Key Points
Genomic disruption of CD7 prior to CAR transduction allows generation of CD7 CAR T cells without extensive self-antigen-driven fratricide.
CD7 CAR T cells have robust activity against T-cell malignancies in vitro and in vivo.
Abstract
Extending the success of chimeric antigen receptor (CAR) T cells to T-cell malignancies is problematic because most target antigens are shared between normal and malignant cells, leading to CAR T-cell fratricide. CD7 is a transmembrane protein highly expressed in acute T-cell leukemia (T-ALL) and in a subset of peripheral T-cell lymphomas. Normal expression of CD7 is largely confined to T cells and natural killer (NK) cells, reducing the risk of off-target-organ toxicity. Here, we show that the expression of a CD7-specific CAR impaired expansion of transduced T cells because of residual CD7 expression and the ensuing fratricide. We demonstrate that targeted genomic disruption of the CD7 gene prevented this fratricide and enabled expansion of CD7 CAR T cells without compromising their cytotoxic function. CD7 CAR T cells produced robust cytotoxicity against malignant T-cell lines and primary tumors and were protective in a mouse xenograft model of T-ALL. Although CD7 CAR T cells were also toxic against unedited (CD7+) T and NK lymphocytes, we show that the CD7-edited T cells themselves can respond to viral peptides and therefore could be protective against pathogens. Hence, genomic disruption of a target antigen overcomes fratricide of CAR T cells and establishes the feasibility of using CD7 CAR T cells for the targeted therapy of T-cell malignancies.
Introduction
Many patients with refractory or relapsed B-cell malignancies have achieved complete remission after receiving T cells that are redirected with chimeric antigen receptors (CARs) targeting the pan-B-cell antigen CD19.1-3 A number of factors have contributed to the clinical success of CAR T cells in this patient cohort, including the relative accessibility of malignant B cells, the presence of costimulatory molecules on normal and malignant CD19+ B cells, and the ability to effectively manage the loss of normal B cells—a common on-target/off-tumor side effect.
Broadening the success of CAR T cells to treat T-cell malignancies has proven challenging because of the shared expression of many targetable antigens between normal and malignant T cells. This shared antigenicity can cause fratricide in CAR-transduced T cells, inhibiting their proliferation and viability, and in the clinic may result in eradication of normal peripheral T cells. Such an on-target/off-tumor effect would be more profound and less treatable than is found after depletion of normal B cells with CD19 CAR T cells.
We previously reported that the fratricide in T cells expressing a CD5-specific CAR is limited and does not impair their expansion.4 This phenomenon was attributed to the rapid downregulation of CD5 from the cell surface of CAR T cells, reflecting the property of CD5 to internalize upon binding to a specific antibody.5 CD5, however, is not expressed by many T-cell tumors, and even when present, expression is often dim, emphasizing the need to broaden the range of target antigens. Moreover, it is now clear that tumor immune editing leads to frequent antigen-escape relapses following CAR T-cell and other immunotherapies.1,6,7 Hence, the ability to target more than one T-cell-associated antigen may be critical for the effective long-term treatment of malignancies arising from these cells.
CD7 is a transmembrane glycoprotein expressed by T cells and natural killer (NK) cells and their precursors8,9; it is also expressed in >95% of lymphoblastic T-cell leukemias and lymphomas and in a subset of peripheral T-cell lymphomas.10,11 CD7 plays a costimulatory role in T-cell activation upon binding to its ligand K12/SECTM1.12-14 However, it appears not to make a pivotal contribution to T-cell development or function because genomic disruption of CD7 in murine T-cell progenitors permits normal T-cell development and homeostasis and only minor alterations in T-cell effector function.15,16 Notably, CD7 is internalized on ligation17 and was previously evaluated as a target for immunotoxin-loaded antibodies in patients with T-cell malignancies.18 Although there were no severe CD7 antibody-related permanent adverse reactions, tumor responses were limited.18 We hypothesized that enhancing the potency of CD7-directed cytotoxicity by substituting autologous CAR T cells for a monoclonal antibody would augment the efficacy of CD7-targeted therapy in patients with T-cell malignancies.
Here, we investigated the feasibility of targeting T-lineage malignancies by using CD7 CAR T cells. We found that unlike CD5, the internalization of CD7 from the T-cell surface following CAR expression is incomplete and leads to extensive fratricide of CD7 CAR T cells. We therefore developed a means to permit the generation of functional CAR T cells using genome editing to eliminate persisting self-target antigens in T cells. In this study, we demonstrate that targeted disruption of the CD7 gene using CRISPR/Cas9 prior to CAR expression minimizes fratricide in T cells and allows the expansion of the CD7-knockout (CD7KO) CD7 CAR T cells with robust antitumor activity for preclinical and potential clinical application.
Materials and methods
CAR design and transduction
Three CD7-specific single-chain variable fragments derived from 3A1e, 3A1f,19 and TH-6920 clones of CD7-specific antibodies were created using commercial gene synthesis (Bio Basic, Inc) and cloned into a second-generation backbone CAR containing CD28 and CD3z endodomains and the CH3 domain from IgG1 Fc as a spacer region. A truncated CD7 CAR lacking signaling endodomains was used as a control. After initial efficacy studies, the best-performing clone (3A1e) was used for the remaining experiments presented here. T cells were activated by plate-bound OKT3 and anti-CD28 antibodies, transduced with gammaretroviral vectors, and expanded in the presence of interleukin-7 (IL-7) and IL-15 in cytotoxic T-lymphocyte (CTL) media, as has been described before.4
Genomic disruption of CD7 in T cells
Two guide RNAs (gRNAs) for the CD7 gene (gRNA-85: GGAGCAGGTGATGTTGACGG and gRNA-72: GGGGTCAATGTCTACGGCTC) were designed using CRISPRscan and COSMID algorithms.21,22 The CD7 gene was genomically disrupted in T cells according to the published protocol.23 Briefly, the 20-nt sequence complementary to the specific gene locus was incorporated into an oligonucleotide primer and used to amplify the gRNA scaffold from PX458 plasmid (a gift from Feng Zhang; Addgene 48138). gRNAs were generated through in vitro transcription with High-Yield RNA Synthesis Kit (NEB Bio Labs) from the DNA template, following the manufacturer’s instructions, and purified using the RNA Clean & Concentrator-25 kit (Zymo Research). We electroporated 0.4 μg of gRNA and 1 μg of cas9 protein (PNA Bio) with 0.25 × 106 of activated T cells by using the Neon Transfection System (Thermo Fisher Scientific) in 10 μL of buffer T and using 3 1600-V 10-ms pulses. Following electroporation, T cells were incubated in CTL media supplemented with 20% fetal bovine serum in the presence of IL-7 and IL-15 overnight after electroporation. T cells were then expanded in normal media.
