ABSTRACT
A major pathway for the detoxification of organic hydroperoxides, such as cumene hydroperoxide (CHP), involves the MarR family transcriptional regulator OhrR and the peroxidase OhrA. However, the effect of these peroxides on the global transcriptome and the contribution of the OhrA/OhrR system to bacterial virulence remain poorly explored. Here, we analyzed the transcriptome profiles of Chromobacterium violaceum exposed to CHP and after the deletion of ohrR, and we show that OhrR controls the virulence of this human opportunistic pathogen. DNA microarray and Northern blot analyses of CHP-treated cells revealed the upregulation of genes related to the detoxification of peroxides (antioxidant enzymes and thiol-reducing systems), the degradation of the aromatic moiety of CHP (oxygenases), and protection against other secondary stresses (DNA repair, heat shock, iron limitation, and nitrogen starvation responses). Furthermore, we identified two upregulated genes (ohrA and a putative diguanylate cyclase with a GGDEF domain for cyclic di-GMP [c-di-GMP] synthesis) and three downregulated genes (hemolysin, chitinase, and collagenase) in the ohrR mutant by transcriptome analysis. Importantly, we show that OhrR directly repressed the expression of the putative diguanylate cyclase. Using a mouse infection model, we demonstrate that the ohrR mutant was attenuated for virulence and showed a decreased bacterial burden in the liver. Moreover, an ohrR-diguanylate cyclase double mutant displayed the same virulence as the wild-type strain. In conclusion, we have defined the transcriptional response to CHP, identified potential virulence factors such as diguanylate cyclase as members of the OhrR regulon, and shown that C. violaceum uses the transcriptional regulator OhrR to modulate its virulence.
KEYWORDS: oxidative stress, organic hydroperoxides, CHP stimulon, OhrA/OhrR system, MarR family, bacterial virulence, cumene hydroperoxide, diguanylate cyclase, transcription factors
INTRODUCTION
Bacteria are exposed to reactive oxygen species (ROS) generated endogenously or produced by phagocytic cells from the mammalian immune system (1–3). Other harmful oxidants are organic hydroperoxides (OHPs) produced either enzymatically during eicosanoid metabolism (4) or by the free radical-catalyzed oxidation of polyunsaturated fatty acids (PUFAs) during lipid peroxidation (5). In the lipid peroxidation process, multiple toxic breakdown molecules are generated, including lipid hydroperoxides (LHPs) and short-chain aldehydes such as malondialdehyde (MDA) (5, 6). Because bacterial membranes are usually devoid of PUFAs and contain mainly saturated fatty acids and monounsaturated fatty acids (UFAs) (7), lipid peroxidation in bacteria is a controversial issue. However, the findings that bacteria are able to incorporate exogenous PUFAs into their membranes (8) and accumulate PUFA-derived reactive aldehydes upon challenge with oxidizing agents (9, 10) suggest the relevance of this process in prokaryotes.
While the one-electron reduction of OHP in the presence of transition metals promotes the propagation of lipid peroxidation, the two-electron reduction catalyzed by enzymes generates the unreactive corresponding alcohols (5, 6). These detoxifying enzymes are mainly glutathione peroxidases (Gpx) in mammalian cells (6, 11) or the Cys-based peroxidase Ohr (organic hydroperoxide resistance protein) in bacteria (12, 13). Numerous data from in vitro and in vivo analyses using cumene hydroperoxide (CHP), tert-butyl hydroperoxide (TBHP), and LHP as OHP molecules indicate that Ohr(A) and its negative regulator OhrR compose a dedicated system for OHP detoxification (14, 15). The oxidation of OhrR upon exposure to these OHP compounds disrupts its DNA binding activity, causing its reversible inactivation and the derepression of ohrA. After OhrR oxidation, an N-terminal cysteine residue undergoes thiolation by low-molecular-weight thiols (1-Cys subfamily) or reacts with a C-terminal cysteine residue forming an intermolecular disulfide bond (2-Cys subfamily) (14, 15). Recently, bacilliredoxins and thioredoxins were shown to be the reducing systems for the 1-Cys OhrR from Bacillus subtilis (16) and for the 2-Cys OhrR from Chromobacterium violaceum (17), respectively. OhrR belongs to the MarR family of transcription factors, which contains many other thiol-based redox-sensing regulators that respond to ROS or to reactive electrophilic species (14, 18).
Chromobacterium violaceum is a betaproteobacterium belonging to the family Neisseriaceae that is found in soil and water of tropical and subtropical areas around the world (19, 20). As a saprophytic microorganism, this Gram-negative bacterium encounters natural and xenobiotic aromatic compounds present in soils and is potentially able to use them as carbon sources or deal with their toxic effects (21, 22). The aromatic peroxide CHP can be released in these environments as an intermediate product from lignin catabolism (23) or as a pollutant derived from the chemical industry (24). In addition to its free-living lifestyle, C. violaceum can also act as an occasional opportunistic pathogen of animals and humans, infecting individuals mainly through skin lesions (25). Clinical manifestations include abscess formation in the skin, liver, and lung and, in many cases, progression to death (19, 25). A type III secretion system (T3SS) is essential for C. violaceum virulence in mice (26, 27), and the T3SS needle protein CprI can be recognized by the NLRC4 inflammasome in human macrophages (28). The clearance of C. violaceum infection is mediated by the NLRC4 inflammasome that triggers pyroptosis in macrophages or natural killer cytotoxicity in liver cells to expose C. violaceum to neutrophil killing (29). NADPH oxidase-deficient mice or patients with chronic granulomatous disease (CGD) are highly susceptible to C. violaceum infection (25, 29, 30), indicating the pivotal role of ROS derived from phagocytes in the control of C. violaceum.
In this study, we determined the transcriptional profile of the adaptive response of C. violaceum to CHP insult and characterized the OhrR regulon. CHP caused increased expression levels of genes related to multiple stress response pathways, including the OhrR-repressed genes ohrA and ohrR, as expected, and CV_0208, which encodes a putative diguanylate cyclase-containing GGDEF domain. Furthermore, the deletion of ohrR resulted in the decreased expression of three genes (CV_0362, CV_1440, and CV_2001) encoding putative virulence factors. Remarkably, we found that an ohrR mutant strain is attenuated for virulence in a mouse acute-infection model, which might be related to the increased production of the second messenger cyclic di-GMP (c-di-GMP) and the decreased expression of extracellular enzymes required for tissue dissemination in this mutant strain. These data led us to propose that the C. violaceum OhrA/OhrR system is required for bacterial defense against OHP and to modulate virulence in the host.
RESULTS
Global transcriptional profile of C. violaceum in response to CHP. (i) Overview of the CHP stimulon.
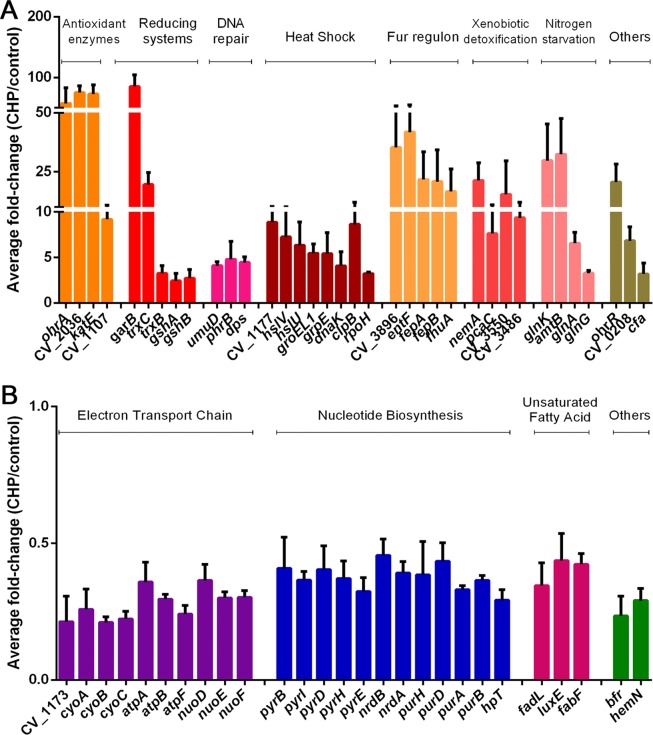
To characterize the processes involved in the adaptive response of C. violaceum to CHP, we performed genome-wide transcriptome analyses of mid-log-phase bacterial cells exposed to 100 μM CHP for 10 min, which is a sublethal dose that causes the maximum expression of ohrA (17). These conditions resulted in the upregulation of 255 genes (see Table S1 in the supplemental material) and the downregulation of 175 genes (Table S2) compared to their expression levels in the untreated control, using the criterion of a minimum of a 2-fold expression change as a cutoff. These differentially expressed genes were grouped by functional categories (Tables S1 and S2), and the most representative genes of the main biological processes are shown in Fig. 1. Among the upregulated genes, highlighted categories include antioxidant enzymes, xenobiotic degradation pathways, DNA repair, the heat shock response, and iron limitation and nitrogen starvation responses (Fig. 1A), while for downregulated genes, some of the categories were the electron transport chain, nucleotide biosynthesis, and the unsaturation of fatty acids (Fig. 1B). Overall, we observed a switch in the global gene expression pattern from biosynthetic and maintenance functions to stress protection and detoxification cellular functions upon exposure to CHP.
FIG 1.
Transcriptome analysis of the CHP stimulon in C. violaceum. Data are average ratios with standard deviations from three biological replicates comparing cultures from wild-type strain ATCC 12472 exposed to 100 μM CHP for 10 min to cultures of untreated cells. (A) Upregulated genes in the presence of CHP. (B) Downregulated genes in the presence of CHP. Here, we show only some genes representative of the main functional categories discussed in the text (all genes with altered expression levels are shown in Tables S1 and S2 in the supplemental material).
