ABSTRACT
Acinetobacter baumannii is a major cause of antibiotic-resistant nosocomial infections worldwide. In this study, several rifampin-resistant spontaneous mutants obtained from the A. baumannii ATCC 17978 strain that differed in their point mutations in the rpoB gene, encoding the β-subunit of the RNA polymerase, were isolated. All the mutants harboring amino acid substitutions in position 522 or 540 of the RpoB protein were impaired in surface-associated motility and had attenuated virulence in the fertility model of Caenorhabditis elegans. The transcriptional profile of these mutants included six downregulated genes encoding proteins homologous to transporters and metabolic enzymes widespread among A. baumannii clinical isolates. The construction of knockout mutants in each of the six downregulated genes revealed a significant reduction in the surface-associated motility and virulence of four of them in the A. baumannii ATCC 17978 strain, as well as in the virulent clinical isolate MAR002. Taken together, our results provide strong evidence of the connection between motility and virulence in this multiresistant nosocomial pathogen.
KEYWORDS: Acinetobacter baumannii, surface-associated motility, rpoB, rifampin resistance
INTRODUCTION
Acinetobacter baumannii is a Gram-negative bacterium often responsible for nosocomial infections, based on its capacity to acquire and develop antimicrobial resistance (1). It is one of the most frequently encountered pathogens in intensive care, neonatal, and burn units, where it causes pneumonia and infections involving the central nervous system, skin, soft tissues, and bone (1). However, despite the increasing clinical importance of A. baumannii, little is known about the virulence factors that contribute to its pathogenetic properties.
In previous studies of DNA damage-mediated mutagenesis in A. baumannii (2–5), we observed an atypical motility pattern in several spontaneous rifampin-resistant (Rifr) mutants derived from the wild-type (WT) strain ATCC 17978 growing on semisolid medium. Although Acinetobacter spp. lack flagella, they exhibit surface-associated motility (6, 7). In a recent study, hypermotile A. baumannii derivatives with increasing virulence were isolated, but whether a lack of motility negatively affects the virulence and pathogenesis of A. baumannii is unclear (8, 9).
The point mutations conferring rifampin resistance are located in the rpoB gene (encoding the β-subunit of the RNA polymerase), and they produce substantial changes in the transcriptional profile of bacterial cells by affecting several promoters (3, 10, 11). Nonetheless, not all mutations conferring rifampin resistance give rise to global modifications in the transcriptional profile of the respective bacterium (12, 13). In this study, A. baumannii rifampin-resistant mutants with altered surface-associated motility were isolated. Using DNA microarray technology, several genes involved in the motility and virulence of the bacterial species were identified.
RESULTS
Motility and virulence of A. baumannii rpoB mutants.
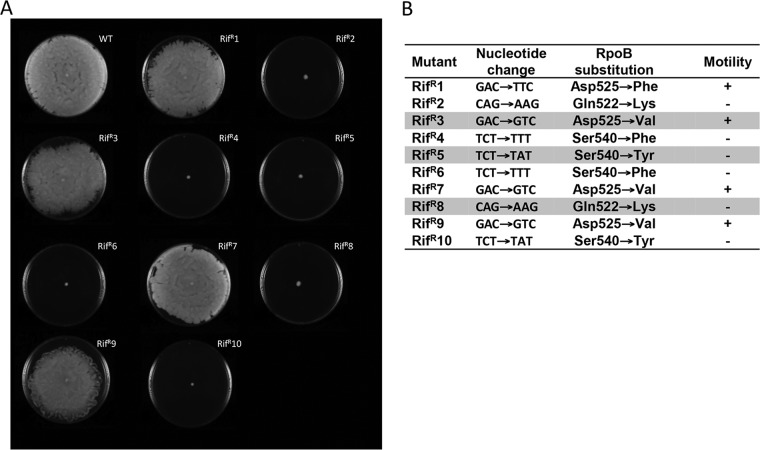
By plating saturated cultures of A. baumannii ATCC 17978 in the presence of rifampin (50 mg/liter), we isolated 10 spontaneous mutants (Rifr1 to Rifr10) resistant to the antimicrobial. In subsequent motility assays, impaired surface-associated motility was confirmed in 60% of the isolated rifampin-resistant clones (Fig. 1A). Sequencing data for the rpoB gene revealed that rifampin-resistant clones with altered motility patterns had an amino acid substitution at either Gln522 or Ser540 of the wild-type RpoB protein (Fig. 1B). Moreover, wild-type motility was detected in rifampin-resistant mutants harboring an amino acid change at position Asp525 of RpoB (Fig. 1B). In several additional experiments, independent rpoB mutants that contained an amino acid substitution at either Gln522 or Ser540 were isolated; all of the mutants were similarly impaired in their surface-associated motility (data not shown).
FIG 1.
(A) Surface-associated motility assays of A. baumannii ATCC 17978 (WT) and 10 rifampin-resistant mutant derivatives (Rifr1 to Rifr10). (B) RpoB amino acid substitutions in the rifampin-resistant mutant strains and effects on motility (+, presence of motility; −, absence of motility). Mutants used in subsequent experiments are shaded.
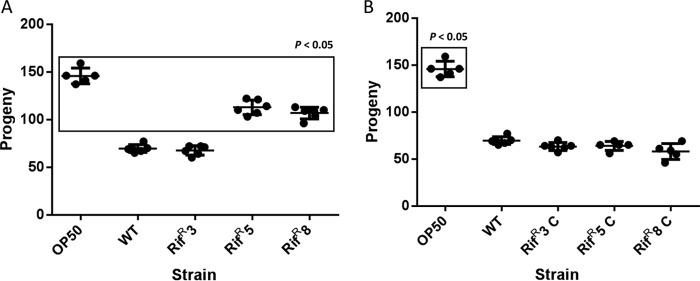
To analyze whether motility plays a role in the pathogenesis of A. baumannii, the virulence of three different rifampin-resistant mutants, two with altered motility (Rifr5 and Rifr8) and the third with wild-type motility (Rifr3), was determined in the Caenorhabditis elegans fertility model, recently validated for studies of the virulence of this bacterial species (14). The results showed that the virulence of the Rifr5 and Rifr8 mutant strains (both with impaired motility), but not of the Rifr3 mutant (wild-type motility), was significantly lower than that of the wild-type strain (Fig. 2A). Since the growth kinetics of the Rifr5 (growth rate [μ] = 0.86 ± 0.03 h−1) and Rifr8 (μ = 0.86 ± 0.02 h−1) mutants in liquid medium were comparable to those of the wild-type strain (μ = 0.83 ± 0.05 h−1), the defects in the motility and virulence of the two mutants cannot be attributed to any reduction in the bacterial growth rates.
FIG 2.
C. elegans fertility assay with the Rifr3, Rifr5, and Rifr8 strains (A) or the complemented mutants (Rifr3C, Rifr5C, and Rifr8C) carrying the vector pBAV1K-T5-gfp with the wild-type rpoB gene (B). Significant (P < 0.05) differences between the indicated strains and the A. baumannii wild-type strain ATCC 17978 (WT) are boxed. The error bars represent the standard deviations of the means (horizontal lines) from six replicates.