DNA analysis
Genomic DNA was harvested 72 hours posttransfection using a Qiagen DNeasy Blood and Tissue kit, according to the manufacturer’s instructions. For T7EI assays, polymerase chain reaction (PCR) was used to amplify the gRNA target sites in CD7 using Accuprime High Fidelity Taq polymerase (Life Technologies), and the resulting products were purified using magnetic beads, as has been previously described.24 T7EI digestion was performed according to the manufacturer’s instructions, and the resulting cleavage products were analyzed by agarose gel electrophoresis.
GUIDE-Seq
For GUIDE-Seq experiments, gRNAs were generated by cloning annealed oligos containing the target sequence into pX330 (gift from Feng Zhang, Addgene 42230).25 U2OS cells (American Type Culture Collection [ATCC] HTB-96) were maintained in McCoy’s 5a Medium Modified; ATCC), supplemented with 10% fetal bovine serum and 2 mM of L-glutamine. Cells were cultured at 37°C in a 5% CO2 incubator. We nucleofected 200 000 U2OS cells with 1 μg of pX330 Cas9 and gRNA plasmid and with 100 pmol of dsODN by using SE cell line nucleofection solution and the CA-138 program on a Lonza 4D-nucleofector. The nucleofected cells were seeded in 500 μL of media in a 24-well plate, and gDNA was extracted 72 hours postnucleofection using a Qiagen DNeasy Blood and Tissue kit. dsODN integration at the target site was confirmed by restriction fragment length polymorphism assay with NdeI. gDNA was quantified using a qubit high-sensitivity dsDNA assay kit. We sheared 400 ng of gDNA using a Covaris LE220 Ultrasonicator to an average length of 500 bp. The sheared DNA was processed as previously described26 and sequenced on the Illumina Miseq. We analyzed GUIDE-Seq data using the standard pipeline26 with a reduced-gap penalty for better detection of off-target sites containing DNA or RNA bulges.
Bioinformatic off-target identification
Potential off-target sites for the CD7 gRNAs in the human genome (hg38) were identified using the Webtool COSMID21 with up to 3 mismatches allowed in the 19 PAM proximal bases. After off-target site ranking, 17 sites for gRNA-72 and 20 sites for gRNA-85 were selected for off-target screening.
Off-target validation
T cells were electroporated with recombinant Cas9 and CD7-specific gRNA, as indicated above. gDNA was extracted 7 days after ribonucleoprotein delivery, and locus-specific PCRs were performed to amplify off-target sites identified by COSMID and GUIDE-Seq. PCR primers contained adapter sequences to facilitate amplicon barcoding via a second round of PCR, as has been previously described.27 All amplicons were pooled at an equimolar ratio and sequenced on the Illumina Miseq according to the manufacturer’s instructions, except that custom sequencing primers were used for Read 2 and Read Index. Sequencing data were analyzed using a custom indel quantification pipeline.28
Flow cytometry
Anti-human CD7 and CCR7 (Biolegend); CD4, CD8, CD45RA, and CD56 (BD Biosciences); and CD3, CD19, and CD45 (Beckman Coulter) were used to stain cells in phosphate-buffered saline or CTL medium. Anti-IgG Fc (Jackson ImmunoResearch) was used for CAR detection in all assays. All flow cytometry data were obtained in BD fluorescence-activated cell sorter (FACS) Canto II (BD Biosciences) and Gallios (Beckman Coulter) and analyzed with FlowJo software (FlowJo, LLC).
Intracellular staining assay
Tumor cells were stained with eFluor670 (Thermo Fisher Scientific), according to the manufacturer’s instructions. CD7KO CD7 CAR T cells were cultured with stained tumor cells for 5 hours at a 1:4 effector:target ratio. Brefeldin A (BD GolgiPlug) and monensin (BD GolgiStop) were added 1 hour after plating. At the end of coculture, cells were incubated with antibodies for surface antigens and permeabilized for 10 minutes using BD FACS Permeabilizing Solution 2, followed by incubation with anti-tumor necrosis factor-a (anti-TNFa) and anti-interferon-g (IFNg) antibodies.
Cytotoxicity assay
Cell lines Jurkat (ATCC TIB-152), CCRF-CEM (ATCC CCL-119), MOLT-4 (ATCC CRL-1582), Hut 78 (ATCC TIB-161), SupT1 (ATCC CRL-1942), and Raji (ATCC CCL-86) were purchased from ATCC. NALM6 cells were a gift from Stephen Gottschalk. The cells were expanded according to ATCC recommendations. Tumor cells were stained with eFluor670 and incubated with CD7KO CD7 CAR T cells for 3 days without exogenous cytokines, unless stated otherwise. Cells from individual wells were collected at indicated time points. We added 7-AAD to discriminate dead cells, and we obtained absolute cell counts with CountBright Absolute Counting Beads (Thermo Fisher Scientific). In some experiments, autologous peripheral blood mononuclear cells (PBMCs) were labeled with eFluor670 and incubated with CD7KO CD7 CAR T cells in the presence of IL-2 (100 U/mL), IL-7, and IL-15. Cells were collected 24 hours later and stained with specific antibodies for subsequent flow cytometric analysis of eFluor670+ cells.
Primary T-ALL blasts
Deidentified fresh or frozen peripheral blood samples from acute T-cell leukemia (T-ALL) patients were thawed and used immediately for analysis. Fresh cells only were used in cytotoxicity assays because of the lower viability of frozen cells on thawing. The protocol for collection of peripheral blood from T-ALL patients was approved by the institutional review board at Baylor College of Medicine.
ELISPOT
T cells (2 × 105 cells in triplicates) were plated in 96-well MultiScreen HTS IP plates (EMD Millipore, MA) previously coated with anti-human IFNγ mAb 1-D1K (Mabtech, Cincinnati, OH), and kept overnight at 4°C. The cells were stimulated with pepmixes (15mers overlapping by 11 aa peptide libraries; JPT Technology) spanning EBV-EBNA-1, LMP-1, LMP-2, BARF-1, EBNA-3a,3b,3c., BZLF1; Adv-Hexon, Penton; and CMV-pp65, IE-1, and with staphylococcal enterotoxin B (Sigma-Aldrich Corporation) as a positive control. After 20 hours at 37°C, the cells were incubated with anti-human IFNγ mAb 7-B6-1-biotin (Mabtech) for 2 hours at 37°C, and avidin-peroxidase-complex (Vector Laboratories, Burlingame, CA) was added for 1 hour at room temperature. The plates were then developed with 3-amino-9-ethylcarbazole (Sigma, St. Louis, MO) substrate, dried, and sent to Zellnet Consulting (Fort Lee, NJ) for quantification. The spots formed were counted as spot-forming cells per 105 cells as a measure of the number of cells releasing IFNγ in response to viral antigen pepmixes.
