ABSTRACT
Similar to other intracellular pathogens, Leishmania parasites are known to evade the antimicrobial effector functions of host immune cells. To date, however, only a few virulence factors have been described for Leishmania major, one of the causative agents of cutaneous leishmaniasis. Here, we have characterized the expression and function of an L. major phosphatase, which we termed LmPRL-1. This enzyme shows a strong structural similarity to the human phosphatases of regenerating liver (PRL-1, -2, and -3) that regulate the proliferation, differentiation, and motility of cells. The biochemical characterization of the L. major phosphatase revealed that the enzyme is redox sensitive. When analyzing the subcellular localization of LmPRL-1 in promastigotes, amastigotes, and infected macrophages, we found that the phosphatase was predominantly expressed and secreted by promastigotes via the exosome route. Finally, we observed that ectopic expression of LmPRL-1 in L. major led to an increased number of parasites in macrophages. From these data, we conclude that the L. major phosphatase LmPRL-1 contributes to the intracellular survival of the parasites in macrophages.
KEYWORDS: Leishmania, exosome, macrophage, tyrosine phosphatase
INTRODUCTION
Leishmaniasis is an infectious disease prevalent worldwide that is caused by kinetoplastid protozoan parasites belonging to the genus Leishmania (1). In nature, this heteroxenous parasite is transmitted by the bites of sand flies. Following the inoculation of flagellated promastigotes into the dermis of the host, the parasites are rapidly endocytosed by phagocytic cells, in which they differentiate into amastigotes, multiply, and reach inner compartments and organs, such as draining lymph nodes, spleen, liver, and bone marrow. The life cycle of the parasite is completed after ingestion of amastigote-infected cells by sand flies during their blood meal. In the digestive tract of their vector, parasites transform back into extracellular promastigotes that develop into infectious metacyclics (2). Depending on the status of the host immune system and the Leishmania species, the infection of mammals leads either to mostly self-healing skin lesions (cutaneous leishmaniasis [CL]) or to a systemic disease, termed visceral leishmaniasis (VL) or kala-azar, that is lethal if untreated (3–5).
To survive in their hosts, Leishmania parasites need to quickly adapt their growth, metabolism, and mechanisms of protection to their new environment. Interestingly, only a few genes are differentially expressed during this adaptive process, suggesting an important role for the regulation of protein translation (6). Leishmania species-specific gene diversity also participates in the adaptation of the parasite to its organ-specific microenvironment in the host (7). The stress response protein A2 is an example of such a gene, expressed only by Leishmania species causing VL. A2 appears to mediate heat shock resistance and thereby to support parasite survival and visceralization in inner and warmer organs (8). Other Leishmania genes were found to regulate the promastigote-to-amastigote transformation and the intracellular growth of amastigotes; examples include the genes encoding the cation transporter-like protein MGT2 (9), the MPK7 protein kinase (10), and the homologue of the proliferation-associated 2G4 protein Ebp1, named LmaPA2G4 (11).
In addition to these adaptive responses, Leishmania parasites have also developed strategies to manipulate the phagocytic cells in which they primarily reside (reviewed in references 12–16). Recent studies revealed that homologues of the macrophage migratory inhibitory factor expressed by Leishmania major, LmMIF-1 and -2, can alter the leishmanicidal activity of macrophages, leading to T cell exhaustion and disease progression during in vivo infection (17, 18). Similarly, another well-studied immunomodulatory molecule of Leishmania is its promastigote surface molecule lipophosphoglycan (LPG) (19, 20), which delays the fusion of the phagosome with late endosomes or lysosomes in macrophages (21, 22). In addition, parasites secrete virulence factors, such as the zinc metalloprotease Gp63 (also known as leishmanolysin) or the elongation factor EF-1α, to inhibit the production of leishmanicidal nitric oxide or of proinflammatory cytokines by macrophages (15, 23, 24). These two parasite-derived factors lead to the rapid activation of src homology 2 domain-containing tyrosine phosphatase 1 (SHP-1) and other host phosphatases (25, 26), the modulation of protein kinase C (PKC) activities (27, 28), the inhibition of mitogen-activated protein kinase (MAPK) (29, 30), and the disruption of the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway after Leishmania infection (14, 25, 31). Altogether, the changes in host kinase and phosphatase activity caused by these virulence factors contribute to the immune silencing induced by Leishmania.
Although these host cellular pathways all rely on phosphorylation events, so far no Leishmania virulence factor has been reported to directly modulate the phosphorylation status of host proteins (32, 33). Despite the presence of secreted or membrane-bound acid phosphatases in different Leishmania species, such as histidine acid phosphatases (34–36), there is only limited evidence to support a function of these enzymes in the virulence of the parasite (37–39). Moreover, to date, neither their mechanisms of action nor their modes of access to the host cell cytoplasm have been elucidated.
In our present study, we focused on L. major, one of the causative agents of CL, which is prevalent in the Near and Middle East, northern Africa, the northwestern states of India, and presumably also in northwestern China (40, 41). Here, we identify a new tyrosine phosphatase encoded by L. major. First, we characterize the biochemical activity of this Leishmania phosphatase, which we named LmPRL-1 due to its similarity to the family of mammalian phosphatases of the regenerating liver (PRLs). PRLs are thought to participate in the control of various cellular processes, such as proliferation, differentiation, and motility (42). Second, we provide evidence that the LmPRL-1 phosphatase can be secreted during infection of macrophages and that it contributes to the survival of the parasite in these host cells.
RESULTS
The Leishmania phosphatase LmPRL-1 is a homologue of the human PRLs.
Our research on new virulence factors of L. major focuses on proteins that are putatively released by parasites and that might affect the host cell signaling machinery. Leishmania parasites secrete proteins mainly via exosomes. The analysis of the protein content of these exosomes (43, 44) combined with the genome analysis (data not shown) of our L. major strain (MHOM/IL/81/FEBNI) (45) allowed us to identify a gene on chromosome 16 (LmjF.16.0230) that is identical to the one annotated in the L. major Friedlin strain (46) and encodes a protein tyrosine phosphatase (PTP) here called LmPRL-1. A BLAST search with this gene revealed the presence of a paralogous gene (LmjF.16.0250) on the same chromosome 16 of L. major. However, the protein product of this gene (which we termed LmPRL-2) was not reported in the published exosome analyses (43, 44), excluding the enzyme from the list of putative new secreted virulence factors. In addition, the BLAST results revealed that LmPRL-1 belongs to the PRL family (Fig. 1A and B) and that there are numerous homologues in the family Trypanosomatidae.
FIG 1.
Homology between L. major phosphatases, LmPRL-1 and LmPRL-2, and their homologues from the PRL family in mammals and Trypanosomatidae. (A) Phylogenetic comparison between the mammalian PRLs and their homologues in Trypanosomatidae. Available gene sequences belonging to the PRL family from all Trypanosomatidae and from mouse and human, for the mammals, were used to construct a phylogenetic topology by the minimum-evolution method. The circles indicate nodes supported in ≥70% (open), ≥80% (gray), or ≥90% (black) of 500 random bootstrap replicates of all neighbor-joining trees. The evolutionary distance (scale) is measured in numbers of base substitutions per site. Sequences are labeled in red for mammalian, black for Leishmania, cyan for Endotrypanum, violet for Crithidia, pink for Leptomonas, and orange for Trypanosoma. The red stars indicate LmPRL-1 (LmjF.16.0230) and LmPRL-2 (LmjF.16.0250). (B) Comparison of LmPRL-1 (Q4QEZ7) and LmPRL-2 (Q4QEZ5) protein sequences with those of the three members of the phosphatase of the regenerating liver family, PRL-1 (Q93096), PRL-2 (Q12974), and PRL-3 (O75365) (the references in parentheses refer to the UniProtKB database). The CLUSTAL W program (86) was used for the alignment. The asterisks indicate amino acid residues identical in all proteins, and the colons indicate similarity. The catalytic cysteine is indicated in red and the regulatory cysteine in orange. The different loops involved in the catalytic site, the polybasic region, and the putative farnesylation site are indicated with black lines. (C) Alignment of the three-dimensional (3D) structures of LmPRL-1 (PDB no. 3S4O) (blue) and PRL-1 (PDB no. 1XM2) (gray). PyMOL software was used to align the proteins, represented in cartoon form. The vicinity of the catalytic cysteines of LmPRL-1 and PRL-1, C114 and C104, respectively, is illustrated in the box, revealing their close proximity to the regulatory cysteines of the two proteins C53 and C49, respectively. (D) Conservation of loop sequences. The sequences of the conserved amino acids of the four loops described in panel B are listed for the mammalian PRLs (PRL group; red branch in panel A), the LmPRL-1-like phosphatases of Leishmania (panel A, blue branch), and the LmPRL-2-like phosphatases of Leishmania (panel A, green branch).
The phylogenetic analysis of all available sequences of Trypanosomatidae that are homologues of mammalian PRL sequences uncovered a peculiar gene organization for the genus Leishmania. All 17 sequenced species of Leishmania except L. panamensis include two PRL-related phosphatases in their genomes, as seen in L. major (strain Friedlin). Moreover, these paralogous genes always segregate separately in the phylogenetic tree, creating two subfamilies of genes, one related to LmjF.16.0230 (Fig. 1A, blue branch) and the other related to LmjF.16.0250 (Fig. 1A, green branch). Interestingly, this pairwise organization has also been observed in the genera Leptomonas, Endotrypanum, and Crithidia, whereas the gene number can vary from one to four copies in the genus Trypanosoma (Fig. 1A). Our overall knowledge about this phosphatase family in Trypanosomatidae is still very limited. So far, only one study has reported the expression of a family member, termed TcPRL-1. This protein, which is encoded by the gene TcCLB.503851.24 (Fig. 1A, arrow), was detected in amastigote extracts of Trypanosoma cruzi, but its function has not yet been studied (47). A high-throughput RNA interference (RNAi) screen performed in Trypanosoma brucei led to the discovery of the gene Tb927.8.5780 (Fig. 1A, arrowhead), which might be involved in the survival of the parasite during in vitro differentiation (48). However, one has to be cautious with extrapolations, as T. cruzi has four paralogues of PRL-like genes whereas T. brucei carries only one.
The two L. major phosphatases, LmPRL-1 and LmPRL-2, show 58.5% identity, 26.2% similarity, and only a 15.3% difference in their amino acid sequences. The LmPRL-1 and LmPRL-2 proteins also share strong sequence identity (between 38.9 and 41.1%) with the three proteins of the human PRL phosphatase family (PRL-1, -2, and -3) (Fig. 1B), which themselves form a homogeneous group of proteins with a high degree (>75%) of amino acid sequence identity (Fig. 1B). This strong protein sequence relationship translates into comparable structural organizations, as shown by the alignment of the human PRL-1 crystal structure (Protein Data Bank [PDB] no. 1XM2) (49) with the crystal structure of L. major LmPRL-1 (PDB no. 3S4O) (Fig. 1C). The structures of both proteins consist of α-helices surrounding a five-stranded β-sheet, which represents the canonical structure of tyrosine phosphatases. This alignment also illustrates the highly conserved orientation of the amino acids that are critical for their phosphatase activity (Fig. 1C). The P loop and the WDP loop are maintained in all mammalian PRL and Leishmania PRL-like sequences (Fig. 1D). Interestingly, the catalytic cysteine 114 located in the C-X5-R P loop of LmPRL-1 (C107 for LmPRL-2) neighbors cysteine 53 in loop 2 (C52 for LmPRL-2) (Fig. 1C). In PRL-1, this proximity between cysteines was previously described as essential for the regulation of catalytic activity, since the two residues could form an inactivating disulfide bond (50, 51). Another important feature shared by all these phosphatases is the presence of a farnesylation site (C-A-A-X) at the C terminus of the protein, which suggests possible membrane localization for the Leishmania phosphatases (Fig. 1B). Membrane localization is also supported by the interaction between a nearby polybasic sequence (Fig. 1B) and the negatively charged phospholipids present in the cellular membrane (52).
