Abstract
The phenotypic and biochemical similarities between caspase-mediated apoptosis and cellular differentiation are striking. They include such diverse phenomenon as mitochondrial membrane perturbations, cytoskeletal rearrangements and DNA fragmentation. The parallels between the two disparate processes suggest some common ancestry and highlight the paradoxical nature of the death-centric view of caspases. That is, what is the driving selective pressure that sustains death-inducing proteins throughout eukaryotic evolution? Plausibly, caspase function may be rooted in a primordial non-death function, such as cell differentiation, and was co-opted for its role in programmed cell death. This review will delve into the links between caspase-mediated apoptosis and cell differentiation and examine the distinguishing features of these events. More critically, we chronicle the evolutionary origins of caspases and propose that caspases may have held an ancient role in mediating the fidelity of cell division/differentiation through its effects on proteostasis and protein quality control.
Facts
Caspase and caspase-like proteases (termed metacaspases) are a conserved group of proteins that are present in multicellular and unicellular eukaryotes, as well as several prokaryotes.
Caspases, which have primarily been associated with apoptotic cell death, are involved in numerous non-apoptotic processes including cell differentiation, cell cycle progression, proteostasis and cellular remodeling.
There are overt similarities – both in phenotype and cell signaling – between caspase-mediated cell death and cell differentiation.
Open Questions
Does caspase-mediated apoptosis engage the same substrates as those involved in cell differentiation?
Are the origins of caspase-mediated signaling rooted in non-apoptotic functions as opposed to facilitating cell death?
What determines the choice between cell death and non-death adaptation once caspase proteases are activated?
The role of caspases, a class of cysteine-dependent aspartate-directed proteases, in initiating and executing apoptotic programmed cell death (PCD) has been substantiated by a plethora of studies conducted over the past several decades.1 Although several caspases (e.g., caspase-1 and -11) were originally discovered as non-death proteases, the majority of these enzymes were characterized from their action in inducing apoptotic cell death (details of both the intrinsic and extrinsic apoptotic pathways are reviewed by Elmore2). Nevertheless, parallel research efforts have demonstrated that caspase activity is indispensable to many other cellular processes independent of inducing cell death.3 This apparent paradox is best exemplified in the study of cell differentiation, where caspase activity has been shown to modify the differentiation of virtually all somatic cell types tested, across a diverse spectrum of metazoan organisms.4, 5
In the following review, we will discuss the non-canonical role for caspase proteases in cell differentiation and the evolutionary origin of this protein activity. We will present evidence that the death and non-death roles of caspase proteins are equally conserved across the phyla and that this duality of function may have co-evolved from homologous proteins in single-cell eukaryotes. The corollary to this hypothesis is that caspase control of cell differentiation is not a recent co-option of a death-centric protein, rather the non-death role of this protease clad may be its primordial function.
Phenotypic Similarities Between Apoptosis and Differentiation
Apoptotic PCD is typically characterized by a number of biochemical and morphological changes to the cell. For example, one of the defining characteristics of apoptosis is the endonuclease-driven hydrolysis of the DNA into small fragments.6 This is usually accompanied by chromatin and nuclear condensation and eventually fragmentation of the nucleus.7 In addition, several other organelles are subject to destruction, including the Golgi apparatus, endoplasmic reticulum and mitochondria.8, 9, 10 The cellular superstructure is also extensively cleaved upon the activation of apoptosis, with microfilaments, actin-associated proteins, microtubules, microtubule-associated proteins and intermediate filaments all being targeted for degradation.11 A consequence of cytoskeletal destruction is dynamic membrane blebbing. This is characteristic of apoptosis and typically leads to the formation of vesicles, termed apoptotic bodies, which are subsequently phagocytosed by surrounding immune cells.12, 13 Critical to this process appears to be the immune cell recognition of cell membrane phosphatidylserine, which characteristically translocates from the inner leaflet to the outer leaflet of the membrane during apoptosis.14, 15
Interestingly, many of the above apoptosis-related events are observed during the differentiation of varied cell types. For instance, the exposure of phosphatidylserine in the outer leaflet of myoblast cell membranes appears necessary for mediating myoblast fusion and the formation of myotubes.16 Recent evidence suggests that the expression of the phosphatidylserine receptor, stabilin-2, increases during myoblast differentiation and is critical for this fusion process.17 Furthermore, akin to the process of membrane blebbing and apoptotic body formation, neural progenitor cells appear to release membranous particles during the onset or early stages of neurogenesis.18
Surprisingly, some differentiating cells undergo organelle degradation similar to that observed during apoptosis. This is especially true for erythroblasts, lens fiber cells and keratinocytes, where nuclei and other organelles are degraded in an apoptosis-like event as the cells proceed toward terminal differentiation.19, 20 Significantly more common in differentiating cells is the occurrence of DNA strand breaks, which is akin to apoptotic DNA fragmentation. Evidence for DNA cleavage during differentiation appears in granulocytes,21 monocytes,22 myoblasts,23, 24, 25 lymphocytes,26 keratinocytes27 and erythroleukemic cells.28 The multitude of phenotypic similarities between apoptotic cell death and the differentiation of a number of stem/precursor cells suggest that a common signaling cascade may direct both processes.
Conservation of biochemical signaling pathways that initiate apoptosis and differentiation
Although being well established as mediators of cell death, the TNF superfamily of ligands and receptors are important to a number of intracellular processes including initiation of cell differentiation. For instance, several studies have indicated that signaling through TNFR is critical to the differentiation of osteoclasts,29, 30 and monocytes.31 Osteoclastogenesis provides a particularly apt example, with differentiation being mediated in part through the TNF receptor, RANK, and its ligand, RANKL.29 Subsequent examination of this receptor–ligand combination elucidated that it is also responsible for the initiation of apoptosis in osteoclast precursor cells,32 thereby highlighting the coincident signaling of apoptotic and differentiating cells.
One of the key initiating events that trigger the intrinsic apoptotic pathway is the disruption of the MTP. Interestingly, a similar event appears to occur during erythroid differentiation, where a reduction in MTP and an activation of downstream caspases is critical for erythroid maturation.33 Further, evidence exists that suggests alterations in MTP occur during osteogenesis34 and the differentiation of erythroleukemia cells,35 although the research has not linked these changes in MTP to caspase activation. In addition to the changes in MTP, the intrinsic apoptotic pathway is characterized by the release of cytochrome c from the mitochondrial intermembrane space. This has been observed in the differentiation of human peripheral blood monocytes undergoing differentiation into mature macrophages.36
Caspase proteases: conserved inductive cues for cell Differentiation
Caspase proteases have long been typecast as the initiators and executers of apoptotic cell death; however, evidence, especially over the past two decades, has revealed a surprising functional diversity for these enzymes that is entirely independent of cell death. The physiologic activity of caspase proteins has been most studied in the context of cell differentiation and involves a plethora of observations linking both initiator and executioner caspases with the induction and management of this cell fate (Table 1). This alternate biology for caspase function was first presented in four independent studies. Work from the laboratory of Raff and colleagues provided the first evidence that caspase-3 activity was essential for cell differentiation, specifically in lens epithelial cells.37 In a subsequent article, intrinsic pathway-mediated caspase-3 activity was shown to be indispensable for erythropoiesis, specifically the terminal differentiation and maturation of red blood cells.33 This stance was later modified by a study that determined caspase-3 to be critical to the early stages of erythroid development, and not in terminal enucleation.38 That being said, at the time of the earlier publications, the prevailing consensus in the field viewed this non-canonical role of caspase-3 as simply engaging a modified or attenuated form of cell death, as both keratinocytes and red blood cells are short lived. However, this death-centric view of caspase-3 in cell differentiation was challenged by two complimentary, yet independent observations. First, transient caspase-3 activity was shown to be indispensable for murine skeletal muscle differentiation, managing the fusion of myoblasts to form fully differentiated myofibers, whereas concurrently engaging a pro-differentiation gene expression program.39 Unlike red blood cells and keratinocytes, vertebrate skeletal muscle fibers are long-lived cell types that continue to grow and adapt in response to external and internal cues. Accordingly, this study provided the first demonstration that effector caspase activity could be transiently engaged to alter cell fate, whereas maintaining all organelle structures. The second observation to refute the attenuated apoptosis concept of caspase-mediated differentiation was provided by examining caspase function in Drosophila. In an elegant study, Arama et al.40 demonstrated that activation of the traditional mitochondrial death pathway was a conserved feature in spermatid differentiation and that activation of drICE (the Drosophila homolog of caspase-3) was essential to complete the process of maturation. Reminiscent of the structural reorganization that caspase-3 engineered in developing myoblasts, drICE proteolytic activity promoted the removal of superfluous cytoplasmic content, allowing for the emergence of mature individualized spermatids.
