A key discovery in the field of programmed cell death (PCD) is the realization that apoptosis, the most abundant form of PCD in metazoa, is executed by an evolutionary conserved family of proteases called caspases (for cysteine aspartic acid-specific proteases).1 Since then, the combined efforts of many research groups have led to a deep understanding of how caspases are activated and regulated during apoptosis, and a conserved model of the biochemical and morphological features of apoptosis has materialized (Figure 1). However, a constantly growing body of research, mainly performed in the past decade and a half, indicates that caspase activation may not necessarily lead to cell death. This special issue of CDD brings a comprehensive collection of reviews highlighting diverse non-lethal functions of caspases in a variety of organisms, cell types and cellular processes, such as signaling, proliferation, differentiation, remodeling and neuronal plasticity. Furthermore, this issue also features intriguing tangentially related topics, concerning viral hijacking of host caspases, biochemical and biological functions of metacaspases, the possible origin of caspases, non-lethal functions of BCL-2 family proteins and alternative (mostly developmental) caspase-independent cell death pathways in metazoa. Collectively, we hope that this collection of articles will attract the attention of many scientists to this growing research area, for which we suggest the term CDP, for caspase-dependent, non-lethal cellular process. The plethora of reports and constant discoveries of new CDPs call for creating a sense of community, among which exciting new findings and future directions could be discussed and communicated.
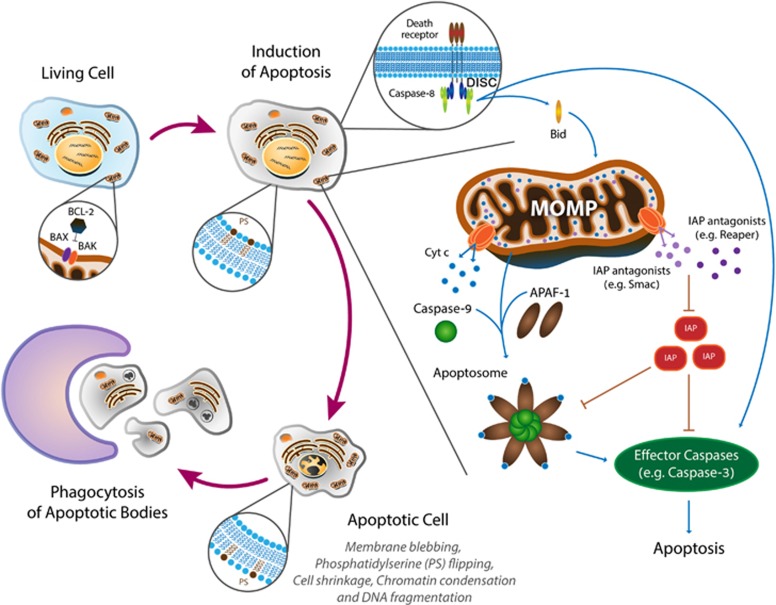
Figure 1.