Despite all these similarities between mammalian and Leishmania PRLs, substantial differences can be observed in the sequences of loops 1 and 2 that are crucial to the substrate specificity of a PTP. As illustrated in Fig. 1D, LmPRL proteins share only 2 out of 9 amino acid residues with loop 1 of mammalian PRL, namely, proline 29/28 (LmPRL-1/2) and leucine 34/33. Regarding loop 2, the divergence is also remarkable, as only 4 out of 7 residues are conserved, namely, arginine 51/50, cysteine 53/52, threonine 56/55, and tyrosine 57/56. Interestingly, there are also differences in the sequences of the two loops in the different Leishmania PRL-like proteins (Fig. 1D). Asparagine 33 of loop 1 of LmPRL-1 is replaced by serine 32 in LmPRL-2. Similarly, valine 52 of loop 2 of LmPRL-1 is replaced by alanine 51 in LmPRL-2. These differences discriminate between the LmPRL-1 like and LmPRL-2 like proteins described in the phylogenetic tree. Altogether, these amino acid sequence disparities suggest a distinct substrate specificity of Leishmania and human PRLs.
Characterization of the enzymatic activity of recombinant His6-LmPRL-1.
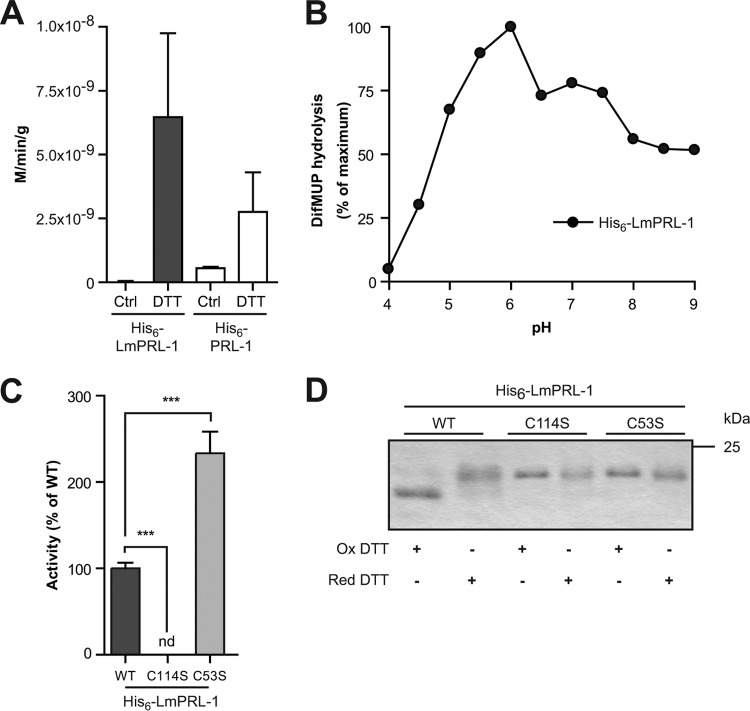
As the protein LmPRL-1 was the only PTP detected in Leishmania exosomes (43, 44), we focused our further investigation on that enzyme. To assess their enzymatic functions, the Leishmania protein LmPRL-1 and the human PRL-1 were overexpressed in E. coli cells with a His6 tag and purified by affinity chromatography using a Ni-nitrilotriacetic acid (NTA) matrix. Hydrolysis of para-nitrophenyl phosphate (pNPP), a classical PTP substrate, was monitored to determine their enzymatic activities. Despite purification under reducing conditions (5 mM β-mercaptoethanol), hydrolysis was hardly detectable unless the enzymes were preincubated with 10 mM dithiothreitol (DTT) for 1 h (Fig. 2A). Under both nonreducing and reducing conditions, the activities detected for His6-LmPRL-1 were in the same order of magnitude as those measured for the human His6-PRL-1 (Fig. 2A). Since PRL-1 has higher affinity for the fluorogenic substrate 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP), this substrate was chosen for the further catalytic study of His6-LmPRL-1 (50). The optimal pH for the activity of His6-LmPRL-1 turned out to be slightly acidic (pH 6.0) (Fig. 2B). In addition, the kinetic characteristics of the Leishmania phosphatase were determined. A Vmax of 2.6 × 10−5 M/min/g and a Km of 9.32 × 10−6 M were measured for the dephosphorylation of DiFMUP. Considering the requirement for a reducing milieu, we also investigated the roles of the putative catalytic and regulatory cysteines. First, the activity of a serine mutant of the catalytic cysteine C114 was tested. As expected, the phosphatase activity of the His6-LmPRL-1-C114S mutant was fully abolished (Fig. 2C). In contrast, a mutant of the putative regulatory cysteine, His6-LmPRL-1-C53S, showed markedly higher activity toward DiFMUP (>2-fold increase compared to wild-type [WT] enzyme) (Fig. 2C). The latter finding suggested that a disulfide bond formed between the regulatory and the catalytic cysteine needs to be reduced to trigger the LmPRL-1 phosphatase activity. To further validate this hypothesis, the migration behaviors of the WT protein and the different mutants of the His6-tagged phosphatase were analyzed with nonreducing SDS-PAGE, as it allows the indirect visualization of disulfide bond formation. Under oxidative conditions, a protein containing an oxidized disulfide bond runs at a lower molecular weight than predicted because the sequence is more compact (53). The WT phosphatase, pretreated with 10 mM oxidized DTT, migrated as two bands. The predominant lower band corresponds to the oxidized form, whereas the minor upper band corresponds to the reduced form of His6-LmPRL-1 (Fig. 2D). Only the latter form was detectable when the protein was preincubated with 10 mM reduced DTT, leading to the reduction of the disulfide bond. Similar treatment of mutants of either the catalytic (His6-LmPRL-C114S) or the regulatory (His6-LmPRL-1-C53S) cysteine revealed that these proteins could no longer convert into an oxidized, fast-migrating form. These data support a regulatory mechanism for the phosphatase activity of LmPRL-1 based on a pair of cysteines that sense the redox potential of the milieu.
FIG 2.
Biochemical characterization of His6-tagged LmPRL-1. (A) Phosphatase activity of WT His6-LmPRL-1 against pNPP compared to the activity of the human His6-PRL-1. The phosphatase activity was measured by incubating His6-tagged LmPRL-1 (black bars) and PRL-1 (white bars) with pNPP for 1 h at 37°C and pH 6.0. Some reactions were carried out with previously reduced proteins. (B) Phosphatase activity of WT His6-LmPRL-1 against DiFMUP as a function of pH. His6-tagged LmPRL-1 was reduced before its phosphatase activity was measured by incubating it with DiFMUP for 20 min at 37°C and pHs varying from 4.0 to 9.0. (C) Effects of the catalytic (C114) and regulatory (C53) cysteines on the phosphatase activity of His6-LmPRL-1. The phosphatase activities of His6-LmPRL-1-C114S and His6-LmPRL-1-C53S (light-gray bar) against DiFMUP at pH 6.0 after 20 min at 37°C were measured and compared to the activity of the wild-type His6-LmPRL-1 (dark-gray bar). (D) Change of the electrophoretic migration of LmPRL-1 induced by oxidation is controlled by the catalytic and regulatory cysteines. The wild type and the catalytic and regulatory mutants of LmPRL-1 were either oxidized with 10 mM oxidized (Ox) DTT or reduced with 10 mM reducing (Red) DTT for 1 h at 4°C. The proteins were separated on nonreducing SDS-PAGE and dyed by Coomassie staining. The data represent the means and SD of the results of one of three experiments with similar results. ***, P < 0.001; nd, not determined.
LmPRL-1 expression and subcellular localization in L. major promastigotes and amastigotes.
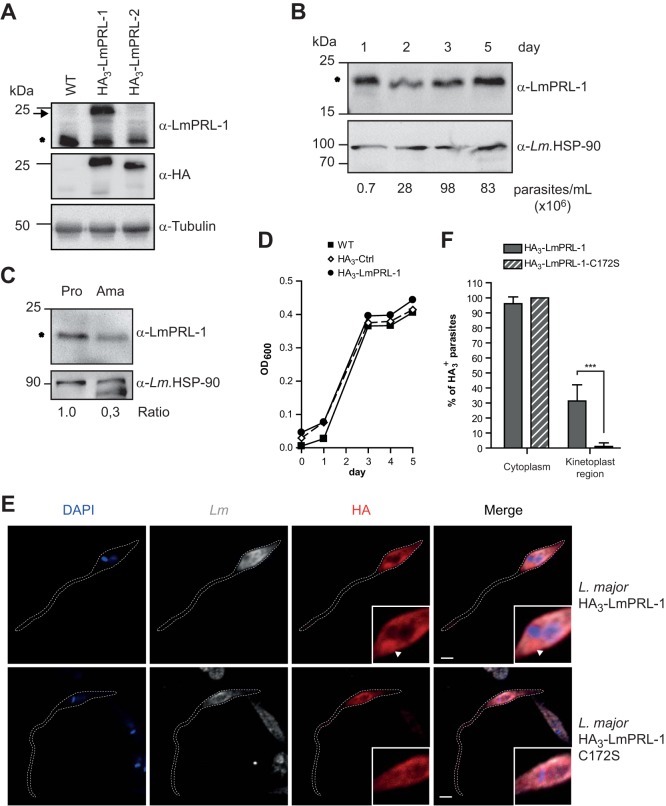
As an initial step toward its functional characterization, we investigated whether the LmPRL-1 phosphatase shows a parasite stage-dependent expression pattern. To this end, we obtained a rabbit anti-LmPRL-1 peptide antiserum and generated L. major parasites that ectopically expressed hemagglutinin (HA3)-tagged LmPRL-1 (23.1 kDa) following electroporation of pCLN-3xHA-based vectors (see Table S1 in the supplemental material). As a control, we also expressed HA3-LmPRL-2 in L. major to ascertain the specificity of the rabbit anti-LmPRL-1 antiserum and to exclude the detection of this paralogous protein. Western blot analysis of lysates of these parasites revealed that both HA-tagged proteins could be expressed (Fig. 3A, α-HA) and that the anti-LmPRL-1 antiserum selectively detected HA3-LmPRL-1 (Fig. 3A, α-LmPRL-1, arrow); no cross-reaction with HA3-LmPRL-2 was observed. Furthermore, endogenous LmPRL-1 (19.4 kDa) was detectable in the lysates of the two strains ectopically expressing HA3-tagged proteins, as well as in the lysate of the WT L. major strain (Fig. 3A, α-LmPRL-1, asterisk).
FIG 3.
