Abstract
Background:
Mitochondria are considered relevant players in many tumour entities and first data indicate beneficial effects of mitochondria-targeted antioxidants in both cancer prevention and anticancer therapies. To further dissect the potential roles of mitochondria in NSCLC we comprehensively analysed somatic mitochondrial mutations, determined the spatial distribution of mitochondrial DNA within complete tumour sections and investigated the mitochondrial load in a large-scale approach.
Methods:
Whole mitochondrial genome sequencing of 26 matched tumour and non-neoplastic tissue samples extended by reviewing published data of 326 cases. Systematical stepwise real-time PCR quantification of mitochondrial DNA covering 16 whole surgical tumour sections. Immunohistochemical determination of the mitochondrial load in 171 adenocarcinoma and 145 squamous cell carcinoma.
Results:
Our results demonstrate very low recurrences (max. 1.7%) and a broad distribution of 456 different somatic mitochondrial mutations. Large inter- and intra-tumour heterogeneity were seen for mitochondrial DNA copy numbers in conjunction with a correlation to the predominant histological growth pattern. Furthermore, tumour cells had significantly higher mitochondrial level compared to adjacent stroma, whereas differences between tumour entities were negligible.
Conclusions:
Non-evident somatic mitochondrial mutations and highly varying mitochondrial DNA level delineate challenges for the approach of mitochondria-targeted anticancer therapies in NSCLC.
Keywords: somatic mtDNA mutations, mtDNA copy number variations, spatial distribution, heterogeneity, NSCLC
Lung cancer is the leading cause of cancer-related mortality. Although the patients’ prognosis is still limited, the successful translation of targeted therapies based on tissue-derived biomarkers into the clinical setting, for example EGFR mutations or ALK and ROS1 translocations, has paved the way for future strategies to significantly improve the patients’ outcome. However, it became evident that tumours develop resistance mechanisms against targeted therapies, usually based on selection of resistant clones (Warth et al, 2016). Therefore, the better understanding of the complex interactions and mechanisms that take place during tumorigenesis and tumour progression seems to be of utmost importance for further therapeutical improvements.
Mitochondria are indispensable for respiratory energy metabolism, apoptosis regulation, cellular senescence, immune response, and cell signalling. The role of mitochondria in cancer in general and specifically the impact of somatic mitochondrial DNA (mtDNA) alterations as well as of quantitative mitochondrial changes on cancer progression are poorly understood. However, there is growing evidence that mitochondria are relevant players in this context and might also serve targets for anticancer therapies. In solid tumours, such as breast cancer, a high mitochondrial mass was identified in a sub-population of chemo-resistant stem-like cancer cells (Farnie et al, 2015); however, low mtDNA content in general was reported to be associated with a poor outcome (Weerts et al, 2016). Furthermore, mitochondrial reprogramming was shown to regulate tumour progression (Kannan et al, 2016). In contrast to normal mitochondria, mitochondria in cancer cells are characterised by reactive oxygen species (ROS) overproduction, which promotes cancer development, recurrence, and therapy resistance by inducing genomic instability, and modifying gene expression and signalling pathways. Mitochondrial genetic changes are therefore considered as a molecular initiator of mutations in various cancers (Reznik et al, 2016; Yan et al, 2016; Yang et al, 2016).
To shed more light on the potential roles of mitochondria in non-small cell lung cancer (NSCLC), we performed whole mtDNA sequencing of tumour and corresponding non-neoplastic tissue samples to systematically assess somatic mitochondrial mutations, we spatially quantified the amount of mtDNA across whole surgical specimen sections, and determined the mitochondrial load on protein level in a large-scale approach discriminating between tumour cells and tumour stroma.
Materials and methods
Samples
All analysed tumours were resected at the Thoraxklinik at Heidelberg University and were diagnosed in the Institute of Pathology at Heidelberg University, according to the criteria of the 2015 WHO Classification for lung cancer (Travis et al, 2015).