Introduction of the wild-type rpoB gene into Rifr mutants completely restores the wild-type phenotype.
It had been reported that a wild-type rpoB gene present in a high-copy-number plasmid restored rifampin susceptibility in a resistant mutant (15). For this reason, and to rule out the possibility that a secondary mutation different from those encountered in the rpoB gene was responsible for the phenotypes of the Rifr5 and Rifr8 mutants, the A. baumannii wild-type rpoB was cloned into the high-copy-number expression vector pBAV1K-T5-gfp (16) and introduced into Rifr3, Rifr5, and Rifr8 A. baumannii mutants. As expected, the presence of the wild-type rpoB gene in the three resistant mutants restored rifampin susceptibility, evidenced by a reduction in the MIC from >512 mg/liter to 4 mg/liter in all three complemented mutants.
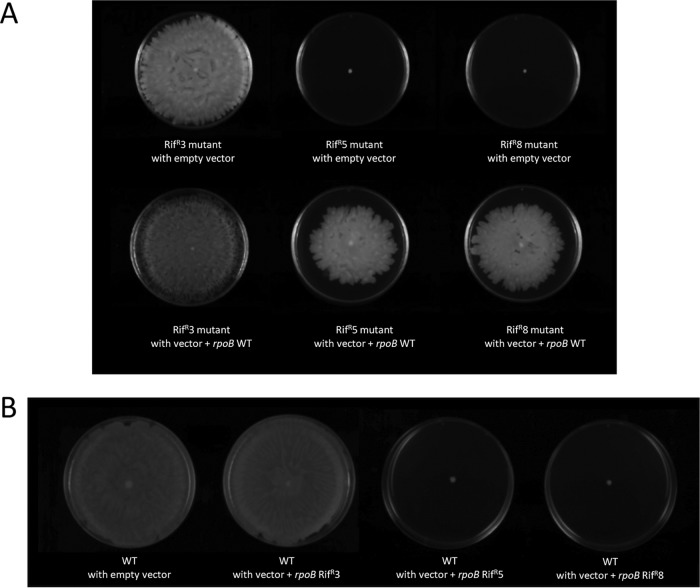
In addition, expression of the wild-type rpoB gene was able to restore both the virulence (Fig. 2B) and the surface-associated motility (Fig. 3A) of the Rifr5 and Rifr8 A. baumannii mutant strains. The vector pBAV1K-T5-gfp without the rpoB gene had no effect on rifampin susceptibility (MIC, >512 mg/ml in all cases), virulence (data not shown), or surface-associated motility (Fig. 3A) when introduced into the three Rifr mutants of A. baumannii strain ATCC 17978.
FIG 3.
(A) Surface-associated motility assays of the indicated A. baumannii ATCC 17978 Rifr derivative mutants carrying either the empty expression vector pBAV1K-T5-gfp (empty vector) or the same vector containing the wild-type rpoB gene (vector + rpoB WT). (B) Surface-associated motility assays of the wild-type A. baumannii ATCC 17978 (WT) strain carrying the empty expression vector pBAV1K-T5-gfp (empty vector) or the same vector containing the rpoB gene amplified from the indicated Rifr strain.
Introduction of the pBAV1K-T5-gfp vector containing the rpoB gene isolated from any of the three Rifr mutants into the wild-type strain conferred rifampin resistance (MIC, >512 mg/ml in all cases). However, only the pBAV1K-T5-gfp plasmid constructions containing the rpoB genes amplified from the Rifr5 and Rifr8 mutants, and not that from the Rifr3 mutant, were able to impair motility when introduced into the A. baumannii ATCC 17978 wild-type strain (Fig. 3B). Together, these findings definitively demonstrated that the rpoB gene mutations of A. baumannii strains Rifr5 and Rifr8 were responsible for the lack of both surface-associated motility and virulence in the nosocomial pathogen.
Global transcriptional analysis of rifampin-resistant mutants of A. baumannii with impaired motility and virulence.
As mentioned above, mutations in the rpoB gene alter gene expression patterns in several bacterial species (17). To obtain insights into the effect of the rpoB mutations on A. baumannii motility and virulence, the gene expression profiles of A. baumannii strains Rifr5 and Rifr8 were examined in cDNA microarrays (see Materials and Methods). Among the 3,431 cDNA sequences examined, the expression of 30 from strain Rifr5 and 221 from strain Rifr8 differed by more than 2-fold compared to the parental strain. In Rifr5, 12 sequences were upregulated and 18 were downregulated, while in Rifr8, 59 were upregulated and 162 were downregulated.
Six downregulated genes were shared by the two strains: two encoding putative membrane transporters and four encoding metabolism-related enzymes (Table 1). Decrease in the expression of these genes was validated by reverse transcription-quantitative real-time PCR (RT-qPCR) (Fig. 4). Furthermore, introduction of the expression vector pBAV1K-T5-gfp carrying a copy of the wild-type rpoB gene was able to increase the expression of these six downregulated genes in both the Rifr5 and Rifr8 A. baumannii mutants to levels similar to those of the wild-type parental strain (Fig. 4).
TABLE 1.
Downregulated genes in the Rifr5 and Rifr8 mutants of A. baumannii
| Gene | Product description | Fold changea |
|
|---|---|---|---|
| Rifr5 | Rifr8 | ||
| RS14730 | Taurine ATP-binding transport system component | −2.7 | −2.2 |
| RS16805 | Aldehyde dehydrogenase | −5.6 | −4.7 |
| RS17030 | Aldehyde dehydrogenase | −5.6 | −5.6 |
| RS17035 | Fumarylacetoacetate hydrolase | −6.6 | −5.4 |
| RS17040 | Major facilitator superfamily permease | −4.7 | −4.3 |
| RS17045 | Dihydroxy-acid dehydratase | −5.4 | −5.5 |
Fold change in gene expression in the indicated mutants with respect to the wild-type parental strain (ATCC 17978).
FIG 4.
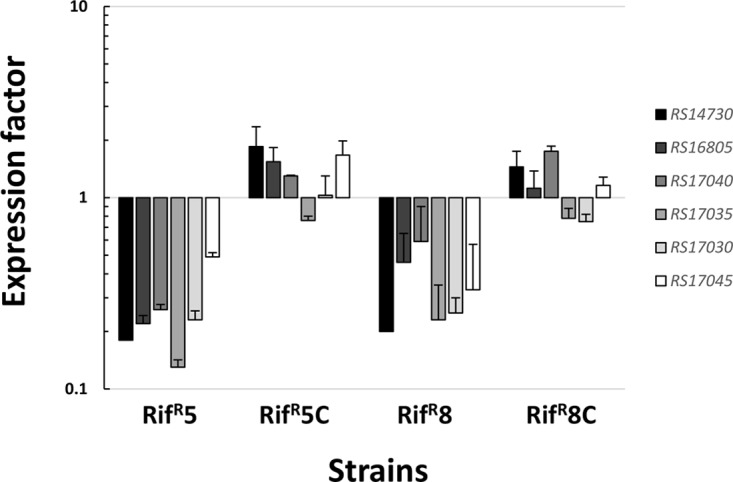
Expression factors of the indicated genes in A. baumannii strains. Rifr5C and Rifr8C represent the complemented rifampin-resistant strains Rifr5 and Rifr8, respectively, carrying the vector pBAV1K-T5-gfp with the wild-type rpoB gene. The expression factor is the ratio of the mRNA concentration of each gene from the indicated strain with respect to the wild-type A. baumannii parental strain (ATCC 17978). The error bars represent the standard deviations of the mean of at least two independent experiments, each carried out in duplicate.
