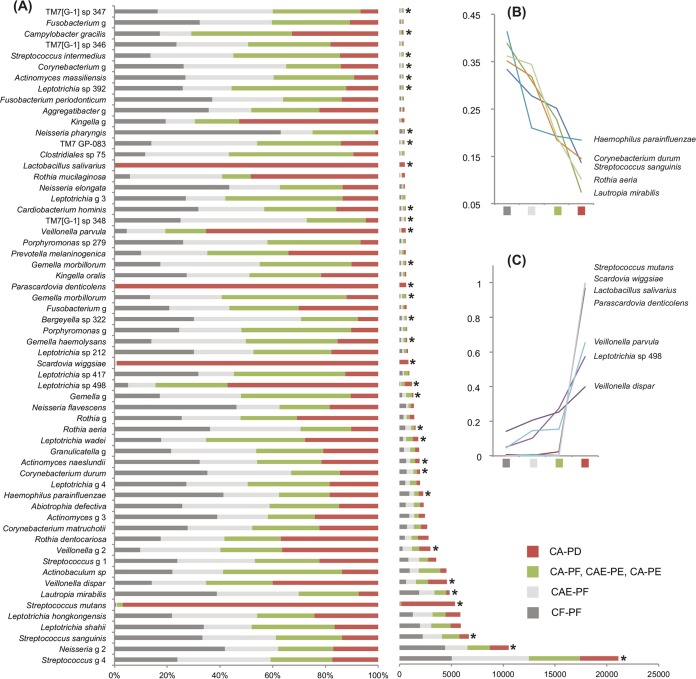
Due to the lack of significant differentiation among CA-PF, CAE-PE, and CA-PE, these three types of plaque samples were grouped in all subsequent analyses. Out of the 492 OTUs assigned, 115 OTUs showed significant differences in abundance among the four remaining plaque groups, CF-PF, CAE-PF, CA-PF/CAE-PE/CA-PE, and CA-PD. Table S2 in the supplemental material shows the normalized taxon/OTU counts (means for each of the four groups) for those taxa that showed a significant difference in abundance among the plaque groups, and Table S3 shows the normalized counts for all taxa/OTUs. Figure 2 shows the 60 most frequent OTUs (out of the 492 OTUs assigned) among the plaque groups. Out of these 60 OTUs, 32 showed significant differences in frequencies of occurrence when the groups were compared.
FIG 2.
(A) Distribution of the 60 most abundant taxa (out of 492 taxa) among the four plaque groups. *, taxon showing a significant difference in abundance among the groups; sp, species probe; g, genus probe. The left chart shows the mean taxon counts for each of the plaque groups expressed as a proportion of the sum of the means. The right chart shows the sum of the means (see Table 1 for a description of the taxa captured by the genus probes). (B and C) Two line charts showing the frequencies of certain taxa that, in addition to showing significant differences in abundances among the groups, also showed strong and progressive increases or decreases in frequency as caries progressed. Frequency is expressed as a proportion of the sum of the means.
Overall, Streptococcus genus probe 4 and Neisseria genus probe 2 were the two most frequently detected taxa among the four groups (Fig. 2A; for a description of genus probes, see Table S4). The third most frequently detected taxon was Streptococcus sanguinis, which was significantly more abundant in CF-PF than in the other groups. Five additional taxa were also significantly more abundant in CF-PF and occurred at relatively high frequencies: Lautropia mirabilis, Haemophilus parainfluenzae, Corynebacterium durum, Actinomyces naeslundii, and Rothia aeria. With the exception of Actinomyces naeslundii, these species showed consistent decreases in frequency through the groups toward CA-PD (Fig. 2B). Seven different Leptotrichia species and two distinct genus probes were also particularly frequent among the groups. In order of frequency, they were Leptotrichia shahii, Leptotrichia hongkongensis, Leptotrichia genus probe 4, Leptotrichia wadei, Leptotrichia sp. 417, Leptotrichia sp. 498, Leptotrichia sp. 212, Leptotrichia genus probe 3, and Leptotrichia sp. 392. Of note, Leptotrichia sp. 498, Leptotrichia wadei, and Leptotrichia sp. 392 were detected at significantly higher frequencies in disease states.
