Abstract
Initially found to be critically involved in inflammation and apoptosis, caspases have since then been implicated in the regulation of various signaling pathways in animals. How caspases and caspase-mediated processes evolved is a topic of great interest and hot debate. In fact, caspases are just the tip of the iceberg, representing a relatively small group of mostly animal-specific enzymes within a broad family of structurally related cysteine proteases (family C14 of CD clan) found in all kingdoms of life. Apart from caspases, this family encompasses para- and metacaspases, and all three groups of proteases exhibit significant variation in biochemistry and function in vivo. Notably, metacaspases are present in all eukaryotic lineages with a remarkable absence in animals. Thus, metacaspases and caspases must have adapted to operate under distinct cellular and physiological settings. Here we discuss biochemical properties and biological functions of metacaspases in comparison to caspases, with a major focus on the regulation of developmental aspects in plants versus animals.
Facts
Metacaspases and paracaspases are ancestors of caspases.
Although both metacaspases and paracaspases possess the caspase-hemoglobinase fold, they exhibit distinct substrate specificity and activation mechanism from caspases.
Despite differences in substrate specificity, some targets of C14 proteases seem to be evolutionary conserved across kingdoms.
In plants, metacaspases cooperate with autophagy to regulate aging, immune responses, terminal differentiation of cells and post-mortem cell clearance.
Open Questions
Is the repertoire of metacaspase substrates larger than that of caspases, considering that metacaspases have less stringent substrate specificity than caspases?
How did substrates and physiological functions of metacaspases evolve from bacteria to eukaryotes?
Which molecular mechanisms determine the pro-death versus the pro-survival functions of metacaspases?
How do metacaspases crosstalk with autophagy to regulate cell fate?
How do metacaspases act in the clearance of protein aggregates and is it the only mechanism whereby metacaspases can control aging?
“Nothing in Biology Makes Sense Except in the Light of Evolution”
Theodosius Dobzhansky, 1973
The discovery of caspases in the early 90s1, 2, 3 gave a major impetus to elucidating the biochemical mechanisms of apoptotic cell death in animals. However, it also became clear that apart from apoptosis, caspases can control other developmental processes, including cell proliferation, differentiation and migration.4, 5, 6, 7 As the early attempts to identify caspase homologues in non-metazoan genomes were not successful, it was assumed that caspase-dependent mechanisms were relatively recent evolutionary innovations restricted to animals. How then caspases and their functional divergence evolved?
The paradigm shift occurred in year 2000, when Uren and colleagues8 reported the discovery of two groups of proteins distantly related to caspases and named metacaspases and paracaspases, in the genomes of various organisms spanning about 4 billion years of evolution. To date, caspases, metacaspases and paracaspases constitute a large family (C14) within CD clan cysteine proteases, all sharing tertiary structure but exhibiting variation in structural topology, substrate specificity and activation mechanisms.9
The phylogenetic distribution of the family C14 proteases up to date is best compatible with the idea that metacaspases were acquired by eukaryotes through primary mitochondrial and plastidic endosymbiosis.10, 11, 12 The origin of paracaspases is more obscure, but considering that they could simply be a subgroup of the metacaspase family, the possibility that paracaspases evolved several times independently from metacaspases cannot be ruled out.13 Finally, canonical caspases are thought to originate from ancient metacaspase-like proteins, conceivably through horizontal gene transfer from α-proteobacteria to early multicellular organisms.11
Clearly, evolution of the C14 family proteases encompassed a long and complex series of speciation and duplication events that resulted in significant variation in biochemical properties and hence a high degree of functional divergence. This notion is particularly true for metacaspases which exhibit broader structural variation than caspases and paracaspases.9, 14 In this review we first compare the C14 proteases from structural and biochemical point of view and then discuss their biological functions. As plant biologists, we put a major focus on metacaspase-dependent regulation of plant development, aging and cell fate specification. We anticipate that comparing caspase- and metacaspase-dependent processes across kingdoms will increase understanding of the molecular and functional evolution of these important enzymes.
Molecular Structure and Biochemical Properties of C14 Family Proteases
The name caspase stands for cysteine-dependent aspartate-specific protease. It describes biochemical characteristics of these enzymes, which require cysteine residue for their proteolytic activity and cleave a polypeptide chain after an aspartate residue. Metacaspases and paracaspases were originally identified as cysteine proteases similar to caspases,8 but were later discovered to lack aspartate specificity,15 making the naming choice rather unfortunate.
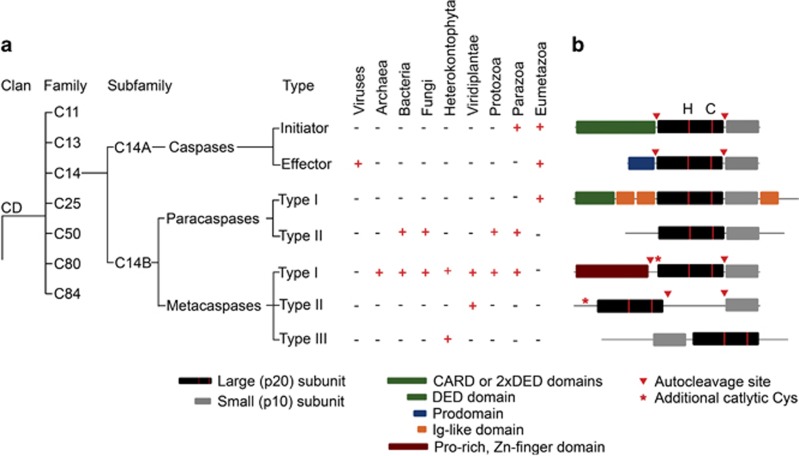
According to the current MEROPS classification,16 all known cysteine proteases are grouped into 14 different clans based on their tertiary structure. Within each clan, proteases are clustered into families according to their sequence similarity. Caspases, metacaspases and paracaspases belong to the same clan of proteases (CD clan) and also to the same family (C14) based on the sequence similarity (Figure 1a). The CD clan comprises cysteine-dependent proteases with a unique α/β-fold called caspase-hemoglobinase (CHF) fold which consists of a large (p20) subunit containing the catalytic histidine/cysteine dyad and a small (p10) subunit17 (Figures 1b and 2). The most conserved parts of the CHF fold are three motifs: one at the N-terminal β-strand, one before the catalytic histidine and one before the catalytic cysteine.10, 13
Figure 1.
Domain structure and representation of C14 proteases in various taxonomic groups. (a) CD clan of cysteine-dependent proteases comprises endopeptidases containing CHF. Based on their primary structure members of CD clan can be clustered into 7 families with various substrate specificity: C11 peptidases cleave their substrates after Arg; C13- after Asn; C14- after Asp, Arg or Lys; C25 -after Arg or Lys; C50- after Arg; C80- after Leu; C84- after Arg. Members of the C14 family can be further divided into two subfamilies C14A, consisting of caspases (Asp-specific cysteine proteases); and C14B consisting of paracaspases (Arg-specific cysteine proteases) and metacaspases (Arg or Lys-specific proteases). C14 proteases are present in all taxonomic groups. Caspases have been so far identified only in Eu- and Parazoa and viruses, albeit most of viral caspases do not have proteolytic activity. Type I paracaspases have so far been identified only in Eumetazoa, while type II paracaspases have been found in a large variety of taxonomic groups spanning both eu- and prokaryotic domains. The most wide-spread type of C14 proteases is type I metacaspases, which so far have not been identified only in genomes of viruses and Eumetazoa. Up to date type II metacaspases are known to be specific for green plants (Viridiplantae), while type III metacaspases have been found only in phytoplankton species (Heterokontophyta, Haptophyta and Cryptophyta) (b) Caspases, paracaspases and metacaspases can be divided into several types according to their typical domain architecture. All C14 proteases contain the typical C14 peptidase domain (pfam00656) that forms the CHF and consists of a large (p20) and a small (p10) subunits. The p20 subunit carries the catalytic His/Cys dyad (annotated with red stripes and H/C letters, correspondingly). The additional catalytic Cys is capable of carrying out proteolytic activity of an activated processed metacaspase. C14 peptidases are produced as zymogens and undergo modifications for activation. Initiator and inflammatory caspases contain a CARD (caspase recruiting domain) or DED (death effector domain) that are necessary for interaction with activating molecules, oligomerization and activation of these enzymes; these domains are removed upon activation. Effector caspases contain a short N-terminal prodomain that is important for stabilizing the zymogen, inhibiting its activity and initiation of dimerization; this domain is removed upon caspase activation. All characterized type II metacaspases, most of caspases, some of the type I metacaspases also need to undergo autocleavage of the loop connecting p20 and p10 subunits. Paracaspases do not require autoprocessing for activation. Paracaspases of type I contain an N-terminal DED domain, and several Ig-like domains, while only C14 peptidase domain was predicted to be present in type II paracaspases. Metacaspases type I have an N-terminal domain, often containing a Zn-finger motif and Pro-rich motif. For type II metacaspases it is typical to have a large linker separating p20 and p10 domain and no N-terminal domain. Metacaspases of type III are unique among C14 peptidase, as they have a reversed positioning of p10 and p20 subunits
Figure 2.