Schematic diagram of the conserved biochemical machinery and the corresponding morphological changes during apoptosis
The idea that apoptosis is largely regulated at the post-translational level leads to an ‘all-or-nothing’ model of caspase activation (Figure 1). This model has been initially characterized in model organisms, such as Caenorhabditiselegans and Drosophila, consequently creating a genetic framework for subsequent biochemical studies in mammalian cell-free and cell culture systems, which filled in the (physical) interaction gaps (Figure 1). In brief, apical (initiator) caspases, such as caspase-8 and -9, are activated in large multimeric protein complexes called the death-inducing signaling complex (DISC) and the apoptosome, respectively.2 Once activated, the initiator caspases proteolytically cleave and activate the executioner (effector) caspases, such as caspases-3 and -7, which in turn proteolytically cleave hundreds of cellular proteins, ultimately leading to apoptosis.3 In vertebrates, the mitochondria intrinsically mediate caspase activation by releasing to the cytosol pro-apoptotic proteins, such as cytochrome c and Smac/Diablo, in a process called mitochondrial outer membrane permeabilization (MOMP). MOMP is in turn regulated by pro- and anti- apoptotic proteins of the BCL-2 protein superfamily.4 Apoptosis can also be triggered by extrinsic signals modulated by death receptors.5 Finally, both the initiator and effector caspases are regulated by members of the inhibitor of apoptosis protein family and their antagonists, such as the Drosophila Reaper-family proteins and the mammalian Smac/Diablo.6
While intellectually satisfying on the biochemical side, classical physiological studies, mainly performed in model organisms, implied that this model of caspase activation is highly generalized and oversimplified, as it could not explain how different cell types, or even the same cell at different developmental stages, display high variability in their sensitivities to apoptotic stimuli. Furthermore, the realization that some cells may harness caspase activity for non-lethal cellular processes while avoiding cell death, strongly implies that additional regulatory layers and mechanisms must exist in order to tune down caspase activity levels in these cells. To date, more than 50 distinct CDPs have been described in a variety of high metazoan organisms (Figure 2; also see in this issue). Perhaps the most important questions emerging from these studies are concerned with the mode of regulation and versatile functions of caspases in such diverse CDPs, while avoiding excessive caspase activity and cell death. The significance of understanding CDP mechanisms goes beyond the importance of studying cell type-specific developmental or signaling processes, as one of the hallmarks of cancer cells is their ability to resist cell death, implying that these normal and pathological cells may share some mechanisms of cell death resistance.
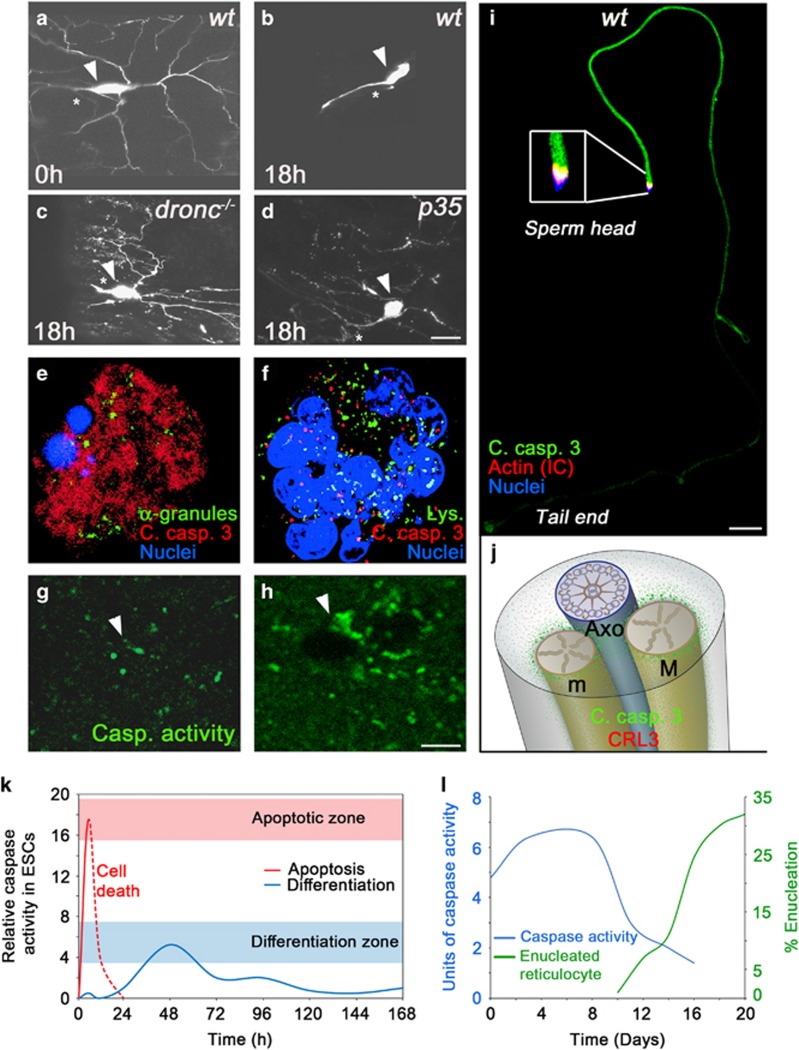
Figure 2.