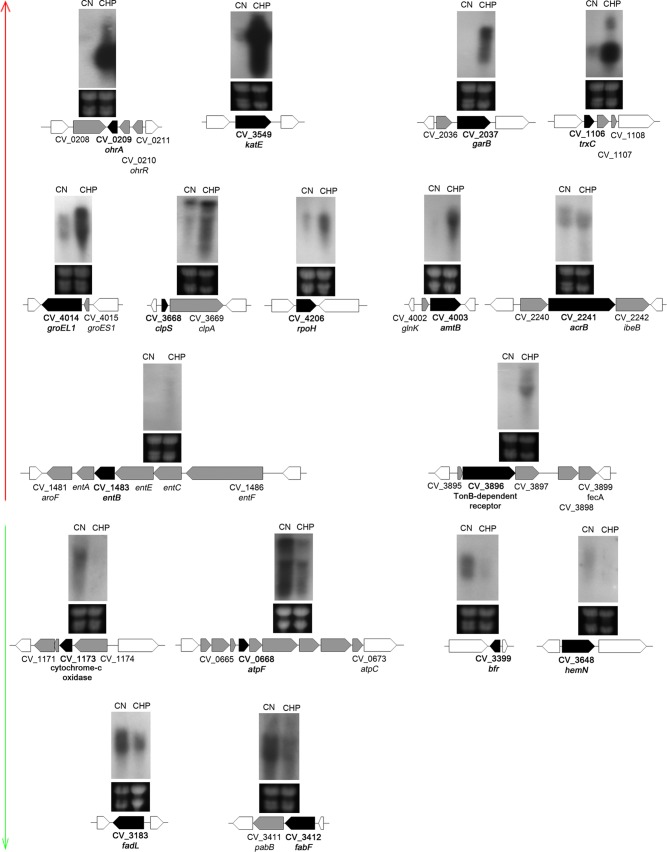
To validate our microarray results, we performed Northern blot assays for differentially expressed genes representative of the main functional categories altered by CHP stress (Fig. 2). For all tested target genes, the microarray and Northern blot data were consistent with each other, allowing us to identify the CHP stimulon of C. violaceum. Noteworthy, considering that most of the 17 genes selected for validation are organized into putative operons, we potentially confirmed the microarray data for several other genes (Fig. 2). Additional validation was obtained for the katE gene by a catalase assay. The strong upregulation of the katE gene correlated with high catalase activity under CHP stress (Fig. S1). Below are some considerations on each one of the categories altered by CHP.
FIG 2.
Northern blot validation of selected genes with altered expression in the presence of CHP in C. violaceum. Total RNA was extracted from wild-type strain ATCC 12472 grown in LB medium (control [CN]) or after exposure to 100 μM cumene hydroperoxide for 10 min (CHP). RNA was separated on a 1.5% agarose gel, transferred onto a nylon membrane, and hybridized with radiolabeled probes specific for each indicated gene (black arrows). Genes indicated in black or gray arrows were differentially expressed in the microarray analysis. White arrows indicate neighboring genes. We validated 11 upregulated genes (red vertical arrow) and six downregulated genes (green vertical arrow). Levels of rRNA were used as a loading control (bottom gel).
(ii) Upregulation of antioxidant enzymes and xenobiotic detoxification pathways.
Many genes encoding antioxidant enzymes showed increased expression in the presence of CHP, such as the glutathione amide-dependent peroxidase CV_2036 (75-fold), the monofunctional catalase KatE (73-fold), the lipoyl-dependent peroxidase OhrA (61-fold), the glutathione peroxidase CV_1107 (9-fold), the cytochrome c peroxidase CV_0300 (3-fold), the peroxiredoxin BCP/PrxQ (2-fold), and the superoxide dismutase SodB1 (2.4-fold) (Fig. 1A and 2; see also Table S1 in the supplemental material). Among these enzymes, OhrA, glutathione amide peroxidase, and glutathione peroxidase were previously described as OHP-scavenging enzymes (12, 13, 31). Interestingly, besides ohrA and ohrR, which compose a dedicated system for OHP detoxification in C. violaceum (17), we also observed increased expression levels of trxC (19-fold) and trxB (3-fold). Enzymes of the thioredoxin system were shown to be involved in OhrR reduction in vitro (17).
The thioredoxin system operates in parallel with the glutathione-dependent glutaredoxin system to repair thiol groups from oxidation (32, 33). Accordingly, upon CHP stress, we detected the upregulation of genes encoding glutathione synthetase enzymes (CV_4275 and CV_4276) and glutathione S-transferases (GSTs) (CV_1164 and CV_2424) (Table S1). We also detected a strong upregulation of CV_2037 (85-fold), which encodes a putative glutathione amide reductase (GAR), a flavoprotein disulfide reductase that probably supports the peroxidase activity of the glutathione amide-dependent peroxidase CV_2036 (Fig. 1A and 2 and Table S1) (31). GSTs participate in xenobiotic degradation and protection against oxidative stress (34). The induction of many genes encoding glyoxalase domain-containing proteins (CV_0197, CV_0969, CV_1319, CV_2341, and CV_3486), quinone oxidoreductase (CV_2043), and the NADH-dependent oxidoreductase NemA (CV_3501) (21-fold induction) suggests a response to electrophilic stress. This stress could arise from aldehydes derived from CHP-dependent lipid peroxidation (5, 6). Indeed, the nemRA-gloA operon is critical for electrophile detoxification in Escherichia coli (35). Genes encoding an aromatic ring-opening extradiol dioxygenase (CV_3550) (15-fold induction), a carboxymuconolactone decarboxylase (pcaC), and a hydroxybenzoate transporter (pcaK) were also induced (Fig. 1A and Table S1), suggesting a meta-cleavage pathway (36) for the degradation of the aromatic moiety of CHP in C. violaceum.
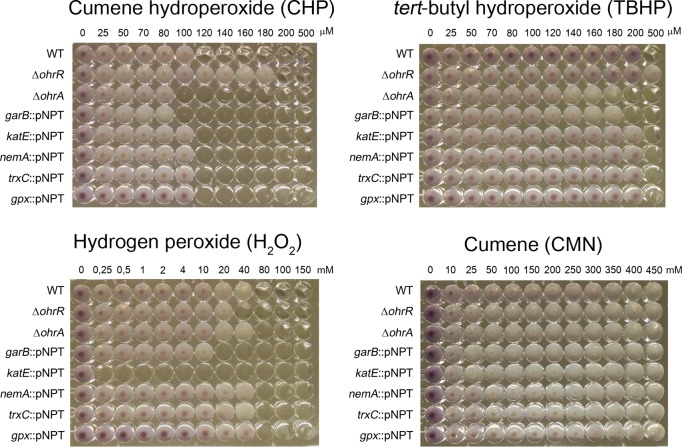
To access the contribution of CHP-induced genes to C. violaceum resistance to CHP and other oxidant agents, we performed phenotype characterization of mutant strains for five selected genes (garB, katE, nemA, trxC, and gpx) as well as for ΔohrA and ΔohrR mutants as controls (Fig. 3). Compared to the wild-type (WT) strain, the ΔohrA and garB::pNPT mutant strains were more susceptible to CHP and TBHP, whereas the ΔohrR mutant was more resistant. These data are consistent with the role of the OhrA/OhrR system in OHP detoxification for many bacteria (14, 15, 17) and are the first description of OHP susceptibility for a garB mutant, reinforcing biochemical evidence of the GAR/Prx/Grx system (encoded by the CV_2036-CV_2037 [garAB] operon) as enzymes for OHP detoxification (31). Although the katE, nemA, trxC, and gpx genes were highly induced by CHP, none of the mutant strains showed a phenotype for CHP and TBHP (Fig. 3), perhaps due to the high redundancy observed for antioxidant enzymes and reducing systems (13, 32). As expected, the katE::pNPT mutant strain was highly susceptible to H2O2 (Fig. 3) and showed no catalase activity (Fig. S1). As a control, wild-type C. violaceum and all mutant strains were able to grow even with high concentrations of cumene (CMN) in comparison to CHP (Fig. 3).
FIG 3.
Susceptibility of C. violaceum strains to CHP, TBHP, and H2O2. An MIC assay was performed to assess the sensitivities of C. violaceum mutant strains to distinct hydroperoxides. The identity of each tested strain is shown on the left. The concentrations of hydroperoxides used are shown at the top of each panel. MIC assays were done at least in biological triplicates. CMN was used as a control, and no growth inhibition was detected for all tested strains.
(iii) Upregulation of DNA repair enzymes.
CHP also induced the expression of genes encoding DNA repair systems, including the photolyase PhrB and a photolyase-related protein (phrB and CV_3229), an Smr domain-containing protein (CV_2814), and translesion DNA polymerase V (Pol V) (umuD) (Fig. 1A and Table S1), which suggests that DNA damage may be caused by CHP-derived aldehydes (5, 37), as suggested above. Two nucleoid-associated proteins (dps and CV_1363) were also induced, which is consistent with the role of Dps in protection against iron and peroxide-mediated DNA damage (38).
(iv) Upregulation of the σ32-dependent heat shock response.
Exposure to CHP also resulted in the induction of a classical heat shock response, including chaperones (htpG, grpE, dnaK, groES, groEL, groES1, and groEL1), proteases (hslV, hslU, clpB, lon, clpS, and clpA), and a small heat shock protein (CV_1177) (Fig. 1A and 2 and Table S1). The induction of the heat shock sigma factor σ32 (rpoH), confirmed by Northern blotting (Fig. 2), suggests that these heat shock genes belong to the σ32 regulon in C. violaceum, as described previously for E. coli (39). The stimulus that induced this heat shock response could be derived from the aromatic moiety of the CHP molecule, as observed for phenol and catechol compounds (40), or from the cumyl alcohol group as a consequence of a two-electron CHP reduction, as described previously for TBHP and its corresponding alcohol in B. subtilis (41).
(v) Upregulation of the Fur-dependent iron limitation response.