Formalin fixed and paraffin embedded (FFPE) NSCLC specimens were handled with the support of the tissue bank of the National Center for Tumor Diseases (NCT; project: # 1746, # 2015). Tissues were used in accordance with the ethical regulations of the NCT tissue bank established by the local ethics committee.
For the analysis of somatic mtDNA mutations FFPE tissues from 26 pulmonary adenocarcinomas (ADC) and corresponding non-neoplastic lung tissues from the same patient, histologically confirmed to be tumour free, were tested (Supplementary 1).
For the determination of the spatial distribution of mitochondria, central tumour sections in largest tumour diameter of 16 ADC were analysed. Following formalin fixation, the tumour sections were systematically cut into segments according to a 5 × 5 mm grid and labelled with ink marks to enable the reconstruction of the full section (Supplementary 2). Each segment was embedded separately in paraffin and the respective predominant histological growth pattern was determined by an experienced pathologist (A.W.) on consecutive H&E-stained sections (see Supplementary 1 for clinicopathological data).
For the quantification of mitochondrial protein levels eight TMA slides representing FFPE material (two punch samples for each tumour) of 171 ADC and 145 squamous cell carcinomas (SQCC) were immunohistochemically analysed. Construction of the TMA and comprehensive clinicopathological assessment of the cases was described previously in detail (Warth et al, 2012).
DNA extraction and quantification
Genomic and mitochondrial DNA were extracted together from unstained 10 μm tissue slides. Following deparaffinisation all samples were digested with proteinase K overnight and DNA was extracted automatically using a Maxwell 16 Research system and the Maxwell 16 FFPE Tissue LEV DNA Purification Kit (both Promega, Madison, WI, USA), in accordance to the manufacturer’s instructions. Total DNA concentrations were determined using the Qubit HS DNA assay (Thermo Fisher Scientific, Waltham, MA, USA), whereas genomic DNA yields were assessed using the RNaseP Detection system on a StepOne qPCR system (both Thermo Fisher Scientific), according to the manufacturer’s protocols, respectively.
Library preparation and massive parallel sequencing
The complete mitochondrial genome was sequenced on an Ion Torrent PGM System applying the Precision ID mtDNA Whole Genome Panel (Thermo Fisher Scientific) according to the manufacturer’s protocol. For each sample two amplicon libraries were prepared using two different primer pools containing 81 primer pairs each and the Ion AmpliSeq Library Kit 2.0 (Thermo Fisher Scientific) with 0.5 ng DNA input. The thermal cycler (Biometra, Göttingen, Germany) conditions were as follows: 99 °C for 2 min (initial denaturation), 18 cycles of 99 °C for 15 s and 60 °C for 4 min. Subsequently, both libraries of one sample were processed separately. PCR products were partly digested using FuPa reagent (Thermo Fisher Scientific) and ligated to barcoded sequencing adapters (Ion Xpress Barcode Adapters, Thermo Fisher Scientific). In the final preparation step the libraries were purified with Agencourt AMPure XP magnetic beads (Beckman Coulter, Krefeld, Germany) and quantified using the Ion Library Quantitation kit on a StepOnePlus qPCR system (both Thermo Fisher Scientific).
All libraries were adjusted to a concentration of 100 pM and the two libraries of each sample were combined. Up to 25 samples were pooled for library amplification on Ion Spheres using the Ion OneTouch 2 instrument with the 200 bp chemistry. Ion Sphere quality control measurement for unenriched libraries was performed on a QuBit 2.0 instrument, before the libraries were enriched using the Ion OneTouch ES system. Finally, the barcoded libraries were processed using the Ion Torrent 200 bp sequencing v2 chemistry and loaded onto a 318v2 chip.
NGS data analysis
To generate sequences from the raw data and align them to the revised Cambridge reference sequence (rCRS; NC_012920) the Torrent Suite software (version 4.2, Thermo Fisher Scientific) was used. Variations to the reference sequence were detected by the variant caller plug-in package and confirmed in the IGV browser. Cutoffs for mutations were an allele frequency greater than 3% and a minimum coverage of 100 reads. In accordance to the guidelines of the mitochondrial DNA population database of the European DNA Profiling Group of the International Society for Forensic Genetics (EMPOP) length variants around positions 309, 445, 463, 573, 960, 5899, 8276, 8285 and 16 193 involving C- or T-stretches were ignored. Somatic mutations were identified by comparing the semiconductor sequencing results of tumour samples and corresponding non-neoplastic lung tissue samples. Variations present in the tumour and the non-neoplastic tissue were referred to as polymorphism and variations that were solely detected in the tumour were designated as somatic mutations.