Different CDP paradigms. (a–d), Dendritic pruning in ddaC neurons in the Drosophila pupa requires caspase activity. Eighteen hours into metamorphosis all the ddaC neuron’s dendrites are eliminated (a,b). In flies mutant for the initiator caspase (dronc−/−) pruning is completely blocked (c). However, overexpression of the effector caspase inhibitor protein p35 only attenuates the elimination of the severed dendrites, suggesting an additional role for the initiator caspase unrelated to its role in the activation of the effector caspases (d). Arrowheads indicate the neuronal cell bodies, while asterisks indicate the unpruned major axon. Scale bar, 20 μm. (e,f), Activated caspases are confined to cytoplasmic granules of unknown nature during platelets formation in maturing megakaryocytes (MKs). Proplatelet-displaying MKs induced to undergo apoptosis by staurosporine display diffused active caspase-3 throughout the cytoplasm (e). In contrast, active caspase-3 was confined to cytoplasmic granules that do not co-localize with lysosomes (Lys.) or α-granules in these maturing MKs (not shown; f). (g,h), Dendritic spines in the auditory forebrain of adult zebra finches during song habituation training. Sections from the auditory forebrain taken from a bird hearing only silence (g) or a bird immediately after listening to ten novel songs (h). Arrowheads indicate the areas of the dendritic spines where a transient increase in the density of caspase-3 active sites is detected after song habituation training. Scale bar, 4 μm. (i), Drosophila spermatids at the onset of the individualization process. A cyst of 64-spermatid bundle displaying caspase activation at a graded pattern descending from sperm head to tail end. Note, that the cytoplasmic contents together with the active caspases are gradually removed at the same direction as that of caspase activation, such that the regions that are the first to individualize encounter the highest levels of active caspases but for the shortest time. Scale bar, 50 μm. (j), An illustration of a cross section through an elongated spermatid depicting the axoneme (Axo; blue) attached to the two mitochondrial derivatives (M and m; gold). The CRL3 complex is expressed throughout the cytoplasm (red dots). However, once CRL3 binds to the mitochondrial surface (through a protein called A-Sβ), it gets activated, and in turn activates effector caspases in the vicinity of the mitochondria (green dots), which then diffuse in the cytoplasm. (k), Changes in the intensity and duration of caspase activity can determine cell death versus cell differentiation of mESCs into cardiomyocytes. Caspase-3/7 activity was monitored in mESC lysates at different time points showing a rapid and intense elevation in caspase activity following a death signal (red line), and delayed and reduced caspase activity levels associated with differentiation (blue line). (l), Casapase-3 activity precedes enucleation of red blood cells. The graphs describe erythropoiesis induced in culture. A peak in caspase-3 is recorded several days before the enucleation process begins. The figures were adopted with permission from the following papers: (a–d) from ref. 7; (e,f) from ref. 12; (g,h) from ref. 13; (i) from ref. 9; (j) was modified from ref. 8; (k) is a different presentation of the data in Figure 5 from ref. 14; (l) is a combined presentation of the data in Figures 1 and 3 from ref. 10
Perhaps the most common characteristic of many CDPs appears to be the involvement in cytoskeletal remodeling and associated cellular shaping. This may not come as a surprise, as the process of apoptosis itself is associated with extensive cytoskeletal and morphological changes of the doomed cell. To give a few examples for such CDPs, caspases were reported to promote major changes in cell morphology during dendritic pruning in some of the Drosophila sensory neurons (Figures 2a–d),7 the cytoplasmic extrusion during spermatid terminal differentiation in Drosophila (Figure 2i)8, 9 and during nuclear extrusion in mammalian red blood cells (Figure 2l).10 Another emerging feature of CDPs is the multiple functions of caspases in modulating learning, memory and behavior by shaping neurons and affecting neurite remodeling and synaptic plasticity (Figures 2a–d, g and h). These and additional CDPs involving cell morphology changes and/or neuroplasticity are thoroughly reviewed in this issue.