Table 1. Eukaryotic caspases involved in cellular differentiation.
| Organism | Tissue/cell | Caspase(s) involved | Reference(s) |
|---|---|---|---|
| Mammals | Lens epithelial cells | Caspase-3 | 37 |
| Erythrocytes | Caspase-3 | 33 | |
| Skeletal muscle | Caspase-3, -9 | 39, 57 | |
| Bone cells | Caspase-2, -8 | 41 | |
| Bone marrow stromal cells | Caspase-3 | 42 | |
| Monocytes | Caspase-2, -3, -8, -9 | 36 | |
| Neurons | Caspase-3 | 44, 45, 46 | |
| Keratinocytes | Caspase-3 | 47 | |
| Embryonic stem cells | Caspase-3 | 48 | |
| Hematopoietic stem cells | Caspase-3 | 49 | |
| Cardiomyocytes | Caspase-3 | 50, 51 | |
| Dental hard tissues | Caspase-7 | 52 | |
| Drosophila | Spermatids | drICE (caspase-3) | 40 |
| C. elegans | Seam cells | CED-3 (caspase-3, -9) | 55 |
Following these definitive studies, the role of caspase activity in promoting differentiation was demonstrated across a broad spectrum of cell lineages, from invertebrates to mammals. These studies include characterization of individual initiator and/or effector caspases, in addition to observations that fully intact extrinsic and/or intrinsic cell death pathways are employed to engage differentiation programs. Following these hallmark studies, osteogenesis,41 bone marrow stromal cells,42 monocytes,36 were determined to involve the activation of initiator and/or executioner caspases without the occurrence of apoptosis. Subsequently, the differentiation of a broad range of both invertebrate and vertebrate neuronal cell lineages was shown to be dependent on caspase-3 activity, including astrocytes, oligodendrocytes, striatal and olfactory neurons.43, 44, 45, 46 Still further, caspases have been shown to be critical in the differentiation of keratinocytes,47 embryonic stem cells (ESCs),48 hematopoietic stem cells,49 cardiomyocyte progenitors,50, 51 and pre-odontoblasts and -ameloblasts during the development of dental hard tissues.52 Most fittingly, the conservation of caspase-mediated differentiation has now been extended to and verified in the round worm C. elegans, the very model system where caspase-dependent apoptotic signal pathways were originally discovered.53, 54 Here, a recent study involving the C. elegans initiator and executioner caspase, CED-3, identified this protease as a novel regulator of stem cell-like seam cells. CED-3 was found to proteolytically process key pluripotency factors, such as LIN-14, LIN-28 and DISL2, which are necessary for the progression of these cells through their differentiation program.55
Many of these studies focused on effector caspase activity alone, yet parallel literature has emerged to show that the differentiation-specific function of effector caspases is largely derived from a fully reconstituted activation of the intrinsic cell death pathway. The conservation and re-deployment of this death pathway was first suggested in the pioneering study on Drosophila spermatogenesis, where Arama et al.40 indicated that drICE engagement was dependent on a distal cytochrome c signal. In a series of elegant experiments, Menko and colleagues also demonstrated that lens fiber epithelial cell differentiation was dependent on a pulsatile release of mitochondrial cytochrome c, engaging the intrinsic pathway.56 This study also demonstrated that the differentiation process was concurrent to elevated expression of anti-apoptotic genes, perhaps as a regulatory loop to limit caspase activation levels, preventing the transition to fulminant apoptosis. Subsequently, the role of the intrinsic pathway as the priming source for caspase-mediated differentiation was confirmed in a number of cell types including skeletal muscle, neurons and other epithelial derivatives.5, 45, 57
Given the phyla conservation for caspase-mediated cell differentiation there is a compelling interest to identify the substrate(s) that are targeted by these proteases. For example, in apoptotic scenarios, caspase-3 directed cleavage activates a number of protein kinase substrates by removing their N-terminal regulatory domain, freeing these enzymes to propagate a signaling cascade. The mitogen-activated protein kinase kinase kinase (MEKK1), which is a stress-activated protein kinase, provides an example of this phenomenon. MEKK1 is known to be cleavage activated by caspases in response to a number of proapoptotic stimuli such as Fas ligation,58 genotoxin insult,59 and possibly, when a cell loses integrin contact with the extracellular matrix (termed anoikis).60 The first caspase substrate to be definitively identified in a pro-differentiation context was the Ste20-like kinase MST1. Like other MAPKs, MST1 has a catalytic domain that when cleaved and released from its nascent regulatory region, assumes maximal activity and stimulates apoptosis.61 During skeletal muscle differentiation, caspase-3 cleaves and activates MST1 and this activated form of the kinase can partially rescue the differentiation deficit in caspase-3 null myoblasts.39 A number of other examples exist where cleavage activation of a particular protein kinase occurs during both apoptosis and differentiation, including the cleavage activation of MEKK1,62 p21-activated kinase 1 and 2 (PAK1 and PAK2),43, 63, 64 ROCK1,65 and PKCδ.47, 66, 67
Apoptotic activation of caspase-3 leads to a plethora of downstream events that results in cell death. One such event is the activation of the endogenous endonuclease, CAD, which mediates DNA fragmentation and promotes cell death. Interestingly, CAD becomes active following caspase-3 activation during skeletal muscle myoblast differentiation, and it proceeds to cleave DNA at specific sites throughout the genome. One well-studied example of CAD action during this process occurs when CAD targets the promoter region of p21, a cyclin-dependent kinase inhibitor.25 This transient cleavage promotes p21 expression and exit of the cell cycle, thereby promoting myoblast differentiation. Continued research into CAD targeting of genes within the differentiation program is necessary to elucidate the full spectrum of genes affected, but recent work does support the supposition that this phenomenon broadly targets the genome.68 Specifically, XRCC1, a DNA repair protein that mends CAD-induced DNA strand breaks, is present in numerous locations within the nuclei of differentiating myoblasts and myoblast specific loss of XRCC1 leads to unresolved DNA strand breaks with a catastrophic failure in differentiation.68 The general conservation of caspase-3 activation during cell differentiation suggests other cell lineages are likely to utilize a similar damage-mediated genome-reprogramming event.
Although a complete discussion of the caspase substrates cleaved during apoptosis and differentiation has not been conducted, the above evidence suggests that there may be little variation in the substrate selection between these divergent cell fates. Examples of context-dependent caspase substrates exist, yet the overwhelming number of caspase substrates that have been identified in differentiating cell types were previously identified as bona fide substrates in apoptotic conditions.