Determination of mtDNA copy number variations
Copy numbers of mtDNA were determined using an adapted real-time PCR assay for the absolute quantification of mtDNA (Kavlick et al, 2011) on a StepOnePlus qPCR system (Thermo Fisher Scientific). This assay utilises a synthetic double stranded oligonucleotide for standard curve generation. A 105 bp long region of the mitochondrial genome corresponding to positions 13 288–13 392 of the rCRS is amplified and detected for quantification. In order to take varying cell numbers of the analysed samples into account, we normalised the mtDNA copy numbers (mtDNA-CN) to the total DNA content of a sample (see above for DNA quantification).
Immunohistochemical quantification of mitochondria
For the quantification of mitochondria on protein level a Mitochondria Ab-2, mouse monoclonal antibody, clone MTC02, ready-to-use solution (Thermo Fisher Scientific, Dreieich, Germany) was applied according to the manufacturer’s instructions using an autostainer (BenchMark ULTRA, Ventana Medical Systems Inc, Tucson, AZ, USA). The antibody recognises a 60 kDa non-glycosylated protein component localised in human mitochondria. The stainings were evaluated digitally on the HALO platform (v.2.0.1038, Indica Labs, Corrales, NM, USA) applying an algorithm (Supplementary 3) to count cells subdivided by three intensities (weak, moderate, strong). H-scores were calculated according to the percentage of cells stained at a respective intensity (1 × weak, 2 × moderate, 3 × strong), yielding scores from 0 to 300. Tumour and stroma cells were differentiated using the HALO program together with a specific classifier that was set-up and verified by a pathologist (H.A.).
Statistical analysis and plot generation
Shapiro–Wilk test (normality test) and Mann–Whitney U test (nonparametric hypothesis test) were carried out using R (v.3.3.0) (R Core Team, 2016) and the ‘coin’ (v1.1–2) package. Plots were generated either using R (v.3.3.0) in combination with the ‘ggplot2’ (v2.1.0) and the ‘GGally’ (v.1.2.0) packages or by using Microsoft Excel 2013 (Microsoft, Redmond, WA, USA) together with the ‘Daniel’s XL Toolbox NG’ (7.1.4) add-in (https://www.xltoolbox.net/).
Literature research
Since only few data were available on somatic mtDNA mutations in lung cancer, we screened PubMed (http://www.ncbi.nlm.nih.gov/pubmed) for published studies on lung cancer with whole mtDNA mutation analysis to complement our own data. Studies covering only parts of the mitochondrial genome, for example, the D-loop, were not considered. Finally we included nine publications (Fliss et al, 2000; Liu et al, 2004; Jakupciak et al, 2005; Jin et al, 2007; Jakupciak et al, 2008; Dasgupta et al, 2009, 2012; Ju et al, 2014; Stewart et al, 2015).