Expression and subcellular localization of LmPRL-1 in L. major parasites. (A) Detection of LmPRL-1 protein in promastigotes. Protein extracts were prepared from stationary-phase promastigote cultures of WT L. major and parasites ectopically expressing HA3-LmPRL-1 or HA3-LmPRL-2. The proteins were analyzed by immunoblotting with rabbit serum anti-LmPRL-1, as well as anti-HA antibody to estimate the specificity of the rabbit serum and with anti-tubulin antibody (loading control). Endogenous LmPRL-1 (expressed by WT parasites) is indicated by the asterisk and HA3-tagged proteins of transfected L. major by the arrow. (B) Expression of LmPRL-1 in logarithmic- or stationary-growth-phase promastigotes in vitro. A culture of WT L. major promastigotes in modified complete Schneider's medium inoculated with 1.0 × 105 parasites/ml was analyzed daily for its concentration (parasites per milliliter) until it reached stationary phase (day 5). Expression of endogenous LmPRL-1 (marked by the asterisk) was analyzed by anti-LmPRL-1 immunoblotting, using the HSP-90 housekeeping gene as a control. (C) Expression of LmPRL-1 in promastigotes and amastigotes. Protein extracts prepared from the same number (20 × 106) of in vitro-cultivated promastigotes (Pro) and ex vivo-isolated amastigotes (Ama) were loaded onto SDS-PAGE. Expression of endogenous LmPRL-1 (marked by the asterisk) and of the HSP-90 housekeeping gene was analyzed by immunoblotting. The ratios of the band intensities are indicated below. (D) Ectopic expression of HA3-LmPRL-1 has no effect on in vitro growth of L. major promastigotes. WT promastigotes and parasites either carrying the empty pCLN-3xHA vector or expressing HA3-LmPRL-1 were cultivated in modified complete Schneider's medium starting at 1.0 × 105 parasite/ml until stationary phase. Growth was monitored by measuring the OD600. (E and F) Subcellular localization of LmPRL-1 in L. major promastigotes depends on its C-terminal farnesylation. (E) Parasites expressing HA3-LmPRL-1 or HA3-LmPRL-1-C172S were analyzed by confocal microscopy after staining with an anti-HA antibody (red), an anti-Leishmania serum (gray), and DAPI (blue). The arrowheads indicate the region of the kinetoplast. Bars, 2 μm. (F) Means and standard deviations of more than 250 promastigotes per strain of L. major. ***, P < 0.001. The data presented are from one of three experiments with similar results.
Next, we tested whether the flagellated promastigote form of L. major parasites expressed different amounts of LmPRL-1 depending on the growth phase. In their logarithmic growth phase (days 1 to 3 of culture), promastigotes were clearly positive for LmPRL-1 (Fig. 3B, asterisk). The stationary-phase promastigotes (day 5 of culture), which represent the more infectious stage of the parasite, maintained the expression of LmPRL-1 (Fig. 3B). When analyzing the expression of the phosphatase in 20 × 106 amastigotes prepared from the skin lesions of infected BALB/c mice, we found that LmPRL-1 was still expressed in this developmental stage but to a considerably lesser extent per cell than in the same number of stationary-phase promastigotes (Fig. 3C, asterisk).
To characterize the localization and fate of LmPRL-1 inside Leishmania parasites, rabbit anti-LmPRL-1 serum was tested on WT L. major promastigotes. Unfortunately, the quality of the rabbit anti-LmPRL-1 serum was not sufficient for reliable detection of LmPRL-1 by confocal laser scanning fluorescence microscopy (CLSFM). Therefore, we used parasites ectopically expressing the HA3-tagged protein. During a culture period of 5 days, the growth rates of the HA3-LmPRL-1-expressing promastigotes of the WT parent strain and of a parasite line carrying the empty HA vector (HA3-Ctrl) were comparable (Fig. 3D). The subcellular localization of HA3-LmPRL-1 was analyzed by CLSFM. The cytosol of more than 97% of the promastigotes analyzed stained positively for HA3-LmPRL-1 (Fig. 3E and F). In addition, the protein appeared to be enriched around the kinetoplast in 30% of the parasites (Fig. 3F). Strikingly, the accumulation of HA3-LmPRL-1 in the kinetoplast region was dependent on its C-terminal farnesylation motif. As illustrated in Fig. 3E and F, the C172S mutation of the C-A-A-X sequence resulted in diffuse cytosolic staining and loss of staining around the kinetoplasts of the parasites. This result demonstrates that the farnesylation motif of LmPRL-1 is functional in L. major promastigotes. Since the kinetoplast region is in close proximity to the flagellar pocket in promastigotes, which has been described as a region actively involved in protein secretion and in the production of exosomes (54, 55), these data are indicative of secretion of the LmPRL-1 phosphatase by Leishmania parasites during infection.
LmPRL-1 is secreted by L. major promastigotes via the exosome pathway.
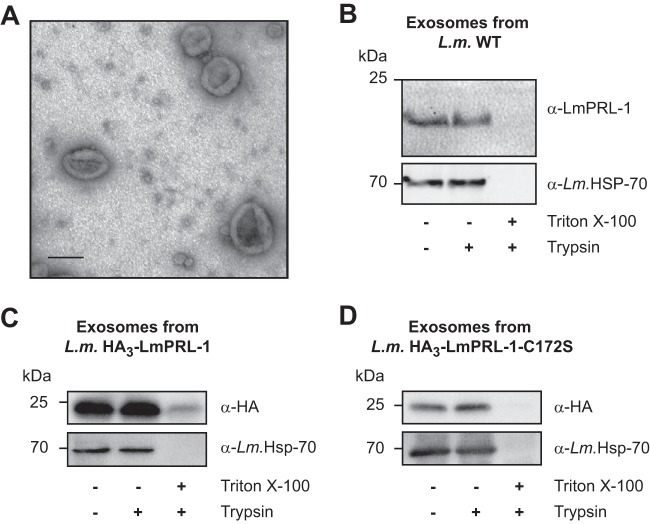
To validate the possible association of LmPRL-1 with exosomes of L. major, exosomes from a culture of WT stationary-phase promastigotes were purified by ultracentrifugation following established protocols (43). Using electron microscopy, a homogeneous population of vesicles with sizes ranging from 70 to 150 nm was observed, confirming that the integrity of the exosomes was maintained during purification (Fig. 4A). The protein content of these exosomes was analyzed by Western blotting. LmPRL-1, as well as HSP70, a protein enriched in Leishmania exosomes (56), was detected in the exosome preparation (Fig. 4B, top, left lane). In addition, the purified exosomes were subjected to trypsin digestion. LmPRL-1 was not degraded by trypsin alone (Fig. 4B, top, middle lane), whereas in the presence of the detergent Triton X-100, both the LmPRL-1 band and the HSP70 protein disappeared (Fig. 4B, top, right lane). This indicates that LmPRL-1 and the known exosome marker HSP70 were protected from the action of the protease by the exosome phospholipid membrane. Similar results were obtained with exosomes produced by parasites expressing HA-tagged WT LmPRL-1, indicating that this N-terminal tag does not impair the release of the phosphatase in exosomes (Fig. 4C). Surprisingly, HA-tagged LmPRL-1 with a mutated farnesylation motif (HA3-LmPRL-1-C172S) was also detected inside exosomes (Fig. 4D), suggesting that the loading of exosomes with LmPRL-1 does not require a functional farnesylation site. Finally, to exclude direct secretion of the phosphatase by the parasite through the classical secretory pathway, we also estimated the quantity of LmPRL-1 secreted outside exosomes. The protein content of the supernatants (SN) resulting from the ultracentrifugation step during the preparation of L. major exosomes was precipitated and analyzed by Western blotting. The majority of LmPRL-1 was detected in the pelleted exosomes, whereas only a minor proportion of the protein was found in the soluble supernatants (see Fig. S1 in the supplemental material). Overall, these results clearly show that WT or HA-tagged LmPRL-1 is secreted by L. major promastigotes inside exosomes.
FIG 4.
LmPRL-1 is secreted by L. major promastigotes via exosomes. (A) Electron micrograph of exosomes produced by WT L. major promastigotes. Exosomes were purified from a culture of metacyclic promastigotes incubated overnight in serum-free RPMI 1640 medium at 28°C and then fixed and analyzed by electron microscopy. Bar, 100 nm. (B) Endogenous LmPRL-1 is secreted in exosomes of WT L. major (L.m.) promastigotes. Purified exosomes from WT promastigotes were analyzed by immunoblotting for the presence of LmPRL-1 and the exosome marker HSP-70. Untreated exosomes were compared to exosomes digested with trypsin or with trypsin in the presence of the detergent Triton X-100. (C) Ectopic expression and HA tagging does not alter LmPRL-1 secretion via exosomes. Purified exosomes from transfected L. major ectopically expressing HA3-LmPRL-1 were analyzed by immunoblotting for the presence of HA-tagged protein and the exosome marker HSP-70 following treatment similar to that described for panel B. (D) HA-tagged LmPRL-1 is secreted in exosomes independently of its farnesylation motif. Purified exosomes from transfected L. major ectopically expressing HA3-LmPRL-1-C172S were analyzed as described for panel C. The data presented are from one of three experiments with similar results.
LmPRL-1 is expressed during macrophage infection by L. major promastigotes.
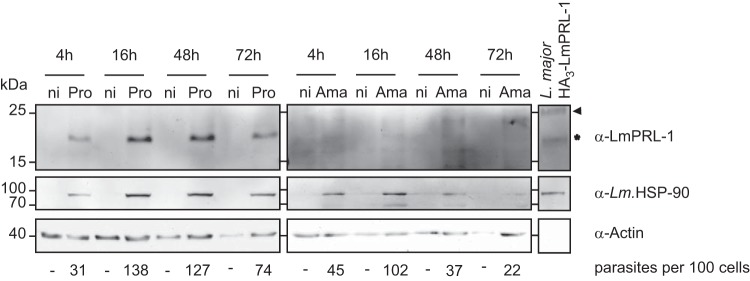
Having established that LmPRL-1 can be secreted by L. major promastigotes, its expression and fate during infection of macrophages were analyzed. Bone marrow-derived macrophages (BMM) were infected with WT stationary-phase L. major promastigotes at a multiplicity of infection (MOI) of 5. The infection was ascertained by the specific detection of the L. major HSP90 protein. Increasing amounts of HSP90 were detected during the course of the experiment (Fig. 5). The slight decrease in HSP90 observed after 72 h of infection reflects the reduced number of intracellular parasites due to spontaneous killing by BMM. LmPRL-1 was clearly present in the lysates of promastigote-infected BMM throughout the in vitro infection (Fig. 5, asterisk). We also directly infected BMM with amastigotes purified from skin lesions of infected and nonhealing BALB/c mice. Unexpectedly, LmPRL-1 was hardly detectable in amastigote-infected BMM throughout the observation period (Fig. 5). These data show that LmPRL-1 is present in promastigote-infected macrophages for at least 72 h but remains downregulated in lesional amastigotes even after in vitro uptake by cultured macrophages.
FIG 5.
Expression of LmPRL-1 during macrophage infection by L. major. BMM were infected by L. major WT promastigotes (Pro) or amastigotes (Ama) at an MOI of 5. Noninfected BMM (ni) were used as controls. At the indicated time points, protein samples were analyzed by immunoblotting for the expression of LmPRL-1, Leishmania HSP-90, and BMM actin. Lysates of parasites ectopically expressing HA3-LmPRL-1 were used to control the reactivity of the LmPRL-1 antiserum. WT LmPRL-1 protein is marked with an asterisk and HA3-LmPRL-1 with an arrowhead. The numbers of parasites per 100 cells of the culture are shown below the Western blots. BMM were infected with L. major promastigotes or amastigotes at an MOI of 5. Extracellular parasites were washed away with PBS (2×) after 4 or 16 h. At the indicated time points, Diff-Quik staining was performed, and the number of amastigotes per 100 cells was determined (a total of 300 cells/per sample). The data presented are from one of three experiments with similar results.
HA3-LmPRL-1 is secreted by L. major during macrophage infection.
