Abstract
Protease biology is intimately linked to the functional consequences of substrate cleavage events. Human caspases are a family of 12 fate-determining cysteine proteases that are best known for driving cell death, either apoptosis or pyroptosis. More recently, caspases have been shown to be involved in other cellular remodeling events as well including stem cell fate determination, spermatogenesis, and erythroid differentiation. Recent global proteomics methods enable characterization of the substrates that caspases cleave in live cells and cell extracts. The number of substrate targets identified for individual caspases can vary widely ranging from only a (few) dozen targets for caspases-4, -5, -9, and -14 to hundreds of targets for caspases-1, -2, -3, -6, -7, and -8. Proteomic studies characterizing the rates of target cleavage show that each caspase has a preferred substrate cohort that sometimes overlaps between caspases, but whose rates of cleavage vary over 500-fold within each group. Determining the functional consequences of discrete proteolytic events within the global substrate pool is a major challenge for the field. From the handful of individual targets that have been studied in detail, there are only a few so far that whose single cleavage event is capable of sparking apoptosis alone, such as cleavage of caspase-3/-7 and BIMEL, or for pyroptosis, gasdermin D. For the most part, it appears that cleavage events function cooperatively in the cell death process to generate a proteolytic synthetic lethal outcome. In contrast to apoptosis, far less is known about caspase biology in non-apoptotic cellular processes, such as cellular remodeling, including which caspases are activated, the mechanisms of their activation and deactivation, and the key substrate targets. Here we survey the progress made in global identification of caspase substrates using proteomics and the exciting new avenues these studies have opened for understanding the molecular logic of substrate cleavage in apoptotic and non-apoptotic processes.
Facts
The human caspase family regulates crucial biological functions, such as cell death in apoptosis and pyroptosis, as well as non-cell death functions in inflammation, dendrite trimming, and cell differentiation.
Thousands of potential caspase substrates have now been identified, but each is cleaved at different rates, and preferentially by different family members.
Open Questions
What substrate pool does each caspase target and what cellular functions do these cleavage events drive?
Are there critical and non-critical substrate subsets within individual caspase target pools?
Which caspase-cleaved substrates have gain-of-function, loss-of-function, or non-functional phenotypes?
What are the mechanisms present during cell differentiation that prevent activated caspases from promoting apoptosis?
What are the mechanisms that lead to specific caspase activation during non-apoptotic functions?
Can we develop more selective means to activate or inhibit specific caspases either with small molecules or genetic manipulation?
Caspases are a family of cysteine proteases whose functions are inextricably linked with the process of programmed cell death, or apoptosis, in all metazoans, including C. elegans, Drosophila, mouse and humans.1, 2, 3 The enzymes predominantly cleave their substrates on the C-terminal side of aspartate,4, 5 less frequently after glutamate and in rare cases following phosphoserine residues.6 Proteolytic cleavage leads to important changes in cell morphology such as membrane blebbing, DNA fragmentation, phosphatidylserine exposure at the cell surface, and formation of apoptotic vesicles.7 Recent proteomics approaches allowing the identification of caspase substrates in apoptosis8, 9, 10, 11, 12 have advanced the field, but our understanding of the role of caspases in other biological processes such as cell differentiation and other programmed forms of cell death like pyroptosis (inflammation) and necroptosis (necrosis)13, 14 is lagging behind. A deeper understanding of caspase biology relies on many facets, including finding out how each caspase gets activated, what substrates they cleave, what biological functions they each drive, and in what cellular context. This review will focus on the technologies for identifying and characterizing caspase substrates using modern proteomics, and how this information has been applied to shed light on the role of caspases in biology.
Caspase classification
The human caspase family is divided into three main groups, primarily based on sequence similarity and biological function.15 Group I comprises the inflammatory caspases-1, -4, and -5 (caspase-11 in mouse), based on the commonalities of having a long caspase-recruitment domain and a preference for a large aromatic or hydrophobic residue at the P4 position. Within this group, caspase-1 is the enzyme that has been best characterized and well known for processing of IL-1β involved in inflammation. Group II is formed by the apoptotic effector caspases-3, -6, and -7 that share a similar short pro-domain, and are classically described as the ‘executors of apoptosis’. Finally, group III includes the human initiator caspases-8, -9, and -10, which all contain a long pro-domain and prefer substrates with a leucine or valine at the P4 position. These broad group classifications are admittedly imperfect. For example, although caspase-2 has been characterized as an initiator caspase (group III) because of its long pro-domain, its substrate specificity is more similar to ‘executioner-like’ group II caspases.16, 17, 18, 19 Conversely, caspase-6 has been characterized as an executioner caspase because of its short pro-domain and sequence recognition motif, but caspase-6 activation alone is not sufficient to lead to apoptosis in all cells.20 The group classification also does not accurately highlight the specialized cellular contexts in which some caspases may function. Some notable examples include the observation that only subsets of the human population detectably express caspase-12 and the highly specific role proposed for caspase-14 in the differentiation process of human keratinocytes.21
Biochemistry
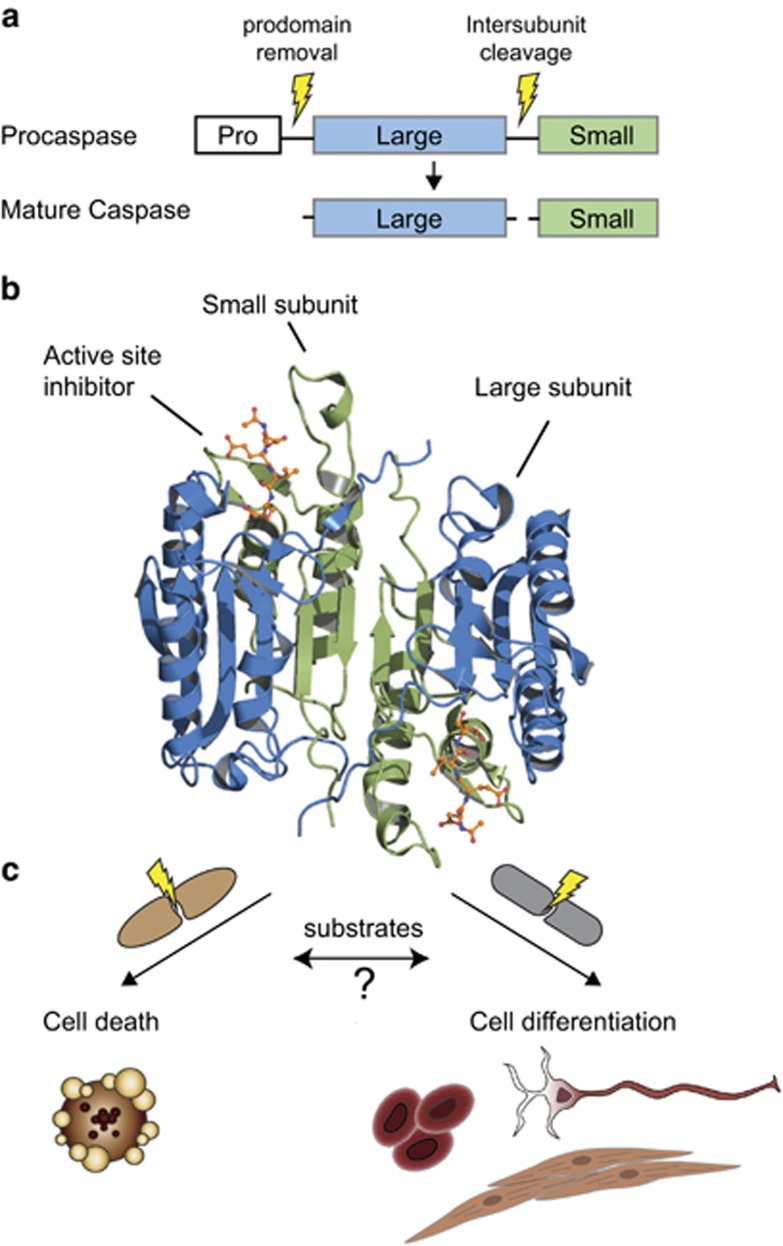
As is the case for most proteases, caspases are expressed in cells as inactive zymogens also known as procaspases (Figure 1a). All mature caspases form a head-to-tail dimer with each unit composed of a large and small subunit containing one active site, as illustrated by the crystal structure of caspase-7 (Figure 1b). Some procaspases, such as procaspase-1 and the initiator procaspases-8 and -9, are monomeric.22, 23, 24 These caspases become activated via dimerization after recruitment onto a protein scaffold and/or proteolytic cleavage of the inter-subunit linker between the small and large subunits.22, 25 Other procaspases, such as procaspases-3, -6, and -7, are expressed in the form of constitutive dimers and cleavage within in the inter-subunit linker region, either by initiators or autoproteolysis, induces activation.26, 27 Upon activation, caspases begin to cleave a set of pre-defined substrates, ranging from a few to several hundred targets, at rates that can vary by over 500-fold between targets. Caspase activation can lead to cell death via apoptosis or pyroptosis, or cell differentiation. Specific phenotypic outcomes are likely dependent on multiple factors including (i) which caspases are activated, (ii) which substrates are cleaved, (iii) the cellular context, and (iv) the post-activation regulatory machinery in the cell. Currently, many important gaps exist in our understanding of the interplay between these factors and how they control death versus non-death phenotypes (Figure 1c).