Guiding caspase deployment to determine cell fate: restraint is the key
The conservation of caspase signaling between apoptosis and differentiation strongly suggests that these proteases achieve phenotypic variation by being actively managed. Clearly, differentiation would require a more tightly scripted caspase activation pattern, which would subdue the transition to apoptosis. Several examples exist that demonstrate that modulating the intensity or duration of caspase signaling may be critical in the death/non-death cell fate decision. For instance, cytochrome c release and caspase activity in mouse ESCs rose rapidly following the induction of apoptosis, but was more slow to develop during differentiation of ESCs into the cardiomyocyte lineage; maximum caspase activity being delayed by 24 h when differentiation was induced.69 Moreover, many differentiating cell types display a transient increase in caspase activity, including skeletal muscle myoblasts, osteoblasts, bone marrow stromal cells and neurons.39, 41, 42, 43 This is in stark contrast to the continuous and unregulated increase in caspase activity during apoptosis.39, 70
In addition to differences in the timing of signaling events during apoptosis and differentiation, these processes appear to be distinguished by the intensity of the associated signal. Almost universally, the signal associated with differentiation is dampened when compared with that observed during apoptosis. Typically this is observed as lower levels of caspase activity, as is seen for differentiating lens epithelial cells56 and ESCs,51, 69 or the transient activation of differentiation/apoptosis-related kinases, as found for differentiating keratinocytes71 and myoblasts39
One may envision a number of mechanisms whereby post-translational modifications may limit caspase activity in cell differentiation. Indeed, phosphorylation of caspases (i.e., caspase-2, -3, -6, -7, -8 and -9) acts to dampen proteolytic action.72 One specific example is the negative regulation of caspase-9 activity by phosphorylation at several specific sites,73, 74 which suppresses the transition to full-blown apoptosis. Indeed, there is evidence that the phosphorylation of caspase substrates may compete with caspase proteolytic action, and may have an important role in fine-tuning caspases during differentiation. Systems biology experiments have revealed that casein kinase 2 (CK2) phosphorylation sites and caspase-3 cleavage sites are identical, and that CK2 phosphorylation precludes cleavage across this large substrate cohort while also phosphorylating and inhibiting cleavage activation of caspase-3 itself.75, 76 Interestingly, CK2 activity competes with and limits caspase-3 targeting of the paired box transcription factor Pax7, an event that is critical for promoting self-renewal of skeletal muscle progenitor/stem cells.77, 78 For skeletal muscle progenitor cells to commit to differentiation, CK2-mediated phosphorylation of Pax7 must be reduced, allowing for cleavage inactivation of Pax7.77 These observations do not preclude other vital kinase caspase interactions, however, the ubiquitous expression of CK2 strongly suggests that the countervailing balance between this kinase and effector caspases may be a common regulatory theme in managing metazoan cell differentiation.
Post-translational phosphorylation is by no means the sole modification that affects caspase function, with ubiquitination (caspase-3, -7, -8 and -9) affecting both protease activity and protein stability/turnover.72 To date, with respect to the differentiation process, most of the work has been derived from Drosophila models. Beginning with the discovery of the REAPER/HID/GRIM family of proteins that target and disrupt the interactions between the apical caspase and its nascent inhibitors from the DIAP family (XIAP in mammals), it was noted that these interactions were essential for Drosophila spermatid individualization and differentiation.79 Following this discovery, a cullin 3-based ubiquitin ligase complex was shown to target effector caspases to regulate its engagement during spermatid maturation.80 Similarly, a cullin 1-engaged F-box protein, nutcracker, has also been implicated in Drosophila sperm development.81 The cullin 3-based control of caspase activation appears to be regulated by the Krebs cycle protein succinyl-CoA synthase, which sits on the mitochondrial membrane and spatially restricts effector caspase activation.82 These observations suggest that proteosomal control of caspase inhibitors and initiators serves as a titratable mechanism for tuning non-death caspase signaling, whether it is operational in other examples of caspase-mediated differentiation is not yet known.
The evolutionary origin and utility of caspase-mediated signaling events
The conservation of caspase-mediated biochemical signals in both death and differentiation raises the question as to the evolutionary origin of this protease family. Did caspase proteins and their ancestral equivalents simply evolve from a primordial apoptotic function? Alternatively, did these enzymes co-evolve from ancestral proteins that retained both death and non-death functions? Clearly, the re-deployment of death-centric proteins over evolutionary timescales would lessen selection pressures to remove genes and proteins that were exclusively detrimental to cell survival.
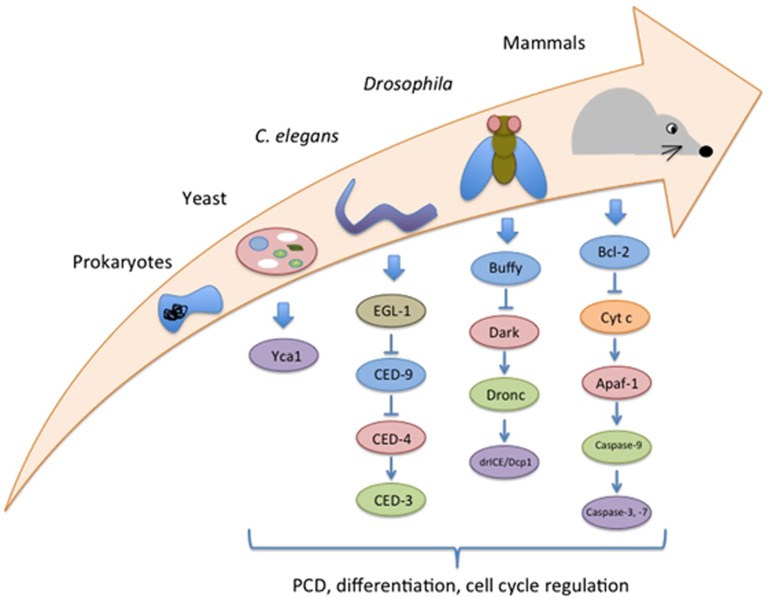
As noted previously, specific cell lineages in Drosophila and C. elegans rely on caspase-mediated control of cell differentiation. These observations support the hypothesis that dual caspase function may be as old as the advent of multicellular animals (Figure 1). Indeed, one could posit based on the available evidence that caspase proteases may have been critical to the very emergence of multicellular life forms. Specifically, caspases are central to two key requirements for all multicellular life forms, elimination of superfluous or damaged cells and cellular specialization, which is the end stage of differentiation. Extension of this logical thread would argue that caspases and their protein equivalents were/are present in single-cell organisms and were/are engaged in both cell death and differentiation-like activities.
Figure 1.
Conservation of caspase signaling cascades through eukaryotic evolution. The effector caspases (purple), caspase-3 and -7 (mammals), drICE and Dcp1 (Drosophila), and CED-3 (functions as both initiator and effector caspase in C. elegans) dictate a diverse set of physiological outcomes including caspase-mediated PCD, differentiation and cell cycle regulation. The lone caspase-like protein within budding yeast, Yca1, acts in a similar manner as the above effector caspases and is involved in yeast cell death, cell cycle progression and proteostasis. Upstream of the effector caspases there are homologous proteins that partake in caspase signaling, including initiator caspases (green), caspase activators (red) and related anti-apoptotic proteins (blue)
Caspase-like proteins – termed metacaspases – exist in unicellular organisms, such as protists, fungi and bacteria, although the precise relationship between these proteases and caspases is still being debated.83, 84 The critical argument against accepting the homology of caspases and metacaspases lies in the differences in protease activation. Specifically, metacaspase enzymes do not require dimerization or the presence of adaptor proteins, as is the case for metazoan caspases.83 Moreover, active metacaspases cleave proteins at arginine/lysine residues as opposed to the aspartate residues typical of metazoan caspases.85, 86, 87 Despite these differences, other key structural features are conserved between caspases and metacaspases. For example, metacaspases contain the caspase-specific catalytic diad,88, 89 and structural analyses by X-ray crystallography have confirmed the presence of a caspase fold in the yeast metacaspase.90
The role of caspases and metacaspases in mediating a death-inducing cellular programs in their respective organisms/cells argues for a significant functional homology between caspases and metacaspases.91 Metacaspase involvement in cell death processes extends to many lower eukaryotes including several fungi,92, 93, 94, 95, 96 protists97, 98 and plants.99 Despite the strong functional overlap between metacaspase and caspase enzymes, enough doubt remains to question whether these proteases are in a direct evolutionary lineage, that is, are caspases the evolved metazoan equivalent of metacaspases? Certainly, gathering and sequencing more rudimentary metazoan species may reveal the existence of a transitional protease that shares features of both metacaspase and caspase proteases. However, one can also envision a number of experiments to more directly address this issue, all of which should be priorities for the field. The most straightforward experiment is to test functional equivalency, which could be readily accomplished in the yeast model by expressing individual caspase genes in the endogenous loci of the yca1 gene. A yeast strain expressing a metazoan caspase (in place of yca1) able to maintain both apoptosis and proteostasis control would provide strong support to the contention that caspase and metacaspase proteases are evolutionarily related. The experiment could also be performed in reverse, expressing a metacaspase in a metazoan system and asking whether this protease can be a functional substitute for caspase activity in the standard apoptotic and differentiation phenotypes. Clearly there are caveats to such an approach, yet a demonstration of functional substitution would provide a compelling argument for evolutionary conservation between metacaspase and caspase enzymes.