Four species occurred almost exclusively in CA-PD: Streptococcus mutans, Scardovia wiggsiae, Parascardovia denticolens, and Lactobacillus salivarius. Other taxa that were detected at significantly high frequencies in CA-PD were Veillonella parvula, Veillonella dispar, Veillonella genus probe 2, and Leptotrichia sp. 498. Veillonella parvula, Veillonella dispar, and Leptotrichia sp. 498 showed a strong and progressive pattern of increasing frequency from CF-PF to CA-PD (Fig. 2C). The pattern for the increasing frequency of Veillonella genus probe 2 was not quite as linear, as this taxon was more frequently detected in CAE-PF than in the CA-PF/CAE-PE/CA-PE group.
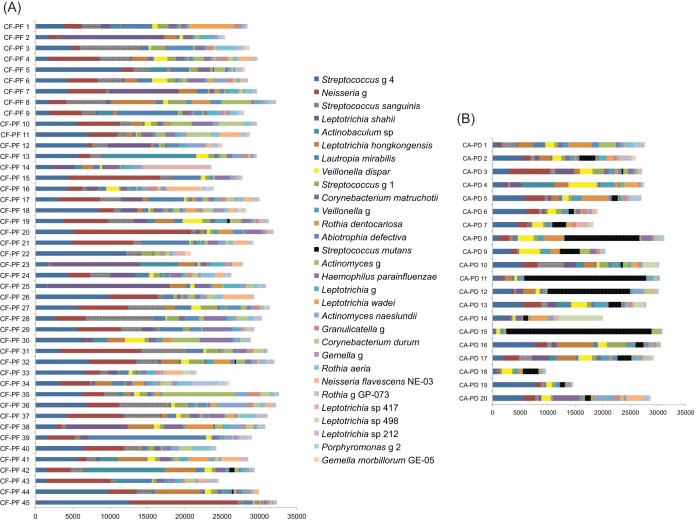
Significant differences in beta diversity values appeared to reflect the levels of alpha diversity among the plaque groups. Specifically, groups separated by large beta distances also had significant differences in alpha diversity values (Fig. 3A). There was no significant difference in alpha diversities between CF-PF and CAE-PF and between CA-PF/CAE-PE/CA-PE and CA-PD. Median levels of alpha diversity for plaque samples increased progressively from CF-PF to CA-PD (Fig. 3B). The CA-PD category had the widest distribution of alpha diversity values among the groups despite the fact that several CA-PD samples presented very low alpha diversity values. Figure 4 shows the distribution of the 30 most frequently detected taxa within each individual CF-PF and CA-PD sample.
FIG 3.
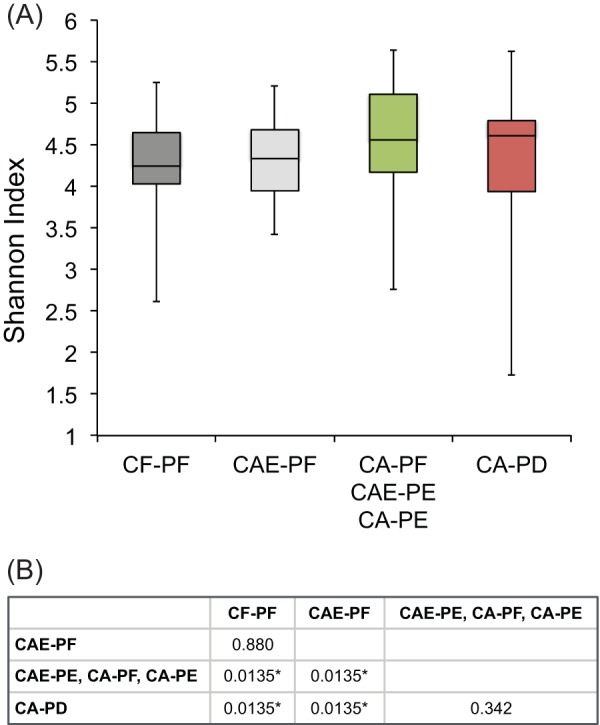
Levels of alpha diversity for the different groups of site-specific plaque samples. (A) Box-and-whisker plots showing alpha diversity levels for the different groups of site-specific plaque samples. (B) Pairwise t tests for significant differences in alpha diversity values among the different groups of site-specific plaque samples. P values were corrected by using FDR analysis; *, significant P values.
FIG 4.
Distribution of the 30 most frequently detected taxa among the CF-PF samples (A) and CA-PD samples (B).