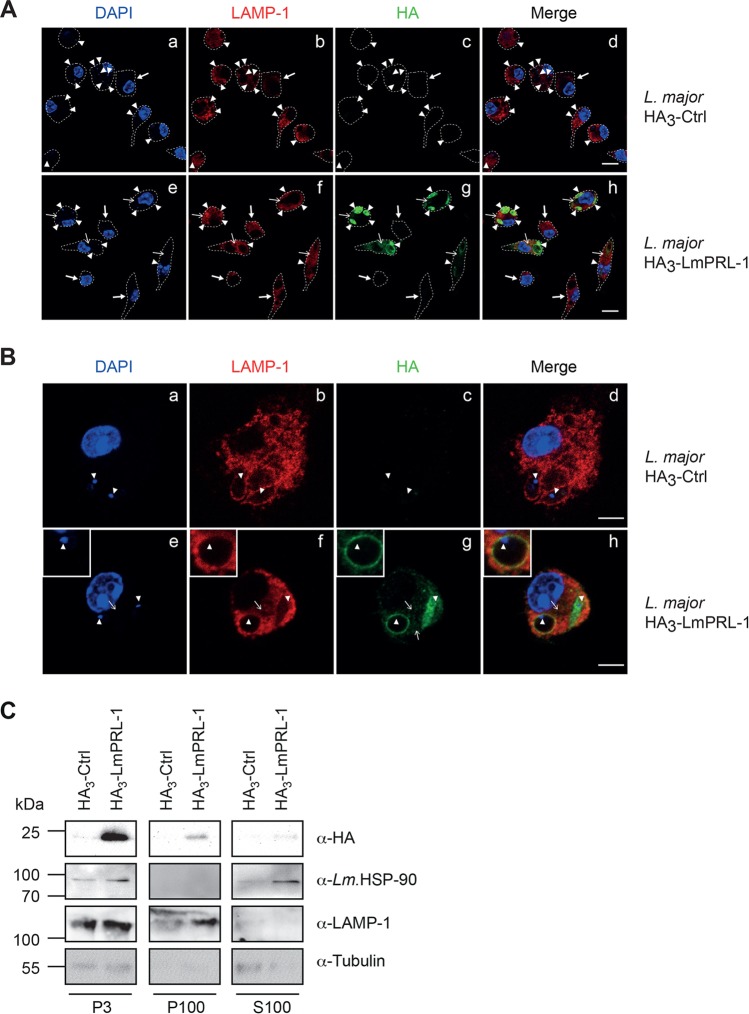
To follow the fate of Leishmania LmPRL-1 after infection, BMM were infected with L. major promastigotes expressing the HA3-tagged phosphatase. After 72 h of infection, the subcellular localization of the proteins was analyzed by CLSFM. Whereas no HA-derived staining was seen in BMM infected with L. major carrying the empty control vector pCLN-3xHA (Fig. 6A, c), L. major parasites expressing HA3-tagged LmPRL-1 were readily detectable as amastigotes (Fig. 6A, e to h) inside parasitophorous vacuoles (PV) that were labeled by an antibody (Ab) against the lysosome-associated membrane protein 1 (LAMP-1) (Fig. 6A, f, arrowheads). In addition, a HA signal was also consistently detected outside the PV when the macrophages were infected with HA3-LmPRL-1 parasites (Fig. 6A, g, thin arrows), whereas no signal was detected in noninfected cells (Fig. 6A, g, thick arrows). This distribution pattern indicates that HA3-LmPRL-1 leaves the PV and enters the cytoplasm of host cells, which is in line with the detected exosome-dependent secretion of LmPRL-1 by WT Leishmania (Fig. 4).
FIG 6.
Subcellular localization of HA3-LmPRL-1 during infection of BMM by L. major. (A and B) BMM were infected for 72 h by L. major promastigotes carrying the empty plasmid pCLN-3xHA (HA3-Ctrl) or the plasmid pCLN-3xHA-LmPRL-1 (HA3-LmPRL-1). Localization of HA-LmPRL-1 (green), the phagosomal marker LAMP-1 (red), and the DNA of the BMM and the parasite (blue) was detected by CLSFM. (A) Overview. (B) Single-cell analyses. The dashed white lines delineate the BMM cell shapes. The arrowheads highlight the DNA of the L. major kinetoplast. The thin arrows mark the HA-derived signal inside the cytoplasm of the BMM. The thick arrows mark noninfected BMM. Bars, 5 μm. (C) HA3-LmPRL-1 localization in the cytosolic fraction of BMM during infection. BMM were infected for 72 h with the same parasites as for panel A. The fractions P3 (nucleus and PV), P100 (cytoplasmic membrane and debris), and S100 (cytosol) were analyzed by immunoblotting for the presence of HA-tagged protein, Leishmania HSP-90, LAMP-1, and tubulin (from parasites and BMM). The data presented are from one of three experiments with similar results.
A closer microscopic analysis of macrophages infected with L. major expressing HA3-LmPRL-1 confirmed its diffuse intraparasitic localization and its presence outside the PV (Fig. 6B, g, thin arrow). We also observed LAMP-1-positive compartments that were decorated with HA3-LmPRL-1 and contained amastigote parasites (Fig. 6B, e to h, arrowheads). To confirm the secretion of HA3-LmPRL-1 during macrophage infection, subcellular fractions were prepared from BMM infected with either HA3-Ctrl or HA3-LmPRL-1 for 72 h. Most of the HA signal was detected in the heaviest fraction pelleted at 3,000 × g (Fig. 6C, P3). This fraction comprises the BMM nuclei and large organelles, such as PV containing amastigotes that express the HA3-tagged protein. The latter account for the presence of Leishmania HSP90, Leishmania tubulin, and the phagolysosome marker LAMP-1. LAMP-1, but not tubulin or HSP90, was also detected in the P100 fraction containing small vesicular structures and the cytoplasmic membrane. HA3-LmPRL-1 was probably detected in this fraction because of the destruction of some PV vacuoles (Fig. 6C, P100). Strikingly, HA3-LmPRL-1 was also detected in the cytosolic fractions of infected cells (Fig. 6C, S100), in which only tubulin and Leishmania HSP90, but no LAMP-1, were found. Altogether, these data document the fate of HA3-LmPRL-1, which is first secreted by Leishmania parasites during infection and then appears to reach the cytoplasm of infected cells.
HA3-LmPRL-1 expression by L. major increases parasite survival during infection of macrophages.
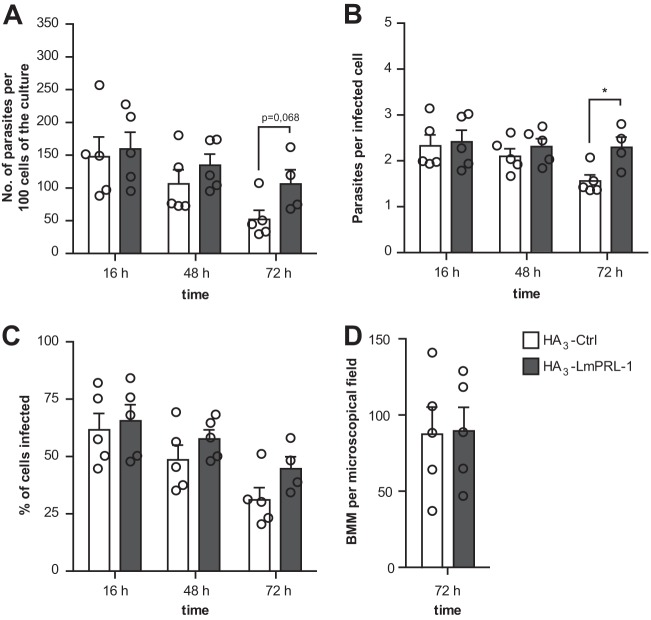
Finally, we tested the effect of HA3-LmPRL-1 overexpression on the intracellular survival of L. major. BMM were infected at an MOI of 5 for 16, 48, or 72 h with either the control strain of L. major (HA3-Ctrl) or the strain ectopically expressing HA3-LmPRL-1. For each time point, the number of intracellular parasites per 100 macrophages of the culture was calculated by measuring the infection rate and determining the number of parasites per infected macrophage. After 72 h of infection, BMM infected with L. major expressing HA3-LmPRL-1 contained 2.1 (±0.4)-fold more parasites per 100 cells than the ones infected with control parasites (Fig. 7A). This phenotype largely resulted from an increased parasite number per infected cell (Fig. 7B) and only to a minor degree from a higher percentage of macrophages infected following expression of HA3-LmPRL-1 (Fig. 7C). In parallel, the total number of BMM per culture was determined after 72 h of infection in order to see whether the Leishmania strains differentially affected host cell survival. The viability of the BMM remained high throughout the course of infection and was independent of the strain of parasite used for infection (Fig. 7D). Altogether, our data suggest that the expression of HA3-LmPRL-1 phosphatase supports the intracellular survival of Leishmania parasites in macrophages.
FIG 7.
Expression of HA3-LmPRL-1 increases the infectivity and survival of L. major during macrophage infection. (A) Numbers of parasites per 100 cells of a culture. BMM were infected with L. major promastigotes (control parasites or parasites expressing HA3-LmPRL-1) at an MOI of 5. Extracellular parasites were washed away with PBS (2×) after 16 h. At the indicated time points, Diff-Quik staining was performed, and the number of amastigotes per 100 cells was determined (a total of 300 cells/sample). (B) Numbers of parasites per infected macrophage. The number of amastigotes per infected cell was determined as described for panel A. (C) Quantification of the percentage of cells infected. The number of BMM containing one or more parasites was calculated as described for panel A. (D) Quantification of macrophage viability. The number of BMM per microscopic field was calculated as described for panel A. Control parasites are depicted in white, and HA3-LmPRL-1 in dark gray. The data represent means and standard errors of the mean (SEM) of five experiments. Each circle represents the result of one independent experiment. *, P < 0.05.
DISCUSSION
Identifying the virulence factors of L. major is crucial for understanding the pathways targeted by the parasite in the mammalian host cell to establish its intracellular habitat. Various surface molecules and secreted proteins of L. major parasites have previously been analyzed (15). In the present study, we report the biochemical and functional characterization of a novel L. major phosphatase, termed LmPRL-1. We established the expression profile of the endogenous phosphatase during the life cycle of the parasite. Interestingly, we observed that LmPRL-1 was secreted by the parasite. In order to characterize its function, we took advantage of an ectopic expression system, since RNA interference cannot be applied in L. major (57, 58). Despite its limitations, this approach revealed the positive effect of LmPRL expression on the survival of L. major amastigotes during macrophage infection.
Our analysis of the genome of L. major (data not shown) for genes related to LmPRL-1 revealed the presence of a paralogous gene that we named LmPRL-2. A phylogenetic analysis demonstrated that this pair of phosphatases has orthologues in other human-pathogenic Leishmania species. This is likely due to a gene duplication event that occurred in a common ancestor of the Leishmania species during evolution. The close proximity (6,946 bp apart) of the respective genes for LmPRL-1 and LmPRL-2, LmjF.16.0230 and LmjF.16.0250, on chromosome 16 further supports this notion. The analysis of their sequences also revealed a strong similarity to the mammalian PRL phosphatase family, as illustrated by the conservation of the classical catalytic site (C-X5-R), a regulatory cysteine at the N terminus, and a farnesylation site at the C terminus. The suspected homology was confirmed by the in vitro biochemical characterization of His6-LmPRL-1 and comparison with the known characteristics of the human PRL-1 (50, 53). First, the catalytic constants (Km) of His6-LmPRL-1 using the DiFMUP substrate (9.32 × 10−6 M) was in line with the previously measured value for its human homologue, PRL-1 (4.6 × 10−6 M) (50). In addition, the activity of the phosphatase was shown to be strictly dependent on the reduction of a disulfide bridge formed between its catalytic cysteine and its regulatory cysteine. This reduction allowed a switch between a closed, oxidized, and inactive form and an open, reduced, and active form of the enzyme. Finally, the sensitivity of His6-LmPRL-1 to oxidation was further confirmed by the enhanced enzymatic activity seen after mutation of the regulatory cysteine. This sensitivity to the redox milieu could be relevant during the oxidative stress encountered by the parasite during infection.