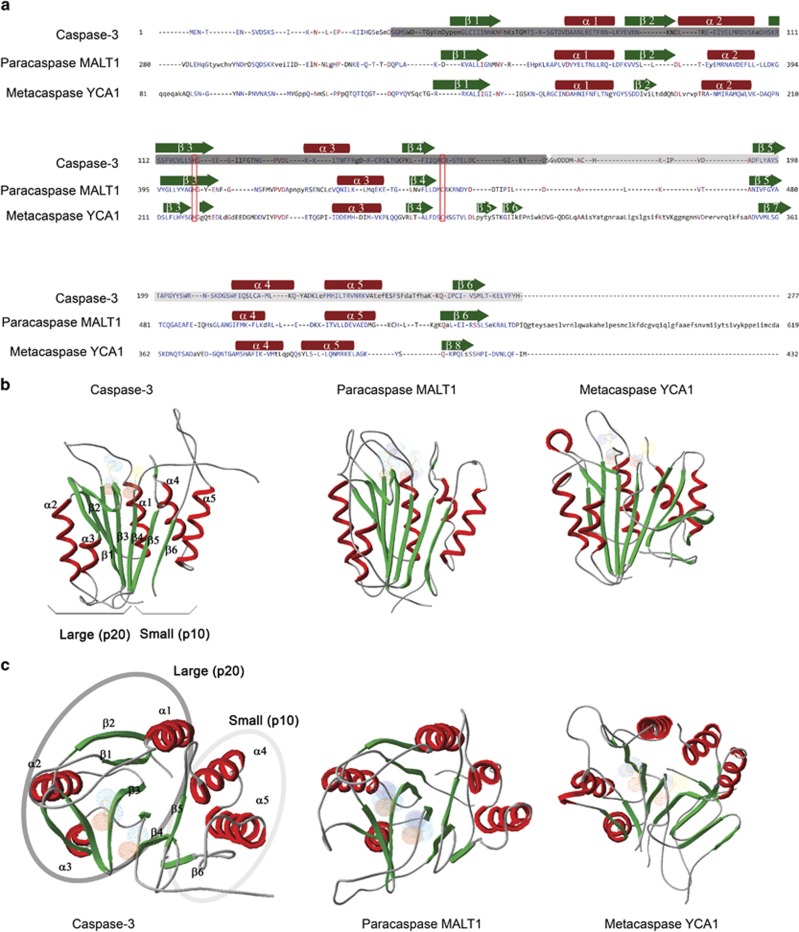
C14 proteases have poor conservation of primary structure, but all possess a highly conserved fold (CHF). (a) Alignment of C14 peptidase domains of caspase-3 (Homo sapiens), paracaspase type I, MALT1 (Homo sapiens) and metacaspase type I, YCA1 (Saccharomyces cerevisiae) reveals low level of amino acid identity. Amino acids forming the large (p20) subunit are marked with dark grey rectangle; small (p10) subunit is marked with light grey rectangle. Position of β-sheets and α-helices are represented with green arrows and red rectangles, respectively. Catalytic His and Cys are marked with red rectangles. (b and c) Ribbon diagrams of crystal structures of the same enzymes as in (a) displayed from two perspectives with 90 °C difference: (B), side view; (c), top view. Surfaces of the catalytic His/Cys are denoted with light blue and red colors. The main structural difference of type I metacaspases is presence of two additional β-sheets within the p20 subunit
There are seven families of proteases belonging to the CD clan: clostripains, C11;18 legumains, C13;19 caspases, C14;20 gingipains, C25;21 separases, C50;22 MARTX cytolysins, C8023 and PrtH-like proteases, C85.24 Notably, only members of the C14 family can be found in all taxonomic groups.16 Our understanding of the phylogeny of the C14 protease family is gradually changing with new members being discovered in recently sequenced genomes.10, 12, 13, 14 Caspases have been found only in the genomes of animals and a few viruses; paracaspases were detected in the genomes of bacteria, slime mold (Dictyostelium) and metazoa, whereas metacaspases can be found in the genomes of virtually all pro- and eukaryotic lineages, with a remarkable absence in animals (Figures 1a and 3).
Figure 3.
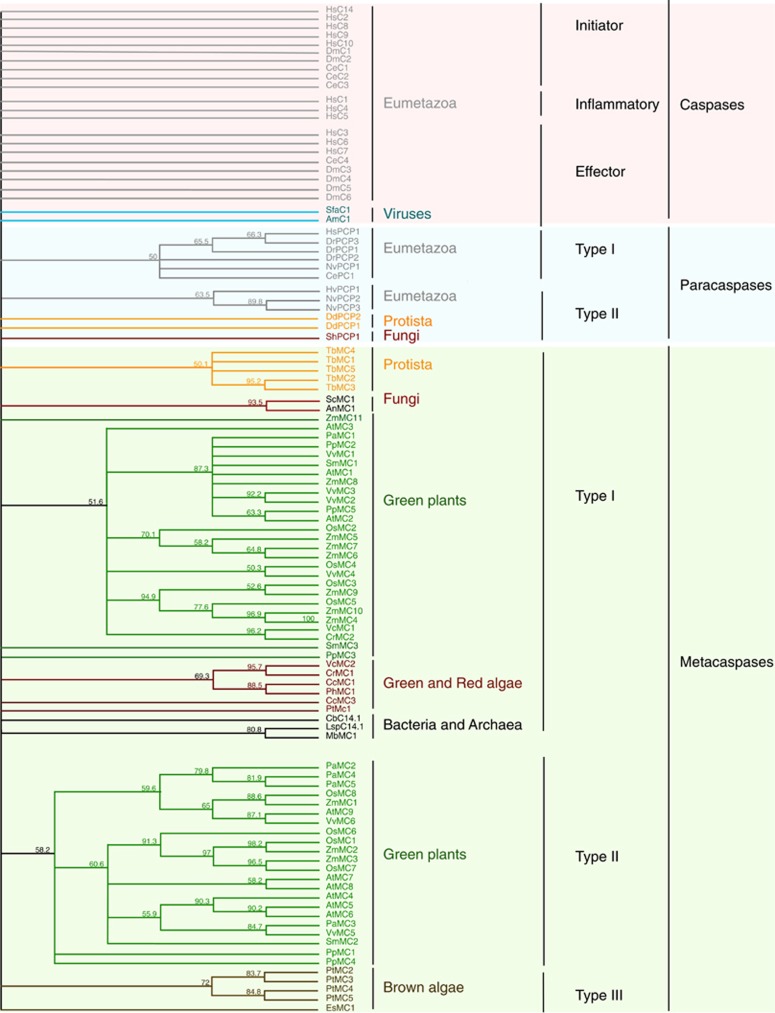
Phylogenetic tree of C14 proteases based on alignment of C14 conserved domain. Position of the C14 domain was predicted for each protein sequence using Conserved Domain search tool.140 Alignment was performed using Clustal Omega tool with default setting and followed by building the tree using neighbor joining method with 2000 bootstraps. ID number of the sequences used for the analysis are provided in the Supplementary Table 1. The comparison revealed that most of C14 proteases form type-specific clades within each kingdom. Interestingly, C14 domains of caspases, do not form any identifiable subgroups, indicating rapid diversification of the primary structure of these proteins
Most of the C14 protease family members are active enzymes with a broad range of substrates, and many of them are constitutively expressed and thus require tight regulation of activity to prevent unwanted proteolysis. C14 peptidases are expressed as zymogens and undergo structural modifications before activation (Table 1). Caspases and paracaspases are activated upon dimerization. In the case of caspases, dimerization presumably enables basal catalytic activity, allowing autoprocessing of the zymogen and freeing the unstructured loop regions for interaction with regions close to active centers. This interaction stabilizes active centers and yields a full protease activity.9, 25, 26, 27 Unlike caspases, paracaspases characterized up to date do not undergo autoprocessing upon dimerization. Dimerization per se seems to be sufficient for stabilization of the active fold of the protease.25, 28 Metacaspases, on the other hand, do not dimerize, but most of them still undergo autoprocessing before activation.15, 29, 30, 31 Autoprocessing activity of vast majority of metacaspases is highly dependent on Ca2+ and thus might be regulated by local changes in the concentration of Ca2+ ions.15, 29, 30, 31, 32 Interestingly, metacaspases contain an additional N-terminal cysteine, which can take over proteolyitiyc activity after the zymogen underwent autoprocessing of the zymogen15, 33 (Table 1).
