ABSTRACT
The mycobacterial cell wall is crucial to the host-pathogen interface, because it provides a barrier against antibiotics and the host immune response. In addition, cell wall lipids are mycobacterial virulence factors. The mycobacterial membrane protein large (MmpL) proteins are cell wall lipid transporters that are important for basic mycobacterial physiology and Mycobacterium tuberculosis pathogenesis. MmpL3 and MmpL11 are conserved across pathogenic and nonpathogenic mycobacteria, a feature consistent with an important role in the basic physiology of the bacterium. MmpL3 is essential and transports trehalose monomycolate to the mycobacterial surface. In this report, we characterize the role of MmpL11 in M. tuberculosis. M. tuberculosis mmpL11 mutants have altered biofilms associated with lower levels of mycolic acid wax ester and long-chain triacylglycerols than those for wild-type bacteria. While the growth rate of the mmpL11 mutant is similar to that of wild-type M. tuberculosis in macrophages, the mutant exhibits impaired survival in an in vitro granuloma model. Finally, we show that the survival or recovery of the mmpL11 mutant is impaired when it is incubated under conditions of nutrient and oxygen starvation. Our results suggest that MmpL11 and its cell wall lipid substrates are important for survival in the context of adaptive immune pressure and for nonreplicating persistence, both of which are critically important aspects of M. tuberculosis pathogenicity.
KEYWORDS: Mycobacterium tuberculosis, biofilm, lipid transport, cell wall, tuberculosis, MmpL11, nonreplicating persistence, granuloma, lipid transporter
INTRODUCTION
Tuberculosis (TB) is one of the leading causes of death due to infectious disease despite the availability of antitubercular drugs. The mycobacterial cell wall is associated with Mycobacterium tuberculosis pathogenesis and provides a barrier against environmental stresses, antibiotics, and the host immune response. The mycobacterial cell wall is unique in architecture and composition. The outer membrane contains an inner leaflet of very-long-chain mycolic acids covalently bound to the arabinogalactan-peptidoglycan layer and an outer leaflet composed of noncovalently associated lipids, such as phthiocerol dimycocerosate (PDIM), sulfolipids, and trehalose 6,6′-dimycolate (TDM) (1, 2). These surface-exposed lipids are immunomodulatory and play a role in host-pathogen interactions (3–9).
The mycobacterial membrane protein large (MmpL) proteins are cell wall lipid transporters that are crucial contributors to mycobacterial physiology and pathogenesis (10–18). MmpL3 is essential and transports trehalose monomycolate (TMM), the TDM precursor, to the mycobacterial surface (19). TDM biosynthesis and incorporation into the mycobacterial cell wall are required for mycobacterial replication and viability (20, 21). MmpL4, MmpL5, MmpL7, MmpL8, MmpL10, and MmpL11 are required for full M. tuberculosis virulence (22, 23). MmpL4 and MmpL5, along with their accessory MmpS proteins, are involved in M. tuberculosis siderophore export (24). MmpL7 and MmpL8 transport PDIM and sulfolipid-1, respectively, to the outer leaflet of the M. tuberculosis cell wall, where these surface-exposed lipids function as virulence determinants (3, 9). MmpL10 is required for transport of diacyltrehalose to the M. tuberculosis periplasm, where it is converted to penta-acyltrehalose (18). The substrates of MmpL11 in M. tuberculosis have not yet been identified.
While all mycobacteria possess genes encoding MmpL proteins, the number in each species can differ. For instance, Mycobacterium leprae possesses 5 intact mmpL genes, whereas there are 31 mmpL genes in M. abscessus. In addition, differences exist in MmpL substrates among species. While MmpL4 is required for siderophore export in M. tuberculosis, it is involved in glycopeptidolipid transport in M. smegmatis and M. bolletii (15, 24–26). Furthermore, the M. smegmatis homologue of MmpL10 transports trehalose polyphleates instead of diacyltrehalose (16). We demonstrated previously that in M. smegmatis, MmpL11 is important for biofilm formation and transports monomeromycolyl diacylglycerol (MMDAG), a specialized triacylglycerol (TAG) molecule, and mycolate wax ester (MWE) (10). Heterologous expression of M. tuberculosis mmpL11 in an M. smegmatis mmpL11 mutant complemented the biofilm phenotype, suggesting that MmpL11 transports similar substrates in the two species.
Two studies independently showed that an M. tuberculosis mmpL11 mutant is attenuated in a mouse model of infection (22, 23). While M. tuberculosis mmpL11 mutants infected mice like wild-type M. tuberculosis, MmpL11 was required for bacterial survival during later stages of chronic infection (22). Neither study identified the mechanism by which the mutant was attenuated or determined the substrate of MmpL11. To further characterize the role of MmpL11 in M. tuberculosis, we constructed and characterized an M. tuberculosis mmpL11 mutant. In agreement with a conserved role in mycobacterial physiology, we show that M. tuberculosis mmpL11 mutants have altered biofilm formation. When grown under biofilm-inducing conditions, surface lipids, including an MWE and long-chain TAGs, are altered in the mmpL11 mutant relative to the wild type, suggesting that these apolar lipids are the MmpL11 substrates in M. tuberculosis. We found that the mmpL11 mutant was able to replicate like the wild type in cultured macrophages, but by capitalizing on an in vitro granuloma model generated from individuals with latent TB infection (LTBI), we show that the mmpL11 mutant is less fit than wild-type M. tuberculosis in the face of adaptive immune pressure. Finally, we show that the mmpL11 mutant is impaired for survival and/or resuscitation relative to wild-type M. tuberculosis when incubated under conditions of nutrient and oxygen starvation. Combined, these data suggest that MmpL11 and its substrates are important for intracellular growth, nonreplicating persistence, and latency.
RESULTS
Mmpl11 plays a conserved role in mycobacterial biofilm formation.
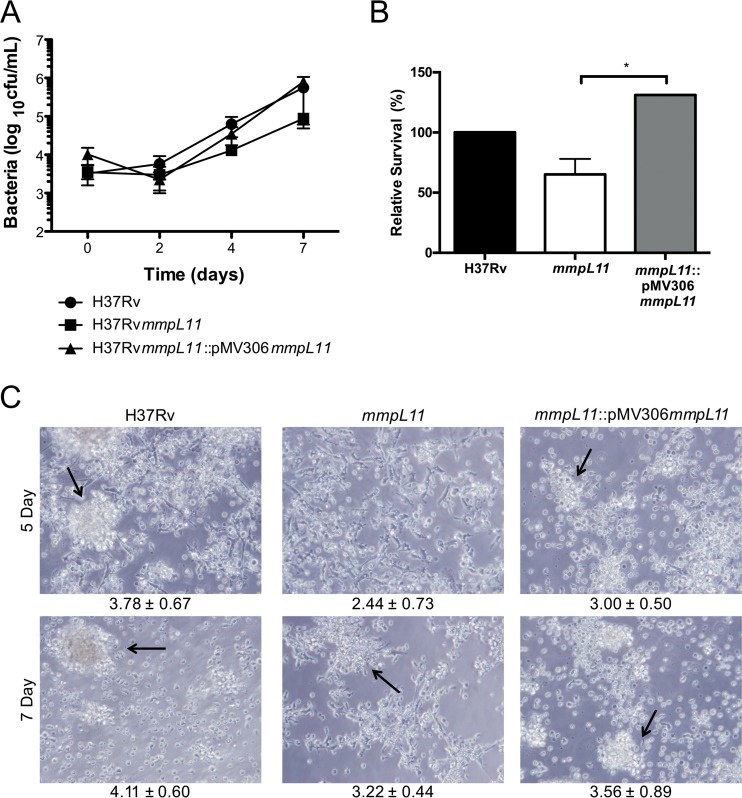
To experimentally determine whether MmpL11 function is conserved, we constructed an allelic exchange mutant in H37Rv to characterize the role and function of MmpL11 in M. tuberculosis. There were no differences in the growth rate between wild-type M. tuberculosis and the mmpL11 mutant when they were grown planktonically in 7H9 medium containing Tween 80 (Fig. 1A). When the mycobacteria were grown in Sauton's medium lacking Tween 80 to induce biofilm formation, we observed that the absence of MmpL11 resulted in a weaker biofilm that was slower to form than that for wild-type bacteria (Fig. 1B). To quantify biofilm formation, we stained the biofilm material that adhered to the wells of a 96-well plate with crystal violet. The level of crystal violet staining for the mutant wells was significantly lower than that for the wild-type wells (Fig. 1C). The impaired biofilm phenotype was complemented via expression of M. tuberculosis MmpL11 (MmpL11Mt). Two complementation strategies were used for the M. tuberculosis mmpL11 mutant. The pVV16mmpL11 plasmid constitutively expressed mmpL11 from an hsp60 promoter. The mmpL11/pVV16mmpL11 strain developed a reticulated biofilm faster than wild-type M. tuberculosis (Fig. 1B, top). We speculate that this occurs because of constitutive overexpression of MmpL11. Our second complementation approach used an integrative pMV306mmpL11 construct where mmpL11 was cloned with the 1,000-bp region upstream such that mmpL11 expression would be under the control of its native promoter. While we anticipated that the levels of mmpL11 would be equivalent to those in wild-type bacteria, expression of the mmpL11 gene in the mmpL11::pMV306mmpL11 strain was 5-fold higher than that of the wild type (5.54-fold ± 0.483-fold over three biological replicates).
FIG 1.