In addition to this biochemical characterization, we also established the expression profile of LmPRL-1 during the growth and stage conversion of L. major parasites. Promastigotes already expressed the LmPRL-1 protein in their logarithmic growth phase and maintained this expression during the stationary growth phase. Moreover, LmPRL-1 was still detectable in macrophage lysates even at 72 h after infection with promastigotes, when the parasites had already transformed into amastigotes. This observation on the protein level corroborates and extends recent mRNA expression profiling experiments (59, 60). The expression of the gene LmjF.16.230, coding for LmPRL-1, was upregulated (1.27-fold) during the transition of procyclic into metacyclic promastigotes (59) and after macrophage infection by promastigotes (1.49-fold) (60). However, LmPRL-1 was hardly detectable in macrophages directly infected with amastigotes purified from mouse skin lesions or in lysates of these amastigotes. Altogether, these expression patterns suggest that endogenous LmPRL-1 might play a role during the development of the infective parasite inside its sand fly vector and/or during the initiation and early stages of infection of mammalian phagocytes. Later, endogenous LmPRL-1 expression might steadily decline following the promastigote-to-amastigote transition, possibly due to the upregulation of other prosurvival mechanisms of the parasite.
In addition to the expression pattern, our study found that LmPRL-1 can be secreted, a key characteristic of many virulence factors. Endogenous LmPRL-1 was detected in exosomes released by WT L. major promastigotes. These exosomes are cargo vesicles produced by promastigotes and amastigotes and used as a main secretory pathway (15, 56). They are loaded with cytoplasmic and membrane proteins of the parasite, including known virulence factors, such as the surface protease leishmanolysin (Gp63) or EF-1α (23, 24, 61). Moreover, exosomes are released within the sand fly gut and are inoculated into the host along with the parasites (62). LmPRL-1 has previously been spotted in a proteomic study that analyzed the content of exosomes produced by Leishmania donovani (43) or gp63-null mutants of L. major (44). These results led us to investigate the expression and fate of LmPRL-1 during the transition of promastigotes into amastigotes inside macrophages. The subcellular localization of LmPRL-1 was addressed in promastigotes, taking advantage of L. major expressing a HA-tagged version of the phosphatase. HA3-LmPRL-1 appeared to spread across the cytoplasm of the promastigotes (Fig. 1A, arrowhead) in a manner similar to that observed for an LmPRL-1 homologue of T. brucei (Tb927.8.5780) that was fused to enhanced yellow fluorescent protein (eYFP) (63). In addition to this diffuse localization, HA3-LmPRL-1 tended to accumulate around the kinetoplasts of promastigotes, a phenotype that was also observed for a homologous enzyme in T. cruzi, TcPRL-1, which, however, was not tested for its potential secretion (47). The observed subcellular localization of LmPRL-1 is in line with its secretion by exosomes, as the flagellar pocket is known as the most active region of that secretory pathway (54, 55). In addition, using a mutant of its C-terminal farnesylation motif, we could demonstrate that the association of LmPRL-1 with the kinetoplast was fully dependent on this sequence. Promastigotes expressing an LmPRL-1 mutant that lacked the terminal farnesylation site (HA3-LmPRL-1-C172S) showed diffuse cytoplasmic staining, as was also observed for a similar mutant of human PRL-1 (64). Interestingly, this mutation did not fully abolish the secretion of the phosphatase by exosomes, suggesting that the exosome loading of LmPRL-1 is independent of its functional farnesylation. One plausible explanation for this observation is the possibility that LmPRL-1 forms trimers, like its human homologue PRL-1. This would allow the HA3-LmPRL-1-C172S mutant to enter exosomes via its trimerization with endogenous WT LmPRL-1. In order to reach a definitive conclusion, the expression of the HA3-LmPRL-1-C172S mutant protein should be tested in an L. major strain with a genetic deletion of LmPRL-1.
When investigating the spatial distribution of the LmPRL-1 phosphatase during infection of macrophages, we found that ectopically expressed HA3-LmPRL-1 was detectable in amastigotes but, strikingly, also appeared outside the PV. Similarly to other exosome markers, such as the heat shock proteins HSP70 and HSP90 (43, 61) and the L. major virulence factor P46 (65, 66), HA3-LmPRL-1 was identified inside the cytoplasm of infected macrophages and colocalized with the late phagosomal marker LAMP-1 on the surfaces of PV, where it is ideally positioned to manipulate host cell signaling pathways in favor of the parasite (67). It is possible that LmPRL-1 directly modulates the phosphorylation status of host signaling molecules and thereby also synergizes with other secreted virulence factors of L. major. This mode of action would explain the positive effect of ectopic HA3-LmPRL-1 expression on L. major survival during macrophage infection. The secreted HA3-LmPRL-1, however, might also interact with its mammalian homologues, the PRL phosphatases, which play important roles in cell division and cell motility (64, 68). As PRLs need to form a homotrimer to be fully active (49, 52), the possibility that LmPRL-1 interferes with the process of mammalian PRL trimerization has to be considered. It is tempting to speculate that this leads to altered phagocyte functions.
In conclusion, we have shown that L. major expresses a PRL-like phosphatase that, based on ectopic expression analyses, is likely to promote parasite survival during infection of macrophages. In addition, LmPRL-1 is actively secreted by the parasite, ends up outside the PV during macrophage infection, and is therefore prone to modulate host cell functions to the advantage of the parasite. Future studies need to address the functional role of LmPRL-1 in vivo using double allelic LmPRL-1 knockout L. major parasites.
MATERIALS AND METHODS
Phylogenetic analysis.
All the gene sequences were extracted from the TritrypDB (69) after a BLAST search for LmjF.16.0230 homologues. The evolutionary history was inferred using the minimum-evolution (ME) method (70). The optimal tree with a sum of branch lengths of 4.67698312 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method (71) and are expressed as the number of base substitutions per site. The ME tree was searched using the close-neighbor interchange (CNI) algorithm (72) at a search level of 1. The neighbor-joining algorithm (73) was used to generate the initial tree. The analysis involved 70 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 305 positions in the final data set. Evolutionary analyses were conducted in MEGA7.0.18 (74).
Bacteria and growth conditions.
Escherichia coli strains XL1-Blue(MRF′) and BL21(DE3) were used for plasmid construction and for overproduction of His6-tagged proteins, respectively. The E. coli strains were grown in LB medium at 37°C with shaking. When required, the LB medium was complemented with 15 g/liter agar-agar, 100 μg/ml ampicillin (Amp), 25 μg/ml kanamycin (Kan), and 15 μg/ml tetracycline (Tet).
Parasites and growth conditions.
L. major promastigotes (strain MHOM/IL/81/FEBNI) (45) were cultivated at 28°C and 5% CO2-95% humidified air in modified complete Schneider′s medium, which was prepared by supplementation of Schneider′s Drosophila insect cell medium (Genaxxon Bioscience; pH adjusted to 6.9 after addition of 4.76 mM NaHCO3, 5.4 mM CaCl2 · 2H2O, 50 mM NaOH, and 35 mM HCl) with 100 U/ml penicillin G, 100 μg/ml streptomycin, 1 mM sodium pyruvate, 2 mM l-glutamine, 0.27 mM l-asparagine, 0.55 mM l-arginine, 10 mM HEPES, 2% (vol/vol) normal human urine, and 10% (vol/vol) heat-inactivated fetal calf serum (FCS) in a modification of a protocol previously published by Lima et al. (75). When required, Geneticin (G418) was added to the culture medium at a concentration of 30 μg/ml. L. major amastigotes were isolated from skin lesions of the footpads of BALB/c mice at day 30 of subcutaneous infection (76). Animal care and experiments were conducted in accordance with German regulations after local governmental approval (Ansbach, Germany).
Cloning of expression vectors.
The genes LmjF.16.0230 and LmjF.16.0250 were amplified by PCR with Phusion Hot Start II DNA Polymerase (Thermo Fisher Scientific) using genomic DNA of L. major (strain MHOM/IL/81/FEBNI) and specific primers to which appropriate restriction sites had been added (see Table S2 in the supplemental material). The PCR product of LmjF.16.0230 was inserted into the pET15b vector for affinity purification of the His6-tagged proteins. For ectopic expression in L. major, the LmjF.16.0230 and LmjF.16.0250 genes were inserted into the pCLN-3xHA vector, where the expression of the gene of interest was under the control of intergenic regions of cbp2 genes of Leishmania mexicana on the 5′ end and lpg1 of Leishmania infantum on the 3′ end (77). A list of the plasmids used in this study is presented in Table S1. The nucleotide sequences of all the synthesized and mutated genes were checked to ensure error-free amplification with the T7 or pCLN-3xHA primer pairs for the related plasmids (see Table S2).
Site-directed mutagenesis.
Site-directed mutagenesis was carried out following a PCR-based amplification protocol as previously described (78). Briefly, pairs of complementary primers carrying the desired mutation were used to amplify whole pET15b- or pCLN-3xHA-LmPRL plasmids (see Table S2 in the supplemental material). Plasmids used as the matrix carrying the wild-type gene were then digested with DpnI for 15 min at 37°C, followed by incubation for 5 min at 80°C to inactivate the DpnI. Mutated plasmids (see Table S1) were transformed into competent E. coli XL1-Blue(MRF′) bacteria. For each mutation reaction, plasmids from clones were prepared and sequenced. The success rate of this method approached 66%.
Overexpression and purification of proteins.
For each overexpression, competent E. coli BL21(DE3) bacteria were freshly transformed with plasmids encoding His6-PRL-1 (79), His6-LmPRL-1 WT, or its related mutants. The bacteria were grown overnight in LB supplemented with Amp and Tet and reinoculated at 1:100 in 500 ml LB with Amp and Tet the following day. At an optical density at 600 nm (OD600) of 0.5, protein overexpression was induced for 3 h by addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The bacteria were centrifuged for 15 min at 8,000 rpm and washed once with purification buffer. Purification with Ni-NTA beads (Qiagen) was performed according to the manufacturer's protocol. Purified fractions were collected and dialyzed overnight against a buffer containing 20 mM Tris-HCl, pH 7.5, 500 mM NaCl, 20% glycerol, 5 mM β-mercaptoethanol and placed at 4°C or −20°C for longer storage (80).
Protein migration assay.
Purified His6-LmPRL was incubated for 1 h with 10 mM either reducing or oxidized DTT (53) and then separated on nonreducing SDS-PAGE. Proteins were detected by Coomassie brilliant blue staining.
Phosphatase assays.
Phosphatase activity against pNPP was measured by incubating 0.2 mg/ml of His6-LmPRL-1 with 10 mM pNPP for 1 h at 37°C in a buffer (pH 6.0) consisting of 100 mM sodium citrate and 1 mM EDTA. The analyzed proteins were previously incubated or not for 1 h at 4°C with 10 mM reducing DTT. The appearance of yellow para-nitrophenol (pNP) was measured on a spectrophotometer (λ = 405 nm) after the reaction was arrested by the addition of 1/3 reaction volume of 4 M NaOH (81). Phosphatase activity against DiFMUP was measured by incubating either 67 μg/mL (activity of the mutant) or 25 μg/ml (optimal pH and kinetic constant determination) of His6-LmPRL-1 with 200 μM DiFMUP (LifeTechnologies) for 20 min at 37°C (50). The appearance of fluorogenic DiFMU was measured on a spectrofluorometer (Fluoroskan Ascent FL; Thermo Fisher Scientific) with the appropriate filter set (excitation [ex]/emission [em], 355 nm/460 nm). For determination of the optimal pH of the enzyme, buffer containing 100 mM sodium citrate and 1 mM EDTA was used at a pH range from 4.0 to 6.5, whereas buffer containing 50 mM HEPES and 1 mM EDTA was used for pH values between 7.0 and 9.0.