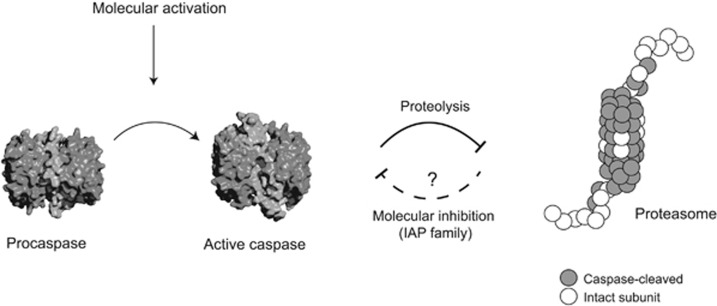
Figure 1.
Activation, structural features, and biological functions of caspases. Caspases are expressed as inactive cysteine proteases. (a) Caspase activation occurs upon cleavage of the pro-domain and inter-subunit linker, the latter of which is most critical. (b) The mature enzyme is formed by a dimer containing a small subunit (green), a large subunit (blue) that generates an active site on each side (inhibitor in sticks shown in orange). The figure shows a cartoon representation of caspase-7 (PDB ID 1F1J). (c) Caspase substrate identification in cells has allowed us to shed light on key biological processes mostly cell death and to a lesser extent cell remodeling and differentiation
Caspase recognition sequences
Caspase substrate recognition sequences are described by the classic protease cleavage nomenclature surrounding the cleavage site (↓) located between P1 and P1′, as Pn-P4-P3-P2-P1↓P1′-P2′-Pn′.28 The first breakthrough in the characterization of substrate specificity of caspases came with the application of the positional scanning synthetic combinatorial library method using fluorogenic tetrapeptide substrates featuring an aspartate at the P1 position.4, 29, 30 Recent advances include the development of mass spectrometry methods that enable unbiased global characterization of native protein protease specificity16, 31 and substrate phage libraries for kinetic characterization of caspase specificity.32, 33 Studies by the Salvesen lab identified the optimal cleavage motifs of caspases using fluorescent peptides5, 34 and the native E. coli proteome.35 A summary of reported caspase sequence specificities for peptides compared with native human proteins (see next section below) is presented in Table 1. Peptide libraries are immensely useful for characterizing short ‘signature’ motifs for proteases and for designing potential peptide-mimetic inhibitors. However, these data are not a substitute for identification of native protein substrates whose folding and domain structure dictate the accessibility of the cleavage site, and in rare cases present exosites that enhance cleavage.36
Table 1. Caspase substrate preference motifs determined from peptide and proteomic studies.
| Enzyme | Peptide substratea | Protein substrateb | References |
|---|---|---|---|
| Caspase-1 | WEHD | YVHD/FESD | Refs 29, 74, 41 |
| Caspase-2 | VDVAD | XDEVD | Refs 30, 19, 16 |
| Caspase-3 | DEVD | DEVD | Refs 4, 42 |
| Caspase-4 | (W/L)EHD, LEVD | − | Refs 4, 30 |
| Caspase-5 | (W/L)EHD | − | Ref. 4 |
| Caspase-6 | VQVD | VEVD | Refs 4, 30, 16 |
| Caspase-7 | DEVD | DEVD | Refs 4, 42 |
| Caspase-8 | LETD | XEXD | Refs 4, 42 |
| Caspase-9 | (W/L)EHD | − | Ref. 4 |
| Caspase-10 | LEHD | LEHD | Ref. 98 |
| Caspase-12 | − | − | |
| Caspase-14 | WEHD | VSQD/HSED | Refs 99, 100 |
Consensus data from positional scanning library studies.
Consensus data from proteomic studies. These represent the dominant motifs, but by no means the only sequence cleaved. For example, DEVD is found in less than 1% of the total protein cleavage sites by caspase-3/-7, but still is the optimal recognition sequence. ‘X’ represents any residue, while ‘–’ remain to be determined.