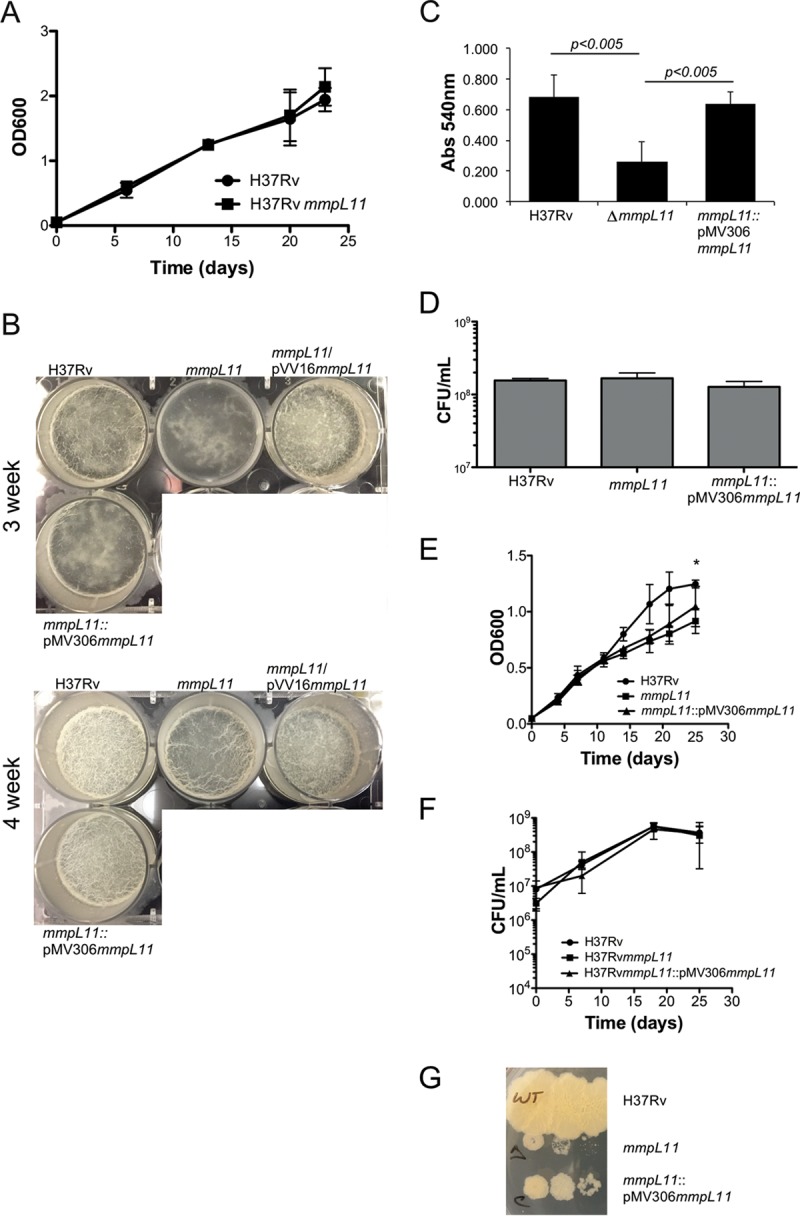
The M. tuberculosis mmpL11 mutant exhibits impaired biofilm formation that does not result from reduced replication in Sauton's medium. (A) Growth of M. tuberculosis wild-type and mmpL11 mutant strains in 7H9 medium. Average values and standard deviations for four biological replicates are shown. (B) Biofilm formation by M. tuberculosis wild-type, mmpL11 mutant, and complemented strains. Biofilms were cultured in 6-well plates for a total of 4 weeks. (C) Quantification of crystal violet staining of M. tuberculosis biofilms. Average values and standard deviations for three independent assays are shown. The difference between the wild type and the mutant was significant (P, <0.005 by Student's t test). (D) The numbers of wild-type, mutant, and complemented M. tuberculosis bacteria present when strains were grown as biofilms were determined by mechanically disrupting 3-week biofilms. Bacteria representing both biofilm and planktonic bacteria were serially diluted and were plated onto 7H11 agar containing ADS and 0.5% glycerol for the determination of the CFU count per milliliter. (E) The growth of wild-type, mutant, and complemented M. tuberculosis strains in Sauton's medium containing Tween 80 was followed by measuring the OD600. An asterisk indicates that the difference between wild-type and mmpL11 mutant bacteria at that time point was significant (P = 0.029). (F) The bacterial CFU counts per milliliter at the indicated time points were determined by plating serial dilutions. The difference between the wild type and the mmpL11 mutant was not significant when they were quantified by this methodology. (G) The mmpL11 mutant forms small colonies on 7H11 agar containing ADS and 0.5% glycerol.
Reduced biofilm formation may result if the mmpL11 mutant grows more slowly than the wild type under biofilm-inducing conditions. To determine if this was the case, biofilms grown in 24-well plates were mechanically disrupted, and serial dilutions of the well contents were plated on 7H11 medium such that both the bacteria in the biofilm and those growing planktonically were represented. There was no difference between the CFU counts of the wild type and the mmpL11 mutant present in the wells (Fig. 1D). This result suggests that the mmpL11 mutant is able to replicate like the wild type under biofilm-inducing conditions but does so planktonically instead of developing a robust biofilm. To formally assess the growth of the M. tuberculosis mmpL11 mutant in Sauton's medium, growth curves were performed and bacterial numbers determined by measuring the optical density at 600 nm (OD600) and by plating serial dilutions on agar plates over a time course. Mycobacterial strains were cultured in Sauton's medium containing Tween 80 to avoid the variability associated with mechanical disruption of biofilm-associated bacteria. Growth curves performed using optical density consistently showed differences between the M. tuberculosis mmpL11 mutant at the late-exponential-growth phase and wild-type M. tuberculosis (Fig. 1E). However, when the CFU per milliliter were enumerated, there was not a significant difference (Fig. 1F). In addition, it is noteworthy that the M. tuberculosis mmpL11 mutant formed smaller colonies than the wild type on 7H11 agar (Fig. 1G). This particular phenotype may be species specific, since the M. smegmatis mmpL11 mutant did not exhibit a growth defect in liquid media or on agar plates (10).
The lipid profiles of the mmpL11 mutant are distinct from those of wild-type M. tuberculosis.
To identify the substrates of the MmpL11 transporter, we examined the lipid profiles of the M. tuberculosis mmpL11 mutant grown under both planktonic and biofilm conditions using thin-layer chromatography (TLC). There were no differences between the lipids isolated from the mmpL11 mutant and those from wild-type M. tuberculosis when the strains were grown planktonically, in agreement with our observation that there were no defects in planktonic growth (data not shown). However, we noted several differences between lipids isolated from wild-type M. tuberculosis and the mmpL11 mutant grown as biofilms. When surface and total lipids were resolved by TLC in the chloroform-methanol-distilled water (dH2O) (90:10:1) solvent system, we saw that the surface levels of TDM in the mmpL11 mutant were lower than those in the wild type, while the levels of free mycolic acids (FM) were increased (Fig. 2A). Previous studies have shown that free mycolic acids are hydrolyzed from TDM and contribute to mycobacterial biofilm formation (27, 28). Therefore, it is possible that the mmpL11 mutant compensates in part for the absence of other biofilm-associated lipids by producing greater amounts of free mycolic acids.
FIG 2.
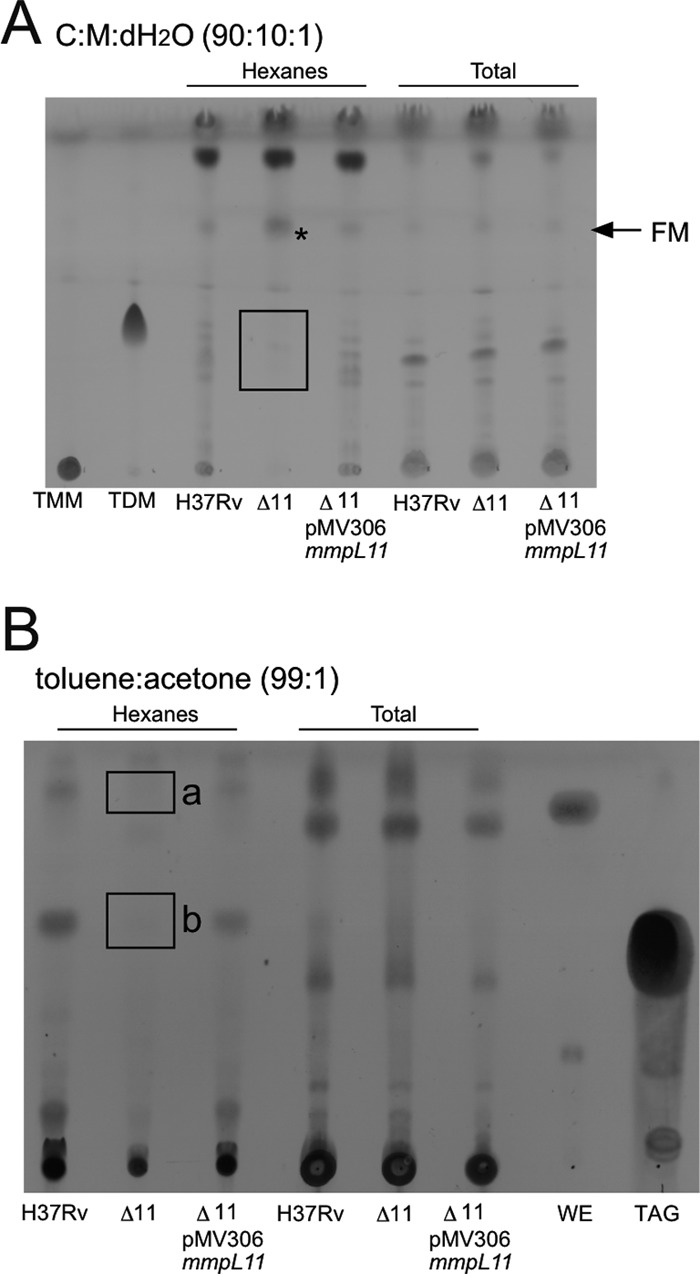
The M. tuberculosis mmpL11 mutant possesses altered cell wall lipids when cultured under biofilm-inducing conditions. (A) Hexane-extractable surface lipids (left) and total apolar lipids (right) were extracted from M. tuberculosis biofilms and were resolved by TLC in chloroform-methanol-dH2O (90:10:1, vol/vol/vol). Free mycolic acids (FM) and the trehalose dimycolate (TDM) standard (Sigma) are indicated. (B) Hexane-extractable surface lipids (left) and total apolar lipids (right) were resolved by TLC in toluene-acetone (99:1, vol/vol). The triacylglycerol (TAG) and wax ester (WE) standards (Sigma) are indicated. Unknown lipids “a” and “b” are noted. In the experiments for both panels, TLC plates were sprayed with MPA and were charred to visualize lipids. Lipids that are absent in the mmpL11 mutant are outlined, and a lipid that appears to accumulate is indicated by an asterisk.