Table 1. Biochemical characteristics of C14 proteasesa.
| Active form | Autocleavage between p20 and p10 subunits | Removal of N-terminal prodomain | Optimal pH | Ca2+ dependency | Additional catalytic Cys | P1 substrate specificity | |
|---|---|---|---|---|---|---|---|
| Caspases | |||||||
| Initiatorb | Dimer | Yes | Yes | Neutral | No | No | Asp |
| Effector | Dimer | Yes | Yes | Neutral | No | No | Asp |
| Paracaspases | |||||||
| Type I | Dimer | No | No | Neutral | No | No | Arg |
| Metacaspases | |||||||
| Type I | Monomer | Some | Some | Neutral | Yes | Yes | Arg or Lys |
| Type II | Monomer | Yes | Not applicable | Neutralc | Yesc | Yes | Arg or Lys |
No information is yet available about biochemical characteristics of type II paracaspases and type III metacaspases.
Initiator and inflammatory caspases.
With the exception of AtMC9.
In addition to the p20 and p10 domains, caspases also have an N-terminal prodomain, which is required for initiation of dimerization and is removed upon activation of the enzyme. Effector caspases have a short, 20–30 residue N-terminal prodomain, while initiator caspases have large, over 90 amino acid N-terminal domains called CARD (Caspase Recruitment Domain) or DED (Death Effector Domain) (Figure 1b). The short prodomains of effector caspases are required for dimerization. Similarly, N-terminal domains of initiator caspases are crucial for interaction with signaling proteins which bring caspase monomers together, stimulating dimerization and triggering the whole cascade of caspase activation.27, 34, 35
Paracaspases are divided into two types based on their domain structure: type I paracaspases contain a DED and immunoglobulin-like domains, while type II paracaspases do not contain them.13 Metacaspases are divided into three types: type I contain N-terminal proline-rich domain with a Zn-finger motif, type II metacaspases do not have an N-terminal domain, but p20 and p10 subunits of their CHF are separated by a long linker region; in type III metacaspases the p10 subunit precedes the p20 subunit12 (Figures 1b and 3). The structural study of a trypanosomal type I metacaspase revealed that N-terminal domain of metacaspases might be inhibiting activity of the protease by folding over the active center.36
Activity of C14 proteases is also regulated by post-translational modifications, such as phosphorylation, ubiquitination, nitrosylation as well as by direct interaction with other proteins;27, 33, 37 by the presence of ions15, 29, 38 and by pH15, 29, 30, 39 (Table 1). An interesting example of evolutionarily conserved mode of C14 protease regulation is by suicide inhibitors serpins, which remain bound to the active sites of proteases upon cleavage.40, 41, 42, 43
The most crucial biochemical difference between caspases and other members of C14 peptidase family is substrate specificity. While caspases cleave polypeptides after an acidic aspartate residue,44 paracaspases cleave after a basic arginine residue28 and metacaspases after basic arginine or lysine residues15, 29, 30, 31, 39 (Table 1). Requirements for measuring proteolytic activity of distinct subfamilies of C14 proteases have been described previously.45
Cellular Substrates of Metacaspases
The evolutionary conservation of C14 protease family members across all taxonomic groups indicates conserved function of these enzymes, but the differences in substrate specificity of caspases, para- and metacaspases suggests the opposite. This juxtaposition spurred a hot debate about clustering of the metacaspases and caspases into different groups.46, 47, 48, 49, 50, 51 Current evidence implies that functional conservation of the C14 proteases is only partial and that establishment of the different molecular machineries for programmed cell death (PCD), immunity and cell differentiation either led to or originated from diversification of the specificity and activity regulation of C14 proteases. In some cases the question about functional conservation of the C14 proteases might be better addressed following the approach adopted by Hulpiau and colleagues,13 which takes into consideration not only a protease and its individual target, but also the whole functional complex of its targets.
Identification of natural substrates of C14 proteases is essential for verifying their proposed phylogeny from the functional perspective. Caspases can be separated into several groups according to their preferences for certain amino acids in the P4 position,52, 53 which reduces redundancy of their activity. So far paracaspases have been shown to have strict preference for arginine in the P1 position and to have restrictions for the minimal length of a potential substrate.28 Metacaspases display arginine- or lysine-P1 specificity, but have loose specificity to amino acids in other positions.14
Although best known for their role in dismantling cells during apoptosis, caspases have key roles in remodeling cells during cell differentiation, synapsis formation, triggering inflammation response and so on.52, 54, 55 Thus known substrates of caspases include a very broad spectrum of targets, several hundreds of which have already been identified.56 Much less information is available about substrates of paracaspases57, 58, 59, 60 and metacaspases.40, 61, 62, 63, 64 Interestingly, while genomes of some organisms contain only one C14 protease gene, the best example being metacaspase YCA1 (also known as MCA1) of budding yeast,65 others have multiple copies,13, 14, 52 suggesting that C14 proteases might be multifunctional and/or redundant.14, 66, 67
The most well studied substrates of the C14 proteases are the proteases themselves. All known caspases,25 and most of the metacaspases,15, 29, 30, 31 excluding some of the type I metacaspases30, 68, 69 require autoprocessing for maturation of the enzyme. Caspases and some type I metacaspases undergo removal of the N-terminal prodomain and cleave the linker between the large and small subunits of the catalytic domain, while type II metacaspases only cleave the linker between the large and small subunits of the catalytic domain (Figure 1b). Caspases can undergo both intra- and intermolecular autocleavage, which is crucial for tuning caspase response according to the strength of the activation stimuli.20, 70 To our knowledge the mechanism of metacaspase autoprocessing has not yet been published. Likewise, possibility for the existence of cascade activation of different metacaspases in a fashion similar to that of caspases remains elusive. Maturation of paracaspases does not seem to involve autocleavage of the zymogen.71 Notably, at least in some cases, autoprocessing is not required for activity of the caspases or metacaspases, but instead has a role in stabilizing the activated protease.72, 73
Despite the differences in the morphology and molecular machinery of PCD74 several of the cell death-related C14 protease substrates are conserved across kingdoms. For example, Tudor Staphylococcal Nuclease (TSN), a protein involved in maintaining cell viability, regulation of gene expression and stress response75, 76 has been found to be a substrate of caspase-3 during apoptosis in animal cells and a substrate of a type II metacaspase during vacuolar PCD in plants.61 Poly (ADP-ribose) polymerase, a classical substrate of caspase-3 during apoptosis77 has been demonstrated to undergo metacaspase-dependent proteolysis during PCD in fungi.63 GAPDH, glyceraldehyde 3-phosphate dehydrogenase, a substrate of caspase-1 during infection or septic shock78 was shown to be cleaved in an NO-dependent manner by YCA1 metacaspase upon H2O2-induced cell death in yeasts.79 Actin, a typical substrate of caspases during apoptosis80 has been found among targets of Arabidopsis thaliana metacaspase AtMC9.62
Another striking common feature of C14 proteases is their involvement in regulating the innate immune responses. Nucleotide-binding leucine rich proteins, NLR, is a large family of intracellular immune receptors found in animals, plants and fungi and involved in defense against microorganisms.81, 82, 83 In mammalian cells NLR proteins trigger an inflammatory response mediated by caspases and nuclear factor κB (NFκB).54 The mammalian paracaspase mucosa-associated-lymphoid tissue lymphoma-translocation gene 1 (MALT1), has essential role in activating the NFκB.84 Arabidopsis metacaspase AtMC9 cleaves the AtCDC48A,62 a negative regulator of NLR-dependent immunity,85 while AtMC1 and AtMC2 have antagonistic roles in establishing an NLR-dependent immune response to pathogen infection68 (Figure 4). Interestingly, fungal homologue of cdc48 was predicted to be also a substrate of metacaspase during farnesol-induced cell death.86
Figure 4.