Ectopic expression of HA3-LmPRL in L. major.
In order to ectopically express HA3-LmPRL, L. major promastigotes were electroporated with pCLN-3xHA-LmPRL vectors following a previously described protocol (77). Briefly, endotoxin-free plasmid DNA was prepared using a NucleoBond Xtra Midi EF plasmid DNA purification kit (Macherey-Nagel). Fifty micrograms of DNA was precipitated with ethanol and rehydrated with 40 μl of sterile electroporation buffer, pH 7.05 (21 mM HEPES, 0.7 mM Na2HPO4, 5 mM KCl, 137 mM NaCl, and 6 mM glucose). Promastigote parasites reaching late log phase in modified complete Schneider's medium were washed with cold phosphate-buffered saline (PBS) and electroporation buffer before being resuspended in electroporation buffer at a density of 1.0 × 108 cells/ml. For each electroporation, 0.4 ml of parasite suspension and the dissolved DNA were mixed in a 4-mm-wide, prechilled, sterile electroporation cuvette. Mock electroporation of the parasites without plasmid was carried out in parallel. Electroporation of the cells was done using a Bio-Rad Gene Pulser with 3 pulses at 3,750 V/cm, 25 μF, and 200 Ω. After electroporation, the cells were kept on ice for 10 min and then transferred into 10 ml of complete Schneider′s medium. After 24 h, G418 was added, and selection of transfected parasites proceeded until the mock-transfected cells died.
Culture and in vitro infection of BMM.
BMM were generated as previously described (82). Briefly, the femur and tibia of a female C57BL/6N mouse were removed and flushed with Dulbecco's modified Eagle's medium (DMEM), and 6.0 × 106 nucleated bone marrow cells were grown in 50 ml conditioned DMEM (supplemented with penicillin-streptomycin, 10% [vol/vol] FCS, 50 μM 2-mercaptoethanol, 1% [vol/vol] 100× nonessential amino acids, and 15% [vol/vol] SN from L929 fibroblast cultures [ATCC clone CCL-1] as a source of macrophage colony-stimulating factor) in Teflon bags for 8 days at 37°C and 10% CO2-90% humidified air. For infection, BMM were seeded in RPMI 1640 (ThermoFisher Scientific no. 21875, supplemented with 10 mM HEPES, 50 μM 2-mercaptoethanol, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% [vol/vol] heat-inactivated FCS) either on glass coverslips in 24-well plates (4.0 × 105 cells/well) or in 6-well plates (3.0 × 106 cells/well). After adherence, the cells were infected with stationary-phase L. major WT or HA3-Ctrl or HA3-LmPRL promastigotes at an MOI of 5. After 16 h, extracellular parasites were removed by three washes with warm PBS. Samples for Diff-Quick staining were taken at 16 h, 48 h, and 72 h after infection. The infection rate (percentage of cells infected) and the number of parasites per infected cell were determined microscopically (100× oil objective; 300 cells in total per condition).
Production of rabbit anti-LmPRL-1 peptide antiserum and immunoblot analysis.
To allow specific detection of LmPRL-1, rabbits were immunized with the peptide CQ156MHWITKYKRRHQG169-amide (Biogenes, Berlin, Germany) coupled to Limulus polyphemus hemocyanin (LPH). The serum of the rabbit was then used for immunoblot detection at a dilution of 1:500. Protein samples were prepared from either 5.0 × 107 L. major parasites or 3.0 × 106 BMM, which were lysed in 200 μl or 60 μl of ice-cold Frackelton buffer (10 mM Tris, 50 mM NaCl, 30 mM sodium pyrophosphate, 1% Triton X-100, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride [PMSF], protease inhibitor cocktail broad range [Roth], and 10 mM phenanthroline for the parasite samples), respectively. In addition, Leishmania lysates were sonicated for 5 min (power, H; interval, 0.5) in a Bioruptor water bath (Diagenode). All the samples were then centrifuged at 14,000 × g for 10 min at 4°C. Protein quantities of the supernatants were determined by the Bradford method (Roti-Quant; Roth). Lysates were separated by SDS-15% PAGE and transferred to a nitrocellulose membrane (pore size, 0.2 μm) using a semidry blotter (Bio-Rad). To block nonspecific binding sites, the blotted membranes were placed in Tris-buffered saline (TBS) with 5% nonfat dry milk and 0.05% Tween 20 for at least 1 h. For detection of the target proteins, the blots were analyzed by enhanced chemiluminescence (RotiLumin; Roth) using different primary Abs and the respective horseradish peroxidase (HRP)-conjugated secondary Ab. Mouse anti-HA Ab (diluted 1:1,000; clone 16B12; Covance), rabbit anti-α-actin Ab (diluted 1:1,000; A2066; Sigma-Aldrich), mouse anti-α-tubulin Ab (diluted 1:1,000; clone B-5-2-1; Sigma-Aldrich), rat anti-LAMP-1 (diluted 1:1,000; clone 1D4B; DSHB), and chicken anti-Leishmania HSP90 and HSP70 sera (diluted 1:500) (83) were used as primary Abs. Anti-mouse IgG Ab (diluted 1:5,000; Roth), anti-rat IgG Ab (diluted 1:10,000; Jackson ImmunoResearch), anti-rabbit IgG Ab (diluted 1:2,500; Roth), and anti-chicken IgG Ab (diluted 1:10,000; Dianova) were used as HRP-conjugated secondary Abs. The intensities of the obtained protein bands were determined with Image J software (84).
CLSFM.
For L. major samples, promastigotes were grown to stationary phase before washing with PBS and subsequent fixation in 4% paraformaldehyde (PFA) (Alfa Aesar) for 20 min at room temperature (RT) in the dark. Fixed promastigotes were seeded on poly-l-lysine (Sigma-Aldrich)-coated glass coverslips (24-well plate; 2.5 × 105 to 5 × 105 cells/well), dried for 15 min, washed with PBS, and permeabilized with ice-cold methanol for 1 min. For BMM samples, the cells that were seeded on glass coverslips (24-well plate; 0.4 × 106 cells/well) were washed, fixed, and permeabilized in a similar way. All the samples were then washed for 15 min with 0.1% Tween 20 in PBS (PBST) and blocked for 30 min with 5% donkey serum (Dianova), 0.1% saponin in PBS. After three washing steps of 5 min each with PBST, the cells were incubated with a primary Ab diluted in 0.5% donkey serum in PBS for 1 to 2 h at RT, followed by another three washing steps and incubation with a related secondary Ab diluted in 0.5% donkey serum in PBS for 1 h at RT in the dark. After three final washings, the cells were mounted with Molecular Probes ProLong Gold Antifade Mountant (Life Technologies), containing DAPI (4′,6-diamidino-2-phenylindole) to stain DNA, on coverslips and cured overnight. Mouse anti-HA Ab (diluted 1:500; clone 16B12; Covance), rat anti-LAMP-1 (diluted 1:600; clone 1D4B; DSHB), and polyclonal human anti-Leishmania serum (diluted 1:1,000; derived from a patient with multilesional cutaneous leishmaniasis due to L. infantum infection [85]) were used as primary Abs. Goat anti-mouse IgG Ab coupled to Alexa 488 (diluted 1:400; Dianova), goat anti-mouse IgG Ab coupled to Alexa 594 (diluted 1:1,600; Dianova), donkey anti-rat IgG Ab coupled to Cy5 (diluted 1:400; Dianova), goat-anti-human IgG coupled to Alexa 488 (diluted 1:1,600; Jackson ImmunoResearch), and goat-anti-human IgG coupled to Alexa 647 (diluted 1:100; Jackson ImmunoResearch) were used as secondary Abs. The slides were analyzed with a confocal microscope (LSM 700; Zeiss) using a 405-nm, 488-nm, 555-nm, or 639-nm laser line. Processing of the images was done with ZEN software 2009 (Zeiss).
Purification of exosomes generated by L. major promastigotes.
Exosome isolation was performed as described previously (61) with minor modification. Briefly, L. major promastigotes were grown to stationary phase (5.0 × 107 cells/ml) in 150 ml modified complete Schneider′s medium supplemented with 30 μg/ml G418 for pCLN-3xHA-containing strains. Twenty-four hours before exosome isolation, the parasites were washed with PBS and kept in RPMI 1640 supplemented with penicillin and streptomycin but without G418 and FCS for 24 h (to avoid contamination by serum proteins). The parasite cultures were then centrifuged for 10 min at 1,000 × g. The supernatants were centrifuged at 1,000 × g for 30 min, followed by another centrifugation at 15,000 × g for 45 min to remove dead parasites and debris. Finally, the supernatants were ultracentrifuged at 110,000 × g for 1 h to pellet the exosomes. All centrifugation steps were done at 4°C. The exosomes were finally resuspended in 50 to 100 μl PBS and subjected to trypsin digestion (50 μg/ml) in the presence of 0.3% Triton X-100 (preincubation for 5 min on ice) at 37°C for 30 min. Samples were then analyzed by immunoblotting.
Electron microscopy.
For electron microscopy, exosomes collected from 50 ml of parasite culture grown under conditions similar to those for exosome preparation were directly resuspended in 2.5% glutaraldehyde in cacodylate buffer (0.1 M, pH 7.3) containing 0.1 M glucose and stored at 4°C until further sample preparation. For negative-contrast staining, Formvar-filmed, carbon-coated, 400-mesh copper grids were prepared and hydrophilized by glow discharge in a high-vacuum coating system (Med 020; Bal-Tec AG, Liechtenstein) immediately before use. Grids were floated with the coated side on a 30-μl drop of the exosome suspension for 10 min, placed on a drop of 10% aqueous uranyl acetate for contrast for 10 min, and briefly dried on filter paper. They were viewed in a FEI Tecnai 12 transmission electron microscopy (TEM) at 80 kV and imaged using electron image film (SO-163; Carestream, USA). The developed film plates were digitized on a scanner (Epson Perfection V700 photo scanner) and processed using Adobe Photoshop CS3.
Subcellular fractionation.
BMM were infected at an MOI of 5 with L. major promastigotes. Extracellular parasites were washed away with PBS (2×) after 16 h. After 72 h, the cells were washed with Hanks' balanced salt solution. The cells were treated with cold hypotonic buffer (20 mM Tris, pH 7.5) containing protease inhibitors and passed 50 times through a 22-gauge needle to disrupt their cytosolic membranes. The nucleus and large organelles were removed by centrifugation at 3,000 × g (P3), and the supernatants were supplemented with NaCl to a final concentration of 0.15 M. The cytosolic fractions were prepared further by centrifuging the supernatants at 100,000 × g for 1 h at 4°C. The supernatants were considered cytosolic fractions (S100), whereas the pellets represented the small organelles and the cytoplasmic membrane (P100).
Statistical analysis.
Data sets were analyzed using the unpaired, two-tailed Student t test. Significance was evaluated by measuring the P value.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Lisa Wirker for preparation of samples and Wolfram Maginot for photographic support for TEM. We also thank Anja Lührmann (UK Erlangen, Germany) for scientific advice and reagents and Jennifer S. Laurence (University of Kansas, USA) for providing reagents.
This study was supported by the German Research Foundation (DFG) (grant SO 1149/1-1 to D.S.; grant CRC1181, project C05, to C.B.), by the Interdisciplinary Center for Clinical Research (IZKF) of the Medical Faculty of the FAU Erlangen-Nürnberg (project grants A61 and A63 to C.B.), and by the Robert Pfleger Stiftung Bamberg (project grant to C.B.). D.S. acknowledges his position as an associated researcher within the CRC796.