PERMANOVAs for differences in beta diversity values were also used to evaluate the microbiomes of PF samples from the five children whose caries status changed from CF to CAE during the study. The microbial profiles of CF-PF collected at baseline were compared to those of CAE-PF collected at 12 months (see Fig. S5 in the supplemental material). Although there was no significant difference in beta diversity values (P = 0.432), several taxa showed clear differences in frequencies of detection between the baseline and 12 months. For example, the frequency of detection of Actinobaculum sp. 183 increased by nearly 100-fold and those of Corynebacterium durum and Gemella genus GP-039 increased by 3-fold, whereas those of Haemophilus parainfluenzae and Leptotrichia shahii decreased by 4-fold and those of Actinomyces genus 3 and Veillonella dispar decreased by 3-fold from the baseline to 12 months.
DISCUSSION
This study reports a prospective microbiome analysis of supragingival dental plaques collected from tooth sites and children of different caries status that substantively enhances the understanding of microbial profile changes with the progressive stages of early childhood caries. A strong pattern of microbial community transition for plaque samples from healthy tooth surfaces to enamel carious lesions and then dentin carious lesions was observed. Plaque communities from dentin carious lesions of caries-active children (CA-PD) showed a very distinctive bacterial profile compared to those of the other communities studied here. Moreover, communities from healthy tooth surfaces of caries-active children (CA-PF) were shown to be more similar to those from enamel carious lesions (CAE-PE and CA-PE) than to those from healthy teeth of caries-free children (CF-PF), suggesting that CA-PF sites appear to be at a greater risk for caries development than CF-PF sites. This finding is concordant with data from previous studies of risk factors for childhood caries (24) and highlights the interconnectedness of plaque communities, in which these communities are part of a larger ecosystem where changes in the structure of one community may eventually affect others. Furthermore, the fact that communities from noncarious tooth sites were shown to be distinctive from those from carious sites also stresses that studies of site-specific dental plaques are far more informative of the caries process than are studies of pooled plaques.
Another important finding of this study is that supragingival plaque communities remained stable for 12 months. Notably, the caries status of the majority of the participating children did not change for the duration of the study, and the sampling and coding methods were the same at baseline and at 12 months. In addition, no significant difference in beta diversity values was observed among the PF samples from children who changed their caries status from CF to CAE after 12 months. Previous studies have shown that microbiomes of different parts of the human body can remain stable for months and even years (25, 26), but they can also be highly variable over short periods of time (27, 28). A tremendous range of compositional variability between individuals and between different oral sites was previously reported (29–32). Recently, the plaque microbiome was described as “highly individualized at the oligotype level and characterized by community stability, with variability in the relative abundance of community members between individuals and over time” (30). In that same study, Corynebacterium, Capnocytophaga, Fusobacterium, Actinomyces, and Streptococcus taxa were shown to be relatively constantly abundant within and between individuals (30). These taxa were also described by that same research group to be the major participants of the “hedgehog” structure, a multigenus microbial consortium characterized by the presence of a mass of Corynebacterium filaments with Streptococcus at the periphery (33). Thus, specific mechanisms of bacterial coadhesion and metabolic cooperation may foster the stability of communities that are characteristic of a particular dental health status in an individual.
Our findings confirm the accepted concepts that S. sanguinis is associated with dental health and that S. mutans is associated with caries development. However, the data also highlight that these associations are neither uniform nor defining of caries status. Previous studies reported either a decrease in community diversity as caries progresses (10, 20, 34) or the opposite (18, 35, 36). Here, communities associated with the most advanced stage of caries studied here (CA-PD) were typically highly diverse, except when S. mutans was the dominant species. Thus, low diversity is not always a signature of disease. In addition, S. mutans and other acidogenic/aciduric species such as Scardovia wiggsiae, Parascardovia denticolens, and Lactobacillus salivarius, which were previously associated with caries (10, 37–42), occurred almost exclusively in CA-PD, suggesting that these species might be more important in the advanced stages of childhood caries.
V. parvula and V. dispar showed consistent increases in frequency from healthy to diseased states, as previously observed (20). The distribution of Veillonella organisms within different oral sites has been correlated with coaggregation with other oral bacteria (43) and their requirement for fatty acids (44). Certain Veillonella species have been associated with the caries process mostly because of the availability of their preferred carbon source, lactic acid, in the environment of carious lesions (44–46). In fact, Edlund et al. (47) used an in vitro multispecies transcriptomic model to show that the metabolic activities of V. dispar and Veillonella atypica were positively stimulated by the presence of lactic acid and low-pH conditions. Furthermore, a recent metatranscriptomic analysis revealed other potential cariogenic features of Veillonella; for example, V. parvula seems to exhibit distinct intracellular pH control mechanisms, which might explain the preponderance of these species in carious lesions (48). Thus, certain Veillonella species may serve as early indicators of caries activity or even indicators of microbial activities that can promote the initiation of a carious lesion. Leptotrichia sp. 498 also showed a consistently increased frequency from health to disease states. Leptotrichia has high saccharolytic potential, is able to ferment a large variety of mono- and disaccharides to lactic acid (49), and is well adapted to thrive under conditions that are conducive to caries formation. Certain Leptotrichia species were shown to be negatively associated with elevated urease activity in plaques, which is correlated with dental health, and therefore positively associated with caries (50).