T-ALL mouse xenograft model
Five- to 7-week-old nonobese diabetic (NOD)–Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) male and female mice (Jackson Laboratory) were engrafted with CCRF-CEM GFP-FFLuc+ cells by IV injection, and 2 × 106 of CD7 CAR+ T cells were injected IV 3 or 6 days later. To measure luminescence, we injected mice with 150 μg/kg of D-luciferin intraperitoneally, and tumor burden was monitored by recording luminesce in an IVIS Imaging system (Caliper Life Sciences) at indicated time points. Living Image software (PerkinElmer) was used to visualize and calculate total luminescence. For analysis of tumor and CAR T cells in peripheral blood of the mice, 100 μL of blood was collected by tail-vein bleeding. After red blood cell lysis, cells were incubated with anti-human CD45, CD4, CD7, and CD8 antibodies for subsequent flow cytometric analysis. All procedures were done in compliance with the Institutional Animal Care and Usage Committee of Baylor College of Medicine.
Statistical analysis
Data points from individual donors are shown in all figures. Statistical significance in pairwise comparisons was determined by an unpaired two-tailed Student t test and in multiple comparisons by a one-way analysis of variance with posttest Bonferroni correction. Statistical significance in Kaplan-Meier survival curves was assessed with the Mantel-Cox log-rank test. All P values were calculated using Prism 6 software (GraphPad).
Results
Expression of CD7 CAR precludes T-cell expansion
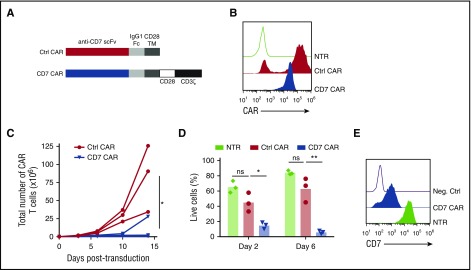
To test whether normal T cells can be redirected to recognize malignant T cells with a CD7-specific CAR, we created a CAR construct with the CD7-specific single-chain variable fragment derived from the antibody clone 3A1e (antigen-binding affinity 8.1 nM)19 fused to a second-generation CAR backbone containing an IgG1 CH3 spacer and cytoplasmic endodomains from CD28 and CD3ζ genes4 (Figure 1A). The CD7 CAR lacking intracellular signaling domains (ΔCAR) was used as a control. Transduction of activated human T cells with the gammaretroviral vectors resulted in efficient expression of the CD7 CAR constructs in T cells (Figure 1B). However, in contrast to control T cells expressing ΔCAR, the T cells expressing full-length CD7 CAR failed to expand (Figure 1C). Lack of expansion was associated with poor viability of CD7 CAR T cells (Figure 1D) and detectable residual expression of CD7 on the cell surface (Figure 1E), suggesting antigen-driven fratricide. Therefore, we hypothesized that ablating CD7 gene expression in T cells would prevent the fratricide and restore CD7 CAR T- cell expansion.
Figure 1.
T cells expressing CD7 CARs fail to expand. (A) Schematic diagram of control (ΔCAR) and full-length CD7 CAR constructs used in the study. (B) Surface expression of the CD7 CAR constructs on retrovirally transduced T cells measured by flow cytometry using anti-human IgG Fc antibodies on day 6 posttransduction. (C) Expansion of T cells transduced with truncated (ΔCAR) of full-length CD7 CAR in vitro for 14 days. (D) Viability of CD7 CAR T cells at days 2 and 6 posttransduction measured by flow cytometry. (E) Expression of CD7 in nontransduced or CD7 CAR-transduced activated T cells. A CD7-negative cell line Raji was used as a negative control. Data represent 2 to 5 independent experiments with n = 3 donors in each. Ctrl, control; Neg., negative; ns, not significant; NTR, nontransduced. *P < .05; **P < .001.
Genetic disruption of the CD7 gene enables expansion of CD7 CAR T cells
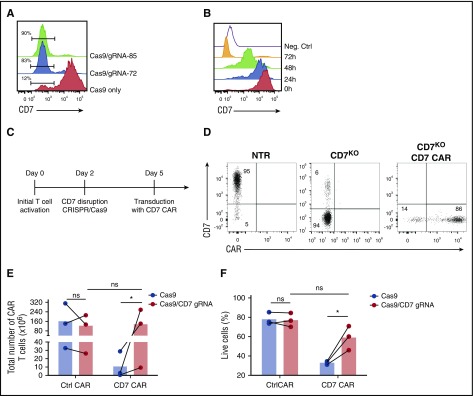
Sustained expansion and activity of CD7 CAR T cells would likely require permanent inhibition or disruption of CD7 gene expression in CAR-transduced T cells and their progeny. Therefore, we disrupted the CD7 gene in T cells prior to transduction by using CRISPR/Cas9. We used CRISPRScan20 and COSMID21 algorithms to select 2 guide RNAs (gRNA-72 and gRNA-85) targeting the CD7 gene with minimal predicted off-target effects. We chose to target exon 2 of the CD7 gene encoding the extracellular domain to reduce the probability of generating a truncated membrane-bound CD7 protein following nonhomologous end joining. Activated PBMCs were electroporated with CRISPR/Cas9 protein complex with either gRNA-72 or gRNA-85; both combinations resulted in efficient disruption of the CD7 gene, reflected by the loss of surface CD7 expression 3 days after electroporation (Figure 2A). After electroporation with CRISPR/Cas9 and a CD7-specific gRNA, T cells only gradually lost cell-surface expression of CD7 protein over 72 hours (Figure 2B), and transducing T cells with CD7 CAR only 24 hours after the gene editing still resulted in impaired expansion of CD7 CAR T cells (supplemental Figure 1, available on the Blood Web site). Therefore, we transduced the cells with CD7 CAR-encoding retrovirus 3 days after electroporation (Figure 2C). This method allowed us to consistently obtain a T-cell product containing >80% CD7-knockout (CD7KO) CD7 CAR T cells (Figure 2D). Most important, the removal of CD7 expression promoted robust expansion of CD7 CAR T cells (Figure 2E) and significantly improved T cell viability (Figure 2F). To verify off-target sites for both gRNAs, we performed unbiased whole-genome screening with the GUIDE-Seq method and validated the identified sites in primary CD7KO T cells (supplemental Figure 2). We detected no off-target activity above the limit of detection (0.1%) for gRNA-72. For gRNA-85, although 5 off-target sites had activity above 0.1%, none were significantly different from mock-treated samples (P = .18-.3; supplemental Table 1). These results indicate that the genomic disruption of the CD7 gene was specific and prevented T-cell fratricide to enable the expansion of CD7 CAR T cells.
Figure 2.
Disruption of CD7 expression with CRISPR/Cas9 restores expansion of CD7 CAR T cells. (A) Representative histogram showing ablation of CD7 expression in T cells after electroporation with CRISPR/cas9 and CD7-specific gRNAs 3 days after electroporation. Numbers denote frequency of CD7-negative cells. T cells electroporated with Cas9 only were used as a negative control. (B) Downregulation of surface CD7 expression in T cells after electroporation with CRISPR/cas9 and gRNA-85. A CD7-negative cell line Raji was used as a negative control. (C) Schematic outline of the optimized protocol for generating CD7-knockout (CD7KO) CD7 CAR T cells. (D) Representative dot plots showing expression of CD7 and CD7 CAR in T cells generated with the optimized protocol. Numbers indicate percentage of cells in each quadrant. (E) Total expansion of CD7 CAR T cells with and without CD7 knockout after 14 days of in vitro culture. (F) Viability of CD7 CAR T cells with and without CD7 gene disruption measured at day 6 after transduction by flow cytometry. Lines denote individual donors. Data represent 3 independent experiments with 3 donors in each. h, hour. *P < .05.