Little work has been conducted on the substrate similarities between caspases and metacaspases, however, one study by Sundström et al.100 indicated that the Tudor staphylococcal nuclease (TSN) is a common substrate for the Norway spruce metacaspase and human caspase-3 during PCD. Further work is necessary to determine the scope of substrate overlap between caspases and metacaspases, yet TSN is unlikely to be the sole example. Future experiments may entail caspase and metacaspase substrate profiling with subsequent in vitro validation using caspase/metacaspase inhibitors. That said, it is clear that differences exist between caspases and metacaspases, yet the conserved structural features and functional homology suggests that an evolutionary relationship exists between these two protease classes.
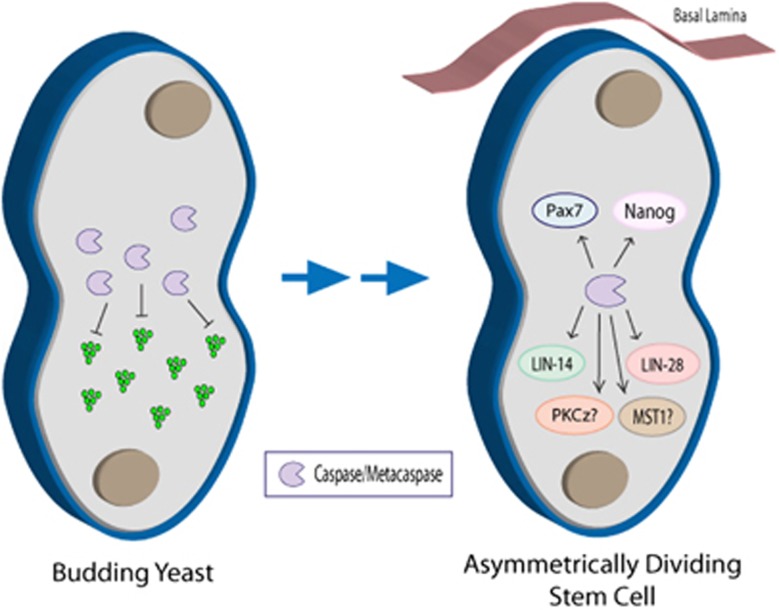
Studies on the unicellular budding yeast, Saccharomyces cerevisiae, may be particularly relevant to explore the evolutionary link between these protease clades as they provide a tractable platform to study protein function in a single-celled eukaryote. Yeast contain a sole caspase-like protein, Yca1, which belongs to the clan C/D metacaspase family, with strong structural homology to caspase proteases.101 Although Yca1 was originally identified in a screen for death-inducing proteins, more recent research has shown that this metacaspase engages a core non-death function, that is, protein quality control. Specifically, work by Lee et al.97 illustrated that Yca1 was essential for protein aggregate disassembly, with deletion/inactivation of Yca1 causing the accumulation of insoluble protein bodies. Clearance of protein aggregates, which accrue following periods of stress or aging, is necessary to prevent PCD. As such, the origin of non-death caspase-like enzyme activity appears as equally conserved in the evolutionary record as the pro-death activity for this protein class. Most intriguingly, Yca1 appears to engage proteostasis control mechanisms in what could be reasonably considered a yeast equivalent of stem cell aging. For example, when stress- or aging-induced protein aggregate accumulation cannot be mitigated sufficiently, budding yeast use another defense mechanism, termed spatial protein quality control, which results in an asymmetrical distribution of protein aggregates during cytokinesis. Yca1 was found to be important in establishing this asymmetry and ensuring that the budding daughter cell was free of insoluble proteins102 (Figure 2).
Figure 2.
The role of caspase in establishing asymmetric cell division: potential primordial link between caspase function and cell differentiation. Caspase-dependent protein processing in the budding yeast, S. cerevisiae, is essential for proteostasis and the destruction of protein aggregates that would be detrimental to cell survival. Upon cytokenesis, the yeast metacaspase, Yca1, is tasked at preventing accumulated protein aggregates from entering the newly formed daughter cell, thereby increasing progeny fitness. This is similar to the emerging role of eukaryotic caspases in regulating stem cell symmetrical/asymmetrical division. To date, caspases have been associated with the degradation of the ESC pluripotency factor, Nanog, as well as the muscle cell lineage-specific factor, Pax7. Caspase targeting of these factors is essential for establishing asymmetric cell division and the initiation of cell differentiation. There is also evidence that suggests that caspase cleavage activation of PKCζ (PKCz in figure) – a known factor in mediating polarity cues in differentiating neuroblasts119 – is critical in initiating asymmetric cell division. Moreover, activation of the mammalian Ste20-like kinase (MST1) via caspase cleavage, which was recently established to be important to the fidelity of muscle cell differentiation, may be involved in polarity signaling during asymmetric cell division. This is based on previous work on Drosophila neuronal cells, where polarity cues relied, in part, on the activated homolog for MST1, Hippo.120, 121 The conservation of caspase involvement in cell polarity signaling is further supported by the caspase-dependent cleavage of LIN-14 and LIN-28 in C. elegans seam cells, with both proteins being critically involved in symmetric/asymmetric cell divisions. Caspase-dependent protein processing during asymmetric cell division may represent the origins of caspase involvement in cell differentiation, and may precede the death-centric functions of these proteases
As noted above, a metazoan equivalent to this ancient mother–daughter cell relationship is the asymmetrical cell division that characterizes the differentiation program in most stem cell populations. In most lineages, stem cells undergo asymmetric cell division to produce a stem cell that repopulates the niche, as well as a daughter cell that engages the differentiation program. A role for effector caspases in general protein aggregation control has not yet been characterized, however, caspase-3 has been implicated in regulating stem cell self-renewal through the targeting of individual protein substrates. Both hematopoietic and ESC fate is controlled by caspase-3, where caspase protease activity is required to limit self-renewal, whereas establishing the milieu for differentiation.48, 49 Although the self-renewal factor targeted by caspase-3 was not identified in the various hematopoietic lineages, it was observed that caspase-3 limits ESC self-renewal/promotes differentiation by cleavage inactivation of the pluripotency factor Nanog.48 Interestingly, this appears to be a conserved phenomenon, with the C. elegans caspase, CED-3, being involved in the cleavage of LIN-14 and LIN-28, which are critical regulators of seam cell symmetric/asymmetric divisions.55 Caspase-3 control of stem cell self-renewal is also evident in lineage restricted stem cell populations such as skeletal muscle satellite cells. Here, caspase-3 targets Pax7, with the ensuing loss of Pax7 function leading to the establishment of a differentiation competent daughter cell.77 Clearly, the yeast mother versus daughter cell asymmetry is not metazoan cell differentiation per se, yet there is broad similarity between each process and each process appears to be dependent on a caspase-type protease activity. Given these observations, it is not unreasonable to conjecture that the origin of caspase control of differentiation is primordial and emerged at or before the death-centric function of caspase-like proteins evolved.