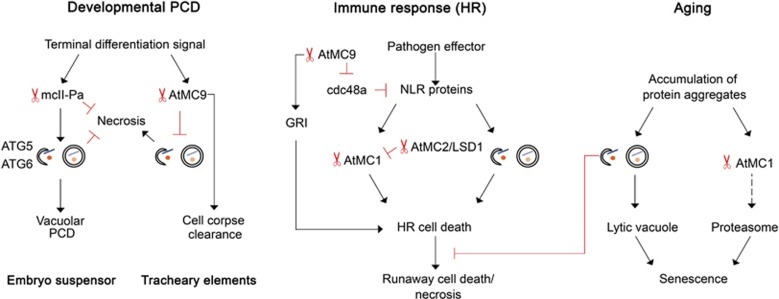
Crosstalk between metacaspases and autophagy. Metacaspase and autophagy are known to be involved in regulating execution of developmental PCD. Depending on the type of tissue and model organism, they may have different roles in the pathway. For instance, type II metacaspase of Norway spruce, mcII-Pa, was demonstrated to be crucial for gradual disassembly of terminally differentiated cells of embryo suspensor via vacuolar PCD. mcII-Pa acts upstream to autophagy, and both metacaspase and autophagy prevent dying cells from early collapse and necrosis. Arabidopsis type II metacaspase AtMC9 is required at the very last stage of vacuolar PCD, post-mortem clearance of root xylem cells. It also has role in suppressing necrosis-inducing signaling from terminally differentiated cells to the surrounding cells by downregulating autophagic activity in the dying cells. Both plant metacaspases and autophagy are activated via NLR-dependent pathway upon pathogen infection. In both cases signaling leads to a hyper-sensitive response (HR) and might result in local lesion or runaway necrotic cell death. Interestingly, autophagy has antagonistic role in developing HR response: it is important for transducing the signal from NLR proteins in infected cells and at the same time aging-related autophagy is important for preventing the spread of runaway death of cells adjacent to the infection site. Likewise, metacaspases do not have a single function in HR response. Different metacaspases have antagonistic roles upon pathogen infection, for example, AtMC1 and AtMC9 have a pro-death role, while AtMC2 in complex with LSD1 acts as an AtMC1 inhibitor. AtMC9 might be involved in stimulating HR on different levels: by processing of cell death inducing peptide GRIM REAPER (GRI) upon oxidative stress and by cleaving cdc48a protein, which is important for removing excess NLR proteins from cells. Aging is associated with accumulation of damaged and/or misfolded proteins that tend to aggregate. Both metacaspases and autophagy participate in removing these aggregates. YCA1 metacaspase of Saccharomyces cerevisiae has been demonstrated to localize to protein aggregates and stimulate their proteasome-dependent degradation. A similar mechanism was proposed for the type I plant metacaspase AtMC1 (dashed arrow). Autophagy removes protein aggregates by delivering them to lytic compartments, such as lytic vacuole. Autophagic flux compensates for a decreased proteasomal activity (and vice versa), linking impacts of autophagy and metacaspase-dependent pathways on aging control
In the past decade high throughput studies provided valuable insights on how strict is substrate specificity of C14 proteases,62, 87 redundancy of proteases,88 and what is the quantitative efficacy of cleavage for individual substrates.89 Mahrus et al.90 demonstrated that during apoptosis, caspases target complexes and networks of proteins. Data from these studies has been used to develop in silico tools to predict caspase substrates.53, 91 Prediction of meta- and paracaspase substrates in silico has been less efficient due to the lower specificity of these proteases and the very sparse information about their preferences to the tertiary structure of substrates, as well as subcellular localization. To our knowledge, up to date only one example of a high throughput identification of metacaspase substrates is available for Arabidopsis metacaspase AtMC9.62
Metacaspases in Terminal Cell Differentiation and Death
The key role of caspases in initiation and execution of apoptosis together with the fact that other members of the C14 family are conserved in all kingdoms of life, makes a compelling case for evolutionary conservation of the molecular mechanism of PCD.8, 10 However, unlike caspases, metacaspases and paracaspases cleave their substrates after the basic residues arginine and/or lysine (Table 1). If the PCD machinery would be conserved in distant taxonomic groups, it would mean that the cleavage sites in the PCD-related targets coevolved in non-animal species to fit specificity of meta- and paracaspases. Alternatively, the role of caspases in these species might be executed by proteases not belonging to the C14 family. Indeed, during PCD in fungi and plants, caspase-like activity was shown to originate from proteases not belonging to the C14 family.74, 92 At the same time, multiple studies with yeasts and plants link PCD to metacaspase activity.14, 93, 94 Growing evidence indicates that PCD regulation in plants, fungi and protozoa drastically differs from PCD regulation in animal organisms,74 with a few interesting examples of coincidence or conservation (see the previous section of this review).
Genomes of both budding yeast and Dictyostelium encode for only one proteolytically active member of C14 proteinase family, metacaspase YCA1 and paracaspase PCP, respectively.65, 95 Interestingly, multiple studies confirm involvement of YCA1 in yeast PCD,65, 66, 96 while PCD of Dictyostelium does not seem to depend on PCP activity.95 Genomes of plants encode for multiple, potentially redundant, metacaspase members;14 thus, investigating their involvement in PCD is more challenging.
Plant metacaspases have been shown to have an important role in some cases of terminal cell differentiation.29, 93, 97, 98, 99 In most cases, overexpression of plant metacaspases does not lead to ectopic cell death phenotype, indicating requirement of an upstream PCD activation signal40 (our unpublished observations on Arabidopsis and spruce).
Plant cells are surrounded by a rigid polysaccharide cell wall, which is often not degraded upon execution of developmental PCD, but is left as a structural component of a tissue (for example, wood, bark, embryo suspensor, seed coat and so on). Developmental PCD in plants coincides with extensive remodeling of cellular architecture, for example, vacuolization, modification of the cell wall, deposition of secondary cell wall structures, gradual removal of organelles and nuclear disassembly. Thus, in contrast to animal cells undergoing apoptosis, terminally differentiated plant cells are maintained in a metabolically active state until remodeling is accomplished and the desired structure is achieved and then degraded from within the cell using lytic vacuoles, without involving extracellular resources.74, 97, 98 Superficially this type of cell death resembles the cornification death typical for epidermal keratinocytes of animals.100
Cell death or remodeling during differentiation often includes changes in the cytoskeleton. Apoptosis involves caspase-dependent cleavage of actin and myosin leading to blebbing.101 Remodeling of the sub-membranous cytoskeleton of trophoblasts during syncytialization and of lens fiber cells during aging require caspase activity.102, 103 Similarly, drastic changes in cytoskeleton morphology were observed during differentiation of plant embryo suspensor cells, coinciding with an increase in metacaspase activity.29, 104, 105 Nuclear disassembly during PCD in Norway spruce embryos depends on the activity of type II metacaspase mcII-Pa, which translocates from the cytoplasm to the nucleus in the terminally differentiated cells.29
Based on the combination of phenotypic features and biochemical and cellular markers, most types of plant cell death can be classified as either vacuolar type or a necrotic type.74 Interestingly, depleting activity of metacaspase mcII-Pa in the embryos of Norway spruce switches the type of cell death in the terminally differentiated cells from vacuolar to necrotic (Figure 4),98 indicating that this metacaspase has a pro-survival role at least at the early stages of cell death. Similarly, caspase-8 is essential for preventing keratinocytes committed to cornification cell death from switching to necrosis.106
Post-Mortem Role of Metacaspases
Both caspases and metacaspases operate mainly in cells that are alive, but post-mortem functions have been reported as well. Caspases can have post-mortem roles if they are secreted into the extracellular space or if they stay intact after phagocytosis. Several studies reported extracellular activities of caspases. For instance, caspase-1 is secreted from dying cells into the extracellular space to propagate inflammation to neighboring cells.107, 108, 109 Caspase-3 is released from apoptotic cells into the extracellular intestinal lumen during Escherichia coli infection to undermine the integrity of the intestinal epithelial cells.110 Common to these extracellular caspase activities is that they affect the fate of the neighboring cells in a non-cell autonomous manner. Recent studies indicate that plant metacaspases might have a cell autonomous, post-mortem role in the clearance of cellular contents. Examples of such processes are cell death in the lateral root cap and in xylem elements. These cells maintain high metabolic activity and intact organelle morphology until the loss of the tonoplast integrity and vacuolar burst (a moment of cell death), after which cellular contents are rapidly hydrolysed post mortem by enzymes released from the intracellular compartments.111, 112, 113, 114 Complete clearance of xylem vessel elements is crucial to allow unobstructed water transport in the empty pipes of the vessels, while clearance of the root cap cells is required for normal root development and especially root branching.115
AtMC9 is the only Arabidopsis metacaspase that is strongly transcriptionally upregulated during xylem and lateral root cap PCD,116 and it is therefore a likely candidate to operate in post-mortem processes. While the role of AtMC9 remains to be established in the lateral root cap cells, it was shown to be required for the rapid autolysis of the xylem vessel elements.97 Considering that complete clearance of xylem vessels is crucial for optimal water transport, it was surprising that the delay in vessel clearance of the AtMC9 mutants did not affect plant growth negatively, and therefore the biological relevance of the AtMC9-mediated post-mortem autolysis remained unclear. Bollhöner et al.97 demonstrated that the AtMC9 mutants are more sensitive to the xylem vessel resident pathogen Ralstonia solanacearum, suggesting that the AtMC9-dependent, efficient autolysis of xylem vessel elements prevents spreading of pathogens in the vasculature. Functional redundancy of AtMC9 with other metacaspases is unlikely considering sequence divergence and unique biochemical properties of AtMC9. The optimal pH of AtMC9 catalysis has been reported to be 5.5 15 and it is around 7 for the Arabidopsis AtMC4, AtMC5 and AtMC8 and the spruce mcII-Pa,29, 30, 117 and most probably for the other type II metacaspases. The low pH optimum of AtMC9 is in line with the low pH of the cells after vacuolar burst and therefore the post-mortem role of this enzyme. Noteworthy, AtMC9 is the only known metacaspase that does not require calcium for its activity.15
Crosstalk Between Metacaspases and Autophagy
Autophagy is a catabolic process, during which the cell content is selectively or in bulk degraded and recycled by the cell. It has a crucial role in maintaining functionality of cells by removing damaged or superfluous molecules and organelles. Autophagy is conserved in all eukaryotes and controlled by the AuTophaGy-related (ATG) genes.118, 119, 120 Autophagy is known as a pro-survival mechanism, but a ‘runaway’ autophagy might lead to the degradation of the whole cell although evidence of existence of such cell death is rather scarce.121 On the contrary, there is ample evidence of crosstalk between autophagy and PCD in various taxonomic groups.121, 122, 123, 124, 125
Suspensor cells of Norway spruce embryos undergo a typical vacuolar cell death,74, 126 which requires both metacaspase mcII-Pa29, 93 and autophagy.98 Interestingly, in this pathway metacaspase acts upstream to autophagy, inducing formation of autohagosomes in terminally differentiated cells and preventing occurrence of early necrotic cell death (Figure 4). It still remains to be identified whether catalytic activity of the metacaspase is required for activation of autophagy and whether the metacaspase cleaves an unknown inhibitor of autophagy upon induction of cell death.98 Recently it was demonstrated that caspase-9 forms a complex with ATG7 protein promoting formation of autophagosomes. This interaction does not depend on caspase-9 proteolytic activity and it does not only promote autophagic flux, but also titrates out caspase-9 from the pool of pro-apoptotic enzymes, thus potentially having a pro-survival role.127
Another plant type II metacaspase, AtMC9 was shown to suppress autophagic activity during development of tracheary elements in Arabidopsis cell cultures99 (Figure 4). As mentioned above, mcII-Pa and AtMC9 differ in the ranges of optimal pH and dependency on Ca2+ for their activity and localization.15, 29, 97 Therefore different modes of their relationship with autophagy are not surprising.