D.S. conceived and coordinated the study. S.L., E.M.L.-T., and D.S. designed and performed experiments and analyzed data. J.C. provided vectors and know-how for generating the transgenic Leishmania strains. C.B. and U.S. provided technical expertise on the macrophage infection experiments and contributed significantly to the conception and design of the study. D.S., S.L., and C.B. wrote the manuscript.
We declare that we have no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00084-17.
REFERENCES
- 1.Akhoundi M, Kuhls K, Cannet A, Votýpka J, Marty P, Delaunay P, Sereno D. 2016. A historical overview of the classification, evolution, and dispersion of Leishmania parasites and sandflies. PLoS Negl Trop Dis 10:e0004349. doi: 10.1371/journal.pntd.0004349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaye P, Scott P. 2011. Leishmaniasis: complexity at the host-pathogen interface. Nat Rev Microbiol 9:604–615. doi: 10.1038/nrmicro2608. [DOI] [PubMed] [Google Scholar]
- 3.Murray HW, Berman JD, Davies CR, Saravia NG. 2005. Advances in leishmaniasis. Lancet 366:1561–1577. doi: 10.1016/S0140-6736(05)67629-5. [DOI] [PubMed] [Google Scholar]
- 4.Bogdan C. 2012. Leishmaniasis in rheumatology, haematology and oncology: epidemiological, immunological and clinical aspects and caveats. Ann Rheum Dis 71(Suppl 2):i60–i66. doi: 10.1136/annrheumdis-2011-200596. [DOI] [PubMed] [Google Scholar]
- 5.Savoia D. 2015. Recent updates and perspectives on leishmaniasis. J Infect Dev Ctries 9:588–596. doi: 10.3855/jidc.6833. [DOI] [PubMed] [Google Scholar]
- 6.Depledge DP, Evans KJ, Ivens AC, Aziz N, Maroof A, Kaye PM, Smith DF. 2009. Comparative expression profiling of Leishmania: modulation in gene expression between species and in different host genetic backgrounds. PLoS Negl Trop Dis 3:e476. doi: 10.1371/journal.pntd.0000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva-Almeida M, Souza-Silva F, Pereira BAS, Ribeiro-Guimarães ML, Alves CR. 2014. Overview of the organization of protease genes in the genome of Leishmania spp. Parasit Vectors 7:387. doi: 10.1186/1756-3305-7-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCall L-I, Matlashewski G. 2010. Localization and induction of the A2 virulence factor in Leishmania: evidence that A2 is a stress response protein. Mol Microbiol 77:518–530. doi: 10.1111/j.1365-2958.2010.07229.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Y, Davis A, Smith BJ, Curtis J, Handman E. 2009. Leishmania major CorA-like magnesium transporters play a critical role in parasite development and virulence. Int J Parasitol 39:713–723. doi: 10.1016/j.ijpara.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 10.Morales MA, Pescher P, Späth GF. 2010. Leishmania major MPK7 protein kinase activity inhibits intracellular growth of the pathogenic amastigote stage. Eukaryot Cell 9:22–30. doi: 10.1128/EC.00196-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norris-Mullins B, VanderKolk K, Vacchina P, Joyce MV, Morales MA. 2014. LmaPA2G4, a homolog of human Ebp1, is an essential gene and inhibits cell proliferation in L. major. PLoS Negl Trop Dis 8:e2646. doi: 10.1371/journal.pntd.0002646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogdan C, Röllinghoff M. 1998. The immune response to Leishmania: mechanisms of parasite control and evasion. Int J Parasitol 28:121–134. doi: 10.1016/S0020-7519(97)00169-0. [DOI] [PubMed] [Google Scholar]
- 13.Bogdan C. 2008. Mechanisms and consequences of persistence of intracellular pathogens: leishmaniasis as an example. Cell Microbiol 10:1221–1234. doi: 10.1111/j.1462-5822.2008.01146.x. [DOI] [PubMed] [Google Scholar]
- 14.Shio MT, Hassani K, Isnard A, Ralph B, Contreras I, Gomez MA, Abu-Dayyeh I, Olivier M. 2012. Host cell signalling and Leishmania mechanisms of evasion. J Trop Med 2012:1–14. doi: 10.1155/2012/819512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambertz U, Silverman JM, Nandan D, McMaster WR, Clos J, Foster LJ, Reiner NE. 2012. Secreted virulence factors and immune evasion in visceral leishmaniasis. J Leukoc Biol 91:887–899. doi: 10.1189/jlb.0611326. [DOI] [PubMed] [Google Scholar]
- 16.Atayde VD, Hassani K, da Silva Lira Filho A, Borges AR, Adhikari A, Martel C, Olivier M. 2016. Leishmania exosomes and other virulence factors: impact on innate immune response and macrophage functions. Cell Immunol 309:7–18. doi: 10.1016/j.cellimm.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 17.Richardson JM, Morrison LS, Bland ND, Bruce S, Coombs GH, Mottram JC, Walkinshaw MD. 2009. Structures of Leishmania major orthologues of macrophage migration inhibitory factor. Biochem Biophys Res Commun 380:442–448. doi: 10.1016/j.bbrc.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holowka T, Castilho TM, Garcia AB, Sun T, McMahon-Pratt D, Bucala R. 2016. Leishmania-encoded orthologs of macrophage migration inhibitory factor regulate host immunity to promote parasite persistence. FASEB J 30:2249–2265. doi: 10.1096/fj.201500189R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Späth GF, Epstein L, Leader B, Singer SM, Avila HA, Turco SJ, Beverley SM. 2000. Lipophosphoglycan is a virulence factor distinct from related glycoconjugates in the protozoan parasite Leishmania major. Proc Natl Acad Sci U S A 97:9258–9263. doi: 10.1073/pnas.160257897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madeira da Silva L, Owens KL, Murta SMF, Beverley SM. 2009. Regulated expression of the Leishmania major surface virulence factor lipophosphoglycan using conditionally destabilized fusion proteins. Proc Natl Acad Sci U S A 106:7583–7588. doi: 10.1073/pnas.0901698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desjardins M, Descoteaux A. 1997. Inhibition of phagolysosomal biogenesis by the Leishmania lipophosphoglycan. J Exp Med 185:2061–2068. doi: 10.1084/jem.185.12.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinet AF, Fukuda M, Turco SJ, Descoteaux A. 2009. The Leishmania donovani lipophosphoglycan excludes the vesicular proton-ATPase from phagosomes by impairing the recruitment of synaptotagmin V. PLoS Pathog 5:e1000628. doi: 10.1371/journal.ppat.1000628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olivier M, Atayde VD, Isnard A, Hassani K, Shio MT. 2012. Leishmania virulence factors: focus on the metalloprotease GP63. Microbes Infect 14:1377–1389. doi: 10.1016/j.micinf.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 24.Nandan D, Yi T, Lopez M, Lai C, Reiner NE. 2002. Leishmania EF-1alpha activates the Src homology 2 domain containing tyrosine phosphatase SHP-1 leading to macrophage deactivation. J Biol Chem 277:50190–50197. doi: 10.1074/jbc.M209210200. [DOI] [PubMed] [Google Scholar]
- 25.Blanchette J, Racette N, Faure R, Siminovitch KA, Olivier M. 1999. Leishmania-induced increases in activation of macrophage SHP-1 tyrosine phosphatase are associated with impaired IFN-gamma-triggered JAK2 activation. Eur J Immunol 29:3737–3744. doi:. [DOI] [PubMed] [Google Scholar]
- 26.Gomez MA, Contreras I, Hallé M, Tremblay ML, McMaster RW, Olivier M. 2009. Leishmania GP63 alters host signaling through cleavage-activated protein tyrosine phosphatases. Sci Signal 2:ra58–ra58. doi: 10.1126/scisignal.2000213. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharyya S, Ghosh S, Sen P, Roy S, Majumdar S. 2001. Selective impairment of protein kinase C isotypes in murine macrophage by Leishmania donovani. Mol Cell Biochem 216:47–57. doi: 10.1023/A:1011048210691. [DOI] [PubMed] [Google Scholar]
- 28.Pingel S, Wang ZE, Locksley RM. 1998. Distribution of protein kinase C isoforms after infection of macrophages with Leishmania major. Infect Immun 66:1795–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nandan D, Lo R, Reiner NE. 1999. Activation of phosphotyrosine phosphatase activity attenuates mitogen-activated protein kinase signaling and inhibits c-FOS and nitric oxide synthase expression in macrophages infected with Leishmania donovani. Infect Immun 67:4055–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soares-Silva M, Diniz FF, Gomes GN, Bahia D. 2016. The mitogen-activated protein kinase (MAPK) pathway: role in immune evasion by trypanosomatids. Front Microbiol 7:183. doi: 10.3389/fmicb.2016.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nandan D, Reiner NE. 1995. Attenuation of gamma interferon-induced tyrosine phosphorylation in mononuclear phagocytes infected with Leishmania donovani: selective inhibition of signaling through Janus kinases and Stat1. Infect Immun 63:4495–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shio MT, Olivier M. 2010. Leishmania survival mechanisms: the role of host phosphatases. J Leukoc Biol 88:1–3. doi: 10.1189/jlb.0210088. [DOI] [PubMed] [Google Scholar]
- 33.Szöör B. 2010. Trypanosomatid protein phosphatases. Mol Biochem Parasitol 173:53–63. doi: 10.1016/j.molbiopara.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gottlieb M, Dwyer DM. 1981. Protozoan parasite of humans: surface membrane with externally disposed acid phosphatase. Science 212:939–941. doi: 10.1126/science.7233189. [DOI] [PubMed] [Google Scholar]
- 35.Remaley AT, Das S, Campbell PI, LaRocca GM, Pope MT, Glew RH. 1985. Characterization of Leishmania donovani acid phosphatases. J Biol Chem 260:880–886. [PubMed] [Google Scholar]
- 36.Shakarian AM, Joshi MB, Yamage M, Ellis SL, Debrabant A, Dwyer DM. 2003. Members of a unique histidine acid phosphatase family are conserved amongst a group of primitive eukaryotic human pathogens. Mol Cell Biochem 245:31–41. doi: 10.1023/A:1022851914014. [DOI] [PubMed] [Google Scholar]
- 37.Remaley AT, Kuhns DB, Basford RE, Glew RH, Kaplan SS. 1984. Leishmanial phosphatase blocks neutrophil O-2 production. J Biol Chem 259:11173–11175. [PubMed] [Google Scholar]
- 38.Papadaki A, Politou AS, Smirlis D, Kotini MP, Kourou K, Papamarcaki T, Boleti H. 2015. The Leishmania donovani histidine acid ecto-phosphatase LdMAcP: insight into its structure and function. Biochem J 467:473–486. doi: 10.1042/BJ20141371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandes ACS, Soares DC, Saraiva EM, Meyer-Fernandes JR, Souto-Padrón T. 2013. Different secreted phosphatase activities in Leishmania amazonensis. FEMS Microbiol Lett 340:117–128. doi: 10.1111/1574-6968.12080. [DOI] [PubMed] [Google Scholar]
- 40.Aoun K, Bouratbine A. 2014. Cutaneous leishmaniasis in North Africa: a review. Parasite 21:14. doi: 10.1051/parasite/2014014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, den Boer M, WHO Leishmaniasis Control Team. 2012. Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rios P, Li X, Köhn M. 2013. Molecular mechanisms of the PRL phosphatases. FEBS J 280:505–524. doi: 10.1111/j.1742-4658.2012.08565.x. [DOI] [PubMed] [Google Scholar]