Methodology for native caspase substrate identification
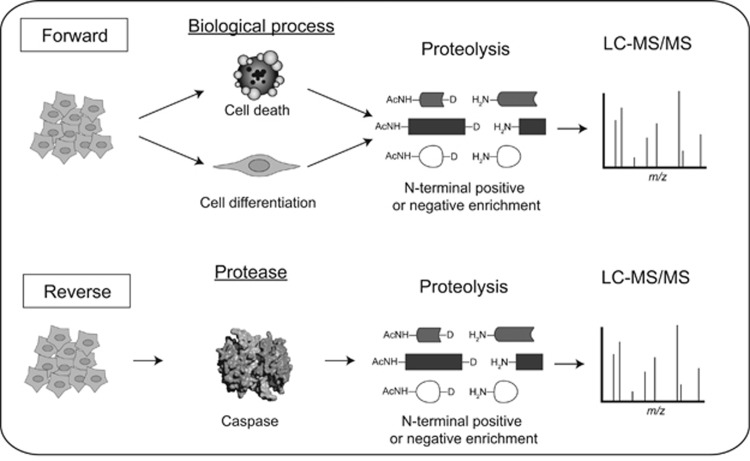
Significant progress has been made using modern proteomics methods to identify native protein substrates either in intact cells by triggering endogenous caspase(s) (the forward approach) or in cell lysates by addition of exogenous caspase (the reverse approach) (Figure 2). To isolate the cleavage products, one can either positively9 or negatively enrich37 for newly formed N termini or separate the fragments by SDS-PAGE.10 The isolated fragments are subjected to trypsin digestion, and peptides corresponding to the native protein targets are identified using tandem mass spectrometry (LC-MS/MS). The advantage of the forward experiment is that one can identify substrates in intact cellular structures with endogenous caspases for the biological process studied, such as cell death. However, one cannot easily determine the specific caspase(s) responsible for the proteolytic events. The reverse experiment, on the other hand, enables identification of potential substrates after addition of a specific caspase to a cell lysate (neutralized of endogenous proteases). This is a good method to link caspases to their protein substrates, but does so in an extract where cell architecture and cellular organelles have been destroyed, and requires controls that ensure cleavage events are not the result of indirect activation of endogenous proteases. Forward and reverse approaches are very complementary and each helps to validate the other in terms of biological relevance and identity of the specific caspase(s) responsible. It should be noted that most of the proteomic approaches involve removal of cellular debris before subjecting samples to mass spectrometry and will therefore usually miss proteolysis that occurs in membrane structures of organelles and the plasma membrane.
Figure 2.
Caspase substrates identification using proteomics approaches. Two main methods are used to identify caspase substrates: the forward and reverse techniques. The forward method involves treating a cell population with an inducer to trigger a biological process that activates endogenous caspases, such as cell death or cell differentiation. Cells are lysed, and cleaved products either captured or separated for identification using mass spectrometry. In the reverse method, one generates a cell lysate in which endogenous proteases are inhibited and adds a purified caspase of interest. After a suitable time(s), proteolytic fragments are captured, identified, and quantified using mass spectrometry
Multiple experimental approaches have been developed to study proteolysis in forward and reverse mode. Gevaert et al.37 developed the COFRADIC method for negative enrichment of N termini generated by proteolysis. COFRADIC was used to study caspase proteolysis during Fas-induced apoptosis (forward approach8), as well as to compare specificity of caspase-3 and -7 specificity (reverse approach38) and to identify caspase-2 substrates.19 Alternatively, our group developed a positive enrichment method in which N termini are tagged using an engineered peptide ligase, subtiligase. This method was employed to identify caspase substrates induced in apoptosis in forward mode.9, 39, 40 It has also been used to identify hundreds of native substrates of a half-dozen caspases in reverse mode.16, 41, 42 Both methods directly tag and identify the site of cleavage. The Cravatt lab devised a method consisting of in-gel digestion of lysates combined with bioinformatics methods called PROTOMAP to identify proteolytic events during apoptosis.10 This method allows for the identification of cleavage targets but unlike the other N-terminomics technologies described above, does not necessarily identify the exact site of cleavage since the peptides surrounding the cleavage site may not be detected. The Lill group has used a method called GASSP, which takes advantage of SILAC labeling to study apoptosis induced by etoposide.11 A commercial method taking advantage of an antibody recognizing specific motifs for enrichment has also been developed (PTMscan)43 and used to identify caspase-cleaved substrates upon induction of DNA damage.44 There are additional proteomic methods developed in the Overall lab (TAILS)45 for studying proteolysis sites, but these have been applied on proteases other than caspases.
Caspase specificity for apoptotic substrates
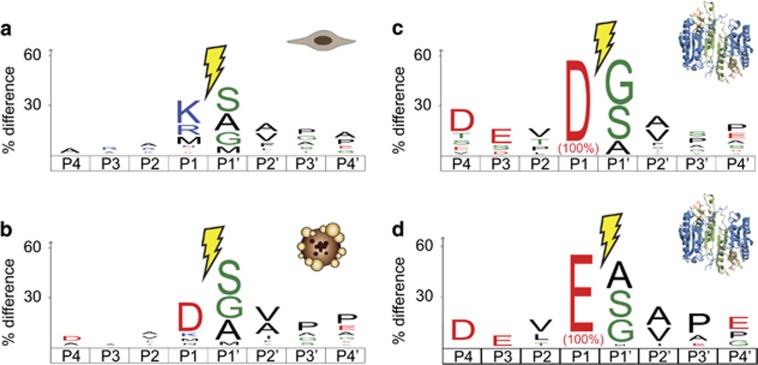
The combination of all these studies has identified over 1500 caspase substrates and has provided a much clearer blueprint of caspase targets and caspase specificity. Several databases that have compiled target data sets are available including: Merops,46 Casbah,12 CutDB,47 TopFind,48 and CaspDB.49 Our group has created the Degrabase, a database of proteolytic events occurring in untreated and apoptotic cells.39 The consensus sequence logo extracted from the Degrabase for many healthy and apoptotic cells are reported in Figures 3a and b, respectively. By extracting only caspase substrates, featuring an aspartate at P1 position, one can clearly see the dominant DEVD motif of the executioner caspases activated during apoptosis. Although this is the prominent consensus motif, this precise sequence is present in only 0.5% of the substrates cleaved after aspartic acid, thereby highlighting the promiscuity of the executioner caspases. Although caspases are named for their prominent cleavage after aspartate (cysteine-aspartic proteases), they can cleave after other acidic residues such as glutamic acid or even phosphor-serine, but the biological significance of these cuts remains to be determined. About 13% of cleavage events occur after glutamate, with the DEVE motif being observed with the highest frequency within that set.6 There is also a small set of substrates potentially cleaved after phosphoserine residues.
Figure 3.