When M. tuberculosis lipids were resolved by TLC in the toluene-acetone (99:1) solvent system, we saw the loss of two surface-extractable lipids in the mmpL11 mutant (Fig. 2B). One was slightly more apolar than the commercial wax ester standard (annotated as “a” in Fig. 2B), and the other migrated alongside the commercial TAG standard (annotated as “b” in Fig. 2B). Complementation with either the integrative or the episomal mmpL11 vector restored all lipids to wild-type levels (Fig. 2 and data not shown). Lipid species “a” was identified by mass spectrometry (MS) as mycolic acid wax ester, which consisted of a tritriacontadienol (33:2) linked to the mycolic acid via an ester bond to form a tritriacontadienyl mycolate ester (see Fig. S1A and B in the supplemental material). This result is consistent with the M. smegmatis MWE reported previously (10). The mycolic acid constituents of the MWE from wild-type M. tuberculosis comprised predominantly methoxy-mycolic acids (marked with an asterisk in Fig. S1A and B), the profile of which resembled that of the methoxy-mycolic acids found in wild-type M. tuberculosis (marked with an asterisk in Fig. S1C). We were unable to detect the corresponding α-mycolic acid- and keto-mycolic acid-containing MWEs, suggesting that the abundances of these species are too low for them to be detected using the present approach. When we examined the mycolic acids present in total lipids, we found that the α-mycolic acid and keto-mycolic acid are also minor species (marked with a number sign and a dagger in Fig. S1C), so the mycolic acid representation of the MWE is consistent with the overall mycolic acid composition of the bacterium. The mycolic acid composition of our biofilm-grown bacteria is consistent with what others have reported recently for H37Rv cultured on Ogawa agar, although we acknowledge that different growth conditions can change the mycolic acid profiles of mycobacteria (29).
Mass spectrometry analysis of spot “b” revealed that it comprised TAGs with very-long-chain (C20 to C38, with 0-1 unsaturation) fatty acid constituents (see Fig. S2 in the supplemental material). The electrospray ionization (ESI) mass spectra of TAGs from the wild type (Fig. S2A) and the mmpL11 mutant (Fig. S2B) appeared similar, although long-chain TAGs with full saturation appeared to be less abundant in the mutant. Combined, these findings suggest that MWE and very-long-chain fatty acid-containing TAGs are substrates of the MmpL11 transporter. The lack of surface-exposed lipids associated with biofilms likely contributes to the reduced biofilm phenotype.
mmpL11 mutant cells are shorter than wild-type M. tuberculosis cells.
Since the M. tuberculosis mmpL11 mutant had altered cell wall-associated lipids when grown as biofilms, we wondered if there were changes to acid fastness or bacterial morphology. Biofilms were mechanically disrupted and bacteria fixed for microscopy. We did not see a difference in acid-fast staining between wild-type and mmpL11 mutant bacteria (data not shown). However, there was a striking difference in cell length between mmpL11 mutant bacteria and wild-type M. tuberculosis (Fig. 3A). Quantification indicated an average cell length of 3.706 ± 0.1119 μm for wild-type bacteria versus 2.287 ± 0.05797 μm for the mmpL11 mutant. Bacterial size was restored upon complementation. Because mmpL11 mutant bacteria grown as biofilms had a reduced cell length, we investigated whether these bacteria were also shorter when grown planktonically in Sauton's medium or 7H9 medium containing Tween 80. The short bacterium phenotype was less pronounced in the planktonically grown mutant, but cell lengths still differed significantly between mutant and wild-type bacteria (Fig. 3B and C), which could explain why the mmpL11 mutant bacteria appeared to have a growth defect when growth curves were acquired using the OD600.
FIG 3.
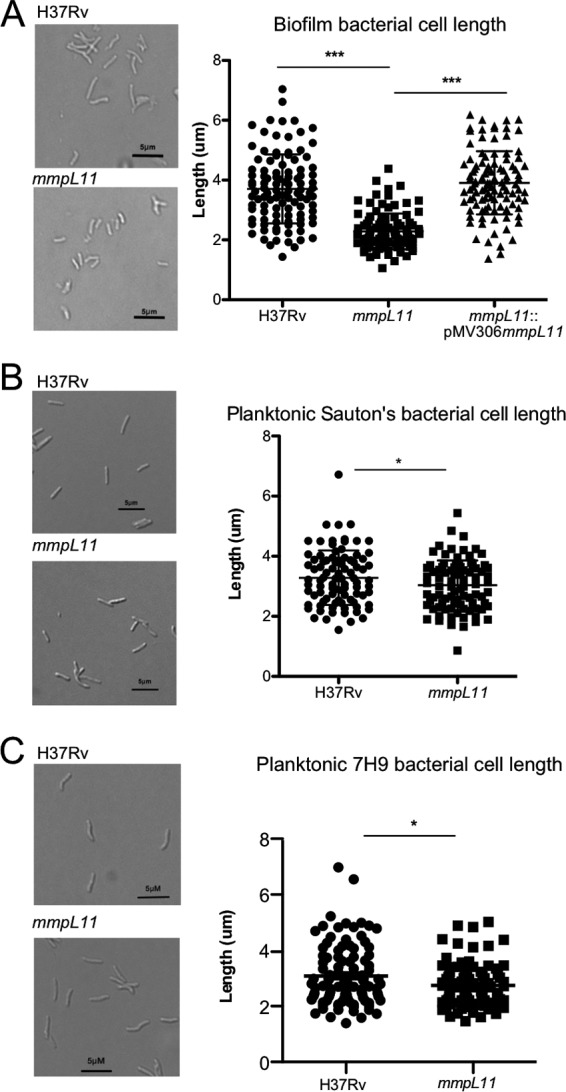
M. tuberculosis mmpL11 mutant bacteria are shorter than the wild type in biofilms. Shown are representative images (left) and quantification of the lengths (right) of M. tuberculosis strains grown in either Sauton's medium lacking Tween 80, to promote biofilm formation (A), Sauton's medium containing Tween 80 (B), or 7H9 medium containing Tween 80 (C). The differences between the wild type and the mutant were significant by Student's t test (***, P < 0.005; *, P < 0.05).
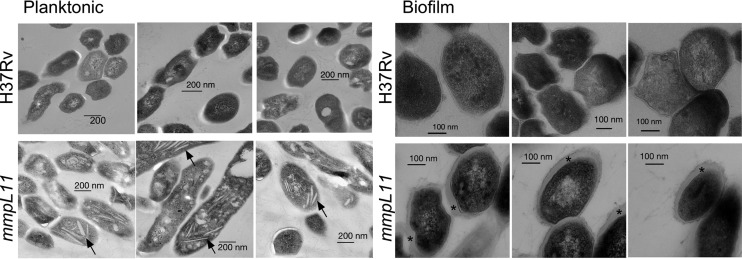
Finally, we performed transmission electron microscopy (TEM) to determine whether there were ultrastructural differences between wild-type and mmpL11 mutant M. tuberculosis bacteria. Intriguingly, planktonic mmpL11 mutant bacteria were distinct from the wild type in that they accumulated electron-light inclusions (Fig. 4, left). While some inclusions were consistent with lipid droplets or vesicles, others had a crystalline appearance (indicated by arrows). Our data indicate that even during planktonic growth, when the mmpL11 mutant lacks an apparent phenotype, loss of the MmpL11 transporter still impacts bacterial physiology. The mmpL11 mutant was also distinct from the wild type when grown as a biofilm: there appeared to be more extracellular material surrounding mmpL11 mutant bacteria than around wild-type bacteria (Fig. 4, right).
FIG 4.
Transmission electron microscopy reveals differences in ultrastructure between the wild-type and mmpL11 mutant M. tuberculosis strains grown in 7H9 medium containing Tween 80 (left) (planktonic) or Sauton's medium without Tween 80 (right) (biofilm). Arrows indicate crystal inclusions in mmpL11 mutants grown planktonically. Asterisks highlight the extracellular material that is enriched in the mmpL11 mutant relative to wild-type M. tuberculosis.
The M. tuberculosis mmpL11 mutant is impaired for survival in an in vitro human granuloma model.