The exact roles of caspases in promoting CDPs are largely unclear, and different CDPs may utilize caspases in distinct functional manners. Furthermore, in some CDPs, the effector caspases appear to be directly involved in the destruction of a subcellular region, such as the destruction of the severed dendrites during dendritic pruning in some Drosophila sensory neurons (Figure 2d).7 In contrast, in other CDPs, such as red blood cell enucleation, caspase activity peaks several days prior to and already declines when the process becomes visible, suggesting an indirect role of caspases in the actual destruction process (Figure 2l).10 The reason why progress in understanding the exact functions of caspases in CDPs has been relatively slow is mainly attributed to the very few examples of identified CDP-specific caspase substrates. Progress in developing more sensitive proteomics technologies could be instrumental in identifying CDP-specific caspase substrates, which would help understand how caspases promote a wide variety of CDPs.
How do some cells avoid cell death in the presence of active caspases that would normally induce apoptosis in other cells? Studies of different CDP paradigms, imply the evolvement of mechanisms that maintain caspase activity levels below a certain threshold required to induce apoptosis.11 In many CDPs, caspase activity appears to be restricted in space and/or time. For instance, caspase activity can be confined to specific subcellular domains, such as the severed dendrites after detachment from the cell body (Figures 2a–d),7 the cytoplasmic granules, of unknown source, during pro-platelets formation in megakaryocytes (Figures 2e and f)12 and the dendritic spines in the auditory forebrain of the zebra finch during song habituation training (Figures 2g and h).13 In terminally differentiating Drosophila spermatids, caspase activation is coordinated with the gradual extrusion of the bulk cytoplasmic contents, such that a ubiquitin ligase complex (CRL3) gradually activates caspases in the same direction throughout the 2 mm long elongated cell (Figure 2i).9 Furthermore, activation of caspases by the CRL3 complex is limited to the surface of the two elongated spermatid mitochondria, thus further reducing the rate of caspase activation throughout these cells (Figure 2j).8 Low and transient caspase activity levels were also reported for other CDPs. During differentiation of cultured mouse embryonic stem cells into cardiomyocytes, a prolonged activation of caspases was recorded with a relatively low activity peak, which was attributed to a delayed apoptosome formation (Figure 2k).14 Caspase activity is also involved in a long-lasting synaptic modification process called long-term depression in NMDA receptor-stimulated hippocampal neurons.15 Interestingly, NMDA stimulation in cultured cortical neurons caused a fast but low and transient 2 to 3 fold increase in active caspase-3 levels, whereas apoptosis of these neurons was associated with slower but more dramatic, 17 fold increase in the level of active caspase-3.15 Whereas the mechanisms responsible for these differences in the intensity and dynamics of caspase-3 activation are not clear, they were at least in part attributed to low levels of activation and duration of the pro-apoptotic BCL-2 family members Bad and Bax.16 Other modes of caspase regulation in CDPs may include tight control over the transcriptional and/or translational levels of caspases and upstream pro-apoptotic regulatory genes,17, 18 post-translational modifications of pro- and anti- apoptotic proteins,19 and spatiotemporal or level modulation of anti-apoptotic genes.13, 20 Finally, effector caspase-independent, non-catalytic (yet unknown) function of the Drosophila initiator caspase-9 ortholog, Dronc, has been recently proposed to induce compensatory proliferation,21 implying that some CDPs may avoid the lethal activity of the effector caspases altogether.