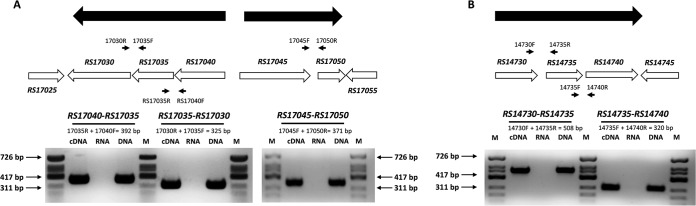
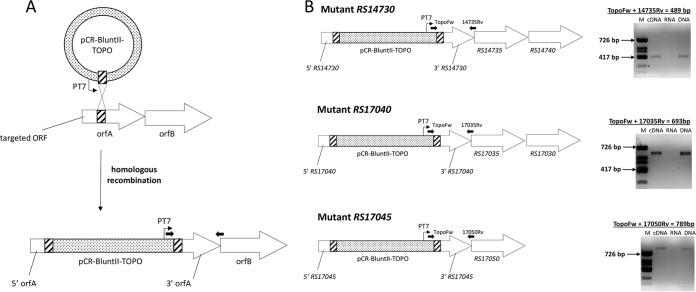
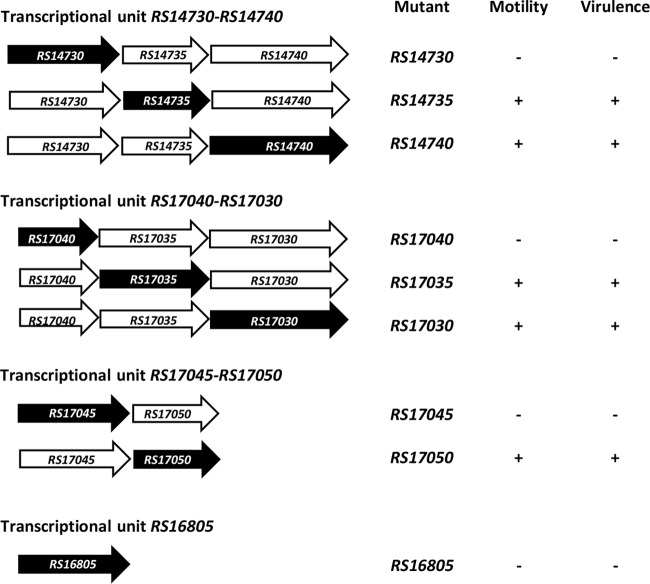
To further understand the genetic organization of the six downregulated genes, a transcriptional analysis of their surrounding regions using RT-PCR was carried out. The six genes are organized into four different transcriptional units, three of them polycistronic (Fig. 5) and one monocystronic. Two of the polycistronic units, designated RS17040-RS17030 and RS14730-RS14740, are constituted of RS17040, RS17035, and RS17030 and RS14730, RS14735, and RS14740, respectively. Another polycistronic unit contains two genes, RS17045 and RS17050, whereas the RS16805 gene is a single monocistronic transcriptional unit.
FIG 5.
Organization and direction of transcription of the downregulated genes in the Rifr3 and Rifr5 mutants inactivated in this work and their surroundings according to the annotation of the genome of A. baumannii strain ATCC 17978. (A) RS17025 to RS17055. (B) RS14730 to RS14745. The small arrows indicate the oligonucleotides used (listed in Table S2 in the supplemental material). The large black arrows represent genes belonging to the same transcriptional unit, determined by RT-PCR analysis. The RT-PCRs were carried out in the presence of cDNA, total RNA, and DNA using the indicated combination of oligonucleotides amplifying a region of overlap between the specified genes of A. baumannii strain ATCC 17978. ϕX174 DNA/HinfI marker (Biotools) was used as a molecular size marker (lanes M). The lengths of the products amplified by the indicated oligonucleotide pairs are reported with respect to some of the marker bands.
Effect of inactivation of the downregulated genes shared by A. baumannii Rifr5 and Rifr8 mutants on motility and virulence.
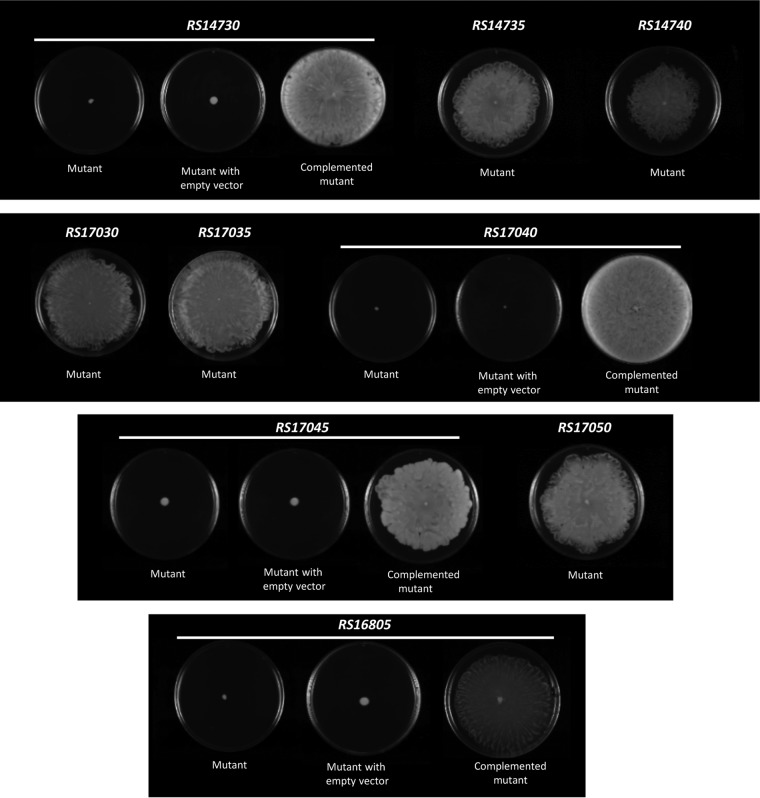
To determine whether all the genes belonging to either the RS17040-RS17030, RS14730-RS14740, or RS17045-RS17050 transcriptional unit are essential for A. baumannii motility, each of the open reading frames (ORFs) of these units was systematically inactivated, and the behavior of the resulting knockout mutants with respect to motility and virulence was studied. The results showed that mutations in four genes (RS14730, RS16805, RS17040, and RS17045) abolished motility, whereas inactivation of the remaining genes did not (Fig. 6). As expected, complementation of any of the four nonmotile mutants with the pBAV1TIC-T5-gfp plasmid carrying the corresponding wild-type gene restored wild-type motility (Fig. 6).