In contrast, a steady decrease in frequency from healthy to diseased states was observed for S. sanguinis, L. mirabilis, H. parainfluenzae, C. durum, and R. aeria, suggesting their possible role in health. S. sanguinis has the ability to raise the pH by catabolizing arginine via the arginine deiminase pathway. Similarly, H. parainfluenzae can raise the pH via urea hydrolysis (51), and Morou-Bermudez et al. (50) showed that this species is significantly more abundant in plaques with high urease activity. Neisseria species also showed a frequent association with health. The second most frequent taxonomic assignment was for the Neisseria genus probe, which was significantly more abundant in CF-PF than in the other groups. Three Neisseria species were detected at high frequencies in CF-PF (Neisseria flavescens, Neisseria elongata, and Neisseria pharynges), with the latter being significant. Previous studies have shown that N. flavescens, N. pharynges, Neisseria flava, and Neisseria mucosa are associated with health (11, 20). N. flavescens is known to be asaccharolytic (52), so it should not contribute to acid production, and therefore caries progression, via fermentation of carbohydrates. It is interesting to note that while S. sanguinis and Neisseria species often dominated CF-PF communities, when they were absent, other species such as Leptotrichia shahii were the dominant species (Fig. 4A). The proportion of Leptotrichia shahii organisms was decreased when the microbial profiles of CF-PF collected at baseline were compared to those of CAE-PF collected at 12 months for the children who changed their caries status from CF to CAE (see Fig. S5 in the supplemental material). Hence, the various oral Leptotrichia spp. identified in this study may differ substantively in their roles in and contributions to health and disease.
Interestingly, most of the taxa detected in plaques of healthy tooth sites are considered commensals or overtly beneficial. However, species deemed caries pathogens (e.g., S. mutans and Veillonella dispar) were also found in these healthy sites albeit at lower proportions than in diseased sites, consistent with the ecological plaque hypothesis. The polymicrobial nature of dental caries and the interdependent functions of different members of the oral microbiome stress the need for further identification of how the species involved in health and disease onset interact with one another and their environment to influence the pathogenic potential and composition of the microbiota at specific sites. Although diet was not a component evaluated in this study, it is likely that differences in the dietary habits of the subjects may help explain the predominance of certain species in different tooth sites. It is also important to consider the established substantial genotypic and phenotypic heterogeneity within given taxa/species of oral bacteria when interpreting microbiome data. Clearly, as knowledge about microbial compositions in healthy and diseased states is acquired, phenotypic and functional studies must also be conducted to establish reliable correlations of specific organisms with health status (12).
Conclusions.
The microbiomes of supragingival dental plaques differ substantially among tooth surfaces and children of different caries status. Importantly, well-known acidogenic and aciduric species, such as S. mutans, S. wiggsiae, P. denticolens, and L. salivarius, were found almost exclusively in plaques collected from dentin carious lesions. Conversely, S. sanguinis and certain species of Neisseria and Leptotrichia were frequently found in plaques collected from healthy tooth surfaces. Moreover, a high diversity of low-frequency “background” species was observed in the transition from health to disease states. Our findings significantly expand the current knowledge of microbial profile changes with stages of childhood caries and the ecological nature of caries etiology. This study provides valuable new insights into the oral microbiome at the community and site-specific levels to support future metabolomic and transcriptomic studies, coupled with functional assays, for the development of novel strategies to identify and manage children at greater risk of developing caries.
MATERIALS AND METHODS
Study group.
A total of 55 children aged 2 to 7 years at baseline were recruited as part of an ongoing longitudinal study and assessed at baseline and at 12 months. Informed consent was obtained from parents or legal guardians of each child under a protocol approved by the Institutional Review Board of the University of Florida Health Science Center (approval number 272-2010).
The selection process for both baseline and 12-month study visits excluded children who were treated with antibiotics within 3 months of either study visit, those who were taking any medication, or those who had orthodontic appliances. At baseline, children were grouped by caries status as caries free (CF) with no clinical or reported evidence of caries experience (no decayed, missing, and filled teeth [DMFT]), caries active with enamel lesions only (CAE) (no decayed teeth [DT] and ≥0 missing and filled teeth [MFT]), and caries active with at least two cavitated, unrestored dentin carious lesions (CA) (≥2 DT and ≥0 MFT). At 12 months, the caries status was reassessed and reassigned, when necessary.