In addition to yeast, many other lower eukaryotes possess metacaspases that function in a similar manner to their more evolutionary advanced protease clad.102 These metacaspases were found to not only partake in an apoptosis-like process, but to have several non-death roles within the cell. Several of the protists studied to date, such as Trypanosoma brucei and Leishmania major, possess metacaspases that are important to cell growth and cell cycle progression.103, 104 Interestingly, the Trypanosoma cruzi metacaspase, TcMCA3, has an essential role in the differentiation of cells into infective metacyclic trypomastigotes; perhaps providing some primordial link to caspase involvement in multicellular eukaryotic differentiation.105
Prokaryotes, which represent the most ancient organisms on the evolutionary time scale, have also been shown to undergo PCD in response to various environmental stresses.106, 107 Moreover, many bacteria studied to date possess orthologous proteins to those present within the conventional apoptotic cascade, such as metacaspases,108 the serine protease Omi/Htr,109 and AIF.110 Several studies have linked the presence of caspase-like proteins to bacterial PCD, such as in the plant pathogen Xanthomonas campestris and the marine phytoplankton Trichodesmium sp., where abrupt cell death is accompanied by the expression of a protein that is detected with an antibody for human caspase-3.111, 112 With respect to Trichodesmium sp., the protein detected was also inhibited by caspase inhibitor Z-VAD-FMK, suggesting that these cyanobacteria contain a bona fide caspase-like enzyme. Although these examples suggest that the prokaryotic metacaspases have an essential role in a primordial form of PCD, there are conflicting reports as to how widespread this phenomenon is. A study by Asplund-Samuelsson et al.108 found that of the 1463 prokaryotic genomes analyzed, <20% of species contained putative metacaspases genes. Interestingly, the prokaryotes that harbor the greatest number of metacaspases – α-proteobacteria, δ-proteobacteria and Cyanobacteria – are some of the most morphologically and developmentally complex prokaryotes. Moreover, it has been suggested that eukaryotic caspase evolution may stem from the presence of metacaspases within α-proteobacteria, which are hypothesized to be predecessors of the eukaryotic mitochondrion organelle.108, 113, 114
Similar to the PCD/metacaspase link, there is a modest relationship between metacaspases and bacterial differentiation. Many bacterial species do undergo cell differentiation as a part of a complex developmental cycle or in response to changing environmental conditions. The bacterial genus Streptomyces is characterized by a developmental cycle that includes mycelium formation and sporulation.115 Bacillus subtilis also undergoes a process of differentiation in response to environmental nutrient limitations, where the bacterial cell asymmetrically divides to produce a long-lived spore and a mother cell that eventually undergoes PCD.116 Although many other examples exist of prokaryotic differentiation,117 few studies, to date, have examined a connection to metacaspase function. However, one relevant example has been noted, where the presence of the Alcanivorax borkumensis SK2 metacaspase gene acts within a larger operon to regulate cell division and cell shape.118 This is a compelling observation and may be representative of other prokaryotic species.
Caspase and caspase-like proteases possess a long evolutionary history in multicellular eukaryotes, and likely derive from some of the earliest eukaryotic ancestors. The necessity for apoptotic cell death in multicellular organisms is readily apparent, yet regulated cell death in unicellular organisms presents an essential paradox. For example, a cell death only protein would be subject to an extraordinarily high rate of natural selection in a single-cell organism, as each cell is a unique genetic entity with its own survival imperative. To balance this conundrum, the field has constructed a prevailing theory where PCD in a single-celled organism is theorized to be an act of altruism for the larger group/colony. Indeed, many unicellular eukaryotes and prokaryotes exist in large communities, such as yeast colonies, slime molds and bacterial biofilms, where altruistic behavior would be plausible. Several studies have chronicled the benefits conferred to a population by PCD events, including releasing nutrients when there are limited environmental resources, ridding the population of unhealthy cells or limiting the spread of viral infections.118 Nevertheless, natural selection is largely considered to occur at the level of individual fitness, rather than through a group or population dynamic. Kin/group selection has been postulated as a driving force for the persistence of PCD in unicellular organisms,117 but the mechanisms behind such a selective pressure have not been identified. Without strong evidence for an altruistic-type selective force, the most reasonable conjecture is that the ‘cell death’ machinery partakes in a variety of functions in unicellular organisms, entrenching a positive selective pressure to maintain and pass on these genes. Domain analysis of primordial metacaspases does support this supposition as metacaspase genes have been associated with diverse functions including transcription, translation and protein modifications, in addition to PCD.108 Indeed, a role for metacaspases anchored in protein quality control would provide a strong retention bias in unicellular organisms, even if these proteins have robust PCD activity. When considering the uniform role of proteostasis in all eukaryotes, it is easy to envision that caspase proteases and their homologs may have been adapted for PCD at a latter evolutionary stage.
Is caspase-mediated apoptosis just cell differentiation taken to the extreme?
The clear relationship between caspase-mediated apoptosis and cell differentiation in many eukaryotic organisms begs the question: is caspase-mediated cell death just hyperactive cell differentiation signaling in response to developmental or environmental signals? In many respects, the research suggests that this is the case. In addition, the work on yeast metacaspases suggest that apoptotic signaling may have been adapted from a previously established set of factors whose main/original function was to regulate protein quality control and establish viable offspring. Thus, it may indeed be the case that caspase-mediated apoptosis originated from cell specification-related signaling, and that a lack of regulation or a purposeful increase in signaling propagates the apoptotic phenotype.
Why do we need to know the evolutionary origin/intent of caspase proteases? Is there any relevance in asking the question other than satisfying the answer to a semantic argument? From our perspective, there is a clear and obvious benefit to be derived in examining this evolutionary question, that is, the discovery of previously unknown function, which may inform or direct knowledge of caspase activity in human biology. The desire to understand yeast metacaspase biology has led to the appreciation that caspases themselves may have broader proteostasis roles, which strongly argues that targeted repression of these proteases for numerous degenerative disease conditions must be approached with an abundance of caution. Indeed, taming the destructive nature of caspase proteases may equally tame the requisite physiologic activity, a consequence that would add to rather than alleviate disease burden.
Acknowledgments
We thank past and present members of the Megeney laboratory for their research contributions and discussions. We apologize to our colleagues for not being able to present all relevant work in the field, as space constraints prevented a complete anthology. Work in LAM's laboratory is supported by grants the Canadian Institutes of Health Research, Heart and Stroke Foundation of Canada, Muscular Dystrophy Association USA and the Ontario Research Fund. LAM held the Mach-Gaensslen Chair in Cardiac Research.
Footnotes
Edited by E Arama
The authors declare no conflict of interest.