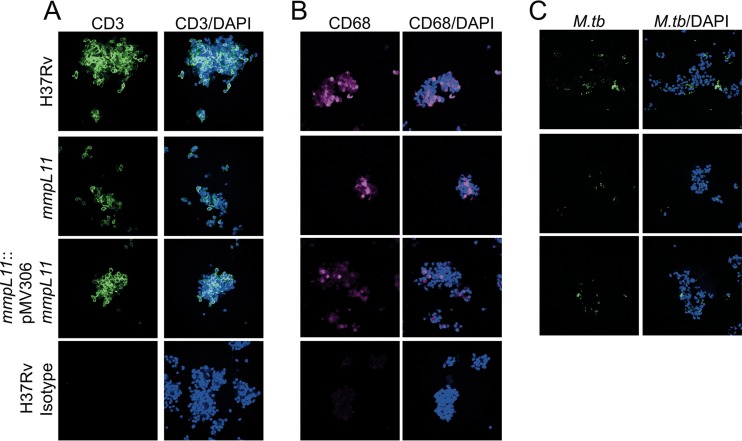
Mice infected with the mmpL11 mutant survive longer than those infected with wild-type M. tuberculosis (22). The M. tuberculosis mmpL11 mutant did not have a defect in the acute stages of mouse infection; however, fewer mmpL11 mutant bacteria than wild-type bacteria were recovered during the chronic stage of infection. In agreement with this observation, the mmpL11 mutant did not have a replication defect in the human THP-1 monocyte-derived macrophage line (Fig. 5A). Although the macrophage is the host niche most studied using in vitro models of infection, natural tuberculosis infections and control of the bacterium involve multiple cell types. To assess the phenotype of the mmpL11 mutant in a more complex system that includes cells involved in the innate and adaptive immune responses, we used an in vitro granuloma model generated from peripheral blood mononuclear cells (PBMCs) from individuals with latent TB infection (LTBI) who had a positive result with the Mantoux tuberculin skin test (TST) and/or gamma interferon (IFN-γ) release assay (IGRA) (TST+ or IGRA+, respectively) (30). Previous characterization of the granulomas showed that these structures include macrophages and multinucleated giant cells (CD11b+), T cells (CD3+, CD4+, CD8+), and B cells (CD19+ cells) and that M. tuberculosis resides within the dense cellular aggregates of the granuloma. These cells are viable, and the T cells are able to proliferate over the time course of infection, as indicated by 5-ethynyl-2′-deoxyuridine (EdU) labeling (30). Granuloma structures generated from PBMCs from individuals with LTBI control M. tuberculosis growth more efficiently than granulomas generated from naïve individuals, a phenomenon related to stronger immune pressure against the bacteria. We observed significantly fewer viable bacteria of the mmpL11 mutant strain than of the complemented strain 7 days following infection (Fig. 5B). The growth of the mmpL11 mutant was also reduced from that of wild-type bacteria, but this difference did not reach significance. In addition, granuloma formation was significantly delayed for the mmpL11 mutant relative to the wild type, and this was partially restored with the complemented strain (Fig. 5C). This was noticeable within just 4 or 5 days of infection, as shown by the representative images and the scoring index, which ranks the granuloma structure on a scale from 1 to 5, with progressive increases in the size and specific cell distribution of granulomas (30). We confirmed that the granuloma structures formed following infection with wild-type, mmpL11 mutant, or complemented M. tuberculosis strains contain CD3+ T cells and CD68+ macrophages (Fig. 6A and B) and that the bacteria are located within the granuloma structures (Fig. 6C). These granuloma structures have lower expression of CD163+ and CD206+ cells than uninfected macrophages (see Fig. S3 in the supplemental material), potentially representing an early granulomatous response (31, 32).
FIG 5.
The M. tuberculosis mmpL11 mutant has a survival defect in an in vitro granuloma model containing human innate and adaptive immune cells. (A) Survival of wild-type, mutant, and complemented M. tuberculosis strains in differentiated THP-1 cells. Average values and standard deviations for a representative experiment performed in triplicate are shown. (B) Survival of wild-type, mutant, and complemented M. tuberculosis strains in an in vitro granuloma model. The relative survival of strains at 7 days (average values ± standard errors of the means for two experiments, performed in triplicate) is shown. The difference between the mutant and the complemented strain was significant by Student's t test (*, P < 0.05). (C) Representative images of granulomas formed in vitro 5 and 7 days after infection with M. tuberculosis. Magnification, ×40. Granuloma formation was scored for each condition; the means ± standard deviations of scores representative of two experiments each are given below the images.
FIG 6.
In vitro granuloma structures contain CD3+ and CD68+ cells and M. tuberculosis. In vitro granuloma structures were fixed at 7 days and were labeled for CD3 (A), CD68 (B), and M. tuberculosis (C) to confirm that granuloma composition was consistent with that found in our previous work (30). Images are representative of two experiments.
To better understand the mechanism underlying delayed granuloma formation, we evaluated the ability of M. tuberculosis-specific CD8+ T cell clones to recognize and respond to autologous dendritic cells (DCs) infected with wild-type or mmpL11 mutant bacteria. We plated CFU of the wild-type and mutant strains at 0 and 18 h and found no significant differences in either the infection rate or the survival of the mutant over this period, in agreement with the phenotype of the mutant in THP-1 monocyte-derived macrophages (data not shown). Despite being infected with equivalent numbers of bacteria, DCs infected with the mmpL11 mutant were less able than those infected with the wild type to stimulate IFN-γ production by four T cell clones that recognize distinct M. tuberculosis antigens (see Fig. S4A in the supplemental material).
CD8+ T cell responses to M. tuberculosis-infected cells require that antigens be secreted from the bacterium and delivered to the major histocompatibility complex type I (MHC-I) processing and presentation pathway. Based on these data, we considered the possibility that lower CD8+ T cell responses to the mutant than to the wild type corresponded to reduced antigen secretion across the mmpL11 bacterial membrane into the host environment because of altered cell wall composition. Western blot analysis of total lysates and culture filtrate proteins (CFP) using antibodies against Antigen 85 and ESAT-6 showed that there are lower levels of these proteins in the liquid growth medium of the mmpL11 mutant than in that of wild-type M. tuberculosis (Fig. S4B). Therefore, we speculate that in the host cell, the mmpL11 mutant is less immunogenic than wild-type M. tuberculosis, resulting in slower T cell activation and delayed granuloma formation.
The differences observed in the secreted protein levels in the mmpL11 mutant may be results of general differences in the cell wall rather than impacts on specific secretion systems. In support of this possibility, we see reduced IFN-γ levels induced by T cell clones that recognize substrates of the ESX (type VII) secretion system, as well as T cell clone 454H1-2, which recognizes Rv1174c (also known as Mtb8.4). Rv1174c has a predicted N-terminal signal sequence suggesting secretion by the Sec system, and proteomic studies have identified both the cleaved signal peptide and mature Rv1174c in the CFP (33). To determine if there were differences in cell wall permeability that may impact protein release, we performed an ethidium bromide uptake assay. The mmpL11 mutant had lower membrane permeability than wild-type M. tuberculosis (Fig. S4C). This result is consistent with the reduced membrane permeability we observed for the M. smegmatis mmpL11 mutant (34). It is reasonable that if small molecules take longer to diffuse into the bacterium, molecules and proteins will take longer to diffuse out of the bacterium into the growth medium and, by extension, into the host cell.
The M. tuberculosis mmpL11 mutant is impaired for survival and resuscitation from nonreplicating persistence.
M. tuberculosis is also subject to nutritional stress during infection and has adapted to utilize multiple host-derived carbon sources. It is thought that M. tuberculosis utilizes host cholesterol during infection, and a genetic screen has indicated that MmpL11 is required for growth on cholesterol (35). To determine if this underlies the attenuation of the M. tuberculosis mmpL11 mutant during persistent infections, we followed bacterial growth in Sauton's medium containing cholesterol as the sole carbon source. For this set of experiments, we utilized the surfactant tyloxapol to avoid the possibility of Tween 80-derived oleic acid complicating the interpretation of cholesterol utility. There was no significant difference between the growth of the mmpL11 mutant and wild-type M. tuberculosis when growth was measured using the OD600 (Fig. 7A) or when CFU per milliliter were enumerated by plating serial dilutions (data not shown).
FIG 7.
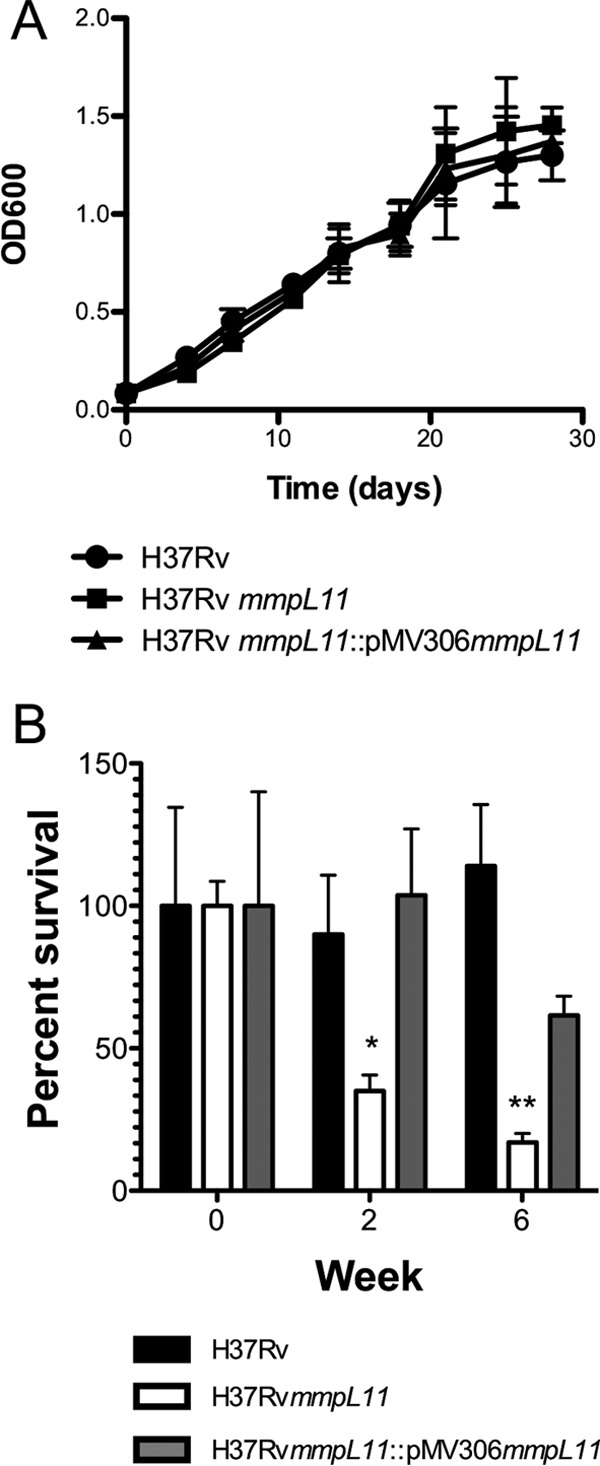
(A) Growth of wild-type, mutant, and complemented M. tuberculosis strains in Sauton's medium containing cholesterol as the sole carbon source. Average values and standard deviations for three biological replicates are shown. (B) Survival of wild-type, mutant, and complemented M. tuberculosis strains in an in vitro model of dormancy. Average values and standard deviations for a representative experiment performed in triplicate are shown. The difference between the wild type and the mutant was significant by Student's t test (*, P < 0.05; **, P < 0.01).