Caries diagnosis.
Carious lesions were detected and diagnosed by a calibrated examiner (M.M.N.) using International Caries Detection and Assessment System II (ICDAS-II) visual criteria (53) and the lesion activity assessment (LAA) scoring system (54), which determines the activity of carious lesions based on clinical appearance, the presence of plaque accumulation, and tactile sensation (55). Teeth were examined before and after the removal of dental plaque as well as before and after being dried with compressed air for 5 s. The ICDAS scores for individual tooth surfaces are defined as follows: 0 for no or a slight change in enamel after air drying, 1 for the first visual change in enamel after air drying, 2 for distinct visual changes in enamel before air drying, 3 for localized enamel breakdown without visual signs of dentin involvement, 4 for an underlying dark shadow from dentin, 5 for a distinct cavity with visible dentin, and 6 for an extensive and distinct cavity with visible dentin affecting more than half of the surface. The range of ICDAS scores as a function of the caries status groups were CF (no activity; ICDAS score = 0), CAE (active lesions; ICDAS score = 0 to 3), and CA (active lesions; ICDAS score = 0 to 6). The threshold to define the CA group was the presence of at least two ICDAS scores of 5 and/or 6; however, CA children could also present with other carious lesions with lower ICDAS scores (noncavitated; enamel and dentin lesions).
Sample collection.
Children were required to refrain from oral hygiene procedures for at least 8 h prior to the collection of dental plaque samples. Supragingival plaque samples were collected separately from (i) tooth surfaces that were caries free (PF) (ICDAS score = 0); (ii) active, enamel carious lesions (PE) (ICDAS score = 1 to 3); and (iii) active, dentin carious lesions (PD) (ICDAS score of ≥4) (55). Each plaque sample was obtained by pooling material from at least two different tooth sites of similar health conditions using sterile periodontal curettes, and more than one type of sample could have been collected from the same subject. PF samples were collected from all participating children, whereas PE samples were collected from CAE and CA (if available) children, and PD samples were collected from CA children. A total of 186 site-specific plaque samples were collected at baseline and at 12 months and grouped as CF-PF (n = 26 and 20, respectively), CAE-PF (n = 15 and 20, respectively), CAE-PE (n = 20 and 14, respectively), CA-PF (n = 13 and 14, respectively), CA-PE (n = 11 and 13, respectively), and CA-PD (n = 13 and 7, respectively).
Bacterial community profiles.
The HOMINGS approach (http://homings.forsyth.org/) was used to survey the bacterial profiles of the plaque samples. Briefly, DNA was purified from plaque samples by using a protocol that includes overnight incubation in the presence of Ready-Lyse lysozyme solution (Epicentre, WI, USA) and the MasterPure DNA purification kit (Epicentre, WI, USA). Purified DNA was sequenced by using the Illumina MiSeq platform (Illumina, San Diego, CA). Sequencing of the 16S rRNA V3-V4 region using primers described previously (56) was performed at the HOMINGS core facility at the Forsyth Institute (Boston, MA, USA). The HOMINGS approach assigns taxonomy by using a customized BLAST program called ProbeSeq, which contains sequences of species- and genus-specific 16S rRNA probes based on data in the Human Oral Microbiome Database (HOMD). Each probe represents a distinct OTU. Bacterial identification is based on the use of 638 oligonucleotide probes (17 to 40 bases) targeting individual oral bacterial species and/or a few closely related species and 129 genus-specific probes that identify closely related species within the same genus (see Table S4 in the supplemental material).
Statistical analyses.
Python scripts within the software package QIIME version 1.9.0 (57) were used to analyze HOMINGS OTU counts. Normalized counts were used to calculate alpha (Shannon index) (58) and beta (Bray-Curtis) (59) diversity measures. Plaque groups were tested for significant differences in alpha diversity values by using Student's t tests and in beta diversity values by using PERMANOVAs. P values were corrected for multiple testing using the false discovery rate (FDR). PERMANOVA components were used to build a dendrogram by the neighbor-joining algorithm implemented in Geneious v7.1.6 (60). The frequencies of OTUs among plaque groups were tested for significant differences by using a Kruskal-Wallis test. P values were generated by using 10,000 permutations and corrected for multiple testing using the FDR. db-RDA was performed by using QIIME.