Interestingly, at least 40 genes involved in iron uptake and transport were upregulated in the presence of CHP, many of which showed strong upregulation (Fig. 1A and Table S1). Several of these genes are organized into large, coregulated clusters or operons, including the two genes (CV_1483 and CV_3896) validated by Northern blotting (Fig. 2). CHP-upregulated iron acquisition systems included outer membrane TonB-dependent receptors (CV_0077, CV_1491, CV_1982, CV_3896, fepA, and fhuA), energy-transducing TonB-ExbB-ExbD protein complexes (CV_0400-CV_0399-CV_0398, CV_1971-CV_1972-CV_1973-CV_1974, CV_1983, CV_1986, and CV_4254), ABC transporters (CV_3895-CV_3896-CV_3897-CV_3898-CV_3899, CV_1491-CV_1490-CV_1489-CV_1488-CV_1487, and CV_2236-CV_2235-CV_2234), and enzymes for the synthesis of iron-chelating molecules such as riboflavin (CV_2387-CV_2388-CV_2389) and the siderophore enterobactin (entFCEBA and CV_2231-CV_2232-CV_2233). As the above-mentioned genes are members of the Fur regulon in many bacteria (42–44) and the C. violaceum genome encodes the Fur protein (21), we hypothesize that CHP is derepressing the Fur regulon. CHP could be causing either direct iron starvation due to a reaction with Fe(II) (45) or the oxidation of the holo-Fur protein, disrupting its DNA binding activity (46).
(vi) Upregulation of the NtrC-dependent nitrogen starvation response.
Several genes belonging to the NtrC regulon that are required for nitrogen assimilation in enteric bacteria (47) were upregulated by CHP in C. violaceum (Fig. 1A and 2 and Table S1). Products of these genes include ammonia (amtB) (32-fold) and glutamate (gltK) transporters, glutamine synthetase (glnA) (6-fold), glutamate synthase (gltB), nitrogen regulatory protein PII (glnK) (39-fold), and the two-component system composed of NtrB (glnL) and NtrC (glnG). The role of this response in CHP stress remains elusive.
(vii) Other CHP-upregulated genes.
Some operons encoding efflux systems were upregulated by CHP stress, including an RND multidrug efflux pump (CV_2240-acrB-ibeB), two putative drug efflux systems (CV_1179-CV_1180-CV_1181 and CV_3922-CV_3923-CV_3924), and two metal exporters (chrBA and zntA) (Fig. 2 and Table S1), suggesting a mechanism for the extrusion of metals or of its own CHP compound. Several genes encoding transcription factors of unknown function belonging to distinct families (AraC, TetR, and GntR) were also induced by CHP (Table S1), suggesting that these regulatory proteins are good candidates to regulate a particular set of genes of the CHP stimulon. Interestingly, two genes (cfa and desD) encoding enzymes required to modify the structure of preexisting fatty acids were induced by CHP (Table S1). The role of front-end Δ6 desaturases (DesD) is to introduce double bonds during the synthesis of PUFA in eukaryotes (48). Cyclopropane fatty acid synthases (Cfa) convert preexisting UFAs to cyclopropane fatty acids in bacteria (7), which could render the C. violaceum membrane less susceptible to oxidation by CHP.
(viii) Genes downregulated by CHP stress.
Energy generation by oxidative phosphorylation is the biological process with more genes downregulated by CHP. Among them are large operons encoding NADH dehydrogenase (nuoDEFGHIJK), fumarate reductase (ducB-frdAB), cytochrome c reductase (petAB), cytochrome c oxidase complexes (CV_1174-CV_1173-CV_1172-CV_1171 and cyoABCDE), and ATP synthase (atpBEFHAGDC) (Fig. 1B and 2 and Table S2). Some of these genes, together with the CHP-downregulated genes bfr (bacterioferritin) and hemN (heme biosynthesis) (Fig. 1B and 2), are activated by Fur in other bacteria (43, 49), reinforcing that CHP could be destructive to the Fur protein.
Another category with several genes downregulated by CHP is nucleotide metabolism, including both purine metabolism (purHD, nrdA-CV_2286-CV_2285-nrdB, CV_2922, purA2, purB, hpT, and prsA) and pyrimidine metabolism (pyrBI, pyrD, pyrH, and pyrE) (Fig. 1B and Table S2). Considering that the synthesis of deoxyribonucleotides by the ribonucleotide reductase NrdAB requires reduction by thioredoxin and glutaredoxin systems (32, 33), the downregulation of nucleotide biosynthesis under CHP stress might be a relevant strategy to direct these thiol-disulfide oxidoreductases for the reduction of antioxidant enzymes.
Some genes that participate in the metabolism of fatty acids were also downregulated by CHP (Fig. 1B and Table S2), two of which were validated by Northern blotting (Fig. 2). Among these genes, fadL encodes a long-chain fatty acid transport protein, fabF and fabH encode enzymes for fatty acids biosynthesis (50), and luxEC encodes a putative fatty acid reductase complex required for aldehyde recycling in bioluminescent bacteria (51). Thus, a diminution in the transport of long-chain fatty acids (FadL) and a decrease in the production of the UFA cis-vaccenate (FabF) may contribute to minimizing the lipid peroxidation process caused by CHP (5, 6). In E. coli, FabF is a temperature-sensitive enzyme that increases the production of cis-vaccenate at low temperatures (50).
Previously, we showed that C. violaceum relies mainly on the OhrA/OhrR system to respond to CHP stress (17). Therefore, we decided to evaluate the transcriptome of a strain that lacks OhrR.
Global analysis of the OhrR regulon in C. violaceum. (i) OhrR has a small regulon, including putative virulence genes.
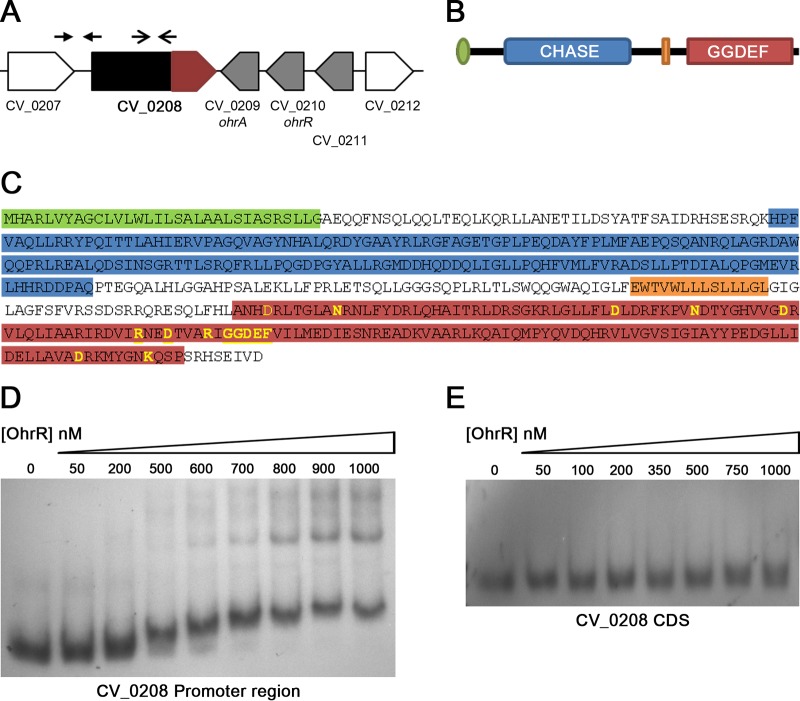
Although the role of the OhrR/OhrA system in OHP detoxification has been studied for several bacteria (14, 15, 17), little is known about other genes whose expression might be regulated by OhrR. To define the global repertoire of genes regulated by OhrR in C. violaceum, we performed DNA microarray analyses by comparing mRNA of wild-type strain ATCC 12472 with that of the ΔohrR mutant strain. Remarkably, only six genes were differentially expressed, with three being upregulated (CV_0208, CV_0209, and CV_0211) and three being downregulated (CV_0362, CV_1440, and CV_2001) in the ohrR mutant (Table 1), and these results were confirmed by Northern blotting (Fig. 4). Because the expressions of ohrA (CV_0209), ohrR (CV_0210), and CV_0208 were also upregulated by CHP (Fig. 1A and 4A; see also Table S1 in the supplemental material), these three genes are probably responsive to CHP through OhrR oxidation in the wild-type strain. While these data confirmed previously reported observations related to the pattern of ohrA and ohrR expression (17), they demonstrate for the first time that a putative diguanylate cyclase (CV_0208) is derepressed by CHP in an OhrR-dependent manner. CV_0208 encodes a protein containing a GGDEF domain that is characteristic of diguanylate cyclases (52), suggesting that, somehow, c-di-GMP production is controlled by OhrR in C. violaceum. With respect to the downregulated genes, they encode putative virulence factors, namely, a thermolabile hemolysin/lecithinase (CV_0362), a putative chitinase (CV_1440), and a microbial collagenase (CV_2001) (Table 1 and Fig. 4B). These enzymes are secreted by C. violaceum, as demonstrated in a previous exoproteome analysis (53).
TABLE 1.
Genes differentially expressed in the ΔohrR mutant strain
| Open reading frame | Gene | Function | Fold change (ΔohrR mutant/WT strain) |
|---|---|---|---|
| Upregulated | |||
| CV_0208 | Putative diguanylate cyclase (GGDEF domain) | 36.66 | |
| CV_0209 | ohrA | Organic hydroperoxide resistance protein | 133.46 |
| CV_0211 | Hypothetical protein | 4.66 | |
| Downregulated | |||
| CV_0362 | Thermolabile hemolysin, lecithin dependent | 0.50 | |
| CV_1440 | Hydrolase transmembrane protein; chitinase | 0.43 | |
| CV_2001 | Microbial collagenase | 0.43 | |
| CV_0210 | ohrR | Transcriptional regulator, MarR family | 0.15 |
FIG 4.