- 43.Silverman JM, Clos J, de'Oliveira CC, Shirvani O, Fang Y, Wang C, Foster LJ, Reiner NE. 2010. An exosome-based secretion pathway is responsible for protein export from Leishmania and communication with macrophages. J Cell Sci 123:842–852. doi: 10.1242/jcs.056465. [DOI] [PubMed] [Google Scholar]
- 44.Hassani K, Shio MT, Martel C, Faubert D, Olivier M. 2014. Absence of metalloprotease GP63 alters the protein content of Leishmania exosomes. PLoS One 9:e95007. doi: 10.1371/journal.pone.0095007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solbach W, Forberg K, Röllinghoff M. 1986. Effect of T-lymphocyte suppression on the parasite burden in Leishmania major-infected, genetically susceptible BALB/c mice. Infect Immun 54:909–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream M-A, Adlem E, Aert R, Anupama A, Apostolou Z, Attipoe P, Bason N, Bauser C, Beck A, Beverley SM, Bianchettin G, Borzym K, Bothe G, Bruschi CV, Collins M, Cadag E, Ciarloni L, Clayton C, Coulson RMR, Cronin A, Cruz AK, Davies RM, De Gaudenzi J, Dobson DE, Duesterhoeft A, Fazelina G, Fosker N, Frasch AC, Fraser A, Fuchs M, Gabel C, Goble A, Goffeau A, Harris D, Hertz-Fowler C, Hilbert H, Horn D, Huang Y, Klages S, Knights A, Kube M, Larke N, Litvin L, Lord A, Louie T, Marra M, Masuy D, Matthews K, Michaeli S, Mottram JC, Müller-Auer S, Munden H, Nelson S, Norbertczak H, Oliver K, O'Neil S, Pentony M, Pohl TM, Price C, Purnelle B, Quail MA, Rabbinowitsch E, Reinhardt R, Rieger M, Rinta J, Robben J, Robertson L, Ruiz JC, Rutter S, Saunders D, Schäfer M, Schein J, Schwartz DC, Seeger K, Seyler A, Sharp S, Shin H, Sivam D, Squares R, Squares S, Tosato V, Vogt C, Volckaert G, Wambutt R, Warren T, Wedler H, Woodward J, Zhou S, Zimmermann W, Smith DF, Blackwell JM, Stuart KD, Barrell B, Myler PJ. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cuevas IC, Rohloff P, Sanchez DO, Docampo R. 2005. Characterization of farnesylated protein tyrosine phosphatase TcPRL-1 from Trypanosoma cruzi. Eukaryot Cell 4:1550–1561. doi: 10.1128/EC.4.9.1550-1561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D. 2011. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res 21:915–924. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeong D, Kim S, Kim J, Son J, Park M, Lim S, Yoon T-S, Ryu S. 2005. Trimeric structure of PRL-1 phosphatase reveals an active enzyme conformation and regulation mechanisms. J Mol Biol 345:401–413. doi: 10.1016/j.jmb.2004.10.061. [DOI] [PubMed] [Google Scholar]
- 50.Sun J-P, Wang W-Q, Yang H, Liu S, Liang F, Fedorov AA, Almo SC, Zhang Z-Y. 2005. Structure and biochemical properties of PRL-1, a phosphatase implicated in cell growth, differentiation, and tumor invasion. Biochemistry 44:12009–12021. doi: 10.1021/bi0509191. [DOI] [PubMed] [Google Scholar]
- 51.Ishii T, Funato Y, Miki H. 2013. Thioredoxin-related protein 32 (TRP32) specifically reduces oxidized phosphatase of regenerating liver (PRL). J Biol Chem 288:7263–7270. doi: 10.1074/jbc.M112.418004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun J-P, Luo Y, Yu X, Wang W-Q, Zhou B, Liang F, Zhang Z-Y. 2007. Phosphatase activity, trimerization, and the C-terminal polybasic region are all required for PRL1-mediated cell growth and migration. J Biol Chem 282:29043–29051. doi: 10.1074/jbc.M703537200. [DOI] [PubMed] [Google Scholar]
- 53.Skinner A, Vartia A, Williams T, Laurence J. 2009. Enzyme activity of phosphatase of regenerating liver is controlled by the redox environment and its C-terminal residues. Biochemistry 48:4262–4272. doi: 10.1021/bi900241k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silverman JM, Reiner NE. 2011. Leishmania exosomes deliver preemptive strikes to create an environment permissive for early infection. Front Cell Infect Microbiol 1:26. doi: 10.3389/fcimb.2011.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McConville MJ, Mullin KA, Ilgoutz SC, Teasdale RD. 2002. Secretory pathway of trypanosomatid parasites. Microbiol Mol Biol Rev 66:122–154. doi: 10.1128/MMBR.66.1.122-154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silverman JM, Chan SK, Robinson DP, Dwyer DM, Nandan D, Foster LJ, Reiner NE. 2008. Proteomic analysis of the secretome of Leishmania donovani. Genome Biol 9:R35. doi: 10.1186/gb-2008-9-2-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson KA, Beverley SM. 2003. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol Biochem Parasitol 128:217–228. doi: 10.1016/S0166-6851(03)00079-3. [DOI] [PubMed] [Google Scholar]
- 58.Kolev NG, Tschudi C, Ullu E. 2011. RNA interference in protozoan parasites: achievements and challenges. Eukaryot Cell 10:1156–1163. doi: 10.1128/EC.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dillon LAL, Okrah K, Hughitt VK, Suresh R, Li Y, Fernandes MC, Belew AT, Corrada Bravo H, Mosser DM, El-Sayed NM. 2015. Transcriptomic profiling of gene expression and RNA processing during Leishmania major differentiation. Nucleic Acids Res 43:6799–6813. doi: 10.1093/nar/gkv656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dillon LAL, Suresh R, Okrah K, Corrada Bravo H, Mosser DM, El-Sayed NM. 2015. Simultaneous transcriptional profiling of Leishmania major and its murine macrophage host cell reveals insights into host-pathogen interactions. BMC Genomics 16:1108. doi: 10.1186/s12864-015-2237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silverman JM, Clos J, Horakova E, Wang AY, Wiesgigl M, Kelly I, Lynn MA, McMaster WR, Foster LJ, Levings MK, Reiner NE. 2010. Leishmania exosomes modulate innate and adaptive immune responses through effects on monocytes and dendritic cells. J Immunol 185:5011–5022. doi: 10.4049/jimmunol.1000541. [DOI] [PubMed] [Google Scholar]
- 62.Atayde VD, Aslan H, Townsend S, Hassani K, Kamhawi S, Olivier M. 2015. Exosome secretion by the parasitic protozoan Leishmania within the sand fly midgut. Cell Rep 13:957–967. doi: 10.1016/j.celrep.2015.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fritz M, Vanselow J, Sauer N, Lamer S, Goos C, Siegel TN, Subota I, Schlosser A, Carrington M, Kramer S. 2015. Novel insights into RNP granules by employing the trypanosome's microtubule skeleton as a molecular sieve. Nucleic Acids Res 43:8013–8032. doi: 10.1093/nar/gkv731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Kirby C, Herbst R. 2002. The tyrosine phosphatase PRL-1 localizes to the endoplasmic reticulum and the mitotic spindle and is required for normal mitosis. J Biol Chem 277:46659–46668. doi: 10.1074/jbc.M206407200. [DOI] [PubMed] [Google Scholar]
- 65.Reiling L, Chrobak M, Schmetz C, Clos J. 2010. Overexpression of a single Leishmania major gene enhances parasite infectivity in vivo and in vitro. Mol Microbiol 76:1175–1190. doi: 10.1111/j.1365-2958.2010.07130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bifeld E, Chrobak M, Zander D, Schleicher U, Schönian G, Clos J. 2015. Geographical sequence variation in the Leishmania major virulence factor P46. Infect Genet Evol 30:195–205. doi: 10.1016/j.meegid.2014.12.029. [DOI] [PubMed] [Google Scholar]
- 67.Liévin-Le Moal V, Loiseau PM. 2016. Leishmania hijacking of the macrophage intracellular compartments. FEBS J 283:598–607. doi: 10.1111/febs.13601. [DOI] [PubMed] [Google Scholar]
- 68.Zeng Q, Dong JM, Guo K, Li J, Tan H-X, Koh V, Pallen CJ, Manser E, Hong W. 2003. PRL-3 and PRL-1 promote cell migration, invasion, and metastasis. Cancer Res 63:2716–2722. [PubMed] [Google Scholar]
- 69.Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, Gardner MJ, Gingle A, Grant G, Harb OS, Heiges M, Hertz-Fowler C, Houston R, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Logan FJ, Miller JA, Mitra S, Myler PJ, Nayak V, Pennington C, Phan I, Pinney DF, Ramasamy G, Rogers MB, Roos DS, Ross C, Sivam D, Smith DF, Srinivasamoorthy G, Stoeckert CJ, Subramanian S, Thibodeau R, Tivey A, Treatman C, Velarde G, Wang H. 2010. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 38:D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rzhetsky A, Nei M. 1992. A simple method for evaluating and testing minimum-evolution trees. Mol Biol Evol 9:945–967. [Google Scholar]
- 71.Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 73.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 74.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lima HC, Bleyenberg JA, Titus RG. 1997. A simple method for quantifying Leishmania in tissues of infected animals. Parasitol Today 13:80–82. doi: 10.1016/S0169-4758(96)40010-2. [DOI] [PubMed] [Google Scholar]
- 76.Bogdan C, Streck H, Röllinghoff M, Solbach W. 1989. Cyclosporin A enhances elimination of intracellular L. major parasites by murine macrophages. Clin Exp Immunol 75:141–146. [PMC free article] [PubMed] [Google Scholar]
- 77.Hombach A, Ommen G, Chrobak M, Clos J. 2013. The Hsp90-Sti1 interaction is critical for Leishmania donovani proliferation in both life cycle stages. Cell Microbiol 15:585–600. doi: 10.1111/cmi.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng L, Baumann U, Reymond J-L. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Laurence JS, Hallenga K, Stauffacher CV. 2004. 1H, 15N, 13C resonance assignments of the human protein tyrosine phosphatase PRL-1. J Biomol NMR 29:417–418. doi: 10.1023/B:JNMR.0000032506.16792.c6. [DOI] [PubMed] [Google Scholar]
- 80.Soulat D, Jault J-M, Duclos B, Geourjon C, Cozzone AJ, Grangeasse C. 2006. Staphylococcus aureus operates protein-tyrosine phosphorylation through a specific mechanism. J Biol Chem 281:14048–14056. doi: 10.1074/jbc.M513600200. [DOI] [PubMed] [Google Scholar]
- 81.Soulat D, Vaganay E, Duclos B, Genestier A-L, Etienne J, Cozzone AJ. 2002. Staphylococcus aureus contains two low-molecular-mass phosphotyrosine protein phosphatases. J Bacteriol 184:5194–5199. doi: 10.1128/JB.184.18.5194-5199.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schleicher U, Bogdan C. 2009. Generation, culture and flow-cytometric characterization of primary mouse macrophages. Methods Mol Biol 531:203–224. doi: 10.1007/978-1-59745-396-7_14. [DOI] [PubMed] [Google Scholar]
- 83.Brandau S, Dresel A, Clos J. 1995. High constitutive levels of heat-shock proteins in human-pathogenic parasites of the genus Leishmania. Biochem J 310:225–232. doi: 10.1042/bj3100225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bogdan C, Stosiek N, Fuchs H, Röllinghoff M, Solbach W. 1990. Detection of potentially diagnostic leishmanial antigens by Western blot analysis of sera from patients with kala-azar or multilesional cutaneous leishmaniasis. J Infect Dis 162:1417–1418. doi: 10.1093/infdis/162.6.1417. [DOI] [PubMed] [Google Scholar]
- 86.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.