Genetic disruption of CD7 does not impair cytotoxicity of CAR T cells
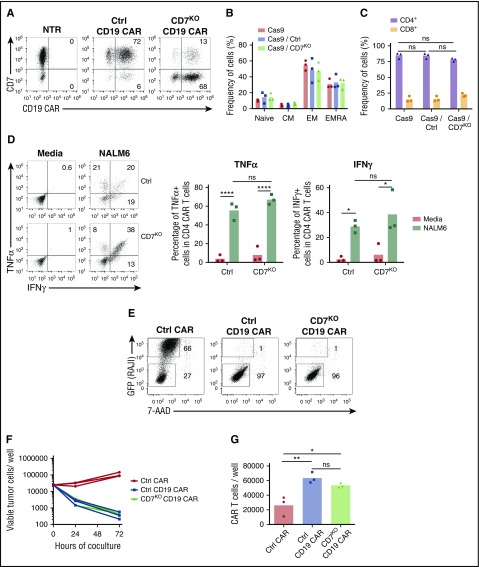
Although the CD7 protein has a costimulatory function in T cells, whether its expression is critical or not for the antitumor activity of CAR T cells is unknown. We therefore determined whether genomic disruption of the CD7 gene using the CRISPR/Cas9 system altered the effector function of CAR T cells. We electroporated T cells with CRISPR/Cas9 and CD7-specific or control gRNA and transduced them with CD19 CAR (Figure 3A), because the activity of this CAR is well documented. Removal of CD7 did not evidently alter the phenotype of CD19 CAR T cells (Figure 3B) and preserved the CD4:CD8 ratio (Figure 3C). Moreover, lack of CD7 did not compromise the ability of CD4+ CD19 CAR T cells to produce the cytokines TNFα and IFNγ on coculture with CD19+ NALM6 cells (Figure 3D). We then measured the cytotoxicity of CD7+ and CD7KO CD19 CAR T cells against a CD19+ cell line, Raji. Both groups of CD19 CAR T cells efficiently eliminated tumor cells after 72 hours of coculture (Figure 3E-F). Loss of CD7 did not reduce overall expansion of CD19 CAR T cells (Figure 3G) and did not affect proliferation of CD4+ and CD8+ subsets (supplemental Figure 3). Therefore, loss of CD7 did not abrogate the short-term survival or cytotoxicity of CAR T cells.
Figure 3.
Loss of CD7 does not alter phenotype or effector function of CAR T cells. (A) T cells were electroporated with Cas9 complexed with CD7-specific or control (CD19 specific) gRNA and transduced with CD19 CAR. Representative dot plots show expression of CD7 and CD19 CAR in T cells electroporated with Cas9+gRNA 7 days posttransduction. Nontreated activated T cells were used as control. Numbers denote frequency of cells in corresponding quadrants. (B) Frequency of naïve-like cells (naïve; CCR7+CD45RA−), central memory (CM; CCR7+CD45RA−), effector memory (EM; CCR7−CD45RA−), and effector memory RA (EMRA; CCR7−CD45RA+) in CD19 CAR T cells assessed by flow cytometry on day 7 posttransduction. (C) Frequency of CD4+ and CD8+ CD19 CAR T cells 7 days posttransduction. (D) CD19 CAR T cells were incubated with CD19+ NALM6 cells, and production of TNFα and IFNγ in CD4+ cells was assessed by intracellular cytokine staining. Dot plots represent cytokine production in CD19 CAR T cells in the presence of NALM6 or in media alone. Summarized data from 3 donors are shown on the right. (E) Control or CD7KO CD19 CAR T cells or control nontransduced T cells were cocultured with GFP+ Raji cells at the effector-to-target ratio 1:1 for 72 hours. Dot plots show representative frequency of gated CAR T cells and GFP+ tumor cells at the end of coculture. Total numbers of live tumor cells (F) and CD19 CAR T cells (G) were counted by flow cytometry at 72 hours using counting beads. Lines denote individual donors. Data represent 2 independent experiments with n = 3 donors in each. *P < .05; **P < .01; ****P < .0001.
CD7KO CD7 CAR T cells eradicate malignant T-cell lines in vitro
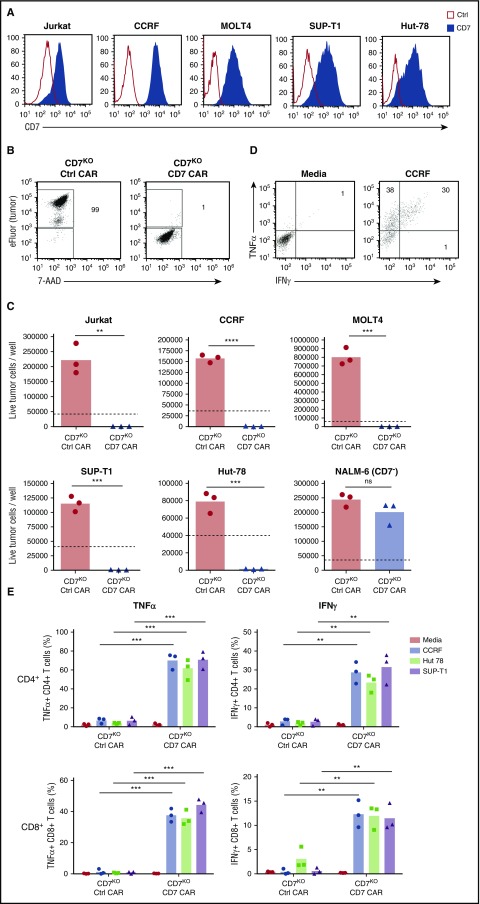
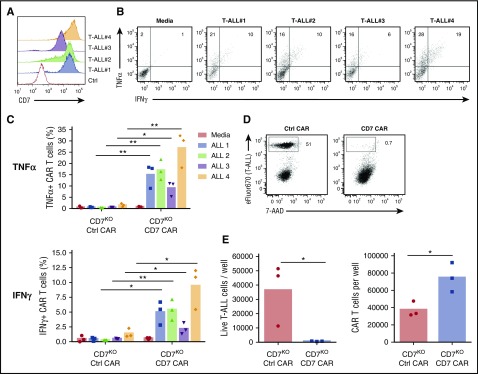
Because CD7 is highly expressed in T-cell malignancies, we assessed the capacity of CD7KO CD7 CAR T cells to kill the T-ALL cell lines Jurkat, CCRF, MOLT-4, and Sup-T1 and a T-cell lymphoma line Hut78, all of which were CD7-positive (Figure 4A). In comparison with control CAR T cells, CD7KO CD7 CAR T cells had robust cytotoxicity against eFluor670-labeled tumor cell lines, with >95% decrease in counts of viable tumor cells after 3 days of coculture at a 1:4 initial effector-to-target (E:T) ratio (Figures 4B-C). Coculture with tumor cells also induced proliferation of CD7KO CD7 CAR T cells (supplemental Figure 4). By contrast, CD7KO CD7 CAR T cells were not cytotoxic against the CD7-negative cell line NALM6 (Figure 4C), indicating that the cytolysis was CD7-specific. Moreover, CD7KO CD7 CAR T cells also effectively eliminated tumor cells at lower E:T ratios 1:5 to 1:20 (supplemental Figure 5). Both CD4+ and CD8+ CD7KO CD7 CAR T cells produced TNFα and IFNγ on coculture with T-ALL cell lines CCRF, and SupT1 and a lymphoma line Hut78 (Figure 4E-F). Therefore, CD7KO CD7 CAR T cells demonstrate potent and specific antitumor activity against malignant T cells.