References
- Kumar S. Caspase function in programmed cell death. Cell Death Diff 2007; 14: 32–43. [DOI] [PubMed] [Google Scholar]
- Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol 2007; 35: 495–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalin S, Dorstyn L, Dawar S, Old KumarS.. new and emerging functions of caspases. Cell Death Diff 2015; 22: 526–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando P, Megeney LA. Is caspase-dependent apoptosis only cell differentiation taken to the extreme? FASEB J 2007; 21: 8–17. [DOI] [PubMed] [Google Scholar]
- Dick SA, Megeney LA. Cell death proteins: an evolutionary role in cellular adaptation before the advent of apoptosis. Bioessays 2013; 35: 974–983. [DOI] [PubMed] [Google Scholar]
- Williams JR, Little JB, Shipley WU. Association of mammalian cell death with a specific endonucleolytic degradation of DNA. Nature 1974; 252: 754–755. [DOI] [PubMed] [Google Scholar]
- Kerr JFR, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 1972; 26: 239–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane JD, Lucocq J, Pryde J, Barr FA, Woodman PG, Allan VJ et al. Caspase-mediated cleavage of the stacking protein GRASP65 is required for Golgi fragmentation during apoptosis. J Cell Biol 2002; 156: 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane JD, Allan VJ, Woodman PG. Active relocation of chromatin and endoplasmic reticulum into blebs in late apoptotic cells. J Cell Sci 2005; 118: 4059–4071. [DOI] [PubMed] [Google Scholar]
- Arounlt D, Akraid K, Grodet A, Petit PX, Estaquier J, Ameisen JC. On the evolution of programmed cell death: apoptosis of the unicellular eukaryote Leishmania major involves cysteine proteinase activation and mitochondrion permeabilization. Cell Death Diff 2002; 9: 65–81. [DOI] [PubMed] [Google Scholar]
- Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol 2008; 9: 231–241. [DOI] [PubMed] [Google Scholar]
- Cotter TG, Lennon SV, Glynn JM, Green DR. Microfilament-disrupting agents prevent the formation of apoptotic bodies in tumour cells undergoing apoptosis. Cancer Res 1992; 52: 997–1005. [PubMed] [Google Scholar]
- Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat Cell Biol 2001; 3: 339–345. [DOI] [PubMed] [Google Scholar]
- Fadok VA, Voelker DR, Campbell PA, Cohen JJ, Bratton DL, Henson PM. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol 1992; 148: 2207–2216. [PubMed] [Google Scholar]
- Martin SJ, Reutelingsperger CP, McGahon AJ, Rader JA, van Schie RC, LeFace DM et al. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J Exp Med 1995; 182: 1546–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Eijnde SM, van den Hoff MJB, Reutelingsperger CPM, van Heerde WL, Henfling MER, Vermeij-Keers C et al. Transient expression of phosphatidylserine at cell-cell contact areas is required for myotube formation. J Cell Sci 2001; 114: 3631–3642. [DOI] [PubMed] [Google Scholar]
- Park SY, Yun Y, Lim JS, Kim MJ, Kim SY, Kim JE et al. Stabilin-2 modulates the efficiency of myoblast fusion during myogenic differentiation and muscle regeneration. Nat Commun 2016; 7: 10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzesco AM, Janich P, Wilscb-Brauninger M, Dubreuil V, Langenfeld K, Corbeil D et al. Release of extracellular membrane particles carrying stem cell marker prominin-1 (CD133) from neural progenitors and other epithelial cells. J Cell Sci 2005; 118: 2849–2858. [DOI] [PubMed] [Google Scholar]
- Bassnett S, Mataic D. Chromatin degradation in differentiating fiber cells of the eye lens. J Cell Biol 1997; 137: 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Yamamoto A, Kuma A, Ohsumi Y, Mizushima N. Organelle degradation during lens and erythroid differentiation is independent of autophagy. Biochem Biophys Res Comm 2006; 339: 485–489. [DOI] [PubMed] [Google Scholar]
- Khan Z, Francis GE. Contrasting patterns of DNA strand breakage and ADP-ribosylation-dependent DNA ligation during granulocyte and monocyte differentiation. Blood 1987; 69: 1114–1119. [PubMed] [Google Scholar]
- Gunji H, Hass R, Kufe D. Internucleosomal DNA fragmentation during phorbol ester-induced monocytic differentiation and G0/G1 arrest. J Clin Invest 1992; 89: 954–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzaneh F, Zalin R, Brill D, Shall S. DNA strand breaks and ADP-ribosyl transferase activation during cell differentiation. Nature 1982; 300: 362–366. [DOI] [PubMed] [Google Scholar]
- Hossain MS, Kurokawa K, Sekimizu K. Induction of fusion-competent myoblast-specific gene expression during myogenic differentiation of Drosphila Schneider cells by DNA double-strand breaks or replication inhibition. Biochim Biophys Acta 2005; 1743: 176–186. [DOI] [PubMed] [Google Scholar]
- Larsen BD, Rampalli S, Burns LE, Brunette S, Dilworth FJ, Megeney LA. Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc Natl Acad Sci USA 2010; 107: 4230–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone AP, Williams GT. Role of DNA breaks and ADP-ribosyl transferase activity in eukaryotic differentiation demonstrated in human lymphocytes. Nature 1982; 300: 368–370. [DOI] [PubMed] [Google Scholar]
- Hartley JA, Gibson NW, Zwelling LA, Yuspa SH. Association of DNA strand breaks with accelerated terminal differentiation in muse epidermal cells exposed to tumor promoters. Cancer Res 1985; 45: 4864–4870. [PubMed] [Google Scholar]
- McMahon G, Alsina JL, Levy SB. Induction of a Ca2+, Mg2+-dependent endonuclease activity during the early stages of murine erythroleukemic cell differentiation. Proc Natl Acad Sci USA 1984; 81: 7461–7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T, Takahashi N, Udagawa N, Jimi E, Gillespie MT, Martin TJ. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocrine Rev 1999; 20: 345–357. [DOI] [PubMed] [Google Scholar]
- Azuma Y, Kaji K, Katogi R, Takeshita S, Kudo A. Tumor necrosis factor-alpha induces differentiation of and bone resorption by osteoclasts. J Biol Chem 2000; 275: 4858–4864. [DOI] [PubMed] [Google Scholar]
- Michishita M, Yoshida Y, Uchino H, Nagata K. Induction of tumor necrosis factor-alpha and its receptors during differentiation in myeloid leukemic cells along the monocytic pathway. A possible regulatory mechanism for TNF-alpha production. J Biol Chem 1990; 265: 8751–8759. [PubMed] [Google Scholar]
- Bharti AC, Aggarwal BB. Ranking the role of RANK ligand in apoptosis. Apoptosis 2004; 9: 677–690. [DOI] [PubMed] [Google Scholar]
- Zermati Y, Garrido C, Amsellem S, Fishelson S, Bouscary D, Valensi F et al. Caspase activation is required for terminal erythroid differentiation. J Exp Med 2001; 193: 247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova SV, Ataullakhanov FI, Globus RK. Bioenergetics and mitochondrial transmembrane potential during differentiation of cultured osteoblasts. Am J Physiol Cell Physiol 2000; 279: C1220–C1229. [DOI] [PubMed] [Google Scholar]
- Levenson R, Macara IG, Smith RL, Cantley L, Housman D. Role of mitochondrial membrane potential in the regulation of murine erythroleukemia cell differentiation. Cell 1982; 28: 855–863. [DOI] [PubMed] [Google Scholar]
- Sordet O, Rébé C, Plenchette S, Zermati Y, Hermine O, Vainchenker W et al. Specific involvement of caspases in the differentiation of monocytes into macrophages. Blood 2002; 100: 4446–4453. [DOI] [PubMed] [Google Scholar]
- Ishizaki Y, Jacobson MD, Raff MC. A role for caspases in lens fiber differentiation. J Cell Biol 1998; 140: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm D, Mazurier C, Giarratana MC, Darghouth D, Faussat AM, Harmand L et al. Caspase-3 is involved in the signaling in erythroid differentiation by targeting late progenitors. PLoS ONE 2013; 8: e62303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA. Caspase-3 activity is required for skeletal muscle differentiation. Proc Natl Acad Sci USA 2002; 99: 11025–11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome c are required for sperm differentiation. Dev Cell 2003; 4: 687–697. [DOI] [PubMed] [Google Scholar]
- Mogi M, Togari A. Activation of caspases is required for osteoblastic differentiation. J Biol Chem 2003; 278: 57577–47482. [DOI] [PubMed] [Google Scholar]