FIG 6.
Surface-associated motility assays of the indicated A. baumannii strain ATCC 17978 derivative mutants and mutants carrying either the empty expression vector pBAV1TIC-T5-gfp (empty vector) or the same plasmid containing the corresponding wild-type gene (complemented mutants).
It is worth noting that the constructions used to obtain the insertional mutations were designed so that the gene immediately downstream of the one to be inactivated was under the T7 promoter present in the pCR-BluntII-TOPO vector (Fig. 7A). To confirm any putative polar effect of the inactivated genes upon distal genes affecting motility, the expression of genes placed downstream of the inactivated gene was confirmed through RT-PCR assays for each strain. The results demonstrated the absence of any polar effect of the genes being inactivated by the insertion of the pCR-BluntII-TOPO plasmid upon the expression of downstream genes (Fig. 7B). Furthermore, all of the A. baumannii gene knockout mutants constructed were stable, as confirmed by their culture for 10 passages without selective pressure (data not shown).
FIG 7.
Strategies for plasmid insertion mutagenesis and assessment of the transcription of downstream genes used in this work. (A) Integration of pCR-BluntII-TOPO plasmid into a target gene. The hatched boxes correspond to the internal segment of the target gene. The vector is integrated into the targeted ORF by a single-crossover event. The direction of transcription from the T7 promoter (PT7) is indicated by the small thin arrows. The small black arrow pair represents the oligonucleotides used to assess the transcription of downstream genes. (B) RT-PCRs were carried out in the presence of cDNA, total RNA, and DNA using the indicated combinations of oligonucleotides amplifying overlapping regions between the T7 promoter of the pCR-BluntII-TOPO plasmid and the specified downstream genes of the indicated A. baumannii ATCC 17978 mutant derivatives. HinfI-digested Φ DNA was used as a molecular size marker (lanes M). The lengths of the products amplified by the indicated oligonucleotide pairs are reported with respect to some of the marker bands.
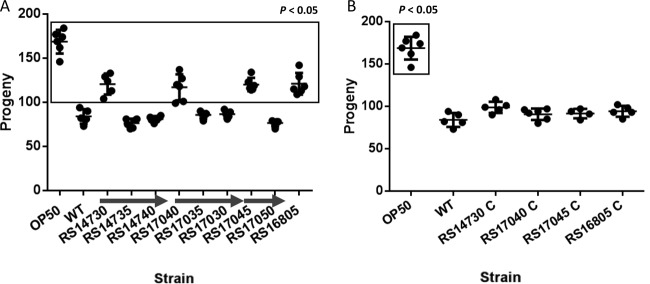
The virulence of four mutants lacking motility (RS14730, RS16805, RS17040, and RS17045) was also examined in the C. elegans fertility assay. The data showed that all of them were significantly less virulent than the wild-type strain (Fig. 8A). Furthermore, the presence in each of the four knockout mutants of the pBAV1TIC-T5-gfp plasmid carrying the corresponding wild-type gene restored virulence to the level of strain ATCC 17978 (Fig. 8B). In concordance with the data obtained in the motility assays (Fig. 6), inactivation of RS17035, RS17030, RS14735, RS14740, and RS17050 did not affect the virulence of strain ATCC 17978 (Fig. 8A). The impaired motility and virulence conferred by these mutations on wild-type A. baumannii strain ATCC 17978 were not attributable to any putative effect on the growth kinetics of the respective mutants, which remained comparable to those of the wild-type parental strain (μ [mean] = 0.83 ± 0.05 h−1).
FIG 8.
C. elegans fertility assay with the indicated A. baumannii strain ATCC 17978 derivative mutants (A) and the corresponding complemented mutants (B). Significant (P < 0.05) differences between the indicated strains with respect to the A. baumannii wild-type strain ATCC 17978 (WT) are boxed. E. coli strain OP50 was also included as a nonvirulent control. The arrows indicate genes belonging to the same transcriptional unit.
The results of the above-described motility and virulence experiments (summarized in Fig. 9), together with the fact that inactivation of the RS14730, RS17040, or RS17045 gene did not give rise to any polar effect on the rest of the genes belonging to their respective polycistronic transcriptional units (Fig. 7), clearly demonstrated that inactivation of any of these three genes alters the motility and virulence of A. baumannii strain ATCC 17978.
FIG 9.
Summary of the effect on A. baumannii motility and virulence of the systematic inactivation of the genes studied in this work. The black arrows represent the inactivated genes.
Downregulated genes of A. baumannii impairing motility are widely distributed among hospital isolates of the species.
Given the significant impact on the behavior of A. baumannii strain ATCC 17978 following the inactivation of genes RS14730, RS16805, RS17040, and RS17045, an in silico search of the sequences of the bacterial species deposited in the NCBI database was carried out to determine the presence of these genes in the genomes of other strains of the microorganism. This search involved 68 A. baumannii strains with complete chromosome sequences (see Table S1 in the supplemental material). The presence of the four genes of A. baumannii ATCC 17978, as well as the genes immediately upstream and downstream, was detected in 61 of the analyzed strains (see Table S1). These data demonstrate the strong conservation of the RS14730, RS16805, RS17040, and RS17045 genes in A. baumannii species.
To further corroborate the relevance of these four genes in other A. baumannii strains, knockout mutants were constructed in A. baumannii strain MAR002, a virulent strain recently isolated from a wound sample collected in the Hospital del Mar in Barcelona, Spain (18). Assays of the mutants indicated that inactivation of RS14730, RS16805, RS17040, and RS17045 homologues in A. baumannii MAR002 also caused a significant reduction in virulence in C. elegans (Fig. 10). In addition, as in strain ATCC 17978, the A. baumannii MAR002 strain defective in either the RS17035 or RS17030 homologue did not exhibit defective virulence (Fig. 10). These results clearly indicate the significant role of the four genes identified in this work in the virulence of A. baumannii.
FIG 10.
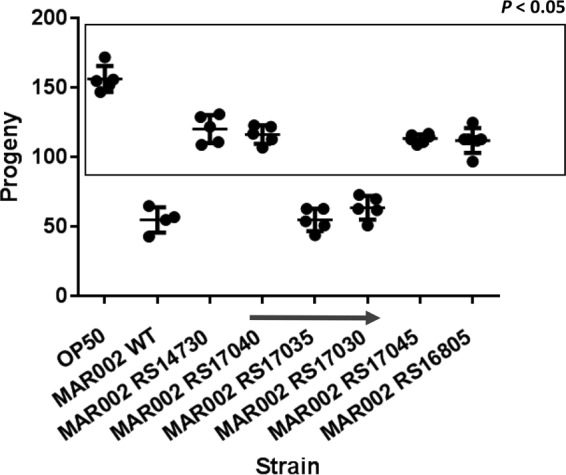
C. elegans fertility assay with the WT A. baumannii strain MAR002 and derivative mutants. Significant (P < 0.05) differences between the specified strains with respect to A. baumannii strain MAR002 (WT) are boxed. The E. coli strain OP50 is also indicated as a nonvirulent control. The arrow indicates genes belonging to the same transcriptional unit.