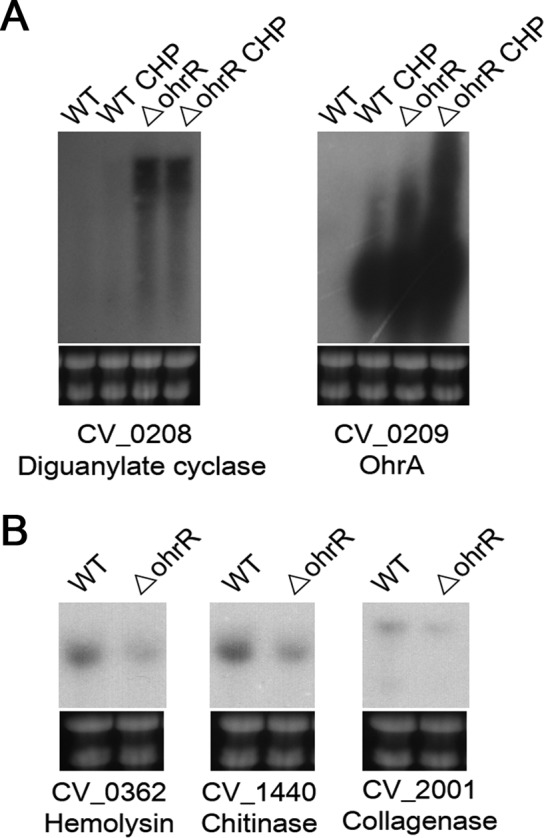
Northern blot validation of the C. violaceum OhrR regulon. Total RNA was extracted from wild-type strain ATCC 12472 (WT) and the ohrR mutant strain (ΔohrR) grown in LB medium without or after exposure to 100 μM CHP for 10 min. These samples were hybridized with specific probes for two upregulated genes (A) and three downregulated genes (B) found by DNA microarray analysis of the WT × ΔohrR mutant (Table 1). Levels of rRNA were used as a loading control (bottom gel).
(ii) The putative diguanylate cyclase CV_0208 is directly repressed by OhrR.
To define whether OhrR binds directly to the promoter region of genes belonging to the OhrR regulon, we performed electrophoretic mobility shift assays (EMSAs) using the purified OhrR protein and PCR-amplified DNA probes (Fig. 5 and Fig. S2). The oxidized or reduced OhrR protein did not bind to the promoter region of the CV_0362, CV_1440, and CV_2001 genes (Fig. S2), indicating that OhrR exerts positive regulation on these virulence genes by an indirect mechanism. As expected, reduced OhrR bound with high affinity to the promoter of ohrA (Fig. S2), confirming previously reported data (17). After a detailed inspection of data in the NCBI Reference Sequence (RefSeq) database, we realized that the CV_0208 gene is larger than previously annotated (21) (Fig. 5A). The new gene annotated in this region (RefSeq accession number CV_RS00965) encodes a protein (RefSeq accession number WP_043595205.1) containing a signal peptide, a CHASE domain, a transmembrane region, and the previously annotated GGDEF domain (Fig. 5A to C). All the amino acid residues required for diguanylate cyclase activity (52) were found within the GGDEF domain (Fig. 5C), suggesting that this protein is a functional enzyme that is able to synthesize the second messenger c-di-GMP. When the EMSAs were performed by using a DNA fragment in the promoter region of the newly annotated CV_0208 gene as a probe (Fig. 5A), we detected the binding of the reduced OhrR protein (Fig. 5D) but not oxidized OhrR (Fig. S2), indicating that CV_0208 is directly repressed by OhrR although with a lower affinity than that observed for the ohrA (Fig. S2) and ohrR (17) promoters. As expected, in a control EMSA, the OhrR protein did not bind to an internal region of the CV_0208 gene (Fig. 5E).
FIG 5.
Analysis of the putative diguanylate cyclase CV_0208. (A) Genomic context of CV_0208. The previous (red) and the new (black and red) annotations for the CV_0208 gene are indicated. Positions of the probes used for EMSAs are indicated by arrows above the genes. (B) Predicted domains found in the reannotated CV_0208 protein. The scheme shows the signal peptide (green), the CHASE domain (blue), the transmembrane region (orange), and the GGDEF domain (red). (C) Amino acid sequence of the reannotated CV_0208 protein highlighting the above-mentioned domains. Key amino acids required for diguanylate cyclase activity are shown in yellow. (D) EMSA using the putative promoter region of the reannotated CV_0208 gene as a probe. The purified OhrR protein was reduced with DTT and used at the indicated concentrations. (E) EMSA using an internal fragment (coding sequence [CDS]) of the CV_0208 gene as a negative control.
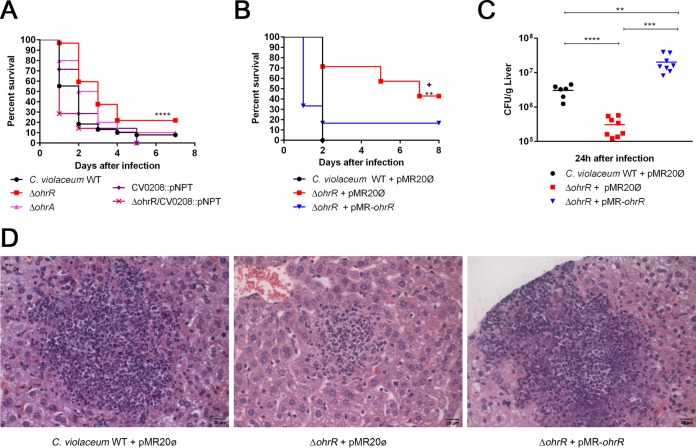
OhrR contributes to C. violaceum virulence in an acute mouse infection model.
Because antioxidant enzymes (OhrA), diguanylate cyclase (CV_0208), and degrading extracellular enzymes (CV_0362, CV_1440, and CV_2001) are all potential virulence factors (54, 55), we verified the contribution of OhrR to C. violaceum virulence in a mouse infection model (Fig. 6). Mice infected with an ΔohrR mutant strain survived longer than did those infected with wild-type strain ATCC 12472, indicating that the ΔohrR mutant is attenuated for virulence. In contrast, the ΔohrA and CV_0208::pNPT mutant strains showed no discernible phenotype with respect to wild-type strain ATCC 12472. Interestingly, a ΔohrR CV_0208::pNPT double mutant strain presented the same virulence phenotype as that observed for the WT strain, suggesting that the increased production of c-di-GMP contributed to the attenuation of virulence of the ΔohrR mutant (Fig. 6A). The attenuated virulence of the ΔohrR mutant strain was fully restored when this mutant was complemented with the ohrR gene in a low-copy-number plasmid (Fig. 6B). Similarly, when we evaluated bacterial dissemination in the liver of mice, we found that the ΔohrR mutant strain showed decreased bacterial burdens (Fig. 6C) and numbers of lesion foci (Fig. 6D; see also Fig. S3 in the supplemental material) for these organs in comparison to the wild-type and complemented strains. Altogether, these results indicated that OhrR is important for virulence control in C. violaceum. Based on the OhrR regulon, we suggest that the synthesis of c-di-GMP and/or the production of enzymes capable of degrading host tissues might play a role in OhrR-mediated virulence control in C. violaceum.
FIG 6.
The transcription factor OhrR controls virulence in C. violaceum. (A) Virulence assays in a mouse infection model. Female BALB/c mice were challenged intraperitoneally with 1 × 106 CFU of each indicated C. violaceum strain. Mice were observed daily, and survival curves were made by using the Kaplan-Meier method (Gehan-Breslow-Wilcoxon test). ****, P < 0.0001 compared to the wild-type strain. At least 7 animals were used for each strain. (B) Complementation tests. The virulence assays were repeated by using the indicated bacterial strains. **, P = 0.0072 compared to the wild-type strain with empty vector pMR20; +, P = 0.0246 compared to the ΔohrR mutant strain with empty vector pMR20. We used at least 6 animals for each strain. (C) Bacterial burden in the liver. Female BALB/c mice were intraperitoneally inoculated with 5 × 105 CFU of each indicated C. violaceum strain. After 24 h of infection, mice were sacrificed for sterile removal of the liver. The liver homogenate was diluted in PBS, and serial dilutions were plated onto LB agar to determine CFU. Data represent the averages of results from at least six biological replicates. Statistical analysis was performed with the Student t test, with a P value of <0.01 being considered significant. ****, P < 0.0001; ***, P = 0.0003; **, P = 0.0044. (D) Liver sections of mice infected as described above for panel C were fixed and stained with hematoxylin and eosin. Images show a larger view of a lesion focus (see Fig. S3 in the supplemental material for counts of lesion foci in the livers). Bar, 20 μm.
DISCUSSION
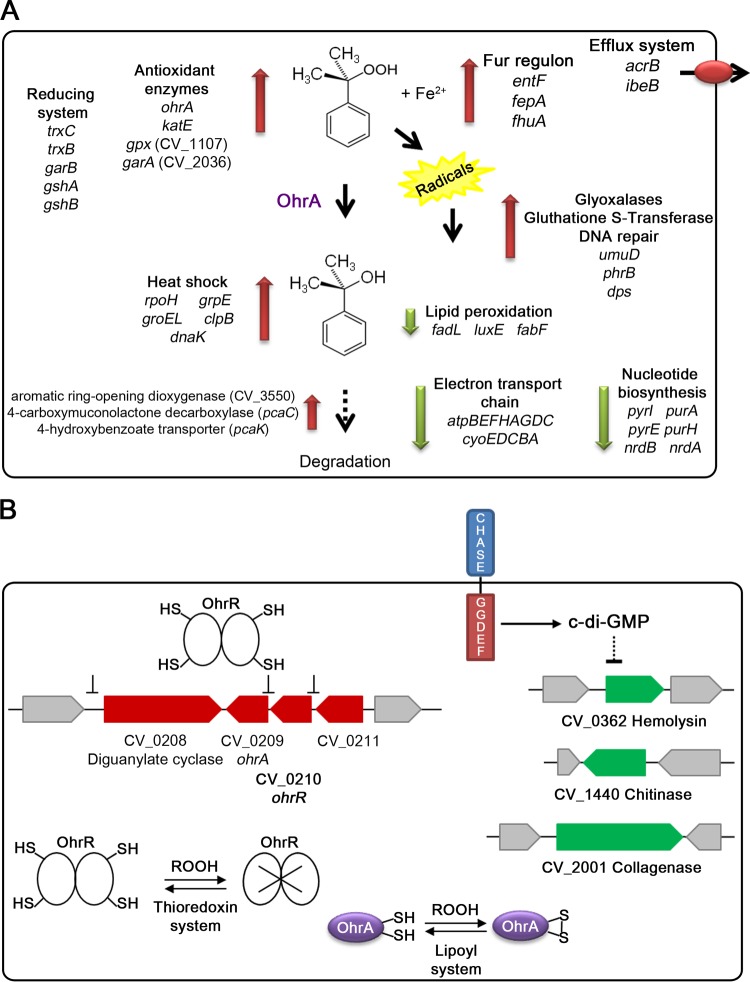
Previous studies on the bacterial response to OHP using TBHP and CHP as model compounds have been focused in the OhrR/OhrA system (14, 15), while genome-wide analysis has been almost completely restricted to hydrogen peroxide (1, 56). In this work, by using transcriptional profiling, we have obtained a global overview of the major transcriptional changes of C. violaceum in response to the aromatic peroxide CHP and have demonstrated that the OhrR regulon includes potential virulence genes, which is in line with our results demonstrating that OhrR controls virulence in this human pathogen. In Fig. 7, we summarize the main adaptive responses and protection mechanisms employed by C. violaceum to deal with CHP and its toxically derived subproducts (Fig. 7A), and we propose a working model to explain how OhrR contributes to virulence control in C. violaceum (Fig. 7B).