Plant autophagy machinery and metacaspases team up during PCD to compensate for the lack of macrophages, which in animal organisms execute the final step of PCD eliminating the remnants of dead cells.128 Multiple studies indicate that autophagy is involved in vacuolar type of cell death in different organisms98, 99, 129, 130 probably having various functional roles: delivering cell content to the lytic compartment for degradation, ensuring lack of toxic waste upon cell collapse and maintaining cell viability until cell differentiation is completed. Metacaspases, on the other hand, regulate autophagic activity98, 99 and degrade the remnants of dead cell after its collapse.97 Interestingly, up-regulation of autophagy in terminally differentiating cultured plant cells results in a runaway cell death,99 presumably caused by an improper disassembly of the dying cell and functionally resembling secondary necrosis occurring in animals in the absence of phagocytosis of apoptotic bodies.128
Metacaspases and Aging
In the last few years a role for metacaspases in the control of aging has started emerging. Initial reports came from the single metacaspase of the budding yeast YCA1, which was shown to contribute to fitness and adaptability through its ability to remodel protein insoluble aggregates.131 YCA1 was shown to target protein aggregates, co-localizing with aggregate remodeling chaperones. Δyca1 mutants were enriched for stress response proteins, vacuolar peptidases and autophagic bodies and had an increased content of insoluble protein aggregates in comparison to wild-type yeast.
These seminal findings were reinforced by the work of Hill et al.,66 who demonstrated that overexpression of YCA1 extended replicative lifespan of yeast and reduced the accumulation of protein aggregates formed during aging or stress. This lifespan extension and aggregate remodeling mediated by YCA1 was dependent on the proteasome and the disaggregase Hsp104. Interestingly, the catalytic YCA1 mutant retained residual lifespan extension132 and aggregate remodeling131 activity, indicating involvement of both proteolytic and non-proteolytic mechanisms in the YCA1 anti-aging function.
In plants, metacaspases can also regulate the process of aging, linked to protein aggregate remodeling. Similar to YCA1, the Arabidopsis type I metacaspase AtMC1 has been shown to localize to insoluble protein aggregates and participate in their clearance during aging133 (Figure 4). AtMC1 protein aggregate localization depends on its N-terminal prodomain, but an intact catalytic site is not strictly required for this localization. The N-terminal domain of AtMC1 contains a zinc finger domain that has been suggested to act as a scaffold to assemble different death regulators.68, 94 Interestingly, protein aggregate localization in yeast was also dependent on the N-terminus of YCA1,131 a prion domain containing a polyQ region, which was previously linked to the aggregate localization propensity of the protein.134 This N-terminus-dependent aggregate targeting of type I metacaspases could represent an ancestral form of the mechanism activating apoptotic initiator caspases. Interestingly, knocking out AtMC1 resulted in accelerated aging. Genetic interaction studies proved that AtMC1 acts additively to autophagy in the control of aging, both acting as protein quality control/homeostatic mechanisms to ensure its correct timing and optimal resource redistribution for seed set.133
In light of the findings in yeast, YCA1 was proposed to contribute to cellular homeostasis in the following contexts:66 (i) the catalytic activity of YCA1 could contribute to cleave proteasomal substrates, making them more accessible for degradation; (ii) the N-terminal domain of YCA1 could facilitate the action of disaggregating chaperones; (iii) YCA1 could aid in the recruitment of certain proteins that contributed in refolding or degradation in protein control compartments. Considering the functional analogies of YCA1 and AtMC1, these scenarios could also apply during plant development.
In contrast to the life-extension functions described above, other plant and fungal metacaspases have been shown to act as catalysts of aging. In the woody fruit tree Litchi chinensis knocking out the type II metacaspase LcMCII-1 results in delayed leaf senescence, whereas its overexpression accelerates the process.135 Similarly, deletion of PaMCA1 or PaMCA2, two metacaspases of the filamentous fungus Podospora anserina, results in lifespan extension.136
Concluding Remarks
Compared with a vast body of knowledge about caspase-dependent processes and numerous examples of translation of this knowledge to medicine, mechanistic understanding of metacaspases remains largely fragmented. This in turn hampers evolutionary analysis of pro-survival and pro-death signaling controlled by C14 proteases. Although it is clear that metacaspases can perform both cytoprotective and cell death functions, it remains an open question whether the assigned pro-death functions are merely compensatory responses to their primordial role as cytoprotective agents.
Cleavage of protease targets can lead not only to the loss of function, as in case of metacaspase-dependent cleavage of TSN during PCD,61 but also to the gain of function62, 64 or even to the change of function.137 Importantly, readiness of caspase substrates for being cleaved has been shown to be greatly influenced by their post-translational modifications138, 139 and ability to form complexes.90 Finding new natural substrates of metacaspases and understanding both structural determinants and functional outcomes of their cleavage is crucial for deciphering mechanistic role of metacaspases in signaling pathways that control cell fate specification, organismal development and aging.
Acknowledgments
Research in our laboratories is supported by grants from Carl Tryggers Foundation (to EAM), the Swedish Research Council (to HT and PVB), Knut and Alice Wallenberg Foundation (to PVB), Olle Engkvist Foundation (to PVB), the Swedish Foundation for Strategic Research (to PVB), the Trees and Crops for the Future (TC4F) programme (to PVB), Spanish Ministry of Economy, Industry and Competitiveness (projects AGL2016-78002, RyC 2014-1658 SEV‐2015‐0533; to NSC) and the CERCA Programme/Generalitat de Catalunya (to NSC). We apologize to those authors whose primary works could not be cited owing to space limitation.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by E Arama
The authors declare no conflict of interest.