DISCUSSION
Bacteria are able to live not only as single-celled organisms exhibiting planktonic growth, but also as communities of sessile cells embedded in extracellular matrices, in which they adhere to one another and to the surfaces on which they reside to form biofilms (19). Bacteria can expand their environments by moving along surfaces through various types of motility that differ in their required components and mechanisms (20). Motility has been linked to the infective capacity of pathogens, such as Pseudomonas aeruginosa (21), Escherichia coli O157:H7 (22), Haemophilus influenzae (23), Neisseria meningitidis (24), Clostridium perfringens (25), Legionella pneumophila (26), and Proteus spp. (27). Motility is also associated with enhanced antibiotic resistance (28), protection against host macrophages (29), and the first stage of biofilm generation (30).
The motility of A. baumannii is responsive to many of the same environmental and cellular signals involved in bacterial virulence (9). Our results clearly demonstrate that A. baumannii mutants with an amino acid substitution at position 522 or 540 of the RpoB protein lack surface-associated motility, which must be attributed to the decreased expression of several genes. The inactivation of some of these genes in A. baumannii also caused a reduction in its virulence. Introduction of the wild-type rpoB copy restored the wild-type phenotype in the Rifr5 and Rifr8 strains (rifampin susceptibility, surface-associated motility, and virulence). Since the wild-type gene encoding the RpoB subunit was cloned in a high-copy-number expression plasmid, recovery of the phenotype in these merodiploids was probably due to the presence of a high number of wild-type RpoB copies, which displaced the mutated RpoB proteins and thereby restored the wild-type phenotype. Similarly, the introduction into the wild-type strain of an expression vector containing the rpoB gene amplified from each rifampin-resistant mutant strain reproduced the same phenotype observed in the strain from which it was amplified.
In E. coli and Streptomyces spp., point mutations in the rpoB gene that confer rifampin resistance give rise to stringent RNA polymerase-like behavior normally associated with nutrient starvation (17). The resulting amino acid substitutions induce a conformational change in the β-subunit of the RNA polymerase that mimics the effect of guanosine tetraphosphate (ppGpp), an effector of the stringent response. The pattern of expression is similar to that seen during this response, but ppGpp is not required, as the same changes are produced in strains defective for the synthesis of the nucleotide (17). Substitutions at position Ser531 of the β-subunit of the E. coli RNA polymerase (which corresponds to Ser540 of the A. baumannii β-subunit reported here) specifically alter the interaction of E. coli RpoB with certain stringent promoters (31). In N. meningitidis, mutations in rpoB cause global transcriptional changes that functionally mimic the stringent response (32); in Pseudomonas spp., a phylogenetic neighbor of Acinetobacter spp., stringent RNA polymerase is involved in twitching motility (33, 34). Thus, by isolating mutants arising from point mutations in the A. baumannii rpoB gene, we were also able to identify genes involved in the surface-associated motility of the bacterium (Table 1).
Six downregulated genes widespread among A. baumannii clinical isolates were shared by the two analyzed Rifr A. baumannii spontaneous mutants. Inactivation of four of these genes (RS14730, RS16805, RS17040, and RS17045) significantly reduced A. baumannii surface-associated motility. Importantly, this loss of motility was associated with attenuation of virulence, not only in A. baumannii strain ATCC 17978, but also in the biofilm-hyperproducing A. baumannii clinical isolate strain (MAR002). The transcriptome response of A. baumannii strain ATCC 17978 during bacteremia was recently analyzed (35). Data obtained in that work confirmed that during this step of the A. baumannii infective process, the bacterial cells are in a planktonic state (35). Moreover, the expression levels of the A. baumannii genes identified in our study (RS14730, RS16805, RS17040, and RS17045) do not increase during bacteremia (35). These results are in concordance with the role of these genes in surface-associated motility. Similar scenarios, in which genes involved in surface-associated motility were not induced under planktonic conditions, have been reported for other bacterial species (36).
In A. baumannii Rifr5 and Rifr8 mutants, all of the downregulated genes described in this work encode putative proteins that in other bacterial species are related to either transporters or metabolic enzymes and play important roles in motility and virulence. For instance, RS17040 is annotated as a major facilitator superfamily (MFS) permease. MFS proteins are involved in chemotaxis, which requires bacterial appendages, such as pili, for directed movement toward or away from attractants and repellents, respectively. In Pseudomonas putida, chemotaxis to the aromatic acid 4-hydroxybenzoate (4-HBA) is mediated by the MFS permease PcaK, a membrane-bound protein that also functions as a 4-HBA transporter (37).
Similarly, Vibrio cholerae possesses a chemoreceptor that mediates the response to taurine, a major constituent of human bile, and thus plays an important role in the chemotaxis and virulence of the pathogen (38). The A. baumannii RS14730 gene encodes a putative taurine ATP-binding transport system component involved in the transport of taurine as a sulfur source. Thus, during the A. baumannii infection process, there may be competition for taurine between the nosocomial pathogen and its host (39).
Two other downregulated genes identified in the Rifr5 and Rifr8 mutants, RS16805 and RS17045, encode a putative aldehyde dehydrogenase and a putative dihydroxy-acid dehydratase, respectively. In enterohemorrhagic E. coli, the aromatic aldehyde dehydrogenase FeaB is involved in chemotaxis to norepinephrine (40), whereas in Pseudomonas syringae, inactivation of the GDP-d-mannose dehydratase decreases the motility of the plant pathogen (41). A previous study showed that, in A. baumannii, genes encoding metabolic enzymes (involved in the synthesis of 1,3-diaminopropane) are also involved in the surface-associated motility of the bacterium (6).
To our knowledge, this is the first report clearly associating loss of motility with the attenuation of virulence in the pathogen A. baumannii. The data obtained in this work provide the basis for further investigations of the cellular components and mechanisms involved in the motility of this multiresistant microorganism.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
A. baumannii ATCC 17978, A. baumannii MAR002, and E. coli (strains DH5α and OP50) were grown in Luria-Bertani (LB) medium and incubated at 37°C with shaking at 180 rpm. When necessary, kanamycin (50 mg/liter) or ticarcillin (80 mg/liter) was added to the growth medium. The plasmids pCR-BluntII-TOPO (Invitrogen) and pBAV1K-T5-gfp (a gift from Ichiro Matsumura; Addgene plasmid 26702), were used for mutant construction and complementation, respectively. Growth was monitored by measuring the absorbance at 600 nm (OD600). The growth rate (μ) was calculated based on the exponential segment of the growth curve and defined as ln 2g−1, where g is the doubling time of an exponentially growing culture. For the isolation of rifampin-resistant mutants, saturated cultures of A. baumannii ATCC 17978 were plated on LB agar containing rifampin (50 mg/liter) and then incubated at 37°C for 24 h. The MICs for rifampin were determined by broth microdilution.