FIG 7.
The CHP stimulon and the role of OhrR in C. violaceum virulence. (A) Proposed mechanisms of CHP detoxification in C. violaceum. Transcriptome changes after CPH exposure suggest the following adaptive responses: two-electron reduction of CHP (antioxidant enzymes and thiol-reducing systems), protection against the denaturing effect of cumyl alcohol (heat shock response), protection against subproducts derived from the one-electron reduction of CHP (glyoxalases, glutathione S-transferases, and DNA repair enzymes), and further catabolism of CHP. Downregulated genes indicate the reduction of energy generation (electron transport chain), biosynthesis of nucleotides, and unsaturation of fatty acids. (B) Proposed mechanisms of virulence control by OhrR in C. violaceum. Exposure of C. violaceum to OHPs such as CHP inactivate OhrR, causing the derepression of its regulon (CV_0208, CV_0209, and CV_0210) (in red). The increased expression of the peroxidase OhrA protects against OHP, while high-level expression of CV_0208, encoding a protein with CHASE and GGDEF domains, could increase the synthesis of c-di-GMP in response to OHP or another unknown signal. High levels of c-di-GMP are associated with a reduction of bacterial virulence, the phenotype observed for the ΔohrR mutant, possibly by downregulating the expression of virulence factors (genes indicated in green). Putative pathways for the reduction of OhrR and OhrA are indicated (12, 17).
With respect to redox homeostasis, CHP induced the upregulation of antioxidant enzymes (OhrA, KatE, the Gpx CV_1107, and the peroxidase CV_2036) and thiol-disulfide oxidoreductases (glutathione synthesis; thioredoxin and GAR). While our data confirmed that ohrA belongs to the OhrR regulon (17), other CHP-induced genes involved in antioxidant protection are under the control of OxyR and RpoS in E. coli (56, 57), two regulators not yet studied in C. violaceum. Considering that according to our data, oxyR was downregulated by CHP, whereas hfq, a positive modulator of RpoS activity, was upregulated, we speculate that RpoS rather than OxyR mediates the response to CHP. Indeed, several genes of the E. coli RpoS regulon (katE, dps, and cfa) were upregulated by CHP in C. violaceum. CHP might activate the RpoS regulon indirectly via the derived product cumyl alcohol, as previously reported for the analog sigma factor sigma B in B. subtilis exposed to tert-butyl alcohol (41). The denaturing effect of the cumyl alcohol may activate the heat shock response through the sigma factor RpoH and its entire regulon (composed mainly of proteases and chaperones), although other aromatic compounds per se activate this response (40). Some enzymes for a meta-cleavage pathway of aromatic compounds (36) were also upregulated by CHP, suggesting that C. violaceum uses CHP and/or products of CHP catabolism as a carbon source (Fig. 7A).
An interesting finding of our work was the altered expression levels of several genes that belong to the Fur regulon (42–44). The CHP-mediated upregulation of Fur-repressed genes and downregulation of Fur-activated genes suggest that CHP may affect the Fur protein. It has been demonstrated in E. coli that the oxidation of Fur by hydrogen peroxide results in the constitutive expression of Fur-repressed genes and the dysregulation of iron uptake (46). The iron-catalyzed one-electron reduction of CHP induces lipid peroxidation, a process that generates other toxic compounds (5, 6). The induction of DNA repair enzymes and aldehyde detoxification pathways and the decrease in the induction of pathways for the production of unsaturated fatty acids seen in our data suggest strategies to avoid or combat CHP-induced lipid peroxidation in C. violaceum (Fig. 7A). Other adaptations of C. violaceum to CHP involved the downregulation of most genes for the electron transport chain and oxidative phosphorylation. A similar response was found in a transcriptome analysis of Saccharomyces cerevisiae exposed to CHP (58). Indeed, the blockage of bacterial respiration was found to be a novel antioxidant strategy to protect Salmonella from oxidative stress (59).
We have previously shown that like most of its orthologs, C. violaceum OhrR acts as an OHP sensor, repressing the ohrR and ohrA genes under reducing conditions (17). Here, we found that, in addition to ohrR and ohrA, C. violaceum OhrR directly repressed CV_0208, encoding a putative diguanylate cyclase, and exerted indirect positive regulation on CV_0362, CV_1440, and CV_2001, all of which are genes that encode putative virulence-related secreted enzymes (53, 55). In our working hypothesis, based on the OhrR regulon and data from our virulence assays, OhrR modulates C. violaceum virulence in response to host-derived oxidants (Fig. 7B). Thus, during host infection, the exposure of C. violaceum to OHP would cause OhrR oxidation, derepressing the genes for OhrA and diguanylate cyclase. While the increased expression of OhrA would render bacteria more resistant to host-derived OHP, a high expression level of CV_0208 could increase the synthesis of c-di-GMP in response to OHP or another unknown signal. By an indirect mechanism, perhaps involving c-di-GMP, the production of some virulence factors (CV_2001, CV_0362, and CV_1440) would be decreased. Indeed, high levels of c-di-GMP have been associated with an overall negative role in virulence or a switch from acute to chronic infection in many bacteria (60–62). It will be interesting to determine whether the CHASE domain modulates the putative diguanylate cyclase activity of the CV_0208 protein. The function of this widespread extracellular sensory domain in signal transduction is still unknown (63).
It has been well established that some redox-sensing transcription factors, such as MgrA, SarZ, and OspR, play a major role in bacterial virulence (18). The contribution of OhrR to virulence is emerging and seems to involve distinct mechanisms (64–66). Thus, while in Bacillus cereus, OhrR restricts toxinogenesis under highly reducing anoxic conditions by regulating all toxin-related genes (65), in Vibrio cholerae, OhrR jump-starts pathogenesis by regulating the virulence activator TcpP during the oxic-to-anoxic transition (66). Our results indicated that in C. violaceum OhrR modulates virulence, whereas the deletion of ohrA did not alter bacterial virulence, at least under the conditions tested. Possibly, the virulence mechanisms mediated by OhrR involve a combination of several factors, such as the increased expression of a putative diguanylate cyclase and the decreased expression of enzymes involved in host tissue degradation. The role of OhrA in C. violaceum virulence remains elusive and requires further investigation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this work are listed in Table 2. C. violaceum wild-type strain ATCC 12472 (21) and derivative mutant strains (17) were grown at 37°C in Luria-Bertani (LB) medium. When needed, LB medium was supplemented with the antibiotics tetracycline (10 μg/ml), kanamycin (50 μg/ml), and ampicillin (100 μg/ml).
TABLE 2.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| C. violaceum strains | ||
| ATCC 12472 | Wild-type strain (sequenced genome) | 21 |
| JF0209 | Wild-type strain with the CV_0209 gene deleted (ΔohrA) | 17 |
| JF0210 | Wild-type strain with the CV_0210 gene deleted (ΔohrR) | 17 |
| JF0208I | Insertion mutant for the CV_0208 gene (CV0208::pNPT) | This study |
| JF1106I | Insertion mutant for the CV_1106 gene (trxC::pNPT) | This study |
| JF1107I | Insertion mutant for the CV_1107 gene (gpx::pNPT) | This study |
| JF2037I | Insertion mutant for the CV_2037 gene (garB::pNPT) | This study |
| JF3501I | Insertion mutant for the CV_3501 gene (nemA::pNPT) | This study |
| JF3549I | Insertion mutant for the CV_3549 gene (katE::pNPT) | This study |
| JF02100208I | Double mutant for CV_0210-CV_0208 (ΔohrR CV0208::pNPT) | This study |
| Plasmids | ||
| pNPTS138 | Suicide vector containing oriT sacB; Kanr | D. Alley |
| pMR20 | Low-copy-number and broad-host-range vector; oriT; Tcr | 67 |
| pMR-ohrR | pMR20 with the complete CV_0210 gene | 17 |
Construction of mutant strains by insertion mutagenesis.
To construct the mutant strains, DNA fragments corresponding to the internal portions of the CV_0208, CV_1106, CV_1107, CV_2037, CV_3501, and CV_3549 genes were amplified by PCR using MutFw and MutRv primers specific for each gene (Table 3). These PCR products were digested with HindIII and BamHI and cloned into the suicide vector pNPTS138. The resulting constructs were introduced into the C. violaceum wild-type strain by conjugation. To obtain a double mutant, the construct of CV_0208 was introduced into the ΔohrR mutant strain. The insertion-disrupted mutant strains (Table 2) were confirmed by PCR using primers MutConf (specific for each gene) and M13Fw (from the integrated vector) (Table 3).
TABLE 3.