Supplementary Material
References
- Cerretti DP, Kozlosky CJ, Mosley B, Nelson N, Van Ness K, Greenstreet TA et al. Molecular cloning of the interleukin-1 beta converting enzyme. Science 1992; 256: 97–100. [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature 1992; 356: 768–774. [DOI] [PubMed] [Google Scholar]
- Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell 1993; 75: 641–652. [DOI] [PubMed] [Google Scholar]
- Ishizaki Y, Jacobson MD, Raff MC. A role for caspases in lens fiber differentiation. J Cell Biol 1998; 140: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam A, Cohen LY, Aouad S, Sekaly RP. Early activation of caspases during T lymphocyte stimulation results in selective substrate cleavage in nonapoptotic cells. J Exp Med 1999; 190: 1879–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy NJ, Kataoka T, Tschopp J, Budd RC. Caspase activation is required for T cell proliferation. J Exp Med 1999; 190: 1891–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Akaike T. Possible involvement of caspase-like family in maintenance of cytoskeleton integrity. J Cell Physiol 1999; 179: 45–51. [DOI] [PubMed] [Google Scholar]
- Uren AG, O'Rourke K, Aravind L, Pisabarro MT, Seshagiri S, Koonin EV et al. Identification of paracaspases and metacaspases: Two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol Cell 2000; 6: 961–967. [DOI] [PubMed] [Google Scholar]
- McLuskey K, Mottram JC. Comparative structural analysis of the caspase family with other clan CD cysteine peptidases. Biochem J 2015; 466: 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Classification of the caspase-hemoglobinase fold: Detection of new families and implications for the origin of the eukaryotic separins. Proteins 2002; 46: 355–367. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ 2002; 9: 394–404. [DOI] [PubMed] [Google Scholar]
- Choi CJ, Berges JA. New types of metacaspases in phytoplankton reveal diverse origins of cell death proteases. Cell Death Dis 2013; 4: e490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulpiau P, Driege Y, Staal J, Beyaert R. MALT1 is not alone after all: identification of novel paracaspases. Cell Mol Life Sci 2016; 73: 1103–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiatsiani L, Van Breusegem F, Gallois P, Zavialov A, Lam E, Bozhkov PV. Metacaspases. Cell Death Differ 2011; 18: 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen D, van de Cotte B, De Jaeger G, Eeckhout D, Casteels P, Vandepoele K et al. Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J Biol Chem 2004; 279: 45329–45336. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Finn R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 2016; 44: D343–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker NPC, Talanian RV, Brady KD, Dang LC, Bump NJ, Ferenz CR et al. Crystal-structure of the cysteine protease interleukin-1-beta-converting enzyme—a (P20/P10)(2) homodimer. Cell 1994; 78: 343–352. [DOI] [PubMed] [Google Scholar]
- Labrou NE, Rigden DJ. The structure-function relationship in the clostripain family of peptidases. Eur J Biochem 2004; 271: 983–992. [DOI] [PubMed] [Google Scholar]
- Dall E, Brandstetter H. Structure and function of legumain in health and disease. Biochimie 2016; 122: 126–150. [DOI] [PubMed] [Google Scholar]
- Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J 2004; 384: 201–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr Protein Pept Sci 2003; 4: 397–407. [DOI] [PubMed] [Google Scholar]
- Moschou PN, Bozhkov PV. Separases: biochemistry and function. Physiol Plantarum 2012; 145: 67–76. [DOI] [PubMed] [Google Scholar]
- Satchell KJF. Structure and function of MARTX toxins and other large repetitive RTX proteins. Annu Rev Microbiol 2011; 65: 71–90. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. Prediction of a caspase-like fold in Tannerella forsythia virulence factor PrtH. Cell Cycle 2009; 8: 1453–1455. [DOI] [PubMed] [Google Scholar]
- Shi YG. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell 2002; 9: 459–470. [DOI] [PubMed] [Google Scholar]
- Oberst A, Pop C, Tremblay AG, Blais V, Denault JB, Salvesen GS et al. Inducible dimerization and inducible cleavage reveal a requirement for both processes in caspase-8 activation. J Biol Chem 2010; 285: 16632–16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish AB, Freel CD, Kornbluth S. Cellular mechanisms controlling caspase activation and function. Cold Spring Harb Perspect Biol 2013; 5. pii: a008672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachmann J, Snipas SJ, van Raam BJ, Cancino EM, Houlihan EJ, Poreba M et al. Mechanism and specificity of the human paracaspase MALT1. Biochem J 2012; 443: 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozhkov PV, Suarez MF, Filonova LH, Daniel G, Zamyatnin AA Jr, Rodriguez-Nieto S et al. Cysteine protease mcII-Pa executes programmed cell death during plant embryogenesis. Proc Natl Acad Sci USA 2005; 102: 14463–14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Lam E. Two Arabidopsis metacaspases AtMCP1b and AtMCP2b are arginine/lysine-specific cysteine proteases and activate apoptosis-like cell death in yeast. J Biol Chem 2005; 280: 14691–14699. [DOI] [PubMed] [Google Scholar]
- Wong AHH, Yan CY, Shi YG. Crystal structure of the yeast metacaspase Yca1. J Biol Chem 2012; 287: 29251–29259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Lam E. Calcium-dependent activation and autolysis of Arabidopsis metacaspase 2d. J Biol Chem 2011; 286: 10027–10040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenghi B, Romero-Puertas MC, Vercammen D, Brackenier A, Inze D, Delledonne M et al. Metacaspase activity of Arabidopsis thaliana is regulated by S-nitrosylation of a critical cysteine residue. J Biol Chem 2007; 282: 1352–1358. [DOI] [PubMed] [Google Scholar]
- Boatright KM, Salvesen GS. Mechanisms of caspase activation. Curr Opin Cell Biol 2003; 15: 725–731. [DOI] [PubMed] [Google Scholar]
- McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol 2013; 5: a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLuskey K, Rudolf J, Proto WR, Isaacs NW, Coombs GH, Moss CX et al. Crystal structure of a Trypanosoma brucei metacaspase. Proc Natl Acad Sci USA 2012; 109: 7469–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelzer C, Cabalzar K, Wolf A, Gonzalez M, Lenz G, Thome M. The protease activity of the paracaspase MALT1 is controlled by monoubiquitination. Nat Immunol 2013; 14: 337–345. [DOI] [PubMed] [Google Scholar]
- Helmersson A, von Arnold S, Bozhkov PV. The level of free intracellular zinc mediates programmed cell death/cell survival decisions in plant embryos. Plant Physiol 2008; 147: 1158–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proto WR, Castanys-Munoz E, Black A, Tetley L, Moss CX, Juliano L et al. Trypanosoma brucei Metacaspase 4 Is a Pseudopeptidase and a Virulence Factor. J Biol Chem 2011; 286: 39914–39925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen D, Belenghi B, van de Cotte B, Beunens T, Gavigan JA, De Rycke R et al. Serpin1 of Arabidopsis thaliana is a suicide inhibitor for metacaspase 9. J Mol Biol 2006; 364: 625–636. [DOI] [PubMed] [Google Scholar]
- Lema Asqui S, Vercammen D, Serrano I, Valls M, Rivas S, Van Breusegem F et al. AtSERPIN1 is an inhibitor of the metacaspase AtMC1-mediated cell death and autocatalytic processing in planta. New Phytol 2017; doi:10.1111/nph.14446. [DOI] [PubMed]
- Ray CA, Black RA, Kronheim SR, Greenstreet TA, Sleath PR, Salvesen GS et al. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme. Cell 1992; 69: 597–604. [DOI] [PubMed] [Google Scholar]
- Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W et al. An overview of the serpin superfamily. Genome Biol 2006; 7: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poreba M, Strozyk A, Salvesen GS, Drag M. Caspase Substrates and Inhibitors. Cold Spring Harb Perspect Biol 2013; 5: a008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozhkov PV, Salvesen G (eds). Caspases, paracaspases, and metacaspases methods and protocols. Methods Mol Biol 2014; 1133. [Google Scholar]
- He R, Rotari V, Willer M, Gallois P. Are plant metacaspases funtional homologues of animal caspases? Comp Biochem Phys A 2005; 141: S287–S288. [Google Scholar]
- Vercammen D, Declercq W, Vandenabeele P, Van Breusegem F. Are metacaspases caspases? J Cell Biol 2007; 179: 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breusegem F, Belenghi B, Beunens T, Vercammen D. Metacaspases are not caspases. Comp Biochem Phys A 2007; 146: S200–S200. [Google Scholar]
- Carmona-Gutierrez D, Frohlich KU, Kroemer G, Madeo F. Metacaspases are caspases. Doubt no more. Cell Death Differ 2010; 17: 377–378. [DOI] [PubMed] [Google Scholar]
- Bozhkov PV, Smertenko AP, Zhivotovsky B. Aspasing out metacaspases and caspases: proteases of many trades. Sci Signal 2010; 3. [DOI] [PubMed] [Google Scholar]
- Enoksson M, Salvesen GS. Metacaspases are not caspases—always doubt. Cell Death Differ 2010; 17: 1221–1221. [DOI] [PubMed] [Google Scholar]
- Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem 2011; 80: 1055–1087. [DOI] [PubMed] [Google Scholar]
- Ayyash M, Tamimi H, Ashhab Y. Developing a powerful in silico tool for the discovery of novel caspase-3 substrates: a preliminary screening of the human proteome. BMC Bioinformatics 2012; 13: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Tschopp J. Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ 2007; 14: 10–22. [DOI] [PubMed] [Google Scholar]
- Yi CH, Yuan JY. The Jekyll and Hyde Functions of Caspases. Dev Cell 2009; 16: 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthi AU, Martin SJ. The CASBAH: a searchable database of caspase substrates. Cell Death Differ 2007; 14: 641–650. [DOI] [PubMed] [Google Scholar]
- Coornaert B, Baens M, Heyninck K, Bekaert T, Haegman M, Staal J et al. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-κB inhibitor A20. Nat Immunol 2008; 9: 263–271. [DOI] [PubMed] [Google Scholar]
- Staal J, Driege Y, Bekaert T, Demeyer A, Muyllaert D, Van Damme P et al. T-cell receptor-induced JNK activation requires proteolytic inactivation of CYLD by MALT1. EMBO J 2011; 30: 1742–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailfinger S, Nogai H, Pelzer C, Jaworski M, Cabalzar K, Charton JE et al. Malt1-dependent RelB cleavage promotes canonical NF-kappa B activation in lymphocytes and lymphoma cell lines. Proc Natl Acad Sci USA 2011; 108: 14596–14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein T, Fung SY, Renner F, Blank MA, Dufour A, Kang S et al. The paracaspase MALT1 cleaves HOIL1 reducing linear ubiquitination by LUBAC to dampen lymphocyte NF-κB signalling. Nat Commun 2015; 6: 8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrom JF, Vaculova A, Smertenko AP, Savenkov EI, Golovko A, Minina E et al. Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat Cell Biol 2009; 11: 1347–1354. [DOI] [PubMed] [Google Scholar]
- Tsiatsiani L, Timmerman E, De Bock PJ, Vercammen D, Stael S, van de Cotte B et al. The Arabidopsis metacaspase9 degradome. Plant Cell 2013; 25: 2831–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel I, Osiewacz HD. Poly(ADP-Ribose) Polymerase Is a Substrate Recognized by Two Metacaspases of Podospora anserina. Eukaryot Cell 2013; 12: 900–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M, Vainonen JP, Stael S, Tsiatsiani L, Help-Rinta-Rahko H, Gauthier A et al. GRIM REAPER peptide binds to receptor kinase PRK5 to trigger cell death in Arabidopsis. EMBO J 2015; 34: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Herker E, Maldener C, Wissing S, Lachelt S, Herlan M et al. A caspase-related protease regulates apoptosis in yeast. Mol Cell 2002; 9: 911–917. [DOI] [PubMed] [Google Scholar]
- Hill SM, Nystrom T. The dual role of a yeast metacaspase: What doesn't kill you makes you stronger. Bioessays 2015; 37: 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien O, Zhuang M, Wiita AP, O'Donoghue AJ, Knudsen GM, Craik CS et al. Quantitative MS-based enzymology of caspases reveals distinct protein substrate specificities, hierarchies, and cellular roles. Proc Natl Acad Sci USA 2016; 113: E2001–E2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL et al. Arabidopsis type I metacaspases control cell death. Science 2010; 330: 1393–1397. [DOI] [PubMed] [Google Scholar]
- Helms MJ, Ambit A, Appleton P, Tetley L, Coombs GH, Mottram JC. Bloodstream form Trypanosoma brucei depend upon multiple metacaspases associated with RAB11-positive endosomes. J Cell Sci 2006; 119: 1105–1117. [DOI] [PubMed] [Google Scholar]
- Kallenberger SM, Beaudouin J, Claus J, Fischer C, Sorger PK, Legewie S et al. Intra- and Interdimeric caspase-8 self-cleavage controls strength and timing of CD95-induced apoptosis. Sci Signal 2014; 7: ra23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal J, Beyaert R. A two-step activation mechanism of MALT1 paracaspase. J Mol Biol 2012; 419: 1–3. [DOI] [PubMed] [Google Scholar]
- Keller N, Grutter MG, Zerbe O. Studies of the molecular mechanism of caspase-8 activation by solution NMR. Cell Death Differ 2010; 17: 710–718. [DOI] [PubMed] [Google Scholar]
- Moss CX, Westrop GD, Juliano L, Coombs GH, Mottram JC. Metacaspase 2 of Trypanosoma brucei is a calcium-dependent cysteine peptidase active without processing. FEBS Lett 2007; 581: 5635–5639. [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Beers EP, Dangl JL, Franklin-Tong VE, Gallois P, Hara-Nishimura I et al. Morphological classification of plant cell deaths. Cell Death Differ 2011; 18: 1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Beltran E, Moschou PN, Smertenko AP, Bozhkov PV. Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell 2015; 27: 926–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Beltran E, Denisenko TV, Zhivotovsky B, Bozhkov PV. Tudor staphylococcal nuclease: biochemistry and functions. Cell Death Differ 2016; 23: 1739–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulares AH, Yakovlev AG, Ivanova V, Stoica BA, Wang G, Iyer S et al. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. Caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. J Biol Chem 1999; 274: 22932–22940. [DOI] [PubMed] [Google Scholar]
- Shao W, Yeretssian G, Doiron K, Hussain SN, Saleh M. The caspase-1 digestome identifies the glycolysis pathway as a target during infection and septic shock. J Biol Chem 2007; 282: 36321–36329. [DOI] [PubMed] [Google Scholar]
- Silva A, Almeida B, Sampaio-Marques B, Reis MIR, Ohlmeier S, Rodrigues F et al. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a specific substrate of yeast metacaspase. Biochim Biophys Acta 2011; 1813: 2044–2049. [DOI] [PubMed] [Google Scholar]
- Mashima T, Naito M, Tsuruo T. Caspase-mediated cleavage of cytoskeletal actin plays a positive role in the process of morphological apoptosis. Oncogene 1999; 18: 2423–2430. [DOI] [PubMed] [Google Scholar]
- Wilmanski JM, Petnicki-Ocwieja T, Kobayashi KS. NLR proteins: integral members of innate immunity and mediators of inflammatory diseases. J Leukocyte Biol 2008; 83: 13–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyrka W, Lamacchia M, Durrens P, Kobe B, Daskalov A, Paoletti M et al. Diversity and variability of NOD-like receptors in fungi. Genome Biol Evol 2014; 6: 3137–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S, Bernoux M, Moncuquet P, Kroj T, Dodds PN. A novel conserved mechanism for plant NLR protein pairs: the ‘integrated decoy’ hypothesis. Front Plant Sci 2014; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome M. Multifunctional roles for MALT1 in T-cell activation. Nat Rev Immunol 2008; 8: 495–500. [DOI] [PubMed] [Google Scholar]
- Copeland C, Woloshen V, Huang Y, Li X. AtCDC48A is involved in the turnover of an NLR immune receptor. Plant J 2016; 88: 294–305. [DOI] [PubMed] [Google Scholar]
- Leger T, Garcia C, Ounissi M, Lelandais G, Camadro JM. The metacaspase (Mca1p) has a dual role in farnesol-induced apoptosis in Candida albicans. Mol Cell Proteomics 2015; 14: 93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GQ, Shin SBY, Jaffrey SR. Global profiling of protease cleavage sites by chemoselective labeling of protein N-termini. Proc Natl Acad Sci USA 2009; 106: 19310–19315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demon D, Van Damme P, Vanden Berghe T, Deceuninck A, Van Durme J, Verspurten J et al. Proteome-wide substrate analysis indicates substrate exclusion as a mechanism to generate caspase-7 versus caspase-3 specificity. Mol Cell Proteomics 2009; 8: 2700–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impens F, Colaert N, Helsens K, Ghesquiere B, Timmerman E, de Bock PJ et al. A quantitative proteomics design for systematic identification of protease cleavage events. Mol Cell Proteomics 2010; 9: 2327–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 2008; 134: 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zhao XM, Tan H, Akutsu T, Whisstock JC, Song J. Cascleave 2.0, a new approach for predicting caspase and granzyme cleavage targets. Bioinformatics 2014; 30: 71–80. [DOI] [PubMed] [Google Scholar]
- Hatsugai N, Yamada K, Goto-Yamada S, Nara-Nishimura I. Vacuolar processing enzyme in plant programmed cell death. Front Plant Sci 2015; 6: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez MF, Filonova LH, Smertenko A, Savenkov EI, Clapham DH, von Arnold S et al. Metacaspase-dependent programmed cell death is essential for plant embryogenesis. Curr Biol 2004; 14: R339–R340. [DOI] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL. Programmed cell death in the plant immune system. Cell Death Differ 2011; 18: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roisin-Bouffay C, Luciani MF, Klein G, Levraud JP, Adam M, Golstein P. Developmental cell death in dictyostelium does not require paracaspase. J Biol Chem 2004; 279: 11489–11494. [DOI] [PubMed] [Google Scholar]