Sequencing of rpoB mutants.
Colony PCR was carried out using the oligonucleotides rpoB-1441F and rpoB-2095R (42) (Invitrogen), which in rpoB amplify a 654-bp region, the site of frequent Rifr-inducing base pair substitutions in the species (42). The PCR products were purified using the GFX PCR DNA and gel band purification kit (GE Healthcare) according to the supplier's protocols and sequenced (Macrogen) using the same oligonucleotide set. The data were analyzed using DNAStar Lasergene software (DNAStar).
Motility assays.
Fresh LB agar motility plates (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 0.5% glucose, and 0.5% Difco agar) were prepared the day of the assay. After sterilization, the medium was deposited on 9-cm petri dishes with constant agitation to ensure the homogeneous spread of the agar. The inoculum was applied with a sterile toothpick. A single colony was picked and inoculated in the center of the plate, avoiding medium penetration. The inoculated plates were incubated at 37°C, usually for 16 to 20 h, previously determined to be the time required for the wild-type strain to reach the plate border under the experimental conditions. All assays were carried out a minimum of three times in independent experiments. Representative images of the motility of each bacterial strain on a semisolid agar surface are shown.
RNA extraction and RT-PCR.
An overnight culture of the corresponding strain in LB medium was diluted 1:50 in fresh medium and then grown until the mid-exponential growth phase (OD600 = 0.6) was reached. The cells in 10 ml of each culture were pelleted by centrifugation at 13,000 × g, resuspended in Tris-EDTA (TE) buffer, and treated with lysozyme (50 mg/ml) for 10 min at 37°C. Total RNA was extracted using an RNeasy minikit (Qiagen). DNA contaminants were removed from the RNA by digestion with DNase Turbo (Ambion). The absence of DNA in the PCR mixtures was confirmed by incubating the RNA samples without reverse transcriptase. RT-PCR was performed using a first-strand cDNA synthesis kit (Nzytech) and the oligonucleotides listed in Table S2 in the supplemental material, following the manufacturer's instructions.
Microarrays.
The microarrays were designed specifically for A. baumannii strain ATCC 17978 by Bioarray Diagnóstico Genético (Alicante, Spain) and were performed using eArray (Agilent). Labeling was carried out following the two-color microarray-based prokaryote analysis Fair Play III Labeling v. 1.3 (Agilent). Three independent RNA extractions per condition (biological replicates) were used for each experiment. The quality of the RNA was determined using Bioanalyzer (Bioarray Alicante, Spain). Statistical analysis was carried out using Bioconductor in the software package RankProd for the R computing environment. A gene was considered to be induced when the ratio of the treated preparation to the nontreated preparation was ≥2 and the P value of the difference compared to the control was <0.05. The resulting microarray data sets (WT versus Rifr5 and WT versus Rifr8) were submitted to the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/). During the revision of the manuscript, the genome of A. baumannii strain ATCC 17978 was updated. Consequently, in this article, the new annotation (NZ_CP018664.1) has been introduced. The correlation between the old and new annotations of the genes described in this work is presented in Table S3 in the supplemental material.
Gene expression.
Gene expression was determined by RT-qPCR using Lightcycler RNA Master SYBR green I (Roche) on a Lightcycler 480 instrument (LC480; Roche) as previously described (43). Specific oligonucleotides (see Table S2 in the supplemental material) were used to validate the expression of the selected genes shown in the microarray to be deregulated. The relative mRNA concentrations of the genes of interest were determined according to a standard curve generated by amplifying an internal fragment of the gyrB gene that was not induced under the tested growth conditions. The expression factor was calculated as the ratio of the mRNA concentration of the target gene as expressed in the wild-type strain versus the studied strain.
Mutant construction.
Plasmids were inserted into the target genes as previously described (44). Briefly, the kanamycin and zeocin resistance plasmid pCR-BluntII-TOPO, which is unable to replicate in A. baumannii, served as a suicide vector. An internal fragment (∼500 bp) of the target gene was PCR amplified with the appropriate primers (see Table S2 in the supplemental material) using genomic DNA from A. baumannii as the template. The PCR products were cloned into the pCR-BluntII-TOPO vector and propagated in E. coli DH5α. The recombinant plasmids (0.1 μg) were then introduced into A. baumannii by electroporation as previously described (44). Mutants were selected on kanamycin-containing plates. Gene inactivation by plasmid insertion via single-crossover recombination was confirmed by sequencing the amplified PCR product using the appropriate oligonucleotides (see Table S2). All the mutants used in this work were assessed for stability after 10 passages (every 24 h) by incubating cultures of the mutants without selective pressure and then counting the colonies plated on LB plates with and without kanamycin.
Mutant complementation.
The plasmid pBAV1K-T5-gfp was used to complement the Rifr3, Rifr5, and Rifr8 mutants by blunt-end cloning the wild-type rpoB gene from the A. baumannii ATCC 17978 wild-type genome into the XbaI site of the vector, whose overhangs were filled with T4 DNA polymerase (Roche). As this plasmid carries a gene encoding kanamycin resistance, complementation of the mutants constructed using the pCR-BluntII-TOPO vector, which also encodes kanamycin resistance, was carried out using a variant of the pBAV1K-T5-gfp vector (pBAV1TIC-T5-gfp), constructed in this work as follows. The β-lactamase-coding region from the pGEM-T vector (Promega) was PCR amplified, and the blunt-ended PCR product was cloned into the pBAV1K-T5-gfp plasmid digested by EcoRV, whose unique site in the plasmid is present in the coding region of the kanamycin resistance gene. All the genes were cloned into the XbaI site of the pBAV1TIC-T5-gfp vector, except rpoB, which contains an XbaI site in its coding region and was cloned into the original vector (pBAV1K-T5-gfp) as described above. The recombinant plasmids were propagated in E. coli DH5α and introduced into the corresponding A. baumannii strain by electroporation as previously described (44). Transformants were selected on plates containing kanamycin (for cells carrying pBAV1K-T5-gfp derivatives) or ticarcillin (for cells carrying pBAV1TIC-T5-gfp derivatives). All of the resulting recombinant plasmids were verified by both PCR and sequencing (Macrogen). The oligonucleotides used in this study are listed in Table S2 in the supplemental material.
In vivo nematode model of virulence.