Since TAGs and wax esters are implicated in M. tuberculosis dormancy (36–39), we investigated whether the M. tuberculosis mmpL11 mutant was able to survive or recover from in vitro dormancy conditions. We washed and resuspended M. tuberculosis bacteria in phosphate-buffered saline (PBS) for 2 and 6 weeks, and then we plated serial dilutions to quantify the surviving bacteria. The mmpL11 mutants recovered very slowly from these conditions, and there was an 85% reduction in countable colonies from the 6-week cultures (Fig. 7B). Taken together, these results indicate that MmpL11 and the MmpL11 substrates are important for survival or resuscitation from a nutrient-starved, hypoxia-induced dormancy model.
DISCUSSION
In this report, we characterized the role of MmpL11 in M. tuberculosis. Our analysis of the M. tuberculosis mmpL11 mutant lipids indicated that the transport of MWE and long-chain TAGs to the bacterial surface is disrupted. The absence of MmpL11 has impacts on bacterial physiology that range from reduced biofilm formation to changes in cell size and attributes, including reduced membrane permeability, which may influence the release of secreted proteins from the bacterium into the environment. We observed that the M. tuberculosis mmpL11 mutants have a morphology and ultrastructural appearance distinct from those of the wild type when grown in broth culture and biofilms, which may result from the inability of the mutant to transport its lipid substrates. Transmission electron microscopy showed that planktonic mmpL11 mutant bacteria accumulated electron-light crystalline inclusions. We were unable to identify these inclusions, but they are reminiscent of the inclusions observed upon tetrahydrolipstatin (THL) treatment of M. kansasii (40). THL inhibits the synthesis of mycolic acids, and the authors of the previous study (40) speculated that these inclusions represented mycolic acid precursors or FAS-1 products esterified to storage molecules. Therefore, we suspect that these inclusions result from the accumulation of MWE or an MWE intermediate in the bacterial cytoplasm.
MmpL11 is required for full virulence of M. tuberculosis in mice (22, 23). While the mutant infected and replicated like the wild type during the acute phase of infection, fewer bacteria were present during the chronic, persistent phase of infection. There was a significant increase in the time to death of mice infected with the mmpL11 mutant (333 days) over that for mice infected with wild-type M. tuberculosis (173 days) (22). The mmpL11 mutant can therefore be classified as an M. tuberculosis “persistence mutant.” Mutants that share this phenotype include those that are less fit in the face of adaptive immune pressure, as well as mutants, such as those lacking Icl and Mce4, that have impaired metabolism or nutrient uptake (41, 42).
The attenuation of the mmpL11 mutant during the chronic phase of infection is likely multifactorial. Our results indicate that the mmpL11 mutant is less fit in the presence of adaptive immune cells than wild-type M. tuberculosis. Granuloma formation is a hallmark of tuberculosis but has been studied primarily in vivo, since it requires the coordination of innate and adaptive immune cells. Several approaches to modeling granuloma formation in vitro using human PBMCs have allowed investigators to gain insight into this aspect of disease. In the context of mycobacteria or mycobacterial antigens, the aggregation of cells occurs within days and can recapitulate the generation of multinucleated cells and epithelioid macrophages that associate with surrounding macrophages and lymphocytes (43). We used an in vitro granuloma model that cultured human PBMCs from individuals with previous exposure to mycobacteria (LTBI; TST+ or IGRA+ subjects). We showed previously that granuloma formation is faster, more robust, and better able to control the microbial burden in TST+ or IGRA+ donors than in their TST− or IGRA− counterparts (30). The results obtained with this model demonstrate that granulomas form less efficiently in response to the mmpL11 mutant, perhaps due to altered cell wall lipid composition and the reduced release of bacterial proteins. The mmpL11 mutant was also more susceptible than the wild type to killing by the cells in the granuloma.
Changes in the lipid composition of the mycobacterial cell wall also likely contribute to the attenuation of the mmpL11 mutant. Specifically, when grown under biofilm-inducing conditions, MWE and long-chain TAGs are absent from the mmpL11 mutant surface-extractable lipids. TAGs and wax esters are associated with M. tuberculosis dormancy, and nonreplicating bacteria are found within human granulomas (36–39). Therefore, we suspected that the ability of the mmpL11 mutant to replicate or recover from the host might also be impaired. Indeed, the survival or resuscitation of the mmpL11 mutant was significantly reduced compared to that of wild-type M. tuberculosis in an in vitro dormancy model. The role of long-chain TAGs and mycolate wax esters remains an active area of research in the lab. We are identifying the enzymes responsible for their biosynthesis with the mindset that the characterization of mutants lacking these individual lipid constituents will refine our understanding of their functions in M. tuberculosis.
Finally, the mmpL11 mutant may also have a defect in nutrient acquisition in the host environment. Both MmpL3 and MmpL11 have been implicated in heme transport (44). Although import of heme or heme complexes may be one function of these transporters, our data and those of others suggest that MmpL3 and MmpL11 are lipid exporters with a primary function in cell wall biosynthesis (10, 19, 45). We do not think that the mmpL11 mutant bacteria are experiencing iron starvation in our studies. The 7H9, 7H11, and Sauton media used in the experiments presented in this report are considered iron-replete and possess sufficient iron concentrations for biofilm formation, and our mmpL11 mutant produces siderophores. It is proposed that during infection, M. tuberculosis utilizes host cholesterol as a carbon source. Mutants lacking cholesterol transporters or catabolic enzymes showed attenuated virulence (42), and a genetic screen indicated that MmpL11 was required for growth on cholesterol (35). Mutants containing transposon insertions in mmpL11 were underrepresented 8-fold following growth on cholesterol as the sole carbon source. However, this phenotype was not as extreme as that observed for mutants containing transposon insertions in genes encoding the Mce4 transporter locus or genes for cholesterol catabolism, which were underrepresented ∼20- to 90-fold. Furthermore, we did not observe a dramatic difference between the mmpL11 mutant and the wild type when grown on cholesterol as the sole carbon source.
Despite evidence that the mmpL11 mutant can grow on cholesterol, we present other data that may suggest that the ability of the mmpL11 mutant to acquire nutrients is impaired. Although 7H11 medium is not considered a nutrient-restricted or minimal medium, the mmpL11 mutant had a small-colony phenotype on 7H11 agar. Nutrient restriction may also explain the shorter cell length of the mmpL11 mutant than of wild-type M. tuberculosis. Previous studies have shown that mycobacterial cell division depends on time, not cell length (46). The cell length of mycobacteria changes depending on the growth stage, with longer bacteria present during logarithmic growth and shorter bacteria present during stationary phase, as nutrients are depleted (47). The mmpL11 mutant may not acquire nutrients as efficiently as the wild type in stationary-phase or biofilm cultures, and perhaps as a result, the mutant does not reach the length of wild-type bacteria before division, resulting in shorter bacilli.
MmpL11 is required for efficient biofilm formation in both M. tuberculosis and M. smegmatis. While this phenotype is similar for the fast-growing nonpathogenic and slow-growing species, there are notable differences in MmpL11 substrates and in the impact of the mmpL11 mutation on mycobacterial growth. We identified MWE and long-chain TAGs as the substrates of MmpL11Mt, whereas MWE and MMDAG are the substrates of MmpL11 in M. smegmatis. MMDAG has been observed in M. kansasii and M. smegmatis; however, we were unable to find MMDAG in lipids isolated from the M. tuberculosis H37Rv or Erdman strain. These results suggest that M. tuberculosis produces very little or none of this mycolic acid-containing lipid. Another difference we observed was that M. tuberculosis mmpL11 mutant bacteria formed colonies smaller than those formed by the wild type on standard 7H11 agar. We did not observe any growth defects in the M. smegmatis mmpL11 mutant (10). M. tuberculosis mmpL11 mutant bacteria appear shorter than wild-type bacteria in stationary cultures, a phenotype that we also have not observed for M. smegmatis. Combined, these results suggest that the loss of MmpL11 function has a greater impact on the physiology of slow-growing or pathogenic mycobacteria than on the nonpathogenic surrogate species M. smegmatis.
MmpL3 and MmpL11 are encoded at a genomic locus that is conserved across pathogenic and nonpathogenic mycobacteria, including M. leprae, suggesting that they are important for the basic biology and physiology of mycobacteria. Our working model is that MmpL3 and MmpL11 are conserved and part of the “core” mycobacterial genes, because they are required for the two phases of mycobacterial existence. MmpL3 is essential for active replication, because exported TMM is required for TDM biosynthesis and cell wall generation. MmpL11 is important for nonreplicating persistence, because it is required for the transport of mycolic acid-containing “storage lipids.” Since the majority of M. tuberculosis infections result in latent TB, where bacteria have altered metabolism and exhibit phenotypic drug tolerance, proteins required for dormancy and resuscitation, including MmpL11, are putative therapeutic targets. In particular, the essential MmpL3 protein represents a target for mycobacterial drug development. In recent years MmpL3 has been identified as the target of several compounds by use of genetic approaches (19, 48–50). Ultimately, these compounds appear to indirectly inhibit MmpL3 by perturbing the proton motive force (PMF) (51, 52). The point-substituted residues identified in MmpL3 that confer resistance likely allow the protein to compensate for a disruption of PMF strength. Further studies on MmpL3, MmpL11, and other virulence-associated MmpL proteins are warranted to facilitate the development of compounds that inhibit these critical proteins and to better characterize the mycobacterial cell wall during various phases of infection.
MATERIALS AND METHODS
Maintenance of bacterial cultures and cells.