To conclude, as new CDPs are constantly being discovered, it is sensible that caspases may play versatile non-lethal functions in many cell types throughout the animal kingdom. These functions require sub lethal levels of effector caspase activity, as opposed to the full-blown activity that triggers apoptosis. Interestingly, maintenance of low/basal caspase activity levels is believed to be a conserved feature of many high metazoan cells.22, 23 Although the significance of this constant basal activity of caspases is currently unclear, it is reasonable to propose that it may fulfill some housekeeping and/or gatekeeping functions. Future studies, as well as the development of sensitive genetic reporters to allow for in situ visualization of the basal caspase activity, will shed light on perhaps some very basic cellular functions of caspases that may be conserved across the metazoan phyla.
Acknowledgments
We are grateful to Najet Debili and David Clayton for the prompt permission to use their figures. We thank Lama Tarayrah for her comments on the manuscript. We warmly thank Genia Brodsky and Noa David Geller from the Weizmann Institute Graphic Design Department for help with the graphic illustrations. EA is supported by grants from the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013)/ERC grant agreement (616088), the Israel Science Foundation (921/13). EA is the Incumbent of the Harry Kay Professional Chair of Cancer Research.
Footnotes
The authors declare no conflict of interest.
References
- Yuan J et al. Cell 1993; 75: 641–652. [DOI] [PubMed]
- Salvesen GS, Riedl SJ. Adv Exp Med Biol 2008; 615: 13–23. [DOI] [PubMed]
- Julien O, Wells JA. Cell Death Differ 2017. (this issue). [DOI] [PMC free article] [PubMed]
- Youle RJ, Strasser A. Nat Rev Mol Cell Biol 2008; 9: 47–59. [DOI] [PubMed]
- Ashkenazi A, Dixit VM. Science 1998; 281: 1305–1308. [DOI] [PubMed]
- Steller H. Cell Death Differ 2008; 15: 1132–1138. [DOI] [PubMed]
- Schoenmann Z et al. J Neurosci 2010; 30: 6375–6386. [DOI] [PMC free article] [PubMed]
- Aram L et al. Dev Cell 2016; 37: 15–33. [DOI] [PubMed]
- Kaplan Y et al. Dev Cell 2010; 19: 160–173. [DOI] [PubMed]
- Carlile GW, Smith DH, Wiedmann M. Blood 2004; 103: 4310–4316. [DOI] [PubMed]
- Florentin A, Arama E. J Cell Biol 2012; 196: 513–527. [DOI] [PMC free article] [PubMed]
- De Botton S et al. Blood 2002; 100: 1310–1317. [DOI] [PubMed]
- Huesmann GR, Clayton DF. Neuron 2006; 52: 1061–1072. [DOI] [PMC free article] [PubMed]
- Akbari-Birgani S et al. J Biol Chem 2014; 289: 16905–16913. [DOI] [PMC free article] [PubMed]
- Li Z et al. Cell 2010; 141: 859–871. [DOI] [PMC free article] [PubMed]
- Jiao S, Li Z. Neuron 2011; 70: 758–772. [DOI] [PMC free article] [PubMed]
- Maurer CW, Chiorazzi M, Shaham S. Development 2007; 134: 1357–1368. [DOI] [PubMed]
- Kang Y, Bashirullah A. Dev Biol 2014; 386: 34–41. [DOI] [PMC free article] [PubMed]
- Gonzalvez F et al. Mol Cell 2012; 48: 888–899. [DOI] [PubMed]
- Ribeiro PS et al. J Cell Biol 2007; 179: 1467–1480. [DOI] [PMC free article] [PubMed]
- Kamber Kaya HE et al. PLoS Genet 2017; 13: e1006438. [DOI] [PMC free article] [PubMed]
- Tang HL et al. Sci Rep 2015; 5: 9015. [DOI] [PMC free article] [PubMed]
- Ding AX et al. Elife 2016; 5: e10936.27058168