Availability of data.
Data have been deposited in the NCBI Sequence Read Archive under accession numbers SRR5643118, SRR5643119, SRR5643121, and SRR5643122.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by National Institute of Dental and Craniofacial Research grant K23-DE023579.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00106-17.
REFERENCES
- 1.Marsh PD. 2006. Dental plaque as a biofilm and a microbial community—implications for health and disease. BMC Oral Health 6(Suppl 1):S14. doi: 10.1186/1472-6831-6-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradshaw DJ, Marsh PD. 1998. Analysis of pH-driven disruption of oral microbial communities in vitro. Caries Res 32:456–462. doi: 10.1159/000016487. [DOI] [PubMed] [Google Scholar]
- 3.Burne RA. 1998. Oral streptococci. Products of their environment. J Dent Res 77:445–452. doi: 10.1177/00220345980770030301. [DOI] [PubMed] [Google Scholar]
- 4.van Houte J, Lopman J, Kent R. 1994. The predominant cultivable flora of sound and carious human root surfaces. J Dent Res 73:1727–1734. [DOI] [PubMed] [Google Scholar]
- 5.van Ruyven FO, Lingstrom P, van Houte J, Kent R. 2000. Relationship among mutans streptococci, “low-pH” bacteria, and lodophilic polysaccharide-producing bacteria in dental plaque and early enamel caries in humans. J Dent Res 79:778–784. doi: 10.1177/00220345000790021201. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi N. 2015. Oral microbiome metabolism: from “who are they?” to “what are they doing?” J Dent Res 94:1628–1637. doi: 10.1177/0022034515606045. [DOI] [PubMed] [Google Scholar]
- 7.Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, Leys EJ, Paster BJ. 2008. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol 46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corby PM, Lyons-Weiler J, Bretz WA, Hart TC, Aas JA, Boumenna T, Goss J, Corby AL, Junior HM, Weyant RJ, Paster BJ. 2005. Microbial risk indicators of early childhood caries. J Clin Microbiol 43:5753–5759. doi: 10.1128/JCM.43.11.5753-5759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. 2003. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol 30:644–654. doi: 10.1034/j.1600-051X.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- 10.Gross EL, Leys EJ, Gasparovich SR, Firestone ND, Schwartzbaum JA, Janies DA, Asnani K, Griffen AL. 2010. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol 48:4121–4128. doi: 10.1128/JCM.01232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crielaard W, Zaura E, Schuller AA, Huse SM, Montijn RC, Keijser BJ. 2011. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics 4:22. doi: 10.1186/1755-8794-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burne RA, Zeng L, Ahn SJ, Palmer SR, Liu Y, Lefebure T, Stanhope MJ, Nascimento MM. 2012. Progress dissecting the oral microbiome in caries and health. Adv Dent Res 24:77–80. doi: 10.1177/0022034512449462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol 192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duran-Pinedo AE, Frias-Lopez J. 2015. Beyond microbial community composition: functional activities of the oral microbiome in health and disease. Microbes Infect 17:505–516. doi: 10.1016/j.micinf.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon-Soro A, Mira A. 2015. Solving the etiology of dental caries. Trends Microbiol 23:76–82. doi: 10.1016/j.tim.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 16.Belstrom D, Holmstrup P, Bardow A, Kokaras A, Fiehn NE, Paster BJ. 2016. Comparative analysis of bacterial profiles in unstimulated and stimulated saliva samples. J Oral Microbiol 8:30112. doi: 10.3402/jom.v8.30112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belstrom D, Holmstrup P, Bardow A, Kokaras A, Fiehn NE, Paster BJ. 2016. Temporal stability of the salivary microbiota in oral health. PLoS One 11:e0147472. doi: 10.1371/journal.pone.0147472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson I, Witkowska E, Kaveh B, Lif Holgerson P, Tanner AC. 2016. The microbiome in populations with a low and high prevalence of caries. J Dent Res 95:80–86. doi: 10.1177/0022034515609554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. 2005. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL. 2012. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One 7:e47722. doi: 10.1371/journal.pone.0047722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanner AC, Kressirer CA, Faller LL. 2016. Understanding caries from the oral microbiome perspective. J Calif Dent Assoc 44:437–446. [PubMed] [Google Scholar]