Figure 4.
Expanded CD7KOCD7 CAR T cells eradicate T-ALL and T lymphoma cell lines. (A) Surface expression of CD7 (solid histograms) in T-ALL and T-lymphoma cell lines measured by flow cytometry in comparison with isotype control (open histograms). (B) Tumor cell lines were labeled with eFluor670 and cocultured with CD7KO CD7 CAR T cells at the effector-to-target ratio 1:4 for 3 days. Dot plots show frequency of gated live tumor cells (CCRF) at the end of coculture. (C) Absolute counts of live tumor cells were measured by flow cytometry using counting beads at the end of coculture with CD7KO CD7 CAR T cells. CD7− cell line NALM6 was used as a negative control. Dashed lines represent the initial number of tumor cells on plating. (D) Representative dot plots showing intracellular cytokine staining for TNFα and IFNγ in CD7KO CD7 CAR T cells upon coculture with CCRF cells or media alone. (G) Mean frequencies of cytokine-positive CD4+ (top) and CD8+ (bottom) CD7KO CD7 CAR T cells on coculture with indicated CD7+ cell lines or media alone. Data represent 2 independent experiments with n = 3 donors in each. **P < .01; ***P < .001; ****P < .0001.
CD7KO CD7 CAR T cells recognize and eliminate primary T-ALL cells
To further evaluate the capacity of CD7KO CD7 CAR T cells to respond to primary tumors, we measured cytokine production on coculture with freshly thawed apheresis samples from T-ALL patients, whose blast cells had a range of CD7 expression (Figure 5A). Both CD4+ and CD8+ CD7KO CD7 CAR T cells produced increased levels of IFNγ and TNFα when cocultured with the primary cells (Figure 5B-C). Similarly, we observed robust cytotoxicity of CD7KO CD7 CAR T cells against freshly isolated primary T-ALL cells, resulting in >97% elimination of malignant T cells after 48 hours of coculture (Figure 5D-E) and concomitant expansion of CD7KO CAR T cells (Figure 5E). These results indicate that CD7 CAR can efficiently redirect CD7KO T cells to eradicate primary T-blast cells.
Figure 5.
Antitumor activity of CD7KOCD7 CAR T cells against primary T-ALL blasts. (A) Surface expression of CD7 in peripheral blasts in 4 T-ALL patient samples. (B) Representative dot plots showing production of TNFα and IFNγ by CD7 CAR T cells on coculture with allogeneic primary T-ALL blasts. (C) Mean frequencies of cytokine-positive CD7KO CD7 CAR T cells on coculture with individual T-ALL samples. (D) Peripheral blasts were freshly isolated from a T-ALL patient by leukapheresis, labeled with eFluor670 and cocultured with control or CD7KO CD7 CAR T cells for 48 hours. Dot plots show the frequency of live tumor cells at the end of coculture. (E) Absolute counts of live tumor cells and CD7KO CD7 CAR T cells quantified by flow cytometry at the end of coculture. Data represent 1 to 2 independent with n = 3 donors in each. *P < .05; **P < .01.
On-target off-tumor effects of CD7KO CD7 CAR T cells
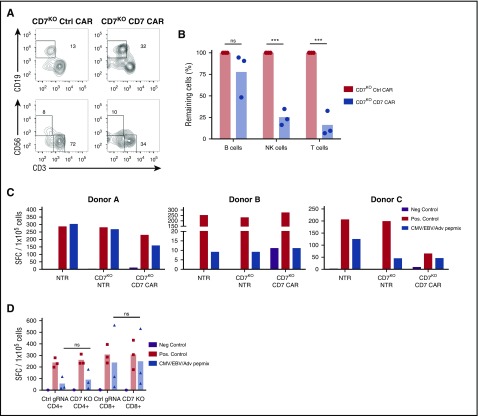
Apart from malignant T cells, CD7 is expressed by the majority of normal T and NK cells, raising the possibility that CD7KO CD7 CAR T cells may produce T- and NK-cell aplasia following infusion. Indeed, we observed a significant reduction in numbers of live T and NK cells on coculture of CD7KO CD7 CAR T cells with autologous PBMC for 24 hours (Figure 6A-B). Depletion of these lymphocytes may result in potentially life-threatening immunodeficiency. However, because lack of CD7 did not affect T-cell effector function (Figure 3), we sought to determine whether CD7KO T cells themselves could protect the host by targeting pathogens through their native receptors. Because the infused product will contain both nontransduced and CD7 CAR-transduced CD7KO T cells, we tested the reactivity of these subsets to viral antigens by incubating cells with a peptide mix derived from CMV, EBV, and adenovirus—the most common causes of viremia in highly immunosuppressed patients—and measured spot-forming units by IFNγ ELISPOT. We found that both nontransduced and CD7 CAR-transduced CD7KO T cells responded to stimulation with viral peptides (Figure 6C), although the response of CD7KO T cells was attenuated in 1 donor. Notably, both CD4+ and CD8+ CD7KO T cells retained their viral reactivity (Figure 6D) suggesting that although CD7KO CD7 CAR T cells can reduce normal T-cell numbers, the infused CAR T-cell product itself retains antiviral activity.
Figure 6.