Caspases recognition motifs. (a) Background proteolysis observed in untreated cells mostly showed tryptic-like protein fragments with K or R at P1 position. (b) Apoptotic cells feature a strong enrichment for protein fragments originating from proteolysis on the C-terminal of D residues. (c) By selecting all proteolytic sites featuring a D at P1 position, a clear ‘DEVD’ consensus sequence can be observed. (d) A similar motif is obtained by selecting only proteolytic sites with an E at P1 position, suggesting caspase cleavage. Data adapted from refs 6, 39
Determining factors guiding the rates of proteolysis of native substrates
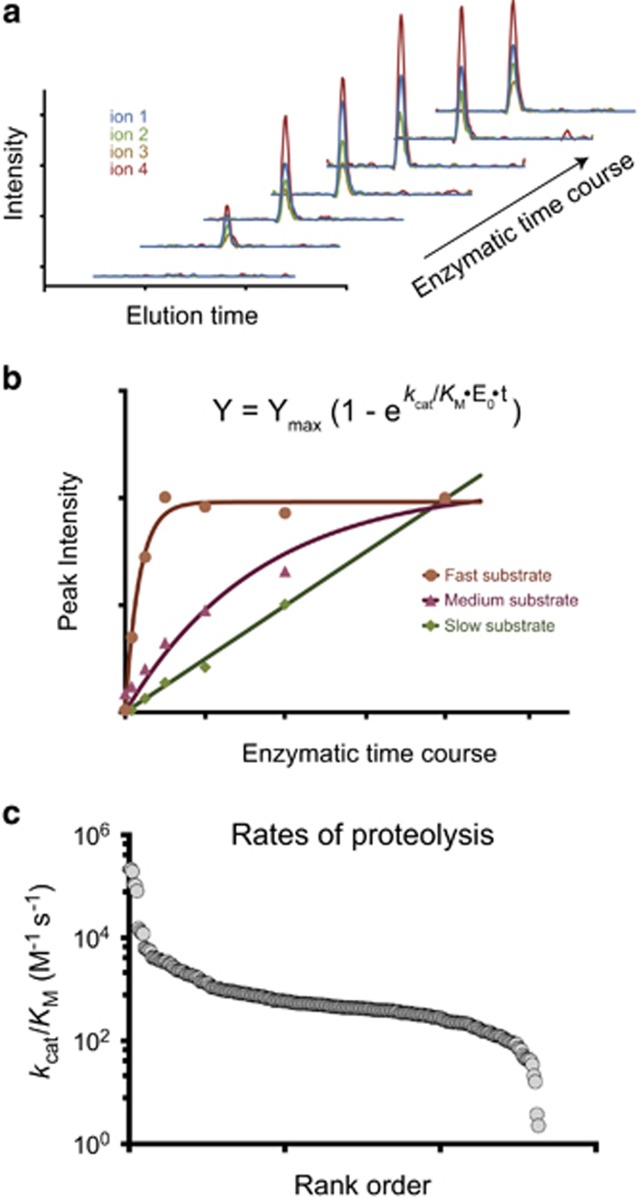
Peptide substrates provide strong evidence of the primary sequence that can be cleaved by caspases. However, caspases cleave natively folded proteins and thus accessibility of the cleavage site is also a critical element of target recognition. Our group and others9, 35 have found that there is a preference for cleavage sites located in particular secondary structure: loops>α-helices>β-strands. Importantly, natural human caspase substrates were found to be superior substrates compared with unnatural caspase sites from E. coli proteins, suggesting that tertiary structures play a key role in proteolysis of protein substrates.35 Supporting this, we found that although caspase-2, -3, and -7 all share very similar primary sequence specificity, each enzyme cleaves a particular set of native substrates at different rates based on factors other than the primary sequence (Figure 4).16, 42 Some of this may be attributed to caspases using exosites to facilitate substrate binding. It has been reported that caspase-7 uses an exosite to enhance cleavage rate for PARP1.36 There is additional suggestive evidence that substrate recognition does not rely solely on the primary sequence of the cleavage site; protein engineering of caspase-7 to acquire caspase-6 primary specificity did not lead to complete overlap of substrate target pools in cell lysates.50
Figure 4.
Rates of proteolysis of natural caspase substrates. With the advances of mass spectrometry, using targeted LC-MS/MS has become standard practice, allowing better characterization of proteolysis in complex mixtures. (a) Chromatograms of targeted transitions in SRM or PRM methods, allowing quantification of peptides captured over a time-course experiment. (b) Rates of proteolysis can be calculated for each peptides originating from proteolytic events by plotting the sum of the peak intensities (or area) as a function of time, assuming pseudo-first-order kinetics. (c) Comparison of the estimated rates of proteolysis for each cleavage sites of each caspase substrates showing up to five order of magnitude difference. Adapted from ref. 16
Functional versus bystander targets
It is possible that some proteins cleaved by caspases are simply bystanders that have little functional impact on the cellular phenotype; this is easy to imagine in apoptosis where active caspases would continue to cleave proteins long after the cell has passed the point of no return. Such post-death proteolysis may help in cellular digestion preceding final engulfment by macrophages and dendritic cells. Time-course experiments that use immunoblotting for specific substrates or targeted proteomics approaches for broader, unbiased coverage can help to identify the rapid and slow events during apoptosis. The Salvesen group determined the catalytic efficiencies (kcat/KM) for caspase-3 cleavage of 11 non-native substrates from E. coli by immunoblotting and calculated rates that ranged between 5 × 101 and 2 × 103/M/s. It is presumed these rates represent the basal level since caspases are not present in E. coli. In contrast, the rates for eight well-known caspase-3 human substrates were greater than 1 × 104/M/s and approached the rate for an optimal DEVD-afc synthetic substrate (~1 × 106/M/s). More recent reverse experiments using targeted proteomics have been applied to caspases-2, -3, -6, -7, and -8 to measure the rates of cleavage for hundreds of substrates simultaneously.16, 42 These studies showed that each caspase cleaves its substrates at wildly different rates that vary by over 500-fold. Moreover, each caspase had a unique set of ‘fast’ (rapid rate) substrates.
Are these fast substrates the most potent drivers of biological outcomes? One clear example of this is Gasdermin D (GSDMD), which has been shown to be cleaved by caspase-1 (FLTD275) using subtiligase reverse N-terminomics.41 In vitro experiments confirmed that GSDMD is cleaved rapidly by caspase-1 at a rate of 1.3 × 105/M/s (kcat/KM). Subsequent in-depth biological studies have shown that GSDMD cleavage triggers pyroptosis by the release of its proteolytic N-terminal domain.51, 52 This is a sterling example of how unbiased proteomics can identify candidate targets worthy of more detailed biological investigation that can uncover important new nodes of proteolytic regulation.
Another proteomic approach to assess the significance of a substrate target is to quantify the extent of proteolysis relative to the total target pool, reasoning that extensively cleaved targets should confer a greater functional impact. The PROTOMAP approach from the Cravatt lab53 is ideal for measuring the extent of cleavage because one can observe not only the target that is cleaved, but also the extent of cleavage in the SDS-PAGE dimension. Indeed, they found that most targets in their data sets are cleaved to near completion in apoptosis of Jurkat cells. However, assuming that the extent of cleavage correlates with impact may have caveats. For example, some cleavage events, especially those that result in a gain-of-function, may require only a small fraction of the pool be cleaved to produce effects.
Gain-of-function and loss-of-function
Proteomic studies show caspase substrates are cleaved only once or a few times, and can create stable domain products in the cell. One of the major challenges in interpreting a native proteolytic event is whether the cleavage results in a gain-of-function or loss-of-function phenotype. These functional differences are particularly evident in apoptosis, where some cleavage events drive apoptosis through gain-of-function, such as the activation of caspases themselves, whereas most of the cleaved substrates, such as inhibitor of the caspase-activated DNAse (ICAD), have undergone a loss-of-function.54 For example, Anania et al.55 used a systems-level approach combining transcriptomics, proteomics, and N-terminomics to study the role of caspases during the unfolded protein response caused by endoplasmic reticulum stress. This study showed that caspase activation not only controls post-translational protein expression of direct caspase substrates, but also regulates transcription of genes involved in cell fate decisions in response to endoplasmic reticulum stress. Although it is attractive to consider the use of CRISPR technology to either inhibit or activate transcription of caspase substrates, in many cases this is not the same as proteolysis. For example, ICAD is cleaved during apoptosis to relieve inhibition of CAD allowing it to cleave DNA, but ICAD also functions as a chaperone for CAD, such that ICAD knockdown will paradoxically result in loss of CAD.56
Cellular engineering is one method that can be used to determine the functional outcomes of specific caspase cleavage events. One example, the chemically induced dimerization system for split TEV protease, the SNIPer, fuses FKBP and FRB to the two parts of the enzyme so that it can be activated by rapamycin.20 By engineering TEV protease sites in place of caspase sites in candidate substrates, it is possible to cleave a single site within a single target, and to monitor the functional outcome. This approach was used to show that activation of caspases-3 or -7, but not caspase-6, was sufficient to drive apoptosis in human cells.20 Recently, a tetracycline inducible expression system (Tet-on) system was used to induce rapid TEV expression and cleave TEV sites engineered into ICAD, providing evidence that activation of CAD was remarkably insufficient to drive cell death.57 Other DNA repair enzymes are known to be cleaved by caspases and when these were inhibited, direct activation of CAD was seen at the single cell level. Other than caspases-3 and -7 themselves, it appears that cleaved caspase substrates are rarely capable of single-handedly sparking apoptosis, except possibly BIMEL in T cells.58 However, while the activation of CAD is insufficient to control the cell death phenotype, the activation of CAD was shown to be required for muscle cell differentiation. For example, Larsen et al.59 showed that caspase-3 cleavage of CAD promotes cell differentiation by directly modifying the DNA/nuclear microenvironment leading to increased expression of critical regulatory genes. These studies are consistent with previous proposals that apoptosis is a massive synthetic lethal process in which sustained caspase activation leads to many cleavage events that work together to drive the cell death.60 In contrast, cell differentiation seems to be a more defined and precise biological process involving transient caspase activation. It is possible the substrates cleaved in these processes are different, owing to differential caspase activation, or selective cleavage of fast substrates due to transient activation. Further experiments will be important to distinguish these possibilities.