- Miura M, Chen XD, Allen MR, Bi Y, Gronthos S, Seo BM. A crucial role of caspase-3 in osteogenic differentiation of bone marrow stromal stem cells. J Clin Invest 2004; 114: 1704–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando P, Brunette S, Megeney LA. Neural stem cell differentiation is dependent upon endogenous caspase 3 activity. FASEB J 2005; 19: 1671–1673. [DOI] [PubMed] [Google Scholar]
- Kanuka H, Kuranaga E, Takemoto K, Hiratou T, Okano H, Miura M. Drosophila caspase transduces Shaggy/GSK-3beta kinase activity in neural precursor development. EMBO J 2005; 24: 3793–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa S, Hamada S, Kuida K, Yoshida H, Igaki T, Miura M. Maturation of the olfactory sensory neurons by Apaf-1/caspase-9-mediated caspase activity. Proc Natl Acad Sci USA 2010; 107: 13366–13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsain N, Barker PA. New views on the misconstrued: executioner caspases and their diverse non-apoptotic roles. Neuron 2015; 88: 461–474. [DOI] [PubMed] [Google Scholar]
- Okuyama R, Nguyen BC, Talora C, Ogawa E, Tommasi di Vignano A, Lioumi M et al. High commitment of embryonic keratinocytes to terminal differentiation through Notch1-caspase-3 regulatory mechanism. Dev Cell 2004; 6: 551–562. [DOI] [PubMed] [Google Scholar]
- Fujita J, Crane AM, Souza MK, Dejosez M, Kyba M, Flavell RA et al. Caspase activity mediates the differentiation of embryonic stem cells. Cell Stem Cell 2008; 2: 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen V, Fleming HE, Riedt T, Karlsson G, Riese MJ, Lo Celso C et al. Hematopoietic stem cell responsiveness to exogenous signals is limited by caspase-3. Cell Stem Cell 2008; 2: 584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdul-Ghani M, Dufort D, Stiles R, De Repentigny Y, Kothary R, Megeney LA. Wnt11 promotes cardiomyocyte development by caspase-mediated suppression of canonical Wnt signals. Mol Cell Biol 2011; 31: 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulatovic I, Ibarra C, Österholm C, Wang H, Beltrán-Rodriguez A, Varas M et al. Sublethal caspase activation promotes generation of cardiomyocytes from embryonic stem cells. PLoS ONE 2015; 10: e0120176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matalova E, Lesot H, Svandova E, Vanden Berghe T, Sharpe PT, Healy C et al. Caspase-7 participates in differentiation of cells forming dental hard tissues. Dev Growth Differ 2013; 55: 615–621. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol 2004; 5: 897–907. [DOI] [PubMed] [Google Scholar]
- Fuchs Y, Stellar H. Programmed cell death in animal development. Cell 2011; 147: 742–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BP, Zabinsky R, Weaver YM, Seung Lee E, Xue D, Han M. CED-3 caspase acts with miRNA to regulate non-apoptotic gene expression dynamics for robust development in C. elegans. eLife 2014; 3: e04265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber GF, Menko AS. The canonical intrinsic mitochondrial death pathway has a non-apoptotic role in signaling lens cell differentiation. J Biol Chem 2005; 280: 22135–22145. [DOI] [PubMed] [Google Scholar]
- Murray TV, McMahon JM, Howley BA, Stanley A, Ritter T, Mohr A et al. A non-apoptotic role for caspase-9 in muscle differentiation. J Cell Sci 2008; 121: 3786–3793. [DOI] [PubMed] [Google Scholar]
- Deak JC, Cross JV, Lewis M, Qian Y, Parrott LA, Distelhorst CW et al. Fas-induced proteolytic activation and intracellular redistribution of the stress-signaling kinase MEKK1. Proc Natl Acad Sci USA 1998; 95: 5595–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann C, Gerwins P, Johnson NL, Jarpe MB, Johnson GL. MEK kinase 1, a substrate for DEVD-directed caspases, is involved in genotoxin-induced apoptosis. Mol Cell Biol 1998; 18: 2416–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone MH, Salvesen GS, Widmann C, Johnson G, Frisch SM. The regulation of anoikis: MEKK-1 activation requires cleavage by caspases. Cell 1997; 90: 315–323. [DOI] [PubMed] [Google Scholar]
- Graves JD, Gotoh Y, Draves KE, Ambrose D, Han DK, Wright M et al. Caspase-mediated activation and induction of apoptosis by the mammalian Ste20-like kinase, Mst1. EMBO J 1998; 17: 2224–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassé E, Bridoux L, Baranek T, Lambert E, Salesse S, Sowa ML et al. Tissue inhibitor of metalloproteinase-1 promotes hematopoietic differentiation via caspase-3 upstream the MEKK1/MEK6/p38α pathway. Leukemia 2007; 21: 595–603. [DOI] [PubMed] [Google Scholar]
- Lee N, MacDonald H, Reinhard C, Halenbeck R, Roulston A, Shi T et al. Activation of hPAK65 by caspase cleavage induces some of the morphological and biochemical changes of apoptosis. Proc Natl Acad Sci USA 1997; 94: 13642–13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathelin S, Rébé C, Haddaoui L, Simioni N, Verdier F, Fontenay M et al. Identification of proteins cleaved downstream of caspase activation in monocytes undergoing macrophage differentiation. J Biol Chem 2006; 281: 17779–17788. [DOI] [PubMed] [Google Scholar]
- Riento K, Ridley AJ. Rocks: multifunctional kinases in cell behavior. Nat Rev Mol Cell Biol 2003; 4: 446–456. [DOI] [PubMed] [Google Scholar]
- Emoto Y, Manome Y, Meinhardt G, Kisaki H, Kharbanda S, Robertson M et al. Proteolytic activation of protein kinase C delta by an ICE-like protease in apoptotic cells. EMBO J 1995; 14: 6148–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghayur T, Huginin M, Talanian RV, Ratnofsky S, Quinlan C, Emoto Y et al. Proteolytic activation of protein kinase C delta by an ICE/CED 3-like protease induces characteristics of apoptosis. J Exp Med 1996; 184: 2399–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khalaf MH, Blake LE, Larsen BD, Bell RAV, Brunette S, Parks RJ et al. Temporal activation of XRCC1-mediated DNA repair is essential for muscle differentiation. Cell Discov 2016; 2: 15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari-Birgani S, Hosseinkhani S, Mollamohamadi S, Baharvand H. Delay in apoptosome formation attenuates apoptosis in mouse embryonic stem cell differentiaton. J Biol Chem 2014; 289: 16905–16913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans C, Megeney LA. How caspase proteases regulate cell fate. InApoptosis: Modern Insights into Disease from Molecules to Man Preedy VR (ed.). CRC Press Taylor & Francis Group: Boca Raton, FL, USA, pp 55–71. [Google Scholar]
- Sayama K, Hanakawa Y, Shirakata Y, Yamasaki K, Sawada Y, Sun L et al. Apoptosis signal-regulating kinase 1 (ASK1) is an intracellular inducer of keratinocyte differentiation. J Biol Chem 2001; 276: 999–1004. [DOI] [PubMed] [Google Scholar]
- Parrish AB, Freel CD, Kornbluth S. Cellular mechanisms controlling caspase activation and function. Cold Spring Harb Perspect Biol 2013; 5: a008672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan LA, Morrice N, Brady S, Magee G, Pathak S, Clarke PR. Inhibition of caspase-9 through phosphorylation at Thr125 by ERK MAPK. Nat Cell Biol 2003; 5: 647–654. [DOI] [PubMed] [Google Scholar]
- Allan LA, Clarke PR. Apoptosis and autophagy: regulation of caspase-9 by phosphorylation. FEBS J 2009; 276: 6063–6073. [DOI] [PubMed] [Google Scholar]
- Duncan JS, Turowec JP, Duncan KE, Vilk G, Wu C, Lüscher B et al. A peptide-based target screen implicates the protein kinase CK2 in the global regulation of caspase signaling. Sci Signal 2011; 4: ra30. [DOI] [PubMed] [Google Scholar]
- Turowec JP, Zukowski SA, Knight JD, Smalley DM, Graves LM, Johnson GL et al. An unbiased proteomic screen reveals caspase cleavage is positively and negatively regulated by substrate phosphorylation. Mol Cell Proteomics 2014; 13: 1184–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick SA, Chang NC, Dumont NA, Bell RAV, Putinski C, Kawabe Y et al. Caspase 3 cleavage of Pax7 inhibits self-renewal of satellite cells. Proc Natl Acad Sci USA 2015; 112: E5246–E5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Moresco JJ, Cabezas F, de la Vega E, Bustos F, Yates JR III et al. Ck2-dependent phosphorylation is required to maintain Pax7 protein levels in proliferating muscle progenitors. PLoS ONE 2016; 11: e0154919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JR, Vernooy SY, Yu H, Yan N, Shi Y, Guo M et al. Multiple apoptotic caspase cascades are required in nonapoptotic role for Drosophila spermatid individualization. PLoS Biol 2004; 2: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arama E, Bader M, Rieckof GE, Steller H. A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol 2007; 5: e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader M, Arama E, Steller H. A novel F-box protein is required for caspase activation during cellular remodeling in Drosophila. Development 2010; 137: 1679–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aram L, Braun T, Braverman C, Kaplan Y, Ravid L, Levin-Zaidman S et al. A Krebs cycle component limits caspase activation rate through mitochondrial surface restriction of CRL activation. Dev Cell 2016; 37: 15–33. [DOI] [PubMed] [Google Scholar]