The M. tuberculosis wild-type strain H37Rv was obtained from the ATCC. Mycobacterial strains were routinely maintained in Middlebrook 7H9 liquid medium (Difco) or on Middlebrook 7H11 agar (Difco) plates supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC; Difco) or albumin dextrose salts (ADS) containing 8.1 mg ml−1 NaCl, 50 mg ml−1 bovine serum albumin (BSA), and 20 mg ml−1 dextrose. Glycerol was added to 7H9 and 7H11 media at a final concentration of 0.5%. Where indicated, bacteria were cultured in Sauton's medium containing 0.5 g liter−1 K2HPO4, 0.5 g liter−1 MgSO4, 4.0 g liter−1 l-asparagine, 0.05 g liter−1 ferric ammonium citrate, 4.76% glycerol, and 1.0 mg liter−1 ZnSO4, with a final pH of 7.0. For growth on cholesterol as the sole carbon source, Sauton's medium that contained 0.5 mM cholesterol and 0.025% tyloxapol (Ty) was prepared. When required, cultures were incubated with kanamycin (25 μg ml−1) or hygromycin (50 μg ml−1).
The M. tuberculosis mmpL11 mutant was constructed by allelic exchange as follows. Roughly 500-bp fragments of the 5′ and 3′ regions of the M. tuberculosis mmpL11 gene were amplified by PCR using primers mmpL11xho (5′-CTCGAGGATCGACCGATAGCACCG-3′) and mmpL11kpn (5′-GGTACCCTGCCGTGCTAGCAACATT-3′) and primers mmpL11BamHI (5′-GATCCGGACCTCCTCCCACTTTG-3′) and mmpL11H3 (5′-AAGCTTGTGTGGATGTGACGGTAAGG-3′), respectively, and were cloned to flank the hygromycin resistance gene in pYUB854-rpsL (53). The resulting plasmid was transformed into electrocompetent M. tuberculosis strain H37Rv. Transformants were selected on 7H11 medium containing OADC supplements and 75 μg ml−1 hygromycin. Individual colonies were streaked onto complete 7H11 agar containing 50 μg ml−1 hygromycin. The mutant was confirmed by PCR. For complementation, primers mmpL11compXba (5′-TTTCTAGACGGCAAGTACTGGTGGTTG-3′) and mmpL11compH3 (5′-GCAAGCTTCGAATGGCGACGGTAGG-3′) were used to amplify M. tuberculosis mmpL11 including 1,000 bp of upstream sequence. This fragment was cloned into the integrative plasmid pMV306Kan (54), which was then transformed into the mmpL11 mutant. The pVV16mmpL11TB plasmid has been described previously (10).
Biofilm growth conditions.
M. tuberculosis biofilm growth conditions were essentially those described previously (27). Briefly, bacteria were cultured in Sauton's medium without Tween 80 in polystyrene bottles (Corning). Flasks were incubated at 37°C under 5.5% CO2 with tightened caps for 2 weeks. After 2 weeks, the caps were loosened to promote biofilm formation. M. tuberculosis was also cultured in Sauton's medium without Tween 80 in 6-well or 24-well plates, where the dishes were kept in a sealed Tupperware container for 3 weeks before the lid was opened and the bacterium was cultured further.
Lipid isolation and analysis.
To harvest total lipids, cultures were harvested by centrifugation and resuspended in chloroform-methanol (2:1, vol/vol). Surface lipids were extracted from M. tuberculosis biofilms as described previously (12). Briefly, biofilms were harvested and were resuspended in 5 ml of hexanes; then they were agitated with glass beads for 3 min. After centrifugation at 3,000 × g for 5 min, the hexane-extracted lipids were removed and an equal volume of chloroform-methanol (2:1, vol/vol) added before removal from the biosafety level 3 (BSL3) laboratory. Following extraction, surface and total lipids were dried under N2 gas. Total and surface lipids were resuspended in chloroform-methanol (2:1, vol/vol), and equivalent amounts were spotted onto thin-layer chromatography (TLC) plates. TMM, TDM, and free mycolic acids were resolved by TLC in the chloroform-methanol-dH2O (90:10:1, vol/vol/vol) solvent system. Triacylglycerol (TAG) was resolved by TLC in the toluene-acetone (99:1, vol/vol) solvent system. The TLC plates were visualized by spraying with 5% molybdophosphoric acid (MPA) in ethanol and were charred. Lipids were purified from silica plates (Analtech) using standard preparative TLC procedures.
MS.
Structural analysis of lipids was conducted on a Thermo Scientific (San Jose, CA) TSQ Vantage EMR mass spectrometer with an Xcalibur operating system. Fractionated lipids from preparative TLC were loop injected, subjected to electrospray ionization (ESI), and analyzed as described previously (10, 55).
Microscopy.
For analysis of bacterial cell length, M. tuberculosis bacteria were fixed overnight at 4°C in 10% buffered formalin. Bacteria were washed with PBS and were heat-fixed to 1.5-mm glass coverslips. After rinsing with deionized (DI) water, coverslips were mounted to slides using Fluoromount-G (Southern Biotech). Slides were visualized at the OHSU Advanced Light Microscopy Core using ×60 magnification on a Deltavision CoreDV Widefield deconvolution system. Cell length was measured using Fiji software (56), and subsequent statistical analysis was performed using GraphPad Prism.
For ultrastructural analysis of M. tuberculosis, bacteria were fixed overnight in 4% formaldehyde–1% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2). Fixed samples were embedded in Spurr's epoxy resin. Transmission electron microscopy was performed at the Multiscale Microscopy Core (MMC) on an FEI Tecnai Spirit TEM system with technical support from the OHSU-FEI Living Lab and the OHSU Center for Spatial Systems Biomedicine (OCSSB). Images were acquired as 2,048-by-2,048-pixel, 16-bit grayscale files using the FEI's TEM Imaging & Analysis (TIA) interface on an Eagle 2K charge-coupled device (CCD) multiscan camera.
For the imaging of in vitro granuloma structures, cells on glass coverslips were first fixed with 4% paraformaldehyde (PFA) for 20 min and then washed twice with PBS. For CD68, CD163, and CD206 staining, cells were treated with ice-cold methanol for 5 min, washed three times with PBS, and incubated in blocking buffer (5% BSA–10% goat serum–PBS) for 1 h at room temperature (RT). Cells were then incubated with primary antibodies against CD3 (UCHT1; Biolegend), CD68 (PG-M1; Dako), CD163 (GH1/61; Thermo Scientific), or CD206 (19.2; BD Biosciences) for 1 h in blocking buffer, washed, and then incubated with secondary antibodies in blocking buffer. After 1 h, cell were washed, stained with 4′,6-diamidino-2-phenylindole (DAPI), and mounted with ProLong Gold antifade reagent. In certain wells, bacteria were stained with TB auramine-rhodamine stain (BD Biosciences), granuloma structures were labeled with DAPI, and coverslips were mounted with ProLong Gold antifade reagent. Cells were imaged with an Olympus FV1000 confocal microscope.
Intracellular survival and bacterial replication in differentiated THP-1 cells.
THP-1 cells were cultured in 90% RPMI 1640 medium (HyClone) with 10% heat-inactivated fetal bovine serum (FBS; Sigma), supplemented with 2 mM l-glutamine (HyClone). THP-1 cells were seeded at a density of 1 × 104/well in 96-well flat-bottom plates in a medium containing 30 ng/ml phorbol-12-myristate-13-acetate (PMA) for 48 h. Differentiated THP-1 cells were infected at a multiplicity of infection (MOI) of 1:1 for 1 h and were then washed with medium to remove extracellular bacteria. At the time points indicated in Fig. 5A, THP-1 cells were lysed with 0.1% Tween 80, serially diluted, and plated on 7H11 agar plates to determine the CFU of surviving M. tuberculosis.
In vitro granuloma model.
Granulomas were generated as described elsewhere (30). Briefly, blood was collected from otherwise healthy individuals who had a positive result with the Mantoux tuberculin skin test (TST) and/or IFN-γ release assay (IGRA) within the previous 12 months following an Ohio State University-approved IRB protocol. PBMCs were isolated on a Ficoll-Paque cushion according to standard protocols (57), and 2 × 106 PBMCs/ml were immediately infected with M. tuberculosis at an MOI of 1:1 in 10% autologous serum–RPMI, in triplicate. Serum was replenished after 4 days, and cells were incubated for a total of 7 days at 37°C under 5% CO2. With this method, the granulomas are stable for up to 12 days. Cells were lysed as described previously (58), diluted, and plated on Middlebrook 7H11 agar plates, in triplicate, for CFU enumeration. The stage of granuloma formation was determined semiquantitatively for each experimental group. Each sample (well) was assessed by light microscopy under a 40× lens (Olympus IX71 DP71 microscope digital camera), and images were taken to establish a scoring index, as described previously (30). Briefly, this index ranks the granuloma structure on a scale of 1 to 5, based on progressive increases in the size and specific cell distribution of granulomas, such that 1 indicates no cell aggregation and 5 indicates large granuloma-like structures with macrophages surrounded by lymphocytes.
IFN-γ ELISPOT assays.
Monocyte-derived dendritic cells (DCs) were isolated as described previously (59) and were infected with wild-type M. tuberculosis or the mmpL11 mutant (MOI, 5) for 18 h. Some infected DCs were lysed to determine bacterial CFU counts. The remaining cells were used as antigen-presenting cells in an IFN-γ enzyme-linked immunosorbent spot (ELISPOT) assay. A total of 10,000 infected DCs were plated in each well, and the IFN-γ response of 20,000 CD8+ T cell clones was measured. The clones used were restricted by HLA-B0801 (D454 B10), HLA-B1501 (D454 H1-2), HLA-B0801 (D480 F6), or HLA-B5701 (D504 F9). D454 B10 recognizes M. tuberculosis peptide EsxG53–61. D454 H1-2 recognizes M. tuberculosis peptide Mtb8.433–43. D480 F6 recognizes M. tuberculosis peptide CFP103–11 (60). D504 F9 recognizes M. tuberculosis peptide EsxJ24–34 (61). The response of the T cell clones to DCs infected with the mmpL11 mutant strain is shown in Fig. S4A as a percentage of the response to DCs infected with wild-type M. tuberculosis.