Caspase substrates in various forms of cell death and differentiation
Extrinsic and intrinsic cell death
It is well established that apoptosis can be induced by outside factors (extrinsic), such as TRAIL and FAS ligand, which lead to activation of caspase-8,61, 62 which triggers activation of executioner caspases-3 and -7 and leads to cell death (for a review see ref. 63). Apoptosis can also be induced by internal factors (intrinsic), such as DNA damage, leading to activation of caspase-9 through mitochondrial cytochrome c release, Apaf1 oligomerization and ultimately activation of caspases-3 and -7.64, 65 Overall, comparison of the final cleaved substrates and their rates of appearance in Jurkat cells show there is a correlation between classic extrinsic and intrinsic inducers.42 Reverse studies further showed that caspases-3 and -7 have unique sets of rapidly cleaved substrates (180 and 58 substrates measured with kcat/KM values >103/M/s) suggesting a set of non-redundant targets for the executioners.42 Some of the most rapidly cleaved substrates for both inducers were those in the miRNA pathway (notably DCGR8 and TARBP2) and targets in the endosomal pathway (especially for TRAIL), consistent with previous work examining several of these targets individually.66, 67, 68 Proteomics was further used to compare apoptotic caspase substrates cleaved in a B-cell line (DB cells, 488 substrates), a T-cell line (Jurkat cells, 505 substrates), and a multiple myeloma cell line (MM1S cells, 520 substrates).40 There was a remarkable overlap of 472 peptides indicating that the apoptotic end states were very similar in these hematopoetic cell lines. However, the abundance of the peptides varied between cell types, and the cleavage rates varied between the apoptosis inducers staurosporine and bortezomib, a proteasome inhibitor. Some of this was due to induction of expression of stress proteins prior to apoptosis that is drug-specific, as bortezomib is known to induce endoplasmic reticulum stress.69 We observed a transient increase in activating transcription factor 4 that preceded cell death but, ultimately cleavage of ATF4 by caspases that may reflect a cell death-resistant stress response followed by capitulation.
It is known that there is a significant time delay of a few hours prior to the induction of caspase activation in cells.70, 71 An extensive study of the transcriptional and translational events that precede induction of caspases in bortezomib-treated MM1S cells was conducted using RNAseq, ribosomal profiling, and quantitative proteomics.72 This work showed extensive mRNA degradation preceding caspase activation. Interestingly, during this mRNA degradation phase there were minimal changes in protein levels, possibly due to the direct inhibition of the proteasome by bortezomib. Ribosomal occupancy and mRNA levels generally tracked, except that translational efficiency (judged by ribosome occupancy/total mRNA) was higher for some proteins in stress-induced pathways, such as ATF4, HSF1, and a few anti-apoptotic proteins among others. Overall, it appeared that the cells were attempting to relieve the consequences of proteasomal inhibition, but ultimately succumb to endoplasmic reticulum stress presumably due to the high burden of immunoglobulin secretion in the MM1S cell. Thus, the initial stages of apoptosis likely depend on the drug-induced mechanism and cellular context, but the final proteolytic outcomes are very similar.
Non-apoptotic cell death mechanisms
The activation of pyroptosis, or inflammatory programmed cell death, is mediated by human caspase-1, as well as caspases-4/-5 (-11 in mouse).73 In fact, caspase-1 was first named interleukin-1β converting enzyme for its role in mediating the production of mature IL-1β, an inflammatory cytokine.74, 75 During bacterial infection, the binding of the cytosolic lipid-A portion of LPS to the caspase-recruitment domains of the inflammatory caspase-4 (and -11) promotes their oligomerization and activation.76 As previously mentioned, GSDMD was identified to be cleaved by caspase-1 (FLTD275)41 but not by either purified caspase-4 or -5. However, it was shown that these two caspases require binding of LPS or lipid-A for cleavage to occur.52 It would thus be interesting to add these cofactors and repeat the reverse experiments with each of the inflammatory caspases to find other potential substrates. It is possible that there are other cofactor molecules (either big or small) that may assist in caspase activation or modify substrate specificity. GSDMD is a key mediator of the host response against Gram-negative bacteria and is essential for mouse caspase-11-dependent pyroptosis and IL-1β maturation.51 Gasdermin family members are capable of binding lipids themselves, such as phosphatidylinositol phosphates and phosphatidylserines. The gasdermin-N domain released from caspase cleavage binds lipids at the cell membrane and promotes the formation of pores, leading to rapid loss of cell membrane integrity and leakage of cytosolic content. The discovery of the role of the gasdermin-N domain is only the latest finding of an essential role for a caspase-generated fragment in inflammation, and highlights the important function of caspase substrates in non-apoptotic cell death processes. It is also well known that phosphorylation of caspases and their substrates can modulate activation, inhibition, or substrate specificity.9, 77, 78
Another non-apoptotic cell death mechanism is necroptosis, or programmed necrosis (see ref. 79 for review). This may seem counter-intuitive, since necrosis has long been considered a non-programmed form of cell death, and also because caspase-8 plays a suppressive role in this process. Caspase-8 is well known for participating in the extrinsic apoptosis pathway by cleaving and directly activating the effector caspases-3 and -7, among others. A lesser-known function of caspase-8 is its pro-survival role in suppressing RIPK1/RIPK3 function, which promotes necroptosis during development.80, 81, 82 One major part of necroptosis signaling is the formation of the necroptosome, formed by RIPK1, RIPK3, FADD, cFLIP, and caspase-8.83, 84 The mechanism by which caspase-8 can suppress necroptosis is not fully understood, but it appears that its catalytic activity is necessary.81 Caspase-8 cleavage of RIPK1/RIPK3 might be preventing oligomerization of these proteins that is required for necroptosis signaling.85 Interestingly, caspase-6 and -8 have been shown to be capable of cleaving RIPK1 in vitro,16, 86 but the identities of other possible caspase substrates and their functional roles in necroptosis remains to be discovered. Finally, as other forms of non-apoptotic cell death emerge, such as a newly described iron-dependent form of cell death named ferroptosis,87 the role played by caspases in promoting or suppressing various cell death mechanisms will have to be addressed.