- 22.Petersen PE. 2008. World Health Organization global policy for improvement of oral health—World Health Assembly 2007. Int Dent J 58:115–121. doi: 10.1111/j.1875-595X.2008.tb00185.x. [DOI] [PubMed] [Google Scholar]
- 23.Kassebaum NJ, Bernabe E, Dahiya M, Bhandari B, Murray CJ, Marcenes W. 2015. Global burden of untreated caries: a systematic review and metaregression. J Dent Res 94:650–658. doi: 10.1177/0022034515573272. [DOI] [PubMed] [Google Scholar]
- 24.Lee HJ, Kim JB, Jin BH, Paik DI, Bae KH. 2015. Risk factors for dental caries in childhood: a five-year survival analysis. Community Dent Oral Epidemiol 43:163–171. doi: 10.1111/cdoe.12136. [DOI] [PubMed] [Google Scholar]
- 25.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ. 2014. Host lifestyle affects human microbiota on daily timescales. Genome Biol 15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. 2013. The long-term stability of the human gut microbiota. Science 341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flores GE, Caporaso JG, Henley JB, Rideout JR, Domogala D, Chase J, Leff JW, Vazquez-Baeza Y, Gonzalez A, Knight R, Dunn RR, Fierer N. 2014. Temporal variability is a personalized feature of the human microbiome. Genome Biol 15:531. doi: 10.1186/s13059-014-0531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, Zhong X, Koenig SS, Fu L, Ma ZS, Zhou X, Abdo Z, Forney LJ, Ravel J. 2012. Temporal dynamics of the human vaginal microbiota. Sci Transl Med 4:132ra52. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nasidze I, Li J, Quinque D, Tang K, Stoneking M. 2009. Global diversity in the human salivary microbiome. Genome Res 19:636–643. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Utter DR, Mark Welch JL, Borisy GG. 2016. Individuality, stability, and variability of the plaque microbiome. Front Microbiol 7:564. doi: 10.3389/fmicb.2016.00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu X, He J, Xue J, Wang Y, Li K, Zhang K, Guo Q, Liu X, Zhou Y, Cheng L, Li M, Li Y, Li Y, Shi W, Zhou X. 2015. Oral cavity contains distinct niches with dynamic microbial communities. Environ Microbiol 17:699–710. doi: 10.1111/1462-2920.12502. [DOI] [PubMed] [Google Scholar]
- 32.Zaura E, Keijser BJ, Huse SM, Crielaard W. 2009. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. 2016. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci U S A 113:E791–E800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Wang W. 2002. Predicting caries in permanent teeth from caries in primary teeth: an eight-year cohort study. J Dent Res 81:561–566. doi: 10.1177/154405910208100812. [DOI] [PubMed] [Google Scholar]
- 35.Luo AH, Yang DQ, Xin BC, Paster BJ, Qin J. 2012. Microbial profiles in saliva from children with and without caries in mixed dentition. Oral Dis 18:595–601. doi: 10.1111/j.1601-0825.2012.01915.x. [DOI] [PubMed] [Google Scholar]
- 36.Thomas RZ, Zijnge V, Cicek A, de Soet JJ, Harmsen HJ, Huysmans MC. 2012. Shifts in the microbial population in relation to in situ caries progression. Caries Res 46:427–431. doi: 10.1159/000339482. [DOI] [PubMed] [Google Scholar]
- 37.Tanner AC, Mathney JM, Kent RL, Chalmers NI, Hughes CV, Loo CY, Pradhan N, Kanasi E, Hwang J, Dahlan MA, Papadopolou E, Dewhirst FE. 2011. Cultivable anaerobic microbiota of severe early childhood caries. J Clin Microbiol 49:1464–1474. doi: 10.1128/JCM.02427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanner AC, Kent RL Jr, Holgerson PL, Hughes CV, Loo CY, Kanasi E, Chalmers NI, Johansson I. 2011. Microbiota of severe early childhood caries before and after therapy. J Dent Res 90:1298–1305. doi: 10.1177/0022034511421201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Modesto M, Biavati B, Mattarelli P. 2006. Occurrence of the family Bifidobacteriaceae in human dental caries and plaque. Caries Res 40:271–276. doi: 10.1159/000092237. [DOI] [PubMed] [Google Scholar]
- 40.Mantzourani M, Gilbert SC, Sulong HN, Sheehy EC, Tank S, Fenlon M, Beighton D. 2009. The isolation of bifidobacteria from occlusal carious lesions in children and adults. Caries Res 43:308–313. doi: 10.1159/000222659. [DOI] [PubMed] [Google Scholar]