- Madeo F, Carmona-Gutierrez D, Ring J, Buttner S, Eisenberg T, Kroemer G. Caspase-dependent and caspase-independent cell death pathways in yeast. Biochem Biophys Res Commun 2009; 382: 227–231. [DOI] [PubMed] [Google Scholar]
- Bollhoner B, Zhang B, Stael S, Denance N, Overmyer K, Goffner D et al. Post mortem function of AtMC9 in xylem vessel elements. New Phytol 2013; 200: 498–510. [DOI] [PubMed] [Google Scholar]
- Minina EA, Filonova LH, Fukada K, Savenkov EI, Gogvadze V, Clapham D et al. Autophagy and metacaspase determine the mode of cell death in plants. J Cell Biol 2013; 203: 917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamez S, Andre D, Zhang B, Bollhoner B, Pesquet E, Tuominen H. METACASPASE9 modulates autophagy to confine cell death to the target cells during Arabidopsis vascular xylem differentiation. Biol Open 2016; 5: 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhart L, Lippens S, Tschachler E, Declercq W. Cell death by cornification. Biochim Biophys Acta 2013; 1833: 3471–3480. [DOI] [PubMed] [Google Scholar]
- Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat Cell Biol 2001; 3: 339–345. [DOI] [PubMed] [Google Scholar]
- Gauster M, Siwetz M, Orendi K, Moser G, Desoye G, Huppertz B. Caspases rather than calpains mediate remodelling of the fodrin skeleton during human placental trophoblast fusion. Cell Death Differ 2010; 17: 336–345. [DOI] [PubMed] [Google Scholar]
- Lee A, Morrow JS, Fowler VM. Caspase remodeling of the spectrin membrane skeleton during lens development and aging. J Biol Chem 2001; 276: 20735–20742. [DOI] [PubMed] [Google Scholar]
- Smertenko AP, Bozhkov PV, Filonova LH, von Arnold S, Hussey PJ. Re-organisation of the cytoskeleton during developmental programmed cell death in Picea abies embryos. Plant J 2003; 33: 813–824. [DOI] [PubMed] [Google Scholar]
- Smertenko A, Franklin-Tong VE. Organisation and regulation of the cytoskeleton in plant programmed cell death. Cell Death Differ 2011; 18: 1263–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko A, Kim JC, Kang TB, Rajput A, Bogdanov K, Dittrich-Breiholz O et al. Caspase-8 deficiency in epidermal keratinocytes triggers an inflammatory skin disease. J Exp Med 2009; 206: 2161–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin BS, Bossaller L, De Nardo D, Ratter JM, Stutz A, Engels G et al. The adaptor ASC has extracellular and 'prionoid' activities that propagate inflammation. Nat Immunol 2014; 15: 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroja-Mazo A, Martin-Sanchez F, Gomez AI, Barbera-Cremades M, Amores-Iniesta J, Martinez CM et al. P2X7 receptor induces the release of NLRP3 inflammasome to propagate inflammation. Purinergic Signal 2014; 10: 817–818. [Google Scholar]
- Baroja-Mazo A, Martin-Sanchez F, Gomez AI, Martinez CM, Amores-Iniesta J, Compan V et al. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat Immunol 2014; 15: 738–748. [DOI] [PubMed] [Google Scholar]
- Wall DM, McCormick BA. Bacterial secreted effectors and caspase-3 interactions. Cell Microbiol 2014; 16: 1746–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avci U, Petzold HE, Ismail IO, Beers EP, Haigler CH. Cysteine proteases XCP1 and XCP2 aid micro-autolysis within the intact central vacuole during xylogenesis in Arabidopsis roots. Plant J 2008; 56: 303–315. [DOI] [PubMed] [Google Scholar]
- Courtois-Moreau CL, Pesquet E, Sjodin A, Muniz L, Bollhoner B, Kaneda M et al. A unique program for cell death in xylem fibers of Populus stem. Plant J 2009; 58: 260–274. [DOI] [PubMed] [Google Scholar]
- Fendrych M, Van Hautegem T, Van Durme M, Olvera-Carrillo Y, Huysmans M, Karimi M et al. Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr Biol 2014; 24: 931–940. [DOI] [PubMed] [Google Scholar]
- Obara K, Kuriyama H, Fukuda H. Direct evidence of active and rapid nuclear degradation triggered by vacuole rupture during programmed cell death in Zinnia. Plant Physiol 2001; 125: 615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Band LR, Kumpf RP, Van Damme D, Parizot B, De Rop G et al. Cyclic programmed cell death stimulates hormone signaling and root development in Arabidopsis. Science 2016; 351: 384–387. [DOI] [PubMed] [Google Scholar]
- Olvera-Carrillo Y, Van Bel M, Van Hautegem T, Fendrych M, Huysmans M, Simaskova M et al. A conserved core of programmed cell death indicator genes discriminates developmentally and environmentally induced programmed cell death in plants. Plant Physiol 2015; 169: 2684–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R, Drury GE, Rotari VI, Gordon A, Willer M, Farzaneh T et al. Metacaspase-8 modulates programmed cell death induced by ultraviolet light and H2O2 in Arabidopsis. J Biol Chem 2008; 283: 774–783. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27: 107–132. [DOI] [PubMed] [Google Scholar]
- Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol 2015; 16: 461–472. [DOI] [PubMed] [Google Scholar]
- Meijer WH, van der Klei IJ, Veenhuis M, Kiel JA. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy 2007; 3: 106–116. [DOI] [PubMed] [Google Scholar]
- Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Bio 2014; 15: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D, Munch D, Bressendorff S, Mundy J, Petersen M. Role of autophagy in disease resistance and hypersensitive response-associated cell death. Cell Death Differ 2011; 18: 1257–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoletopoulou V, Markaki M, Palikaras K, Tavernarakis N. Crosstalk between apoptosis, necrosis and autophagy. Biochim Biophys Acta 2013; 1833: 3448–3459. [DOI] [PubMed] [Google Scholar]
- Minina EA, Bozhkov PV, Hofius D. Autophagy as initiator or executioner of cell death. Trends Plant Sci 2014; 19: 692–697. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Che XR, Zheng QL, Wu A, Pan K, Shao AW et al. Caspases: a molecular switch node in the crosstalk between autophagy and apoptosis. Int J Biol Sci 2014; 10: 1072–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A, Bozhkov PV. Somatic embryogenesis: life and death processes during apicalbasal patterning. J Exp Bot 2014; 65: 1343–1360. [DOI] [PubMed] [Google Scholar]
- Han J, Hou W, Goldstein LA, Stolz DB, Watkins SC, Rabinowich H. A complex between Atg7 and caspase-9A novel mechanism of cross-regulation between autophagy and apoptosis. J Biol Chem 2014; 289: 6485–6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory CD, Devitt A. The macrophage and the apoptotic cell: an innate immune interaction viewed simplistically? Immunology 2004; 113: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosta A, Roisin-Bouffay C, Luciani MF, Otto GP, Kessin RH, Golstein P. Autophagy gene disruption reveals a non-vacuolar cell death pathway in Dictyostelium. J Biol Chem 2004; 279: 48404–48409. [DOI] [PubMed] [Google Scholar]
- Kwon SI, Cho HJ, Jung JH, Yoshimoto K, Shirasu K, Park OK. The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J 2010; 64: 151–164. [DOI] [PubMed] [Google Scholar]
- Lee REC, Brunette S, Puente LG, Megeney LA. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc Natl Acad Sci USA 2010; 107: 13348–13353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SM, Hao XX, Liu BD, Nystrom T. Life-span extension by a metacaspase in the yeast Saccharomyces cerevisiae. Science 2014; 344: 1389–1392. [DOI] [PubMed] [Google Scholar]
- Coll NS, Smidler A, Puigvert M, Popa C, Valls M, Dangl JL. The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: functional linkage with autophagy. Cell Death Differ 2014; 21: 1399–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt M, Wegrzyn RD, Deuerling E. Extra N-terminal residues have a profound effect on the aggregation properties of the potential yeast prion protein Mca1. Plos One 2010; 5: e9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lu P, Zhong S, Chen H, Zhou B. LcMCII-1 is involved in the ROS-dependent senescence of the rudimentary leaves of Litchi chinensis. Plant Cell Rep 2016; 36: 89–102. [DOI] [PubMed] [Google Scholar]
- Hamann A, Brust D, Osiewacz HD. Deletion of putative apoptosis factors leads to lifespan extension in the fungal ageing model Podospora anserina. Mol Microbiol 2007; 65: 948–958. [DOI] [PubMed] [Google Scholar]
- Nakagawa A, Shi Y, Kage-Nakadai E, Mitani S, Xue D. Caspase-dependent conversion of dicer ribonuclease into a death-promoting deoxyribonuclease. Science 2010; 328: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Wang C, Okerberg E, Patricelli MP, Cravatt BF. Functional Interplay between caspase cleavage and phosphorylation sculpts the apoptotic proteome. Cell 2012; 150: 426–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowec JP, Zukowski SA, Knight JD, Smalley DM, Graves LM, Johnson GL et al. An unbiased proteomic screen reveals caspase cleavage is positively and negatively regulated by substrate phosphorylation. Mol Cell Proteomics 2014; 13: 1184–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY et al. CDD: NCBI's conserved domain database. Nucleic Acids Res 2015; 43: D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.