Activation of caspases in differentiation
As reviewed extensively in this issue, caspase activation has been observed in erythroid differentiation,88 lens development,89 spermatogenesis,90 stem cell differentiation,91 neuronal development, and dendrite trimming.92, 93 How these cells transiently activate caspases, and which caspases become activated and ultimately deactivated remains poorly understood. Nonetheless, there are known endogenous mechanisms for turning caspases off through direct binding to members of the inhibitor of apoptosis protein family that include the E3 ligase X chromosome-linked inhibitor of apoptosis protein.94 Other E3 ligases may facilitate caspase deactivation by promoting caspase degradation via the proteasome (Figure 5).20, 95 Proteasomal degradation is an important control point as it has been shown that the rate of apoptosis by direct activation of either caspase-3 or -7 is dramatically accelerated by sublethal inhibition of the proteasome in HEK293 cells.20 Moreover, direct activation of caspase-6 does not lead to apoptosis unless paired with sublethal inhibition of the proteasome in HEK293. Proteasomal activity decreases with caspase activation and 20 of the 33 subunits of the 26s proteasome are cleaved by caspases. Caspase activation, when viewed at the single cell level, is highly precipitous, occurring over about a 10-min period,20, 70, 71, 96 but it is highly stochastic, so that some cells activate caspases early and some late. The basis for this stochasticity is unclear, but this reciprocal negative-feed-back loop between caspases versus the proteasome and E3 ligases could play an important role in stopping the executioner caspases before triggering cell death.
Figure 5.
Regulation of caspase activity. Upon caspase activation, specific proteolytic events occur such as degradation of the proteasome. In non-apoptotic mechanisms, caspase activity must turn off at some point to prevent apoptosis, and inhibitor of apoptosis proteins provide a possible mechanism
Despite the compelling evidence for caspase activation in cell differentiation, there has been little use of proteomic tools to identify caspase substrates in an unbiased fashion. This probably reflects the fact that proteomics studies typically require tens to hundreds of millions of cells, quantities that are not easily obtained for most differentiation models. Moreover, compared to apoptosis, there are likely to be many fewer substrates that may not be cleaved to completion, which further pushes the limits of detection. Immunoblot could be used to sensitively detect cleaved products, but is laborious and requires expensive antibodies to conduct studies on a candidate-by-candidate basis. In the future, we anticipate that proteomics will begin to yield greater insights into cell differentiation as instrumentation improves and the sensitivity and comprehensiveness of coverage increases. As mentioned above, there is a huge range in the rates of cleavage of substrates by any given caspase. It is reasonable to expect that during transient activation of caspases in cell differentiation, only the most highly preferred targets would be cleaved. Thus, one could use existing kinetic data from reverse experiments to construct a candidate list for more targeted proteomics studies. Although little is known about the caspase substrates in differentiation, we believe the future is bright for their identification.
Conclusions and important questions
The identification of caspase substrates using proteomics approaches has exploded in recent years, especially as it applies to apoptosis. It is now clear that hundreds to thousands of substrates can be cleaved by caspases in apoptosis and large databases that catalog these cleavages have emerged. Caspases cleave native proteins sparsely, mostly in exposed loops or dynamic helices, to yield stable domain products that can have both gain-of-function, loss-of-function, or non-functional (bystander) phenotypes. Reverse proteomics has been applied to almost all human caspases except the caspases-10, -12, and -14. Such data are very useful for defining what can be cut by caspases in cell extracts, and in most cases the profiles are not that different from those found in intact cells. Each caspase has highly preferred targets whose rates vary by orders of magnitude. Although many targets can be cut by different caspases, each caspase has a preferred set, suggesting that they have specialized functions.
Many exciting questions remain. For most of the caspase substrates, we do not understand the functional consequences of their cleavage. Shedding light on this area will require detailed biological investigations, such as those conducted on gasdermin and ICAD. Although each caspase has a unique set of preferred substrates, why do some caspases have so many substrates (caspase-1, -2, -3, -6, -7, and -8) while some have so few (caspases-4, -5, -9, -12, and -14)? This is especially true for some of the rarely studied caspases, such as caspase-12 and -14, the latter of which has only one identified substrate, profilaggrin.97 It may also be that caspases have other cofactors and post-translational modifications that are not represented in reverse experiments, as could be the case for LPS/Lipid-A with inflammatory caspases. We have virtually no identification of the caspase substrates cleaved during cell differentiation by proteomics methods, but we believe this will change in the next few years. Although this review has focused on caspase substrates, the biology of cell death and differentiation certainly involves many more players from start to finish. We know there is interplay between kinases and caspases both in phosphorylation of caspases,77 and phosphorylation of caspase substrates to inhibit or enhance cleavage.6, 78 Moreover, other proteases besides caspases are activated during cell death and differentiation, such as calpains, serine proteases (granzymes), and lysosomal proteases, and these could potentially generate almost the same number of cleaved substrates. Clearly, other signaling events are conspiring and cooperating with caspases, and we are only beginning to understand this complex interplay that define cell death and differentiation processes.
Footnotes
Edited by E Arama
The authors declare no conflict of interest.