- McLuskey K, Mottram JC. Comparative structural analysis of the caspase family with other clan cysteine peptidases. Biochem J 2015; 466: 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Fitzgerald P. Just so stories about the evolution of apoptosis. Curr Biol 2016; 26: R620–R627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen D, van de Cotte B, De Jaeger G, Eeckhout D, Casteels P, Vandepoele K et al. Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J Biol Chem 2004; 279: 45329–45336. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Lam E. Two Arabidopsis metacaspases AtMCP1b and AtMCP2b are arginine/lysine-specific cysteine proteases and activate apoptosis-like cell death in yeast. J Biol Chem 2005; 280: 14691–14699. [DOI] [PubMed] [Google Scholar]
- Gonzalez IJ, Desponds C, Schaff C, Mottram JC, Fasel N. Leishmania major metacaspase can replace yeast metacaspase in programmed cell death and has arginine-specific cysteine peptidase activity. Int J Parasitol 2007; 37: 161–172. [DOI] [PubMed] [Google Scholar]
- Uren AG, O’Rourke K, Aravind L, Teresa Pisabarro M, Seshagiri S, Koonin EV et al. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a role in MALT lymphoma. Mol Cell 2000; 6: 961–967. [DOI] [PubMed] [Google Scholar]
- Carmona-Gutierrez D, Fröhlich K-U, Kroemer G, Madeo F. Metacaspases are caspases. Doubt no more. Cell Death Diff 2010; 17: 377–378. [DOI] [PubMed] [Google Scholar]
- Wong AH, Yan C, Shi Y. Crystal structure of the yeast metacaspase Yca1. J Biol Chem 2012; 287: 29251–29259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiatsiani L, Van Breusegem F, Gallois P, Zavialov A, Lam A, Bozhkov PV. Metacaspases. Cell Death Diff 2011; 18: 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Herker E, Maldener C, Wissing S, Lächelt S, Herlan M et al. A caspase-related protease regulates apoptosis in yeast. Mol Cell 2002; 9: 911–917. [DOI] [PubMed] [Google Scholar]
- Hamann A, Brust D, Osiewacz HD. Deletion of putative apoptosis factors leads to lifespan extension in the fungal aging model Podosspora anserine. Mol Microbioql 2007; 65: 948–958. [DOI] [PubMed] [Google Scholar]
- Low CP, Shui G, Liew LP, Buttner S, Madeo F, Dawes IW et al. Caspase-dependent and –independent lipotoxic cell-death pathways in fission yeast. J Cell Sci 2008; 121: 2671–2684. [DOI] [PubMed] [Google Scholar]
- Guérin R, Beauregard PB, Leroux A, Rokeach LA. Calnexin. Calnexin regulates apoptosis induced by inositol starvation in fission yeast. PLoS ONE 2009; 4: e6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Huang S, Dai B, Zhu Z, Lu H, Dong L et al. Candida albicans cells lacking CaMCA1-encoded metacaspase show resistance to oxidative stress-induced death and change in energy metabolism. Fungal Genet Biol 2009; 46: 183–189. [DOI] [PubMed] [Google Scholar]
- Lee REC, Brunette S, Puente LG, Megeney LA. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc Natl Acad Sci USA 2010; 107: 13348–13353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalila H, González IJ, El-Fadili AK, Delgado MB, Desponds C, Schaff C et al. Processing of metacaspase into a cytoplasmic catalytic domain mediating cell death in Leishmania major. Mol Microbiol 2011; 79: 222–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez MF, Filonova LH, Smertenko A, Savenkov EI, Clapham DH, von Arnold S et al. Metacaspase-dependent programmed cell death is essential for plant embryogenesis. Curr Biol 2004; 14: R339–R340. [DOI] [PubMed] [Google Scholar]
- Sundström JF, Vaculova A, Smertenko AP, Savenkov EI, Golovko A, Minina E et al. Tudor staphylococcal nuclease is an evolutionary conserved component of the programmed cell death degradome. Nat Cell Biol 2009; 11: 1347–1354. [DOI] [PubMed] [Google Scholar]
- Hill SM, Hao X, Liu B, Nyström T. Life-span extension by a metacaspase in yeast Saccharomyces cerevisiae. Science 2014; 344: 1389–1392. [DOI] [PubMed] [Google Scholar]
- Kaczanowski S, Sajid M, Reece SE. Evolution of apoptosis-like programmed cell death in unicellular protozoan parasites. Parasites Vectors 2011; 4: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms MJ, Ambit A, Appleton P, Tetley L, Coombs GH, Mottram JC. Bloodstream from Trypanosoma brucei depend upon multiple metacaspases associated with RAB11-positive endosomes. J Cell Sci 2006; 119: 1105–1117. [DOI] [PubMed] [Google Scholar]
- Ambit A, Fasel N, Coombs GH, Mottram JC. An essential role for the Leishmania major metacaspases in cell cycle progression. Cell Death Diff 2008; 15: 113–122. [DOI] [PubMed] [Google Scholar]
- Laverrière M, Cazzulo JJ, Alvarez VE. Antagonic activities of Trypanosoma cruzi metacaspases affect the balance between cell proliferation, death and differentiation. Cell Death Diff 2012; 19: 1358–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramisetty BC, Natarajan B, Santhosh RS. mazEF-mediated programmed cell death in bacteria: “what is this?”. Crit Rev Microbiol 2015; 41: 89–100. [DOI] [PubMed] [Google Scholar]
- Dwyer DJ, Camacho DM, Kohanski MA, Callura JM, Collins JJ. Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol Cell 2012; 46: 561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asplund-Samuelsson J, Bergman B, Larsson J. Prokaryotic caspase homologs: phylogenetic patterns and functional characteristics reveal considerable diversity. PLoS ONE 2012; 7: e49888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Srinivasula SM, Chai J, Li P, Wu JW, Zhang Z et al. Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi. Nat Struct Biol 2002; 9: 436–441. [DOI] [PubMed] [Google Scholar]
- Lorenzo HK, Susin SA, Penninger J, Kroemer G. Apoptosis inducing factor (AIF): a phylogenetically old, caspase-independent effector of cell death. Cell Death Diff 1999; 6: 516–524. [DOI] [PubMed] [Google Scholar]
- Gautam S, Sharma A. Involvement of caspase-3 like protein in rapid cell death of Xanthomonas. Mol Microbiol 2002; 44: 393–401. [DOI] [PubMed] [Google Scholar]
- Berman-Frank I, Bidle KD. The demise of the marine cyanobacterium, Trichodesmium spp., via an autocatalyzed cell death pathway. Limnol Oceangr 2004; 49: 997–1005. [Google Scholar]
- Koonin EV, Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Diff 2002; 9: 394–404. [DOI] [PubMed] [Google Scholar]
- Ferla MP, Thrash JC, Giovannoni SJ, Patrick WM. New rRNA gene-base phylogenies of the Alphaproteobacteria provide perspective on the major groups, mitochondrial ancestry and phylogenetic instability. PLoS ONE 2013; 8: e83383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manteca A, Ye J, Sánchez J, Jensen ON. Phosphoproteome analysis of Streptomyces development revelas extensive protein phosphorylation accompanying bacterial differentiation. J Proteome Res 2011; 10: 5481–5492. [DOI] [PubMed] [Google Scholar]
- Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev 2012; 36: 131–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameisen JC. On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Diff 2002; 9: 367–393. [DOI] [PubMed] [Google Scholar]
- Nedelcu AM, Driscoll WW, Durand PM, Herron MD, Rashidi A. On the paradigm of altruistic suicide in the unicellular world. Evolution 2011; 65: 3–20. [DOI] [PubMed] [Google Scholar]
- Plant PJ, Fawcett JP, Lin DCC, Holdorf AD, Binns K, Kulkarni S et al. A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat Cell Biol 2003; 5: 301–308. [DOI] [PubMed] [Google Scholar]
- Anani S, Bhat S, Honma-Yamanaka N, Krawchuk D, Yamanaka Y. Initiation of Hippo signaling is linked to polarity rather than to cell position in the pre-implantation mouse embryo. Development 2014; 141: 2813–2824. [DOI] [PubMed] [Google Scholar]
- Keder A, Rives-Quinto N, Aerne BL, Franco M, Tapon N, Carmena A. The hippo pathway core cassette regulates asymmetric cell division. Curr Biol 2015; 25: 2739–2750. [DOI] [PubMed] [Google Scholar]