Primers used in this work
| Primer | Sequence (5′→3′)a | Description |
|---|---|---|
| Construction of mutant strains | ||
| CV1106MutFW | CCTAGCAAGCTTAGAAGGGCTGGTCATCCTGG | HindIII/BamHI fragment with 173 bp of the coding region |
| CV1106MutRv | GGCCTAGGATCCGCGTCGGAATGGAGCGGATG | |
| CV1106MutConf | CGACTATCGGCAGGATTG | |
| CV1107MutFW | CCTAGCAAGCTTCACGCCGCAGTTCACGCAAC | HindIII/BamHI fragment with 146 bp of the coding region |
| CV1107MutRv | GGCCTAGGATCCCGCCGTAATTGGCCTGGCAG | |
| CV1107MutConf | GCAATGTGAGACGATAGG | |
| CV2037MutFW | CCTAGCAAGCTTCGATGACCACACCATCGTCC | HindIII/BamHI fragment with 300 bp of the coding region |
| CV2037MutRv | GGCCTAGGATCCAACTGCTCGGACAAGGCCTG | |
| CV2037MutConf | GGCCACACTGACATGTTC | |
| CV3501MutFW | CCTAGCAAGCTTGCTGCAACTGTGGCACACCG | HindIII/BamHI fragment with 411 bp of the coding region |
| CV3501MutRv | GGCCTAGGATCCGAGATGCGGATGCCGACGTG | |
| CV3501MutConf | TGGCCAAGGAATACTACC | |
| CV3549MutFW | CCTAGCAAGCTTAGTTCTACACCGAGGAGGGC | HindIII/BamHI fragment with 480 bp of the coding region |
| CV3549MutRv | GGCCTAGGATCCTGAAGGGACAAGCGTCGGCC | |
| CV3549MutConf | GGCAAAAAGACCGACCTG | |
| CV0208MutFW | CCTAGCAAGCTTCCAACCGCAACCTGTTCTAC | HindIII/BamHI fragment with 157 bp of the coding region |
| CV0208MutRv | GGCCTAGGATCCATCAACTGCAGCACGCGGTC | |
| CV0208MutConf | CAGTTGATCGGTCTGCTG | |
| M13 forward | GTAAAACGACGGCCAGT | |
| Probes for EMSAs | ||
| pCV0208-Fw | TTGGATCCCAGATTCACGCGCTGCGCGC | BamHI/PstI fragment with 233 bp of the promoter region |
| pCV0208-Rv | ATTACTGCAGGAGCGGCTGGCGATGGACAG | |
| CV0208CDS-Fw | TTGGATCCGAAGGCCAGGCGCTGCATCT | BamHI/PstI fragment with 243 bp of the coding region (CDS) |
| CV0208CDS-Rv | ATTACTGCAGCTGTCGCTGCTGCGCACGAA | |
| pCV0209-Fw | TTGGATCCTGCCGGAGCGGAATGCAACT | BamHI/PstI fragment with 212 bp of the promoter region |
| pCV0209-Rv | ATTACTGCAGGCGGTGGCTTCGGCGGTATAC | |
| pCV0362-Fw | TTGGATCCGTCCTGCTTGACGTTTGAGAC | BamHI/PstI fragment with 371 bp of the promoter region |
| pCV0362-Rv | ATTACTGCAGCAAGGGCTTGCCGGCGGAGA | |
| pCV1444-Fw | TTGGATCCCTCTGGACTGATCCGCCCAG | BamHI/PstI fragment with 270 bp of the promoter region |
| pCV1444-Rv | ATTACTGCAGCATCATGAGCGGACTCCCGG | |
| pCV2001-Fw | TTGGATCCGCGAAGCGCGGCATTGAGGT | BamHI/PstI fragment with 435 bp of the promoter region |
| pCV2001-Rv | ATTACTGCAGGGCAGGCCGTGCGGGTAGTC | |
| Probes for Northern blot assays | ||
| CV_0208NB-Fw | GACCATACTAGACAGCTACGCC | Fragment with 115 bp of the coding region |
| CV_0208NB-Rv | CAGCGTGGTGATTTGCGGATAG | |
| CV_0209NB-Fw | TGCAGGTGAAGTTGAGCACCCC | Fragment with 103 bp of the coding region |
| CV_0209NB-Rv | CCGATGAAGCAGGCGGAATAGCC | |
| CV_1106NB-Fw | CAAGATGTTTGGCCCGGTGTTC | Fragment with 115 bp of the coding region |
| CV_1106NB-Rv | GGAGCGGATGTTGAAGTGGGC | |
| CV_1483NB-Fw | GCTGCTGAACGATATGTGGGG | Fragment with 118 bp of the coding region |
| CV_1483NB-Rv | GCTGTAGCGCCATTTGACCAG | |
| CV_2037NB-Fw | GCCTTGTCCGAGCAGTTCTAT | Fragment with 110 bp of the coding region |
| CV_2037NB-Rv | TCGACCTCTTCTCCGTCCAG | |
| CV_2241NB-Fw | TCCAAGACCCATTGAGCCG | Fragment with 123 bp of the coding region |
| CV_2241NB-Rv | CATCGGACGGAATCAGGCC | |
| CV_3549NB-Fw | GGCAACTGGGACCTCGTAG | Fragment with 114 bp of the coding region |
| CV_3549NB-Rv | GTCGCGCATATTGGTGTACG | |
| CV_3668NB-Fw | GACTTCGTGGTGCAGGTTCTC | Fragment with 117 bp of the coding region |
| CV_3668NB-Rv | GTCTTTGGTATAAACGCCACAC | |
| CV_3896NB-Fw | GACCTTCCCCAGCAAGACCTTC | Fragment with 124 bp of the coding region |
| CV_3896NB-Rv | CTTGTAATGGTCCCAGCGCAGC | |
| CV_4003NB-Fw | GACCTTCCAGATGACCTTCGC | Fragment with 121 bp of the coding region |
| CV_4003NB-Rv | CACATAGACCAGCAGAGACCAC | |
| CV_4014NB-Fw | CATCTCCGCCAACTCCGATAG | Fragment with 118 bp of the coding region |
| CV_4014NB-Rv | GTCCAGTTCGTTGTTCAGGC | |
| CV_4206NB-Fw | CACTGGATCAAGGCCGAGA | Fragment with 122 bp of the coding region |
| CV_4206NB-Rv | GAAAAACCGCTCTTCATGCTG | |
| CV_0668NB-Fw | GATCGTGGTTGCCGCTGAAA | Fragment with 121 bp of the coding region |
| CV_0668NB-Rv | CAGCACTTCCTGTTCGGTTTCC | |
| CV_1173NB-Fw | GCGTTCCAGAAATCCGTGAC | Fragment with 120 bp of the coding region |
| CV_1173NB-Rv | GATCATCTGGGTGTGGCAGG | |
| CV_3183NB-Fw | CGGGCGAAGTGATTCCGAA | Fragment with 117 bp of the coding region |
| CV_3183NB-Rv | CGCCCCAGTTGTCGCTATAG | |
| CV_3399NB-Fw | CGACCTGAAGCTGGAAATGGA | Fragment with 115 bp of the coding region |
| CV_3399NB-Rv | CTCGCTCTCCAGGATGTCTTC | |
| CV_3412NB-Fw | AATCTGTTGGCCGGCAAGAG | Fragment with 125 bp of the coding region |
| CV_3412NB-Rv | GCTTCCTTCGGCGAAATGTAGT | |
| CV_3648NB-Fw | GCCTGGACCTGATCTACGGT | Fragment with 117 bp of the coding region |
| CV_3648NB-Rv | GCAGATGGGCGTAGTTGTACA | |
| CV_0362NB-Fw | GAGCAATACCACCCAGCCTT | Fragment with 117 bp of the coding region |
| CV_0362NB-Rv | TTGTCCCAGAACACGTAGGC | |
| CV_1440NB-Fw | GAAACCTTCACCGTGGACAA | Fragment with 113 bp of the coding region |
| CV_1440NB-Rv | CCGTTATTGGGGCAAGACT | |
| CV_2001NB-Fw | GGCGAACGATCCCAATGAAA | Fragment with 119 bp of the coding region |
| CV_2001NB-Rv | TCTCGATGTAGATGCCACCG |
Underlined letters indicate the restriction enzyme recognition sites, used for cloning purposes.
MIC assay.
Cultures of WT and mutant strains (ΔohrR, ΔohrA, garB::pNPT, katE::pNPT, nemA::pNPT, trxC::pNPT, and gpx::pNPT) of C. violaceum were grown on LB medium or LB medium supplemented with kanamycin at 37°C overnight. Each culture was diluted in fresh LB medium to a final optical density at 600 nm (OD600) of 0.01 in 96-well plates. The compounds were added in different concentrations in a final volume of 150 μl. The concentrations of hydroperoxides and cumene used in the MIC assays ranged from 0 to 500 μM for CHP and TBHP, 0 to 150 mM for H2O2, and 0 to 450 mM for CMN. Afterwards, assay plates were incubated at 37°C under agitation (150 rpm), and bacterial growth was observed after 24 h.
RNA isolation.
To investigate the adaptive response of C. violaceum to CHP, wild-type strain ATCC 12472 was grown at 37°C in LB medium until mid-log phase (OD600 of 0.8 to 1.0). The culture was then split into two aliquots in 125-ml flasks and either left untreated or treated with 100 μM CHP for 10 min. After this time, the cells were harvested by centrifugation, and the total RNA was extracted by using TRIzol reagent (Ambion) and purified with the illustra RNAspin Mini RNA isolation kit (GE Healthcare). In another set of experiments, wild-type strain ATCC 12472 and the ΔohrR mutant strain were grown at 37°C in LB medium until mid-log phase (OD600 of 0.8 to 1.0), and the cells were used for RNA extraction, as described above. RNA integrity was evaluated with a formaldehyde-denaturing 1.2% agarose gel. The absence of genomic DNA contamination was verified by PCR. Quantification of total RNA was performed with a NanoDrop spectrophotometer (Thermo Scientific).
DNA microarray analysis.
A 4x44k custom-designed 45- to 60-mer oligonucleotide microarray for the C. violaceum ATCC 12472 strain was produced by Agilent Technologies. The microarray was designed by using the e-Array platform with the 4,407 open reading frames (ORFs) from the former C. violaceum genome annotation (RefSeq accession number NC_005085) (21) plus 270 new ORFs annotated by PATRIC (Pathosystems Integration Resource Center) uploaded. Three distinct oligonucleotide probes were designed for each ORF, and each probe was repeated at least three times on each array. The procedures for cRNA labeling, hybridization, and washing were performed according to the Agilent Two-Color Microarray-Based Exon Analysis protocol, version 1.0 (Agilent Technologies). Briefly, fluorescence-labeled cRNA was generated from total RNA derived under each experimental condition by using the Low Input Quick Amp WT Two-Color kit (Agilent Technologies). Pooled labeled cRNA samples were fragmented and hybridized to the C. violaceum microarray slides. The slides were scanned with an Agilent DNA microarray scanner, and the data were extracted and normalized by using Agilent Feature Extraction software, version 10.5. Data from three independent biological experiments were analyzed under each condition. The values for the relative expression level of each gene were obtained as the averages for at least 27 probes (9 probes from three biological replicates). We considered differentially expressed genes those that had their expression levels altered at least 2-fold relative to the control.