References
- Thornberry NA, Lazebnik Y. Caspases: enemies within. Science 1998; 281: 1312–1316. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Declercq W, Kalai M, Saelens X, Vandenabeele P. Alice in caspase land. A phylogenetic analysis of caspases from worm to man. Cell Death Differ 2002; 9: 358–361. [DOI] [PubMed] [Google Scholar]
- Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell 1993; 75: 641–652. [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem 1997; 272: 17907–17911. [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Renatus M, Meldal M, Salvesen GS. Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem J 2000; 350(Pt 2): 563–568. [PMC free article] [PubMed] [Google Scholar]
- Seaman JE, Julien O, Lee PS, Rettenmaier TJ, Thomsen ND, Wells JA. Cacidases: caspases can cleave after aspartate, glutamate and phosphoserine residues. Cell Death Differ 2016; 23: 1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR. Apoptotic pathways: the roads to ruin. Cell 1998; 94: 695–698. [DOI] [PubMed] [Google Scholar]
- Van Damme P, Martens L, Van Damme J, Hugelier K, Staes A, Vandekerckhove J et al. Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat Methods 2005; 2: 771–777. [DOI] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 2008; 134: 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Cravatt BF. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell 2008; 134: 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VC, Pitti R, Anania VG, Bakalarski CE, Bustos D, Jhunjhunwala S et al. Complementary proteomic tools for the dissection of apoptotic proteolysis events. J Proteome Res 2012; 11: 2947–2954. [DOI] [PubMed] [Google Scholar]
- Lüthi AU, Martin SJ. The CASBAH: a searchable database of caspase substrates. Cell Death Differ 2007; 14: 641–650. [DOI] [PubMed] [Google Scholar]
- Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem 2011; 80: 1055–1087. [DOI] [PubMed] [Google Scholar]
- Yuan J, Kroemer G. Alternative cell death mechanisms in development and beyond. Genes Dev 2010; 24: 2592–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen GS, Ashkenazi A. Snapshot: caspases. Cell 2011; 147: 476–476. [DOI] [PubMed] [Google Scholar]
- Julien O, Zhuang M, Wiita AP, O’Donoghue AJ, Knudsen GM, Craik CS et al. Quantitative MS-based enzymology of caspases reveals distinct protein substrate specificities, hierarchies, and cellular roles. Proc Natl Acad Sci USA 2016; 113: E2001–E2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitevska T, Roberts SJ, Pantaki-Eimany D, Boyd SE, Scott FL, Hawkins CJ. Analysis of the minimal specificity of caspase-2 and identification of Ac-VDTTD-AFC as a caspase-2-selective peptide substrate. Biosci Rep 2014; 34: e00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Wells JA, Arkin MR. Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis. J Biol Chem 2011; 286: 34147–34154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wejda M, Impens F, Takahashi N, Van Damme P, Gevaert K, Vandenabeele P. Degradomics reveals that cleavage specificity profiles of caspase-2 and effector caspases are alike. J Biol Chem 2012; 287: 33983–33995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray DC, Mahrus S, Wells JA. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell 2010; 142: 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippens S, Kockx M, Knaapen M, Mortier L, Polakowska R, Verheyen A et al. Epidermal differentiation does not involve the pro-apoptotic executioner caspases, but is associated with caspase-14 induction and processing. Cell Death Differ 2000; 7: 1218–1224. [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Salvesen GS. Caspases - controlling intracellular signals by protease zymogen activation. Biochim Biophys Acta 2000; 1477: 299–306. [DOI] [PubMed] [Google Scholar]
- Salvesen GS, Riedl SJ. Caspase mechanisms. Adv Exp Med Biol 2008; 615: 13–23. [DOI] [PubMed] [Google Scholar]
- Renatus M, Stennicke HR, Scott FL, Liddington RC, Salvesen GS. Dimer formation drives the activation of the cell death protease caspase 9. Proc Natl Acad Sci USA 2001; 98: 14250–14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennicke HR, Jürgensmeier JM, Shin H, Deveraux Q, Wolf BB, Yang X et al. Pro-caspase-3 is a major physiologic target of caspase-8. J Biol Chem 1998; 273: 27084–27090. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Fuentes-Prior P, Renatus M, Kairies N, Krapp S, Huber R et al. Structural basis for the activation of human procaspase-7. Proc Natl Acad Sci USA 2001; 98: 14790–14795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol 2004; 5: 897–907. [DOI] [PubMed] [Google Scholar]
- Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun 1967; 27: 157–162. [DOI] [PubMed] [Google Scholar]
- Rano TA, Timkey T, Peterson EP, Rotonda J, Nicholson DW, Becker JW et al. A combinatorial approach for determining protease specificities: application to interleukin-1beta converting enzyme (ICE). Chem Biol 1997; 4: 149–155. [DOI] [PubMed] [Google Scholar]
- Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D et al. Substrate specificities of caspase family proteases. J Biol Chem 1997; 272: 9677–9682. [DOI] [PubMed] [Google Scholar]
- O’Donoghue AJ, Eroy-Reveles AA, Knudsen GM, Ingram J, Zhou M, Statnekov JB et al. Global identification of peptidase specificity by multiplex substrate profiling. Nat Methods 2012; 9: 1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S, Pastor R, Sutherlin D, Lowman HB. A substrate-phage approach for investigating caspase specificity. Protein J 2004; 23: 413–425. [DOI] [PubMed] [Google Scholar]
- Tucker MB, MacKenzie SH, Maciag JJ, Dirscherl Ackerman H, Swartz P, Yoder JA et al. Phage display and structural studies reveal plasticity in substrate specificity of caspase-3a from zebrafish. Protein Sci 2016; 25: 2076–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay GP, Salvesen GS, Green DR. Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways. Cell Death Differ 2008; 15: 322–331. [DOI] [PubMed] [Google Scholar]
- Timmer JC, Zhu W, Pop C, Regan T, Snipas SJ, Eroshkin AM et al. Structural and kinetic determinants of protease substrates. Nat Struct Mol Biol 2009; 16: 1101–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher D, Blais V, Denault JB. Caspase-7 uses an exosite to promote poly(ADP ribose) polymerase 1 proteolysis. Proc Natl Acad Sci USA 2012; 109: 5669–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert K, Goethals M, Martens L, Van Damme J, Staes A, Thomas GR et al. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol 2003; 21: 566–569. [DOI] [PubMed] [Google Scholar]
- Demon D, Van Damme P, Vanden Berghe T, Deceuninck A, Van Durme J, Verspurten J et al. Proteome-wide substrate analysis indicates substrate exclusion as a mechanism to generate caspase-7 versus caspase-3 specificity. Mol Cell Proteomics 2009; 8: 2700–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford ED, Seaman JE, Agard N, Hsu GW, Julien O, Mahrus S et al. The DegraBase: a database of proteolysis in healthy and apoptotic human cells. Mol Cell Proteomics 2013; 12: 813–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimbo K, Hsu GW, Nguyen H, Mahrus S, Trinidad JC, Burlingame AL et al. Quantitative profiling of caspase-cleaved substrates reveals different drug-induced and cell-type patterns in apoptosis. Proc Natl Acad Sci USA 2012; 109: 12432–12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agard NJ, Maltby D, Wells JA. Inflammatory stimuli regulate caspase substrate profiles. Mol Cell Proteomics 2010; 9: 880–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agard NJ, Mahrus S, Trinidad JC, Lynn A, Burlingame AL, Wells JA. Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proc Natl Acad Sci USA 2012; 109: 1913–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes MP, Farnsworth CL, Moritz A, Silva JC, Jia X, Lee KA et al. PTMScan direct: identification and quantification of peptides from critical signaling proteins by immunoaffinity enrichment coupled with LC-MS/MS. Mol Cell Proteomics 2012; 11: 187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes MP, Silva JC, Jia X, Lee KA, Polakiewicz RD, Comb MJ. Quantitative profiling of DNA damage and apoptotic pathways in UV damaged cells using PTMScan direct. Int J Mol Sci 2012; 14: 286–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleifeld O, Doucet A, auf dem Keller U, Prudova A, Schilling O, Kainthan RK et al. Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat Biotechnol 2010; 28: 281–288. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Tolle DP, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res 2004; 32: D160–D164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi Y, Eroshkin A, Gramatikova S, Gramatikoff K, Zhang Y, Smith JW et al. CutDB: a proteolytic event database. Nucleic Acids Res 2007; 35: D546–D549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange PF, Overall CM. TopFIND, a knowledgebase linking protein termini with function. Nat Methods 2011; 8: 703–704. [DOI] [PubMed] [Google Scholar]