CD7KOCD7 CAR T cells are cytotoxic against normal T cells and NK cells but can themselves respond to viral antigens. (A) Control or full-length CD7KO CD7 CAR T cells were cocultured with eFluor670-labeled autologous PBMC for 24 hours, and the frequency of T and NK cells was measured by flow cytometry. Dot plots show the frequency of gated CD19+ B cells (top) and CD56+CD3− NK cells and CD3+ T cells (bottom) at the end of coculture. (B) Total numbers of autologous B cells, NK cells, and T cells at the end of coculture with autologous CD7KO CD7 CAR T cells was quantified by flow cytometry using counting beads. Data represent 2 independent experiments with n = 3 donors in each. (C) Nontransduced, CD7KO and CD7KO CD7 CAR T cells were stimulated with pepmixes from cytomegalovirus, Epstein-Barr virus, and adenovirus, and the number of IFNγ+ spot-forming cells was measured by ELISPOT. Individual data from 3 donors are shown as a means of triplicate determinations. (D) CD4+ and CD8+ T cells were MACS-purified and separately assayed for IFNγ production in response to pepmixes. AdV, adenovirus; CMV, cytomegalovirus; EBV, Epstein-Barr virus; Neg., negative; Pos., positive; SFC, spot-forming cells. ***P < .001.
CD7KO CD7 CAR T cells are protective in mouse xenograft models of T-ALL
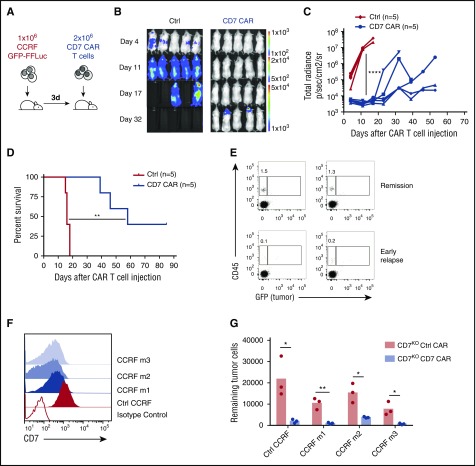
We used a mouse xenograft model of disseminated T-ALL4 by engrafting NOD–severe combined immunodeficiency (SCID) γ chain-deficient (NSG) mice intravenously with CCRF-CEM cells engineered to express GFP-firefly luciferase (Figure 7A). Three days after tumor engraftment, we injected a single dose of CD7KO CD7 CAR T cells or control nontransduced T cells and followed the tumor burden by IVIS imaging. CD7KO CD7 CAR T cells conferred robust protection against leukemia progression (Figure 7B-C) and significantly extended median survival of the mice (16 days in the control group vs 58 days in the CAR T group; P = .0026 by Mantel-Cox log-rank test) (Figure 7D). Similarly, CD7KO CD7 CAR T cells protected mice in a higher tumor burden leukemia model in which CAR T cells were injected 6 days after CCRF administration and systemic engraftment (supplemental Figure 6). In some mice, tumor relapses originated in the periodontal region, a known tumor sanctuary with limited accessibility by human CAR T cells.4,29 Moreover, mice with early tumor relapses had a lower frequency of CD7KO CD7 CAR T cells in peripheral blood at day 32 (Figure 7E), although relapsed tumors retained expression of CD7, albeit displaying moderate downregulation of CD7 surface density (Figure 7F). This downregulation, however, was insufficient to compromise the ability of CD7KO CD7 CAR T cells to recognize and eliminate the relapsed tumor (Figure 7G), indicating that a lack of CAR T-cell persistence rather than antigen escape was the primary culprit for the relapses.
Figure 7.
CD7KOCD7 CAR T cells control the progression of systemic T-ALL in the mouse xenograft model. (A) Schematic outline of the experiment. NSG mice (n = 5 per group) were injected intravenously with 1 × 106 GFP-FFluc CCRF cells followed by a single intravenous injection of 2 × 106 of control or CD7KO CD7 CAR T cells 3 days later. (B) Tumor burden was monitored weekly by measuring luminescence using IVIS imaging. (C) Overall kinetics of systemic tumor progression in mice. Each line denotes an individual animal. (D) Kaplan-Meier survival curve of mice injected with control or CD7KO CD7 CAR T cells. (E) Relative frequency of CAR T cells (hCD45+ GFP−) in peripheral blood of mice in stable remission (top) or during early stages of relapse (bottom) on day 34 after CAR T-cell injection. (F) Surface expression of CD7 on relapsed CCRF tumor cells in peripheral blood of 3 relapsed mice (CCRF m1-m3, blue histograms) in comparison with control in vitro propagated cells (red histogram). Open histogram denotes CD7-negative cell line Raji. Data represent 2 independent experiments. (G) CCRF GFP+ blasts were isolated from spleens of 3 relapsed mice and cocultured with CD7KO control or CD7 CAR T cells from 3 donors for 24 hours at a 1:1 E:T ratio. The numbers of viable tumor cells were counted at the end of coculture by flow cytometry using counting beads. *P < .05; **P < .01, by log-rank Mantel-Cox test; ****P < .0001.
Discussion
Although CD7 is an attractive therapeutic target for T-cell malignancies, effector T cells modified with CD7-specific CARs fail to substantially downregulate CD7 expression, resulting in extensive fratricide and precluding T-cell expansion. Here we show that targeted genomic disruption of the CD7 gene renders CD7 CAR T cells resistant to fratricide, permitting robust expansion without compromising T-cell antigen recognition through their native or chimeric receptors. CD7KO CD7 CAR T cells eliminated malignant T-cell lines and primary tumor cells and protected mice against systemic leukemia progression in a xenograft model. Although CD7 CAR T cells were cytotoxic against normal peripheral T and NK cells, the CD7KO T cells retained their response to virus-derived peptides through their native receptors, which may ameliorate in vivo immunosuppression.
We previously demonstrated that the expression of a CD5-specific CAR on the surface of T cells did not cause complete self-elimination of T cells, because CAR expression coincided with the downregulation of CD5 from the T-cell surface, likely by internalization of CD5 following CD5 CAR engagement.4 In contrast, we found that expression of CD7 CARs promoted only limited downregulation of CD7 expression that was insufficient to mitigate fratricide of CD7 CAR T cells without additional knockout of the CD7 gene. Understanding the mechanisms regulating the differential downregulation of CD5 and CD7 antigens may help extend opportunities for using CAR T cells targeting other T-lineage antigens that internalize upon ligation (eg, CD3).17
The clinical feasibility of using targeted genome editing in T cells prior to their adoptive transfer has been well demonstrated. Genomic disruption of the HIV coreceptor CCR5 in CD4+ T cells using zinc-finger nucleases rendered those cells resistant to infection and enabled CCR5-negative T cells to engraft and persist in HIV-infected patients.30 Ablation of the T-cell receptor (TCR) gene with TALENs in CD19 CAR T cells enabled the use of third-party T cells to successfully induce remission in a patient with B-cell leukemia,31 an encouraging milestone on the way to “universal” off-the-shelf T-cell products with greatly diminished risk of graft-versus-host disease. In the latter study, the CD52 gene was also mutated in the transferred CAR T cells to enable resistance to alemtuzumab. In this study, we used the CRISPR/Cas9 system to ablate CD7 expression, because it allows for fast and efficient disruption of the target gene in T cells.