Northern blot analysis.
Total RNA was extracted as stated above and used for Northern blot analysis as previously described (17). The same quantity (7 μg) of each RNA sample was subjected to electrophoresis in a denaturing formaldehyde–1.5% agarose gel and transferred onto nylon Hybond-XL membranes (GE Healthcare). Specific probes for each gene were PCR amplified by using the primers listed in Table 3. The PCR products were labeled with [α-32P]dCTP (PerkinElmer) by using an Exo-Klenow enzyme DECAprime II kit (Ambion). The membranes were prehybridized in 10 ml of ULTRAhyb (Ambion) at 42°C for 30 min and incubated with the labeled probes at 42°C for 16 h. The membranes were washed twice at 42°C for 5 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.1% SDS and twice at 42°C for 15 min in 0.1× SSC plus 0.1% SDS. The results were detected by autoradiography with various exposure times (18 h to 1 week).
Electrophoretic mobility shift assay.
The promoter regions of the CV_0208, CV_0209, CV_0362, CV_1440, and CV_2001 genes were amplified by PCR using the oligonucleotides listed in Table 3. These DNA fragments were end labeled with [γ-32P]ATP (PerkinElmer) by using T4 polynucleotide kinase (Thermo Scientific) and purified with the NucleoSpin Gel and PCR Cleanup kit (Macherey-Nagel). The DNA binding reactions were performed with a volume of 20 μl containing interaction buffer (20 mM Tris-HCl [pH 7.4], 50 mM KCl, 1 mM EDTA [pH 8.0], 50 μg/ml bovine serum albumin, 5% glycerol), DNA probes, and various amounts of dithiothreitol (DTT)-reduced or CHP-oxidized His-OhrR purified protein (17). All interaction reaction mixtures were incubated at 25°C for 25 min. After the addition of glycerol, the samples were separated by native 5% polyacrylamide gel electrophoresis in Tris-borate-EDTA (TBE) buffer. The gels were dried, and the signal was detected by autoradiography.
Mouse infection assays.
Mice were purchased and maintained for 1 week before experimentation in the Animal Facilities of the Faculdade de Medicina de Ribeirão Preto (FMRP-USP). Six-week-old female BALB/c mice were infected with 1 × 106 CFU of the C. violaceum wild-type and isogenic mutant strains by intraperitoneal (i.p.) injection (26). The injected bacterial doses were monitored by plating serial dilutions of the inoculum onto LB medium. To enumerate animal survival, infected mice were monitored for up to 7 days. For the determination of bacterial burdens in the liver, mice i.p. injected with 5 × 105 bacteria were sacrificed at 24 h postinfection, and the organs were collected aseptically and homogenized in phosphate-buffered saline (PBS). Liver homogenates were serially diluted and plated onto LB medium for bacterial counting. For histopathological analysis, the liver fragments were fixed in formalin and embedded in paraffin to generate 6-μm sections. The sections were stained with a hematoxylin-and-eosin solution. All mouse experimental procedures were performed in accordance with the Ethical Principles in Animal Research approved by the Local Ethical Animal Committee (CEUA) of FMRP-USP (protocol 147/2014).
Accession number(s).
Microarray raw data were deposited in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE90551.
Supplementary Material
ACKNOWLEDGMENTS
We thank Geraldo A. Passos and his laboratory members Nicole Pezzi and Amanda F. Assis for assistance with the microarray scanning and data extraction and Vani M. Alves, Antonio R. Meirelles, and Amilton A. Barrera for technical support in histopathological analysis.
This research was supported by grants from the São Paulo Research Foundation (FAPESP) (grant 2012/20435-9) and the Fundação de Apoio ao Ensino, Pesquisa e Assistência do Hospital das Clínicas da FMRP-USP (FAEPA). During the course of this work, M.P.-M. and D.D.A.M. were supported by FAPESP fellowship grants 2013/25745-9 and 2012/21722-1.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00017-17.
REFERENCES
- 1.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fang FC. 2011. Antimicrobial actions of reactive oxygen species. mBio 2:e00141-11. doi: 10.1128/mBio.00141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurst JK. 2012. What really happens in the neutrophil phagosome? Free Radic Biol Med 53:508–520. doi: 10.1016/j.freeradbiomed.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dennis EA, Norris PC. 2015. Eicosanoid storm in infection and inflammation. Nat Rev Immunol 15:511–523. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayala A, Muñoz MF, Argüelles S. 2014. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev 2014:360438. doi: 10.1155/2014/360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Girotti AW. 1998. Lipid hydroperoxide generation, turnover, and effector action in biological systems. J Lipid Res 39:1529–1542. [PubMed] [Google Scholar]
- 7.Zhang YM, Rock CO. 2008. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol 6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 8.Zhou X, Taghizadeh K, Dedon PC. 2005. Chemical and biological evidence for base propenals as the major source of the endogenous M1dG adduct in cellular DNA. J Biol Chem 280:25377–25382. doi: 10.1074/jbc.M503079200. [DOI] [PubMed] [Google Scholar]
- 9.Pérez JM, Arenas FA, Pradenas GA, Sandoval JM, Vásquez CC. 2008. Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J Biol Chem 283:7346–7353. doi: 10.1074/jbc.M708846200. [DOI] [PubMed] [Google Scholar]
- 10.Belenky P, Ye JD, Porter CB, Cohen NR, Lobritz MA, Ferrante T, Jain S, Korry BJ, Schwarz EG, Walker GC, Collins JJ. 2015. Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep 13:968–980. doi: 10.1016/j.celrep.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liddell JR, Dringen R, Crack PJ, Robinson SR. 2006. Glutathione peroxidase 1 and a high cellular glutathione concentration are essential for effective organic hydroperoxide detoxification in astrocytes. Glia 54:873–879. doi: 10.1002/glia.20433. [DOI] [PubMed] [Google Scholar]
- 12.Cussiol JR, Alegria TG, Szweda LI, Netto LE. 2010. Ohr (organic hydroperoxide resistance protein) possesses a previously undescribed activity, lipoyl-dependent peroxidase. J Biol Chem 285:21943–21950. doi: 10.1074/jbc.M110.117283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishra S, Imlay J. 2012. Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch Biochem Biophys 525:145–160. doi: 10.1016/j.abb.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antelmann H, Helmann JD. 2011. Thiol-based redox switches and gene regulation. Antioxid Redox Signal 14:1049–1063. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubbs JM, Mongkolsuk S. 2012. Peroxide-sensing transcriptional regulators in bacteria. J Bacteriol 194:5495–5503. doi: 10.1128/JB.00304-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaballa A, Chi BK, Roberts AA, Becher D, Hamilton CJ, Antelmann H, Helmann JD. 2014. Redox regulation in Bacillus subtilis: the bacilliredoxins BrxA (YphP) and BrxB (YqiW) function in de-bacillithiolation of S-bacillithiolated OhrR and MetE. Antioxid Redox Signal 21:357–367. doi: 10.1089/ars.2013.5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.da Silva Neto JF, Negretto CC, Netto LE. 2012. Analysis of the organic hydroperoxide response of Chromobacterium violaceum reveals that OhrR is a Cys-based redox sensor regulated by thioredoxin. PLoS One 7:e47090. doi: 10.1371/journal.pone.0047090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen PR, Brugarolas P, He C. 2011. Redox signaling in human pathogens. Antioxid Redox Signal 14:1107–1118. doi: 10.1089/ars.2010.3374. [DOI] [PubMed] [Google Scholar]
- 19.Durán N, Menck CF. 2001. Chromobacterium violaceum: a review of pharmacological and industrial perspectives. Crit Rev Microbiol 27:201–222. doi: 10.1080/20014091096747. [DOI] [PubMed] [Google Scholar]
- 20.Lima-Bittencourt CI, Astolfi-Filho S, Chartone-Souza E, Santos FR, Nascimento AM. 2007. Analysis of Chromobacterium sp. natural isolates from different Brazilian ecosystems. BMC Microbiol 7:58. doi: 10.1186/1471-2180-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brazilian National Genome Project Consortium. 2003. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc Natl Acad Sci U S A 100:11660–11665. doi: 10.1073/pnas.1832124100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carepo MS, Azevedo J, Porto J, Bentes-Souza A, da Silva Batista J, da Silva ALC, Schneider MP. 2004. Identification of Chromobacterium violaceum genes with potential biotechnological application in environmental detoxification. Genet Mol Res 3:181–194. [PubMed] [Google Scholar]
- 23.Hammel KE, Kalyanaraman B, Kirk TK. 1986. Substrate free radicals are intermediates in ligninase catalysis. Proc Natl Acad Sci U S A 83:3708–3712. doi: 10.1073/pnas.83.11.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tice R, Brevard B. 1998. Cumene hydroperoxide [80-15-9]. Review of toxicological literature. Prepared for E. Zeigler of the National Institute of Environmental Health Sciences, p 64 Integrated Laboratory Systems, Research Triangle Park, NC. [Google Scholar]
- 25.Yang CH, Li YH. 2011. Chromobacterium violaceum infection: a clinical review of an important but neglected infection. J Chin Med Assoc 74:435–441. doi: 10.1016/j.jcma.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Miki T, Iguchi M, Akiba K, Hosono M, Sobue T, Danbara H, Okada N. 2010. Chromobacterium pathogenicity island 1 type III secretion system is a major virulence determinant for Chromobacterium violaceum-induced cell death in hepatocytes. Mol Microbiol 77:855–872. doi: 10.1111/j.1365-2958.2010.07248.x. [DOI] [PubMed] [Google Scholar]