The virulences of the different A. baumannii strains used in this work were analyzed in a nematode fertility assay, as previously described (14). Briefly, C. elegans strain N2 was fed on the low-virulence E. coli OP50 strain grown as a lawn on NGM (nematode growth medium) plates. The nematode eggs were recovered and hatched in M9 medium (0.02 M KH2PO4, 0.04 M Na2HPO4, 0.08 M NaCl, and 0.001 M MgSO4), grown to stage L1, and growth arrested overnight at 16°C to physiologically synchronize the worms. The L1 larvae were cultivated to the L4 stage on lawns of the appropriate bacterial strain on NGM plates. A single L4 nematode was then inoculated on a peptone-glucose-sorbitol plate containing the same strain and incubated at 25°C. During the next 3 days, adult nematodes were removed daily to a fresh plate seeded with the same bacterial strain. To determine fertility, nematode progeny were counted daily using a stereomicroscope (Olympus SZ51) 48 h after removal of the parent. Six independent replicates were established for each bacterial strain, and each fertility assay was performed in triplicate. Representative experiments from the C. elegans fertility assay are shown. Statistical analysis consisted of a two-tailed one-way analysis of variance (ANOVA), followed by the Tukey test for post hoc multiple-group comparisons. A P value of < 0.05 was considered to indicate statistical significance.
Accession number(s).
The microarray data sets (wild-type versus Rifr5 and WT versus Rifr8) were submitted to the NCBI GEO database under accession number GSE73193.
Supplementary Material
ACKNOWLEDGMENTS
We thank Pilar Cortés (Universitat Autònoma de Barcelona [UAB]), Joan Ruiz (UAB), and Susana Escribano (UAB) for their excellent technical assistance, as well as our UAB students Pau Obregón and Daniel Quiñones for their helpful support.
This study was supported by grant BIO2016-77011-R from the Ministerio de Economía y Competitividad and 2014SGR572 from the Generalitat de Catalunya. Jesús Aranda is a Serra Húnter Fellow, Generalitat de Catalunya, Spain. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00327-17.
REFERENCES
- 1.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aranda J, Poza M, Shingu-Vázquez M, Cortés P, Boyce JD, Adler B, Barbé J, Bou G. 2013. Identification of a DNA-damage-inducible regulon in Acinetobacter baumannii. J Bacteriol 195:5577–5582. doi: 10.1128/JB.00853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aranda J, López M, Leiva E, Magán A, Adler B, Bou G, Barbé J. 2014. Role of Acinetobacter baumannii UmuD homologs in antibiotic resistance acquired through DNA damage-induced mutagenesis. Antimicrob Agents Chemother 58:1771–1773. doi: 10.1128/AAC.02346-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jara LM, Pérez-Varela M, Corral J, Arch M, Cortés P, Bou G, Aranda J, Barbé J. 2015. Novobiocin inhibits the antimicrobial resistance acquired through DNA damage-induced mutagenesis in Acinetobacter baumannii. Antimicrob Agents Chemother 60:637–639. doi: 10.1128/AAC.01810-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jara LM, Cortés P, Bou G, Barbé J, Aranda J. 2015. Differential roles of antimicrobials in the acquisition of drug resistance through activation of the SOS response in Acinetobacter baumannii. Antimicrob Agents Chemother 59:4318–4320. doi: 10.1128/AAC.04918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skiebe E, de Berardinis V, Morczinek P, Kerrinnes T, Faber F, Lepka D, Hammer B, Zimmermann O, Ziesing S, Wichelhaus TA, Hunfeld KP, Borgmann S, Gröbner S, Higgins PG, Seifert H, Busse HJ, Witte W, Pfeifer Y, Wilharm G. 2012. Surface-associated motility, a common trait of clinical isolates of Acinetobacter baumannii, depends on 1,3-diaminopropane. Int J Med Microbiol 302:117–128. doi: 10.1016/j.ijmm.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Harding CM, Tracy EN, Carruthers MD, Rather PN, Actis LA, Munson RS. 2013. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio 4:e00360-13. doi: 10.1128/mBio.00360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eijkelkamp BA, Stroeher UH, Hassan KA, Elbourne LDH, Paulsen IT, Brown MH. 2013. H-NS plays a role in expression of Acinetobacter baumannii virulence features. Infect Immun 81:2574–2583. doi: 10.1128/IAI.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McConnell MJ, Actis L, Pachón J. 2013. Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol Rev 37:130–155. doi: 10.1111/j.1574-6976.2012.00344.x. [DOI] [PubMed] [Google Scholar]
- 10.Alifano P, Palumbo C, Pasanisi D, Talà A. 2015. Rifampicin-resistance, rpoB polymorphism and RNA polymerase genetic engineering. J Biotechnol 202:60–77. doi: 10.1016/j.jbiotec.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 11.Qi Q, Preston GM, MacLean RC. 2014. Linking system-wide impacts of RNA polymerase mutations to the fitness cost of rifampin resistance in Pseudomonas aeruginosa. mBio 5:e01562-14. doi: 10.1128/mBio.01562-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin DJ, Gross CA. 1989. Characterization of the pleiotropic phenotypes of rifampin-resistant rpoB mutants of Escherichia coli. J Bacteriol 171:5229–5231. doi: 10.1128/jb.171.9.5229-5231.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin DJ, Gross CA. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol 202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 14.Vallejo JA, Beceiro A, Rumbo-Feal S, Rodríguez-Palero MJ, Russo TA, Bou G. 9 July 2015. Optimisation of the Caenorhabditis elegans model for studying the pathogenesis of opportunistic Acinetobacter baumannii. Int J Antimicrob Agents. doi: 10.1016/j.ijantimicag.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 15.Miller LP, Crawford JT, Shinnick TM. 1994. The rpoB gene of Mycobacterium tuberculosis. Antimicrob Agents Chemother 38:805–811. doi: 10.1128/AAC.38.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryksin AV, Matsumura I. 2010. Rational design of a plasmid origin that replicates efficiently in both gram-positive and gram-negative bacteria. PLoS One 5:e13244. doi: 10.1371/journal.pone.0013244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Tozawa Y, Lai C, Hayashi H, Ochi K. 2002. A rifampicin resistance mutation in the rpoB gene confers ppGpp-independent antibiotic production in Streptomyces coelicolor A3(2). Mol Genet Genomics 268:179–189. doi: 10.1007/s00438-002-0730-1. [DOI] [PubMed] [Google Scholar]