Although all genome editing tools are associated with potential off-target activity and mutations outside of the target gene, the off-target activity of CRISPR/Cas9 and other editing systems can in part be predicted by using in silico methods and unbiased genome-wide screening.21,22,26 We used these algorithms to select 2 gRNA with minimal predicted binding sites outside the target CD7 gene and assessed off-target activity by unbiased methods of identification of double-strand breaks in vitro26 and verified in the intended target cells. Further in vivo validation can be achieved using deep sequencing to detect emerging oligoclonality or clonal dominance in patients and thereby identifying mutations that lead to a proliferative or survival advantage of T cells. Although these risks may vary according to the type of genome editing tool used,32 the consequences of off-target activity of genome editing tools should resemble the risks of random insertional mutagenesis by gammaretroviral or lentiviral vectors in T cells.33 Despite the widespread use of these viral vectors over 20 years in more than 2000 patients in clinical trials, no evidence of malignant transformation has been observed when mature T cells are the targets for transduction, although exposure of hematopoietic stem cells to gammaretroviral vectors has led to malignant change following expression of some (but not all) transgenes.34 At the moment, the use of CRISPR/Cas9 for a current good manufacturing practice (cGMP)–compliant production of therapeutic T cells is hampered by limited availability of cGMP-grade Cas9 reagents. However, Cas9 can also be delivered as cGMP messenger RNA, which may circumvent the need for a cGMP-grade recombinant protein; the efficiency of this approach for CD7 gene disruption in T cells remains to be tested.
The direct consequences of CD7 knockdown for residual immune function appear modest. Although CD7 is present in most peripheral T and NK cells, evidence that its expression is critical for mature T-cell function is lacking. CD7-knockout mice display unimpaired development and homeostasis of mature T cells and mount normal T cell–dependent immune responses.15,16 CD7 provides costimulation in T cells by activating PI(3)K and PI(4)K pathways12,13,35 on binding its ligand K12/SECTM, which is expressed by myeloid cells and thymic epithelial and stromal cells.36-38 In contrast, binding of galectin-1 by CD7 can trigger apoptosis,39 suggesting that CD7 may regulate T-cell proliferation and survival in a context-dependent manner. In our study, knocking out the CD7 gene in activated T cells did not affect their expansion or CAR-mediated cytotoxicity, suggesting that loss of CD7 is unlikely to impair the antitumor activity of CAR T cells—at least in the short term—and that costimulatory endodomains embedded in the CAR may compensate for the lack of CD7-derived costimulation. Finally, CD7KO T cells were able to respond to viral antigens through their native receptors in vitro, further demonstrating their maintained functionality.
Because CD7 is normally present on mature T and NK cells and their precursors, administration of CD7KO CD7 CAR T cells may cause long-term ablation of these critical lymphocyte populations. This on-target off-tumor effect may have only limited impact on host immunity. First, CD7 is not expressed by all peripheral T cells40,41 and therefore those CD7-negative subsets may be spared, limiting the extent of T-cell aplasia. Second, the infused CD7KO CD7 CAR T cells are polyclonal and contain both nontransduced and CAR-transduced T cells. If these cells replenish the immune system, they may provide protection to viral (and likely other) pathogens via their native TCRs. Indeed, CD7 CARs could be expressed in multivirus-specific CD7KO T cells to further augment protection.42 Furthermore, CD7KO CD7 CAR T cells could be used as a temporary “bridge-to-transplant” therapy, enabling subsequent stem cell transplant that would in turn terminate CAR T-cell activity. Finally, incorporating a clinically validated suicide switch system to eliminate CAR T cells if persistent toxicity occurs may provide an additional exit strategy.43 One or several of these approaches could be used in combination to mitigate the risk of permanent T-cell aplasia.
In summary, this study demonstrates the feasibility and efficacy of CD7KO CD7 CAR T cells for the targeted therapy of T-cell tumors. Because CD7KO T cells retain activity against viral antigens through their native receptors, it should be feasible to use this approach to eradicate CD7+ malignancies while retaining a functional T-cell immune system. Additionally, our genetic editing approach may enable the generation of CAR T cells to be redirected to other T-lineage antigens to broaden the range of targetable tumors.
Acknowledgments
The authors thank Michael Gundry and Lorenzo Brunetti for technical assistance with genome editing, Stephen Gottschalk for providing NALM6 cells, Haruko Tashiro for technical assistance with ELISPOT assays, Harshavardhan Deshmukh for assistance with cell culture work, Carlos Ramos and Joaquim Cabral for support, and Catherine Gillespie for editing the manuscript.
This work was supported by an American Society of Hematology Scholar Award (M.M.) and by the National Institutes of Health, National Cancer Institute (P50 CA126752), as well as by the Cancer Prevention and Research Institute of Texas (RR140081) (G.B.). D.G.-S. acknowledges Fundação para a Ciência e Tecnologia (Portugal) for financial support (SFRH/BD/52479/2014). The authors also appreciate the support of shared resources from the Cancer Center (support grant P30CA125123).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: D.G.-S., S.S., and C.M.L. planned and performed the experiments, analyzed data, and wrote the manuscript; M.S. planned and performed the experiments; D.L.W. contributed to genome editing experiments; C.M.L., T.H.D., and G.B. performed screening and validation of off-target activity of CD7-specific gRNAs; R.H.R. provided primary T-ALL samples; M.K.B. directed the study and cowrote the manuscript; and M.M. directed the study, planned and performed the experiments, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Maksim Mamonkin, Baylor College of Medicine, Texas Children's Hospital, and Houston Methodist Hospital, 1102 Bates St, Suite FC1770, Houston, TX 77030; e-mail: mamonkin@bcm.edu.