- 27.Miki T, Akiba K, Iguchi M, Danbara H, Okada N. 2011. The Chromobacterium violaceum type III effector CopE, a guanine nucleotide exchange factor for Rac1 and Cdc42, is involved in bacterial invasion of epithelial cells and pathogenesis. Mol Microbiol 80:1186–1203. doi: 10.1111/j.1365-2958.2011.07637.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, Liu L, Shao F. 2011. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 29.Maltez VI, Tubbs AL, Cook KD, Aachoui Y, Falcone EL, Holland SM, Whitmire JK, Miao EA. 2015. Inflammasomes coordinate pyroptosis and natural killer cell cytotoxicity to clear infection by a ubiquitous environmental bacterium. Immunity 43:987–997. doi: 10.1016/j.immuni.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segal BH, Ding L, Holland SM. 2003. Phagocyte NADPH oxidase, but not inducible nitric oxide synthase, is essential for early control of Burkholderia cepacia and Chromobacterium violaceum infection in mice. Infect Immun 71:205–210. doi: 10.1128/IAI.71.1.205-210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vergauwen B, Pauwels F, Jacquemotte F, Meyer TE, Cusanovich MA, Bartsch RG, Van Beeumen JJ. 2001. Characterization of glutathione amide reductase from Chromatium gracile. Identification of a novel thiol peroxidase (Prx/Grx) fueled by glutathione amide redox cycling. J Biol Chem 276:20890–20897. doi: 10.1074/jbc.M102026200. [DOI] [PubMed] [Google Scholar]
- 32.Meyer Y, Buchanan BB, Vignols F, Reichheld JP. 2009. Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu Rev Genet 43:335–367. doi: 10.1146/annurev-genet-102108-134201. [DOI] [PubMed] [Google Scholar]
- 33.Netto LE, de Oliveira MA, Tairum CA, da Silva Neto JF. 2016. Conferring specificity in redox pathways by enzymatic thiol/disulfide exchange reactions. Free Radic Res 50:206–245. doi: 10.3109/10715762.2015.1120864. [DOI] [PubMed] [Google Scholar]
- 34.Allocati N, Federici L, Masulli M, Di Ilio C. 2009. Glutathione transferases in bacteria. FEBS J 276:58–75. doi: 10.1111/j.1742-4658.2008.06743.x. [DOI] [PubMed] [Google Scholar]
- 35.Lee C, Shin J, Park C. 2013. Novel regulatory system nemRA-gloA for electrophile reduction in Escherichia coli K-12. Mol Microbiol 88:395–412. doi: 10.1111/mmi.12192. [DOI] [PubMed] [Google Scholar]
- 36.Fuchs G, Boll M, Heider J. 2011. Microbial degradation of aromatic compounds—from one strategy to four. Nat Rev Microbiol 9:803–816. doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
- 37.Nguyen TT, Eiamphungporn W, Mäder U, Liebeke M, Lalk M, Hecker M, Helmann JD, Antelmann H. 2009. Genome-wide responses to carbonyl electrophiles in Bacillus subtilis: control of the thiol-dependent formaldehyde dehydrogenase AdhA and cysteine proteinase YraA by the MerR-family regulator YraB (AdhR). Mol Microbiol 71:876–894. doi: 10.1111/j.1365-2958.2008.06568.x. [DOI] [PubMed] [Google Scholar]
- 38.Chiancone E, Ceci P. 2010. The multifaceted capacity of Dps proteins to combat bacterial stress conditions: detoxification of iron and hydrogen peroxide and DNA binding. Biochim Biophys Acta 1800:798–805. doi: 10.1016/j.bbagen.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Guisbert E, Yura T, Rhodius VA, Gross CA. 2008. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol Mol Biol Rev 72:545–554. doi: 10.1128/MMBR.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tam LT, Eymann C, Albrecht D, Sietmann R, Schauer F, Hecker M, Antelmann H. 2006. Differential gene expression in response to phenol and catechol reveals different metabolic activities for the degradation of aromatic compounds in Bacillus subtilis. Environ Microbiol 8:1408–1427. doi: 10.1111/j.1462-2920.2006.01034.x. [DOI] [PubMed] [Google Scholar]
- 41.Helmann JD, Wu MF, Gaballa A, Kobel PA, Morshedi MM, Fawcett P, Paddon C. 2003. The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J Bacteriol 185:243–253. doi: 10.1128/JB.185.1.243-253.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.da Silva Neto JF, Lourenço RF, Marques MV. 2013. Global transcriptional response of Caulobacter crescentus to iron availability. BMC Genomics 14:549. doi: 10.1186/1471-2164-14-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fillat MF. 2014. The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch Biochem Biophys 546:41–52. doi: 10.1016/j.abb.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 44.Seo SW, Kim D, Latif H, O'Brien EJ, Szubin R, Palsson BO. 2014. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat Commun 5:4910. doi: 10.1038/ncomms5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rush JD, Koppenol WH. 1990. Reactions of Fe(II)-ATP and Fe(II)-citrate complexes with t-butyl hydroperoxide and cumyl hydroperoxide. FEBS Lett 275:114–116. doi: 10.1016/0014-5793(90)81452-T. [DOI] [PubMed] [Google Scholar]
- 46.Varghese S, Wu A, Park S, Imlay KR, Imlay JA. 2007. Submicromolar hydrogen peroxide disrupts the ability of Fur protein to control free-iron levels in Escherichia coli. Mol Microbiol 64:822–830. doi: 10.1111/j.1365-2958.2007.05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimmer DP, Soupene E, Lee HL, Wendisch VF, Khodursky AB, Peter BJ, Bender RA, Kustu S. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc Natl Acad Sci U S A 97:14674–14679. doi: 10.1073/pnas.97.26.14674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meesapyodsuk D, Qiu X. 2012. The front-end desaturase: structure, function, evolution and biotechnological use. Lipids 47:227–237. doi: 10.1007/s11745-011-3617-2. [DOI] [PubMed] [Google Scholar]
- 49.da Silva Neto JF, Braz VS, Italiani VCS, Marques MV. 2009. Fur controls iron homeostasis and oxidative stress defense in the oligotrophic alpha-proteobacterium Caulobacter crescentus. Nucleic Acids Res 37:4812–4825. doi: 10.1093/nar/gkp509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parsons JB, Rock CO. 2013. Bacterial lipids: metabolism and membrane homeostasis. Prog Lipid Res 52:249–276. doi: 10.1016/j.plipres.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welham PA, Stekel DJ. 2009. Mathematical model of the Lux luminescence system in the terrestrial bacterium Photorhabdus luminescens. Mol Biosyst 5:68–76. doi: 10.1039/B812094C. [DOI] [PubMed] [Google Scholar]
- 52.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castro-Gomes T, Cardoso MS, DaRocha WD, Laibida LA, Nascimento AM, Zuccherato LW, Horta MF, Bemquerer MP, Teixeira SM. 2014. Identification of secreted virulence factors of Chromobacterium violaceum. J Microbiol 52:350–353. doi: 10.1007/s12275-014-3202-5. [DOI] [PubMed] [Google Scholar]
- 54.Kalia D, Merey G, Nakayama S, Zheng Y, Zhou J, Luo Y, Guo M, Roembke BT, Sintim HO. 2013. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem Soc Rev 42:305–341. doi: 10.1039/C2CS35206K. [DOI] [PubMed] [Google Scholar]
- 55.Duarte AS, Correia A, Esteves AC. 2016. Bacterial collagenases—a review. Crit Rev Microbiol 42:106–126. doi: 10.3109/1040841X.2014.904270. [DOI] [PubMed] [Google Scholar]
- 56.Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol 183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sha W, Martins AM, Laubenbacher R, Mendes P, Shulaev V. 2013. The genome-wide early temporal response of Saccharomyces cerevisiae to oxidative stress induced by cumene hydroperoxide. PLoS One 8:e74939. doi: 10.1371/journal.pone.0074939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Husain M, Bourret TJ, McCollister BD, Jones-Carson J, Laughlin J, Vázquez-Torres A. 2008. Nitric oxide evokes an adaptive response to oxidative stress by arresting respiration. J Biol Chem 283:7682–7689. doi: 10.1074/jbc.M708845200. [DOI] [PubMed] [Google Scholar]
- 60.Lamprokostopoulou A, Monteiro C, Rhen M, Römling U. 2010. Cyclic di-GMP signalling controls virulence properties of Salmonella enterica serovar Typhimurium at the mucosal lining. Environ Microbiol 12:40–53. doi: 10.1111/j.1462-2920.2009.02032.x. [DOI] [PubMed] [Google Scholar]
- 61.Zogaj X, Wyatt GC, Klose KE. 2012. Cyclic di-GMP stimulates biofilm formation and inhibits virulence of Francisella novicida. Infect Immun 80:4239–4247. doi: 10.1128/IAI.00702-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen LH, Köseoğlu VK, Güvener ZT, Myers-Morales T, Reed JM, D'Orazio SE, Miller KW, Gomelsky M. 2014. Cyclic di-GMP-dependent signaling pathways in the pathogenic firmicute Listeria monocytogenes. PLoS Pathog 10:e1004301. doi: 10.1371/journal.ppat.1004301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mougel C, Zhulin IB. 2001. CHASE: an extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends Biochem Sci 26:582–584. doi: 10.1016/S0968-0004(01)01969-7. [DOI] [PubMed] [Google Scholar]
- 64.Atichartpongkul S, Fuangthong M, Vattanaviboon P, Mongkolsuk S. 2010. Analyses of the regulatory mechanism and physiological roles of Pseudomonas aeruginosa OhrR, a transcription regulator and a sensor of organic hydroperoxides. J Bacteriol 192:2093–2101. doi: 10.1128/JB.01510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clair G, Lorphelin A, Armengaud J, Duport C. 2013. OhrRA functions as a redox-responsive system controlling toxinogenesis in Bacillus cereus. J Proteomics 94:527–539. doi: 10.1016/j.jprot.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 66.Liu Z, Wang H, Zhou Z, Naseer N, Xiang F, Kan B, Goulian M, Zhu J. 2016. Differential thiol-based switches jump-start Vibrio cholerae pathogenesis. Cell Rep 14:347–354. doi: 10.1016/j.celrep.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roberts RC, Toochinda C, Avedissian M, Baldini RL, Gomes SL, Shapiro L. 1996. Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J Bacteriol 178:1829–1841. doi: 10.1128/jb.178.7.1829-1841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.