- 41.Henne K, Rheinberg A, Melzer-Krick B, Conrads G. 2015. Aciduric microbial taxa including Scardovia wiggsiae and Bifidobacterium spp. in caries and caries free subjects. Anaerobe 35:60–65. doi: 10.1016/j.anaerobe.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 42.Tanner AC. 2015. Anaerobic culture to detect periodontal and caries pathogens. J Oral Biosci 57:18–26. doi: 10.1016/j.job.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes CV, Kolenbrander PE, Andersen RN, Moore LV. 1988. Coaggregation properties of human oral Veillonella spp.: relationship to colonization site and oral ecology. Appl Environ Microbiol 54:1957–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delwiche EA, Pestka JJ, Tortorello ML. 1985. The veillonellae: Gram-negative cocci with a unique physiology. Annu Rev Microbiol 39:175–193. doi: 10.1146/annurev.mi.39.100185.001135. [DOI] [PubMed] [Google Scholar]
- 45.van der Hoeven JS, Toorop AI, Mikx RH. 1978. Symbiotic relationship of Veillonella alcalescens and Streptococcus mutans in dental plaque in gnotobiotic rats. Caries Res 12:142–147. doi: 10.1159/000260324. [DOI] [PubMed] [Google Scholar]
- 46.Mikx FH, van der Hoeven JS, Konig KG, Plasschaert AJ, Guggenheim B. 1972. Establishment of defined microbial ecosystems in germ-free rats. I. The effect of the interactions of Streptococcus mutans or Streptococcus sanguis with Veillonella alcalescens on plaque formation and caries activity. Caries Res 6:211–223. doi: 10.1159/000259801. [DOI] [PubMed] [Google Scholar]
- 47.Edlund A, Yang Y, Yooseph S, Hall AP, Nguyen DD, Dorrestein PC, Nelson KE, He X, Lux R, Shi W, McLean JS. 2015. Meta-omics uncover temporal regulation of pathways across oral microbiome genera during in vitro sugar metabolism. ISME J 9:2605–2619. doi: 10.1038/ismej.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Do T, Sheehy EC, Mulli T, Hughes F, Beighton D. 2015. Transcriptomic analysis of three Veillonella spp. present in carious dentine and in the saliva of caries-free individuals. Front Cell Infect Microbiol 5:25. doi: 10.3389/fcimb.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson J, Pikis A. 2012. Metabolism of sugars by genetically diverse species of oral Leptotrichia. Mol Oral Microbiol 27:34–44. doi: 10.1111/j.2041-1014.2011.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morou-Bermudez E, Rodriguez S, Bello AS, Dominguez-Bello MG. 2015. Urease and dental plaque microbial profiles in children. PLoS One 10:e0139315. doi: 10.1371/journal.pone.0139315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sissons CH, Hancock EM, Perinpanayagam HE, Cutress TW. 1988. The bacteria responsible for ureolysis in artificial dental plaque. Arch Oral Biol 33:727–733. doi: 10.1016/0003-9969(88)90006-4. [DOI] [PubMed] [Google Scholar]
- 52.Boone D, Castenholz R, Garrity GM (ed). 2001. Bergey's manual of systematic bacteriology, 2nd ed Springer, New York, NY. [Google Scholar]
- 53.Ekstrand KR, Martignon S, Ricketts DJ, Qvist V. 2007. Detection and activity assessment of primary coronal caries lesions: a methodologic study. Oper Dent 32:225–235. doi: 10.2341/06-63. [DOI] [PubMed] [Google Scholar]
- 54.Braga MM, Mendes FM, Martignon S, Ricketts DN, Ekstrand KR. 2009. In vitro comparison of Nyvad's system and ICDAS-II with lesion activity assessment for evaluation of severity and activity of occlusal caries lesions in primary teeth. Caries Res 43:405–412. doi: 10.1159/000239755. [DOI] [PubMed] [Google Scholar]
- 55.Nascimento MM, Liu Y, Kalra R, Perry S, Adewumi A, Xu X, Primosch RE, Burne RA. 2013. Oral arginine metabolism may decrease the risk for dental caries in children. J Dent Res 92:604–608. doi: 10.1177/0022034513487907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belstrom D, Holmstrup P, Fiehn NE, Rosing K, Bardow A, Paster BJ, Lynge Pedersen AM. 2016. Bacterial composition in whole saliva from patients with severe hyposalivation—a case-control study. Oral Dis 22:330–337. doi: 10.1111/odi.12452. [DOI] [PubMed] [Google Scholar]
- 57.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shannon CE. 1948. A mathematical theory of communication. Bell Syst Tech J 27:379–423, 623–656. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 59.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 60.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P, Drummond A. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.