Ethidium bromide uptake assays.
To assess membrane permeability, an ethidium bromide uptake assay on intact mycobacteria was performed essentially as described previously (62). Briefly, logarithmic-phase M. tuberculosis strains grown in 7H9 ADS Tween medium (OD600, 0.5 to 1.0) were centrifuged, normalized to an OD600 of 0.5 in uptake buffer (50 mM KH2PO4 [pH 7.0], 5 mM MgSO4), and preenergized with 25 mM glucose for 5 min. One hundred microliters of cells was added per well of black, clear-bottom 96-well microplates (Corning). Ethidium bromide was added to a final concentration of 20 μM. The accumulation of ethidium bromide was followed over time by monitoring emission at 590 nm upon excitation at 530 nm, and results were expressed as relative fluorescence units.
Western blot analysis.
Logarithmic-phase mycobacterial cultures grown in 7H9 medium (OD600, 0.5 to 1.0) were normalized to an OD600 of 1.0, washed once in PBS, and resuspended in Sauton's medium without Tween 80. Standing cultures were maintained at 37°C for 5 days, the bacteria were pelleted by centrifugation, and the supernatant was precipitated by the addition of trichloroacetic acid (TCA) to a final concentration of 10% (vol/vol). Following centrifugation, the pellet containing culture filtrate proteins (CFP) was washed with acetone and was resuspended in 1× SDS-PAGE loading buffer. Total lysates were obtained by bead-beating cell pellets. Proteins were resolved by SDS–15% PAGE, transferred to nitrocellulose membranes, and probed with antibodies against secreted proteins Antigen 85 (NR13800; BEI Resources) and ESAT-6 (NR13803; BEI Resources) and cytosolic protein KatG (NR13793; BEI Resources).
In vitro dormancy model for M. tuberculosis.
We starved M. tuberculosis of nutrients by suspension in PBS with modifications to a previously published protocol (63). We pelleted exponential-phase M. tuberculosis to prepare cultures at a final density of 1 × 108 cells ml−1. M. tuberculosis was washed twice in PBS and was then resuspended in PBS. Dilutions of the final bacterial suspension were plated for enumeration of the starting inoculum. Cultures were incubated in closed tubes at 37°C for durations of 2 and 6 weeks and were then plated to determine the numbers of viable bacteria.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health awards AI113074 and AI087840 to G.E.P. Mass spectrometric analysis was performed in the Washington University Mass Spectrometry Resource, which is supported by National Institutes of Health grants P41GM103422, P30DK020579, P30DK056341, and R21HL120760. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00131-17.
REFERENCES
- 1.Brennan PJ, Nikaido H. 1995. The envelope of mycobacteria. Annu Rev Biochem 64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 2.Bhamidi S, Scherman MS, Jones V, Crick DC, Belisle JT, Brennan PJ, McNeil MR. 2011. Detailed structural and quantitative analysis reveals the spatial organization of the cell walls of in vivo grown Mycobacterium leprae and in vitro grown Mycobacterium tuberculosis. J Biol Chem 286:23168–23177. doi: 10.1074/jbc.M110.210534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilmore SA, Schelle MW, Holsclaw CM, Leigh CD, Jain M, Cox JS, Leary JA, Bertozzi CR. 2012. Sulfolipid-1 biosynthesis restricts Mycobacterium tuberculosis growth in human macrophages. ACS Chem Biol 7:863–870. doi: 10.1021/cb200311s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camacho LR, Constant P, Raynaud C, Lanéelle MA, Triccas JA, Gicquel B, Daffé M, Guilhot C. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis: evidence that this lipid is involved in the cell wall permeability barrier. J Biol Chem 276:19845–19854. doi: 10.1074/jbc.M100662200. [DOI] [PubMed] [Google Scholar]
- 5.Bekierkunst A, Levij IS, Yarkoni E, Vilkas E, Adam A, Lederer E. 1969. Granuloma formation induced in mice by chemically defined mycobacterial fractions. J Bacteriol 100:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perez RL, Roman J, Roser S, Little C, Olsen M, Indrigo J, Hunter RL, Actor JK. 2000. Cytokine message and protein expression during lung granuloma formation and resolution induced by the mycobacterial cord factor trehalose-6,6′-dimycolate. J Interferon Cytokine Res 20:795–804. doi: 10.1089/10799900050151067. [DOI] [PubMed] [Google Scholar]
- 7.Indrigo J, Hunter RL, Actor JK. 2003. Cord factor trehalose 6,6′-dimycolate (TDM) mediates trafficking events during mycobacterial infection of murine macrophages. Microbiology (Reading, Engl) 149:2049–2059. doi: 10.1099/mic.0.26226-0. [DOI] [PubMed] [Google Scholar]
- 8.Geisel RE, Sakamoto K, Russell DG, Rhoades ER. 2005. In vivo activity of released cell wall lipids of Mycobacterium bovis bacillus Calmette-Guérin is due principally to trehalose mycolates. J Immunol 174:5007–5015. doi: 10.4049/jimmunol.174.8.5007. [DOI] [PubMed] [Google Scholar]
- 9.Cambier CJ, Takaki KK, Larson RP, Hernandez RE, Tobin DM, Urdahl KB, Cosma CL, Ramakrishnan L. 2014. Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 505:218–222. doi: 10.1038/nature12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pacheco SA, Hsu F-F, Powers KM, Purdy GE. 2013. MmpL11 protein transports mycolic acid-containing lipids to the mycobacterial cell wall and contributes to biofilm formation in Mycobacterium smegmatis. J Biol Chem 288:24213–24222. doi: 10.1074/jbc.M113.473371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox JS, Chen B, McNeil M, Jacobs WR. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 12.Converse SE, Mougous JD, Leavell MD, Leary JA, Bertozzi CR, Cox JS. 2003. MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc Natl Acad Sci U S A 100:6121–6126. doi: 10.1073/pnas.1030024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domenech P, Reed MB, Dowd CS, Manca C, Kaplan G, Barry CE. 2004. The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J Biol Chem 279:21257–21265. doi: 10.1074/jbc.M400324200. [DOI] [PubMed] [Google Scholar]
- 14.Chalut C. 2016. MmpL transporter-mediated export of cell-wall associated lipids and siderophores in mycobacteria. Tuberculosis (Edinb) 100:32–45. doi: 10.1016/j.tube.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Sondén B, Kocíncová D, Deshayes C, Euphrasie D, Rhayat L, Laval F, Frehel C, Daffé M, Etienne G, Reyrat J-M. 2005. Gap, a mycobacterial specific integral membrane protein, is required for glycolipid transport to the cell surface. Mol Microbiol 58:426–440. doi: 10.1111/j.1365-2958.2005.04847.x. [DOI] [PubMed] [Google Scholar]
- 16.Burbaud S, Laval F, Lemassu A, Daffé M, Guilhot C, Chalut C. 2016. Trehalose polyphleates are produced by a glycolipid biosynthetic pathway conserved across phylogenetically distant mycobacteria. Cell Chem Biol 23:278–289. doi: 10.1016/j.chembiol.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 17.Touchette MH, Holsclaw CM, Previti ML, Solomon VC, Leary JA, Bertozzi CR, Seeliger JC. 2015. The rv1184c locus encodes Chp2, an acyltransferase in Mycobacterium tuberculosis polyacyltrehalose lipid biosynthesis. J Bacteriol 197:201–210. doi: 10.1128/JB.02015-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belardinelli JM, Larrouy-Maumus G, Jones V, de Carvalho LP, McNeil MR, Jackson M. 2014. Biosynthesis and translocation of unsulfated acyltrehaloses in Mycobacterium tuberculosis. J Biol Chem 289:27952–27965. doi: 10.1074/jbc.M114.581199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grzegorzewicz AE, Pham H, Gundi VAKB, Scherman MS, North EJ, Hess T, Jones V, Gruppo V, Born SEM, Korduláková J, Chavadi SS, Morisseau C, Lenaerts AJ, Lee RE, McNeil MR, Jackson M. 2012. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat Chem Biol 8:334–341. doi: 10.1038/nchembio.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belisle JT, Vissa VD, Sievert T, Takayama K, Brennan PJ, Besra GS. 1997. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276:1420–1422. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 21.Bhatt A, Kremer L, Dai AZ, Sacchettini JC, Jacobs WR. 2005. Conditional depletion of KasA, a key enzyme of mycolic acid biosynthesis, leads to mycobacterial cell lysis. J Bacteriol 187:7596–7606. doi: 10.1128/JB.187.22.7596-7606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Domenech P, Reed MB, Barry CE. 2005. Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect Immun 73:3492–3501. doi: 10.1128/IAI.73.6.3492-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamichhane G, Tyagi S, Bishai WR. 2005. Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs. Infect Immun 73:2533–2540. doi: 10.1128/IAI.73.4.2533-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells RM, Jones CM, Xi Z, Speer A, Danilchanka O, Doornbos KS, Sun P, Wu F, Tian C, Niederweis M. 2013. Discovery of a siderophore export system essential for virulence of Mycobacterium tuberculosis. PLoS Pathog 9:e1003120. doi: 10.1371/journal.ppat.1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernut A, Viljoen A, Dupont C, Sapriel G, Blaise M, Bouchier C, Brosch R, de Chastellier C, Herrmann J-L, Kremer L. 2016. Insights into the smooth-to-rough transitioning in Mycobacterium bolletii unravels a functional Tyr residue conserved in all mycobacterial MmpL family members. Mol Microbiol 99:866–883. doi: 10.1111/mmi.13283. [DOI] [PubMed] [Google Scholar]
- 26.Deshayes C, Bach H, Euphrasie D, Attarian R, Coureuil M, Sougakoff W, Laval F, Av-Gay Y, Daffé M, Etienne G, Reyrat J-M. 2010. MmpS4 promotes glycopeptidolipids biosynthesis and export in Mycobacterium smegmatis. Mol Microbiol 78:989–1003. doi: 10.1111/j.1365-2958.2010.07385.x. [DOI] [PubMed] [Google Scholar]