- 18.Álvarez-Fraga L, Pérez Rumbo-Feal A, Merino S, Vallejo M, Ohneck JA, Edelmann EJ, Beceiro RE, Vázquez-Ucha A, Valle JC, Actis J, Bou LA, Poza GM. 2016. Analysis of the role of the LH92_11085 gene of a biofilm hyper-producing Acinetobacter baumannii strain on biofilm formation and attachment to eukaryotic cells. Virulence 7:443–455. doi: 10.1080/21505594.2016.1145335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berlanga M, Guerrero R. 2016. Living together in biofilms: the microbial cell factory and its biotechnological implications. Microb Cell Fact 15:165. doi: 10.1186/s12934-016-0569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarrell KF, McBride MJ. 2008. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6:466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 21.Kazmierczak BI, Schniederberend M, Jain R. 2015. Cross-regulation of Pseudomonas motility systems: the intimate relationship between flagella, pili and virulence. Curr Opin Microbiol 28:78–82. doi: 10.1016/j.mib.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xicohtencatl-Cortes J, Monteiro-Neto V, Saldaña Z, Ledesma MA, Puente JL, Girón JA. 2009. The type IV pili of enterohemorrhagic Escherichia coli O157:H7 are multipurpose structures with pathogenic attributes. J Bacteriol 191:411–421. doi: 10.1128/JB.01306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolappan S, Tracy EN, Bakaletz LO, Munson RS, Craig L. 2012. Expression, purification, crystallization and preliminary crystallographic analysis of PilA from the nontypeable Haemophilus influenzae type IV pilus. Acta Crystallogr Sect F Struct Biol Cryst Commun 68:284–287. doi: 10.1107/S1744309111043910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albiger B, Johansson L, Jonsson A-B. 2003. Lipooligosaccharide-deficient Neisseria meningitidis shows altered pilus-associated characteristics. Infect Immun 71:155–162. doi: 10.1128/IAI.71.1.155-162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varga JJ, Nguyen V, O'Brien DK, Rodgers K, Walker RA, Melville SB. 2006. Type IV pili-dependent gliding motility in the Gram-positive pathogen Clostridium perfringens and other clostridia. Mol Microbiol 62:680–694. doi: 10.1111/j.1365-2958.2006.05414.x. [DOI] [PubMed] [Google Scholar]
- 26.Coil DA, Anné J. 2009. Twitching motility in Legionella pneumophila. FEMS Microbiol Lett 293:271–277. doi: 10.1111/j.1574-6968.2009.01532.x. [DOI] [PubMed] [Google Scholar]
- 27.Hola V, Peroutkova T, Ruzicka F. 2012. Virulence factors in Proteus bacteria from biofilm communities of catheter-associated urinary tract infections. FEMS Immunol Med Microbiol 65:343–349. doi: 10.1111/j.1574-695X.2012.00976.x. [DOI] [PubMed] [Google Scholar]
- 28.Butler MT, Wang Q, Harshey RM. 2010. Cell density and mobility protect swarming bacteria against antibiotics. Proc Natl Acad Sci U S A 107:3776–3781. doi: 10.1073/pnas.0910934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan RM, Kuang Z, Hao Y, Lau GW. 2014. Type IV pilus of Pseudomonas aeruginosa confers resistance to antimicrobial activities of the pulmonary surfactant protein-A. J Innate Immun 6:227–239. doi: 10.1159/000354304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maldarelli GA, Piepenbrink KH, Scott AJ, Freiberg JA, Song Y, Achermann Y, Ernst RK, Shirtliff ME, Sundberg EJ, Donnenberg MS, von Rosenvinge EC. 2016. Type IV pili promote early biofilm formation by Clostridium difficile. Pathog Dis 74:ftw061. doi: 10.1093/femspd/ftw061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou YN, Jin DJ. 1998. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc Natl Acad Sci U S A 95:2908–2913. doi: 10.1073/pnas.95.6.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colicchio R, Pagliuca C, Pastore G, Cicatiello AG, Pagliarulo C, Talà A, Scaglione E, Sammartino JC, Bucci C, Alifano P, Salvatore P. 2015. Fitness cost of rifampin resistance in Neisseria meningitidis: in vitro study of mechanisms associated with rpob H553Y mutation. Antimicrob Agents Chemother 59:7637–7649. doi: 10.1128/AAC.01746-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogt SL, Green C, Stevens KM, Day B, Erickson DL, Woods DE, Storey DG. 2011. The stringent response is essential for Pseudomonas aeruginosa virulence in the rat lung agar bead and Drosophila melanogaster feeding models of infection. Infect Immun 79:4094–4104. doi: 10.1128/IAI.00193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chatnaparat T, Li Z, Korban SS, Zhao Y. 2015. The stringent response mediated by (p)ppGpp is required for virulence of Pseudomonas syringae pv. tomato and its survival on tomato. Mol Plant Microbe Interact 28:776–789. doi: 10.1094/MPMI-11-14-0378-R. [DOI] [PubMed] [Google Scholar]
- 35.Murray GL, Tsyganov K, Kostoulias XP, Bulach DM, Powell D, Creek DJ, Boyce JD, Paulsen IT, Peleg AY. 2017. Global gene expression profile of Acinetobacter baumannii during bacteremia. J Infect Dis 215:S52–S57. doi: 10.1093/infdis/jiw529. [DOI] [PubMed] [Google Scholar]
- 36.Lippolis JD, Brunelle BW, Reinhardt TA, Sacco RE, Thacker TC, Looft TP, Casey TA. 2016. Differential gene expression of three mastitis-causing Escherichia coli strains grown under planktonic, swimming, and swarming culture conditions. mSystems 1:e00064-16. doi: 10.1128/mSystems.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ditty JL, Harwood CS. 1999. Conserved cytoplasmic loops are important for both the transport and chemotaxis functions of PcaK, a protein from Pseudomonas putida with 12 membrane-spanning regions. J Bacteriol 181:5068–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishiyama S, Takahashi Y, Yamamoto K, Suzuki D, Itoh Y, Sumita K, Uchida Y, Homma M, Imada K, Kawagishi I. 2016. Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants. Sci Rep 6:20866. doi: 10.1038/srep20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, Dossat C, Gas S, Kreimeyer A, Lenoble P, Oztas S, Poulain J, Segurens B, Robert C, Abergel C, Claverie J-M, Raoult D, Médigue C, Weissenbach J, Cruveiller S. 2008. Comparative analysis of acinetobacters: three genomes for three lifestyles. PLoS One 3:e1805. doi: 10.1371/journal.pone.0001805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pasupuleti S, Sule N, Cohn WB, MacKenzie DS, Jayaraman A, Manson MD. 2014. Chemotaxis of Escherichia coli to norepinephrine (NE) requires conversion of NE to 3,4-dihydroxymandelic acid. J Bacteriol 196:3992–4000. doi: 10.1128/JB.02065-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiku K, Tsunemi K, Yamamoto M, Ohnishi-Kameyama M, Yoshida M, Ishii T, Taguchi F, Iwaki M, Ichinose Y, Ono H. 2013. Defects in d-rhamnosyl residue biosynthetic genes affect lipopolysaccharide structure, motility, and cell-surface hydrophobicity in Pseudomonas syringae pathovar glycinea race 4. Biosci Biotechnol Biochem 77:505–510. doi: 10.1271/bbb.120736. [DOI] [PubMed] [Google Scholar]
- 42.Norton MD, Spilkia AJ, Godoy VG. 2013. Antibiotic resistance acquired through a DNA damage-inducible response in Acinetobacter baumannii. J Bacteriol 195:1335–1345. doi: 10.1128/JB.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aranda J, Garrido ME, Cortes P, Llagostera M, Barbe J. 2008. Analysis of the protective capacity of three Streptococcus suis proteins induced under divalent-cation-limited conditions. Infect Immun 76:1590–1598. doi: 10.1128/IAI.00987-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aranda J, Poza M, Pardo BG, Rumbo S, Rumbo C, Parreira JR, Rodriguez-Velo P, Bou G, Rodríguez-Velo P, Bou G. 2010. A rapid and simple method for constructing stable mutants of Acinetobacter baumannii. BMC Microbiol 10:279. doi: 10.1186/1471-2180-10-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.