References
- 1.Maude SL, Frey N, Shaw PA, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371(16):1507-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brentjens RJ, Davila ML, Riviere I, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med. 2013;5(177):177ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turtle CJ, Hanafi L-A, Berger C, et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J Clin Invest. 2016;126(6):2123-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mamonkin M, Rouce RH, Tashiro H, Brenner MK. A T-cell-directed chimeric antigen receptor for the selective treatment of T-cell malignancies. Blood. 2015;126(8):983-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kernan NA, Knowles RW, Burns MJ, et al. Specific inhibition of in vitro lymphocyte transformation by an anti-pan T cell (gp67) ricin A chain immunotoxin. J Immunol. 1984;133(1):137-146. [PubMed] [Google Scholar]
- 6.Sotillo E, Barrett DM, Black KL, et al. Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov. 2015;5(12):1282-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rapoport AP, Stadtmauer EA, Binder-Scholl GK, et al. NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat Med. 2015;21(8):914-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haynes BF, Eisenbarth GS, Fauci AS. Human lymphocyte antigens: production of a monoclonal antibody that defines functional thymus-derived lymphocyte subsets. Proc Natl Acad Sci USA. 1979;76(11):5829-5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabinowich H, Pricop L, Herberman RB, Whiteside TL. Expression and function of CD7 molecule on human natural killer cells. J Immunol. 1994;152(2):517-526. [PubMed] [Google Scholar]
- 10.Campana D, van Dongen JJ, Mehta A, et al. Stages of T-cell receptor protein expression in T-cell acute lymphoblastic leukemia. Blood. 1991;77(7):1546-1554. [PubMed] [Google Scholar]
- 11.Gorczyca W. Atlas of Differential Diagnosis in Neoplastic Hematopathology. 3rd ed Boca Raton, FL: CRC Press; 2014. [Google Scholar]
- 12.Ward SG, Parry R, LeFeuvre C, Sansom DM, Westwick J, Lazarovits AI. Antibody ligation of CD7 leads to association with phosphoinositide 3-kinase and phosphatidylinositol 3,4,5-trisphosphate formation in T lymphocytes. Eur J Immunol. 1995;25(2):502-507. [DOI] [PubMed] [Google Scholar]
- 13.Chan AS, Mobley JL, Fields GB, Shimizu Y. CD7-mediated regulation of integrin adhesiveness on human T cells involves tyrosine phosphorylation-dependent activation of phosphatidylinositol 3-kinase. J Immunol. 1997;159(2):934-942. [PubMed] [Google Scholar]
- 14.Lyman SD, Escobar S, Rousseau A-M, Armstrong A, Fanslow WC. Identification of CD7 as a cognate of the human K12 (SECTM1) protein. J Biol Chem. 2000;275(5):3431-3437. [DOI] [PubMed] [Google Scholar]
- 15.Bonilla FA, Kokron CM, Swinton P, Geha RS. Targeted gene disruption of murine CD7. Int Immunol. 1997;9(12):1875-1883. [DOI] [PubMed] [Google Scholar]
- 16.Lee DM, Staats HF, Sundy JS, et al. Immunologic characterization of CD7-deficient mice. J Immunol. 1998;160(12):5749-5756. [PubMed] [Google Scholar]
- 17.Preijers FW, Tax WJ, De Witte T, et al. Relationship between internalization and cytotoxicity of ricin A-chain immunotoxins. Br J Haematol. 1988;70(3):289-294. [DOI] [PubMed] [Google Scholar]
- 18.Frankel AE, Laver JH, Willingham MC, Burns LJ, Kersey JH, Vallera DA. Therapy of patients with T-cell lymphomas and leukemias using an anti-CD7 monoclonal antibody-ricin A chain immunotoxin. Leuk Lymphoma. 1997;26(3-4):287-298. [DOI] [PubMed] [Google Scholar]
- 19.Pauza ME, Doumbia SO, Pennell CA. Construction and characterization of human CD7-specific single-chain Fv immunotoxins. J Immunol. 1997;158(7):3259-3269. [PubMed] [Google Scholar]
- 20.Baum W, Steininger H, Bair H-J, et al. Therapy with CD7 monoclonal antibody TH-69 is highly effective for xenografted human T-cell ALL. Br J Haematol. 1996;95(2):327-338. [DOI] [PubMed] [Google Scholar]
- 21.Cradick TJ, Qiu P, Lee CM, Fine EJ, Bao G. COSMID: a Web-based tool for identifying and validating CRISPR/Cas off-target sites. Mol Ther Nucleic Acids. 2014;3(12):e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno-Mateos MA, Vejnar CE, Beaudoin J-D, et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12(10):982-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gundry MC, Brunetti L, Lin A, et al. Highly efficient genome editing of murine and human hematopoietic progenitor cells by CRISPR/Cas9. Cell Reports. 2016;17(5):1453-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee CM, Zhu H, Davis TH, Deshmukh H, Bao G. Design and validation of CRISPR/Cas9 systems for targeted gene modification in induced pluripotent stem cells. Methods Mol Biol. 2017;1498:3-21. [DOI] [PubMed] [Google Scholar]
- 25.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai SQ, Zheng Z, Nguyen NT, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33(2):187-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee CM, Cradick TJ, Bao G. The Neisseria meningitidis CRISPR-Cas9 system enables specific genome editing in mammalian cells. Mol Ther. 2016;24(3):645-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Y, Cradick TJ, Brown MT, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42(11):7473-7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brentjens RJ, Santos E, Nikhamin Y, et al. Genetically targeted T cells eradicate systemic acute lymphoblastic leukemia xenografts. Clin Cancer Res. 2007;13(18 Pt 1):5426-5435. [DOI] [PubMed] [Google Scholar]
- 30.Tebas P, Stein D, Tang WW, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370(10):901-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qasim W, Amrolia PJ, Samarasinghe S, et al. First clinical application of Talen engineered universal CAR19 T cells in B-ALL. Blood. 2015;126(23):2046. [Google Scholar]
- 32.Cox DBT, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21(2):121-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Recchia A, Bonini C, Magnani Z, et al. Retroviral vector integration deregulates gene expression but has no consequence on the biology and function of transplanted T cells. Proc Natl Acad Sci USA. 2006;103(5):1457-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348(3):255-256. [DOI] [PubMed] [Google Scholar]
- 35.Lee DM, Patel DD, Pendergast AM, Haynes BF. Functional association of CD7 with phosphatidylinositol 3-kinase: interaction via a YEDM motif. Int Immunol. 1996;8(8):1195-1203. [DOI] [PubMed] [Google Scholar]
- 36.Lam GK, Liao H-X, Xue Y, et al. Expression of the CD7 ligand K-12 in human thymic epithelial cells: regulation by IFN-gamma. J Clin Immunol. 2005;25(1):41-49. [DOI] [PubMed] [Google Scholar]
- 37.Slentz-Kesler KA, Hale LP, Kaufman RE. Identification and characterization of K12 (SECTM1), a novel human gene that encodes a Golgi-associated protein with transmembrane and secreted isoforms. Genomics. 1998;47(3):327-340. [DOI] [PubMed] [Google Scholar]
- 38.Wang T, Huang C, Lopez-Coral A, et al. K12/SECTM1, an interferon-γ regulated molecule, synergizes with CD28 to costimulate human T cell proliferation. J Leukoc Biol. 2012;91(3):449-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pace KE, Hahn HP, Pang M, Nguyen JT, Baum LG. CD7 delivers a pro-apoptotic signal during galectin-1-induced T cell death. J Immunol. 2000;165(5):2331-2334. [DOI] [PubMed] [Google Scholar]
- 40.Reinhold U, Abken H, Kukel S, et al. CD7- T cells represent a subset of normal human blood lymphocytes. J Immunol. 1993;150(5):2081-2089. [PubMed] [Google Scholar]
- 41.Reinhold U, Liu L, Sesterhenn J, Abken H. CD7-negative T cells represent a separate differentiation pathway in a subset of post-thymic helper T cells. Immunology. 1996;89(3):391-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papadopoulou A, Gerdemann U, Katari UL, et al. Activity of broad-spectrum T cells as treatment for AdV, EBV, CMV, BKV, and HHV6 infections after HSCT. Sci Transl Med. 2014;6(242):242ra83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Stasi A, Tey S-K, Dotti G, et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N Engl J Med. 2011;365(18):1673-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]