- 27.Ojha AK, Baughn AD, Sambandan D, Hsu T, Trivelli X, Guerardel Y, Alahari A, Kremer L, Jacobs WR Jr, Hatfull GF. 2008. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol Microbiol 69:164–174. doi: 10.1111/j.1365-2958.2008.06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ojha A, Anand M, Bhatt A, Kremer L, Jacobs WR Jr, Hatfull GF. 2005. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123:861–873. doi: 10.1016/j.cell.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 29.Teramoto K, Suga M, Sato T, Wada T, Yamamoto A, Fujiwara N. 2015. Characterization of mycolic acids in total fatty acid methyl ester fractions from Mycobacterium species by high resolution MALDI-TOFMS. Mass Spectrom (Tokyo) 4:A0035. doi: 10.5702/massspectrometry.A0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guirado E, Mbawuike U, Keiser TL, Arcos J, Azad AK, Wang S-H, Schlesinger LS. 2015. Characterization of host and microbial determinants in individuals with latent tuberculosis infection using a human granuloma model. mBio 6:e02537-14. doi: 10.1128/mBio.02537-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattila JT, Ojo OO, Kepka-Lenhart D, Marino S, Kim J-H, Eum S-Y, Via LE, Barry CE, Klein E, Kirschner DE, Morris SM, Lin PL, Flynn JL. 2013. Microenvironments in tuberculous granulomas are delineated by distinct populations of macrophage subsets and expression of nitric oxide synthase and arginase isoforms. J Immunol 191:773–784. doi: 10.4049/jimmunol.1300113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Z, Luo Q, Guo Y, Chen J, Xiong G, Peng Y, Ye J, Li J. 2015. Mycobacterium tuberculosis-induced polarization of human macrophage orchestrates the formation and development of tuberculous granulomas in vitro. PLoS One 10:e0129744. doi: 10.1371/journal.pone.0129744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Målen H, Berven FS, Fladmark KE, Wiker HG. 2007. Comprehensive analysis of exported proteins from Mycobacterium tuberculosis H37Rv. Proteomics 7:1702–1718. doi: 10.1002/pmic.200600853. [DOI] [PubMed] [Google Scholar]
- 34.Purdy GE, Niederweis M, Russell DG. 2009. Decreased outer membrane permeability protects mycobacteria from killing by ubiquitin-derived peptides. Mol Microbiol 73:844–857. doi: 10.1111/j.1365-2958.2009.06801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffin JE, Gawronski JD, DeJesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daniel J, Maamar H, Deb C, Sirakova TD, Kolattukudy PE. 2011. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLos Pathog 7:e1002093. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sirakova TD, Deb C, Daniel J, Singh HD, Maamar H, Dubey VS, Kolattukudy PE. 2012. Wax ester synthesis is required for Mycobacterium tuberculosis to enter in vitro dormancy. PLos One 7:e51641. doi: 10.1371/journal.pone.0051641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim M-J, Wainwright HC, Locketz M, Bekker L-G, Walther GB, Dittrich C, Visser A, Wang W, Hsu F-F, Wiehart U, Tsenova L, Kaplan G, Russell DG. 2010. Caseation of human tuberculosis granulomas correlates with elevated host lipid metabolism. EMBO Mol Med 2:258–274. doi: 10.1002/emmm.201000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garton NJ, Christensen H, Minnikin DE, Adegbola RA, Barer MR. 2002. Intracellular lipophilic inclusions of mycobacteria in vitro and in sputum. Microbiology (Reading, Engl) 148:2951–2958. doi: 10.1099/00221287-148-10-2951. [DOI] [PubMed] [Google Scholar]
- 40.Kremer L, de Chastellier C, Dobson G, Gibson KJC, Bifani P, Balor S, Gorvel J-P, Locht C, Minnikin DE, Besra GS. 2005. Identification and structural characterization of an unusual mycobacterial monomeromycolyl-diacylglycerol. Mol Microbiol 57:1113–1126. doi: 10.1111/j.1365-2958.2005.04717.x. [DOI] [PubMed] [Google Scholar]
- 41.McKinney JD, Höner zu Bentrup K, Muñoz-Elías EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Russell DG. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 42.Pandey AK, Sassetti CM. 2008. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci U S A 105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puissegur M-P, Botanch C, Duteyrat J-L, Delsol G, Caratero C, Altare F. 2004. An in vitro dual model of mycobacterial granulomas to investigate the molecular interactions between mycobacteria and human host cells. Cell Microbiol 6:423–433. doi: 10.1111/j.1462-5822.2004.00371.x. [DOI] [PubMed] [Google Scholar]
- 44.Tullius MV, Harmston CA, Owens CP, Chim N, Morse RP, McMath LM, Iniguez A, Kimmey JM, Sawaya MR, Whitelegge JP, Horwitz MA, Goulding CW. 2011. Discovery and characterization of a unique mycobacterial heme acquisition system. Proc Natl Acad Sci U S A 108:5051–5056. doi: 10.1073/pnas.1009516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varela C, Rittmann D, Singh A, Krumbach K, Bhatt K, Eggeling L, Besra GS, Bhatt A. 2012. MmpL genes are associated with mycolic acid metabolism in mycobacteria and corynebacteria. Chem Biol 19:498–506. doi: 10.1016/j.chembiol.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, Toner M, Fortune SM. 2012. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 335:100–104. doi: 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thanky NR, Young DB, Robertson BD. 2007. Unusual features of the cell cycle in mycobacteria: polar-restricted growth and the snapping-model of cell division. Tuberculosis 87:231–236. doi: 10.1016/j.tube.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 48.La Rosa V, Poce G, Canseco JO, Buroni S, Pasca MR, Biava M, Raju RM, Porretta GC, Alfonso S, Battilocchio C, Javid B, Sorrentino F, Ioerger TR, Sacchettini JC, Manetti F, Botta M, De Logu A, Rubin EJ, De Rossi E. 2012. MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212. Antimicrob Agents Chemother 56:324–331. doi: 10.1128/AAC.05270-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foss MH, Pou S, Davidson PM, Dunaj JL, Winter RW, Pou S, Licon MH, Doh JK, Li Y, Kelly JX, Dodean RA, Koop DR, Riscoe MK, Purdy GE. 2016. Diphenylether-modified 1,2-diamines with improved drug properties for development against Mycobacterium tuberculosis. ACS Infect Dis 2:500–508. doi: 10.1021/acsinfecdis.6b00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tahlan K, Wilson R, Kastrinsky DB, Arora K, Nair V, Fischer E, Barnes SW, Walker JR, Alland D, Barry CE, Boshoff HI. 2012. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob Agents Chemother 56:1797–1809. doi: 10.1128/AAC.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng X, Zhu W, Schurig-Briccio LA, Lindert S, Shoen C, Hitchings R, Li J, Wang Y, Baig N, Zhou T, Kim BK, Crick DC, Cynamon M, McCammon JA, Gennis RB, Oldfield E. 2015. Antiinfectives targeting enzymes and the proton motive force. Proc Natl Acad Sci U S A 112:E7073–E7082. doi: 10.1073/pnas.1521988112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foss MH, Powers KM, Purdy GE. 2012. Structural and functional characterization of mycobactericidal ubiquitin-derived peptides in model and bacterial membranes. Biochemistry 51:9922–9929. doi: 10.1021/bi301426j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bardarov S, Bardarov S Jr, Pavelka MS Jr, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs WR Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- 54.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF, Snapper SB, Barletta RG, Jacobs WR Jr, Bloom BR. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 55.Purdy GE, Pacheco S, Turk J, Hsu F-F. 2013. Characterization of mycobacterial triacylglycerols and monomeromycolyl diacylglycerols from Mycobacterium smegmatis biofilm by electrospray ionization multiple-stage and high-resolution mass spectrometry. Anal Bioanal Chem 405:7415–7426. doi: 10.1007/s00216-013-7179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schlesinger LS. 1993. Macrophage phagocytosis of virulent but not attenuated strains of Mycobacterium tuberculosis is mediated by mannose receptors in addition to complement receptors. J Immunol 150:2920–2930. [PubMed] [Google Scholar]
- 58.Olakanmi O, Britigan BE, Schlesinger LS. 2000. Gallium disrupts iron metabolism of mycobacteria residing within human macrophages. Infect Immun 68:5619–5627. doi: 10.1128/IAI.68.10.5619-5627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Romani N, Gruner S, Brang D, Kämpgen E, Lenz A, Trockenbacher B, Konwalinka G, Fritsch PO, Steinman RM, Schuler G. 1994. Proliferating dendritic cell progenitors in human blood. J Exp Med 180:83–93. doi: 10.1084/jem.180.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lewinsohn DA, Winata E, Swarbrick GM, Tanner KE, Cook MS, Null MD, Cansler ME, Sette A, Sidney J, Lewinsohn DM. 2007. Immunodominant tuberculosis CD8 antigens preferentially restricted by HLA-B. PLoS Pathog 3:e127. doi: 10.1371/journal.ppat.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewinsohn DM, Swarbrick GM, Cansler ME, Null MD, Rajaraman V, Frieder MM, Sherman DR, McWeeney S, Lewinsohn DA. 2013. Human Mycobacterium tuberculosis CD8 T cell antigens/epitopes identified by a proteomic peptide library. PLoS One 8:e67016. doi: 10.1371/journal.pone.0067016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Danilchanka O, Mailaender C, Niederweis M. 2008. Identification of a novel multidrug efflux pump of Mycobacterium tuberculosis. Antimicrob Agents Chemother 52:2503–2511. doi: 10.1128/AAC.00298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li W, Upadhyay A, Fontes FL, North EJ, Wang Y, Crans DC, Grzegorzewicz AE, Jones V, Franzblau SG, Lee RE, Crick DC, Jackson M. 2014. Novel insights into the mechanism of inhibition of MmpL3, a target of multiple pharmacophores in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:6413–6423. doi: 10.1128/AAC.03229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.