- Kumar S, van Raam BJ, Salvesen GS, Cieplak P. Caspase cleavage sites in the human proteome: CaspDB, a database of predicted substrates. PLoS ONE 2014; 9: e110539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill ME, MacPherson DJ, Wu P, Julien O, Wells JA, Hardy JA. Reprogramming caspase-7 specificity by regio-specific mutations and selection provides alternate solutions for substrate recognition. ACS Chem Biol 2016; 11: 1603–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015; 526: 666–671. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015; 526: 660–665. [DOI] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Cravatt BF. Global identification of caspase substrates using PROTOMAP (protein topography and migration analysis platform). Methods Mol Biol 2014; 1133: 61–70. [DOI] [PubMed] [Google Scholar]
- Sakahira H, Enari M, Nagata S. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature 1998; 391: 96–99. [DOI] [PubMed] [Google Scholar]
- Anania VG, Yu K, Gnad F, Pferdehirt RR, Li H, Ma TP et al. Uncovering a dual regulatory role for caspases during endoplasmic reticulum stress-induced cell death. Mol Cell Proteomics 2016; 15: 2293–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakahira H, Enari M, Nagata S. Functional differences of two forms of the inhibitor of caspase-activated DNase, ICAD-L, and ICAD-S. J Biol Chem 1999; 274: 15740–15744. [DOI] [PubMed] [Google Scholar]
- Morgan CW, Diaz JE, Zeitlin SG, Gray DC, Wells JA. Engineered cellular gene-replacement platform for selective and inducible proteolytic profiling. Proc Natl Acad Sci USA 2015; 112: 8344–8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Zhou Q. Caspase cleavage of BimEL triggers a positive feedback amplification of apoptotic signaling. Proc Natl Acad Sci USA 2004; 101: 1235–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen BD, Rampalli S, Burns LE, Brunette S, Dilworth FJ, Megeney LA. Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc Natl Acad Sci USA 2010; 107: 4230–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroh C, Schulze-Osthoff K. Death by a thousand cuts: an ever increasing list of caspase substrates. Cell Death Differ 1998; 5: 997–1000. [DOI] [PubMed] [Google Scholar]
- Muzio M, Chinnaiyan AM, Kischkel FC, O’Rourke K, Shevchenko A, Ni J et al. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death—inducing signaling complex. Cell 1996; 85: 817–827. [DOI] [PubMed] [Google Scholar]
- Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM. An induced proximity model for caspase-8 activation. J Biol Chem 1998; 273: 2926–2930. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science 1998; 281: 1305–1308. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol Cell 1998; 1: 949–957. [DOI] [PubMed] [Google Scholar]
- Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 1997; 90: 405–413. [DOI] [PubMed] [Google Scholar]
- Matskevich AA, Moelling K. Stimuli-dependent cleavage of Dicer during apoptosis. Biochem J 2008; 412: 527–534. [DOI] [PubMed] [Google Scholar]
- Smith LK, Shah RR, Cidlowski JA. Glucocorticoids modulate microRNA expression and processing during lymphocyte apoptosis. J Biol Chem 2010; 285: 36698–36708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin CD, Lawrence DA, Peden AA, Varfolomeev EE, Totpal K, De Mazière AM et al. Death-receptor activation halts clathrin-dependent endocytosis. Proc Natl Acad Sci USA 2006; 103: 10283–10288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson LI, Davies FE. DangER: protein ovERload. Targeting protein degradation to treat myeloma. Haematologica 2012; 97: 1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR. Apoptotic pathways: ten minutes to dead. Cell 2005; 121: 671–674. [DOI] [PubMed] [Google Scholar]
- Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 2009; 459: 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiita AP, Ziv E, Wiita PJ, Urisman A, Julien O, Burlingame AL et al. Global cellular response to chemotherapy-induced apoptosis. Elife 2013; 2: e01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Najafov A, Py BF. Roles of caspases in necrotic cell death. Cell 2016; 167: 1693–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature 1992; 356: 768–774. [DOI] [PubMed] [Google Scholar]
- Alnemri ES, Livingston DJ, Nicholson DW, Salvesen G, Thornberry NA, Wong WW et al. Human ICE/CED-3 protease nomenclature. Cell 1996; 87: 171. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 2014; 514: 187–192. [DOI] [PubMed] [Google Scholar]
- Kurokawa M, Kornbluth S. Caspases and kinases in a death grip. Cell 2009; 138: 838–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Wang C, Okerberg E, Patricelli MP, Cravatt BF. Functional interplay between caspase cleavage and phosphorylation sculpts the apoptotic proteome. Cell 2012; 150: 426–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 2010; 11: 700–714. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature 2011; 471: 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C et al. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 2011; 471: 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhou X, McQuade T, Li J, Chan FK, Zhang J. Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature 2011; 471: 373–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M et al. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell 2011; 43: 449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F et al. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell 2011; 43: 432–448. [DOI] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 2012; 150: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Raam BJ, Ehrnhoefer DE, Hayden MR, Salvesen GS. Intrinsic cleavage of receptor-interacting protein kinase-1 by caspase-6. Cell Death Differ 2013; 20: 86–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012; 149: 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordet O, Rebe C, Plenchette S, Zermati Y, Hermine O, Vainchenker W et al. Specific involvement of caspases in the differentiation of monocytes into macrophages. Blood 2002; 100: 4446–4453. [DOI] [PubMed] [Google Scholar]
- Dahm R. Lens fibre cell differentiation - a link with apoptosis. Ophthalmic Res 1999; 31: 163–183. [DOI] [PubMed] [Google Scholar]
- Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell 2003; 4: 687–697. [DOI] [PubMed] [Google Scholar]
- Li F, He Z, Shen J, Huang Q, Li W, Liu X et al. Apoptotic caspases regulate induction of iPSCs from human fibroblasts. Cell Stem Cell 2010; 7: 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DW, Kondo S, Krzyzanowska A, Hiromi Y, Truman JW. Local caspase activity directs engulfment of dendrites during pruning. Nat Neurosci 2006; 9: 1234–1236. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Zhu S, Younger S, Jan LY, Jan YN. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron 2006; 51: 283–290. [DOI] [PubMed] [Google Scholar]
- Scott FL, Denault JB, Riedl SJ, Shin H, Renatus M, Salvesen GS. XIAP inhibits caspase-3 and -7 using two binding sites: evolutionarily conserved mechanism of IAPs. EMBO J 2005; 24: 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XM, Butterworth M, MacFarlane M, Dubiel W, Ciechanover A, Cohen GM. Caspase activation inhibits proteasome function during apoptosis. Mol Cell 2004; 14: 81–93. [DOI] [PubMed] [Google Scholar]
- Tajon CA, Seo D, Asmussen J, Shah N, Jun YW, Craik CS. Sensitive and selective plasmon ruler nanosensors for monitoring the apoptotic drug response in leukemia. ACS Nano 2014; 8: 9199–9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecker G, Hoste E, Gilbert B, Hochepied T, Ovaere P, Lippens S et al. Caspase-14 protects against epidermal UVB photodamage and water loss. Nat Cell Biol 2007; 9: 666–674. [DOI] [PubMed] [Google Scholar]
- Wachmann K, Pop C, van Raam BJ, Drag M, Mace PD, Snipas SJ et al. Activation and specificity of human caspase-10. Biochemistry 2010; 49: 8307–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolajczyk J, Scott FL, Krajewski S, Sutherlin DP, Salvesen GS. Activation and substrate specificity of caspase-14. Biochemistry 2004; 43: 10560–10569. [DOI] [PubMed] [Google Scholar]
- Hoste E, Kemperman P, Devos M, Denecker G, Kezic S, Yau N et al. Caspase-14 is required for filaggrin degradation to natural moisturizing factors in the skin. J Invest Dermatol 2011; 131: 2233–2241. [DOI] [PubMed] [Google Scholar]