Results
Somatic mutations of the mitochondrial genome
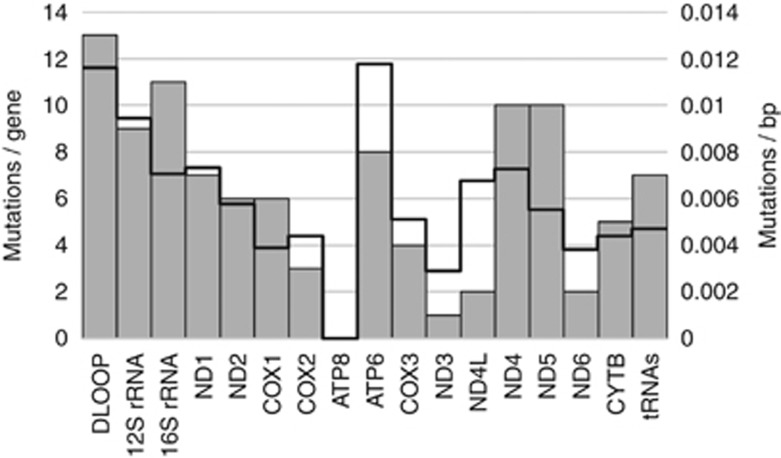
In the examined sample set 24 out of 26 ADC (92%) showed somatic mtDNA mutations with a maximum of nine alterations per sample (Supplementary 4). We identified a total number of 105 mutations, from which only four occurred in more than one sample (n): A8649C (3), G16390A (3), G2701A (2), G10522A (2). The distribution of the mutations spanned widely over the complete mitochondrial genome, with 64, 27 and 14 mutations located in protein coding, RNA coding, and non-coding regions, respectively (Figure 1). The majority (90.6% 58 of 64) of mutations in the protein coding genes caused non-synonymous amino acid changes. Within functional regions, accumulations with more than eight mutations were found in the D-Loop, 12S rRNA, 16S rRNA, ND4, and ND5. The ATP8 region was the only section where no somatic mutation was found. Taking the different region lengths into account the ATP6 and D-Loop showed the highest mutation to base pair ratio, followed by 12S rRNA (Figure 1: bold line).
Figure 1.
Number of somatic mutations in functional regions of the mitochondrial genome found in 26 pulmonary ADCs. Grey bars: absolute numbers. Bold line: mutations per base pair. For absolute numbers see Supplementary 4.
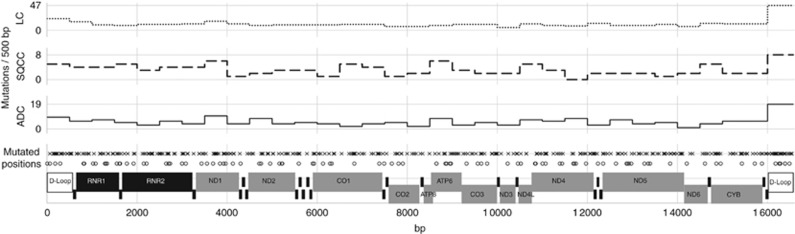
In order to provide a comprehensive overview of mtDNA mutations in NSCLC we reviewed the published literature on somatic mutations in the complete mitochondrial genome. Assessment including our own data resulted in the analysis of 352 lung tumours (Figure 2). In combination with 393 reported somatic mutations (LC subgroup) a total of 498 mutations were identified, whereas 90% of the mutations were non-recurring. Eight of the already reported mutations were also found in our sample set (G1552A, T4752C, T5215C, G5591A, G10522A, T11072C, A16284G and G16390A). With a frequency of 1.7% (6/352) G16390A was the most abundant mutation of the comprehensive cohort. Most of the tumours in the literature were only designated as lung cancer, but 59 were specifically subclassified as ADC and 59 as SQCC. Combined with our own data we found 183 somatic mtDNA mutations in 85 ADCs (Ø 2.15) and 78 in SQCCs (Ø 1.76). The following mutations were found in both groups: C3450T, T4752C, G10522A, A10946+C, G1552A, A16284G, C16290T, G16390A. Determining the numbers of mutations per 500 bp of the mtDNA revealed that most mutations could be found between positions 16 000 and 16 569 in all subgroups (Figure 2, upper graphs).
Figure 2.
Compilation of somatic mutations of the complete mitochondrial genome. Bottom line: functional regions; grey protein coding; black RNA coding (tRNA names are not shown). Mutated positions: o – detected in the present study. x – reported in selected literature. The upper three graphs display the number of mutations per 500 bp section of the mitochondrial genome subdivided by tumour type: ADC (adenocarcinoma, full line; combination of our data and published data); SQCC (squamous cell carcinoma, dashed line; published data); LC (all lung cancer samples of the selective literature).
Spatial distribution of mtDNA copy numbers in ADC
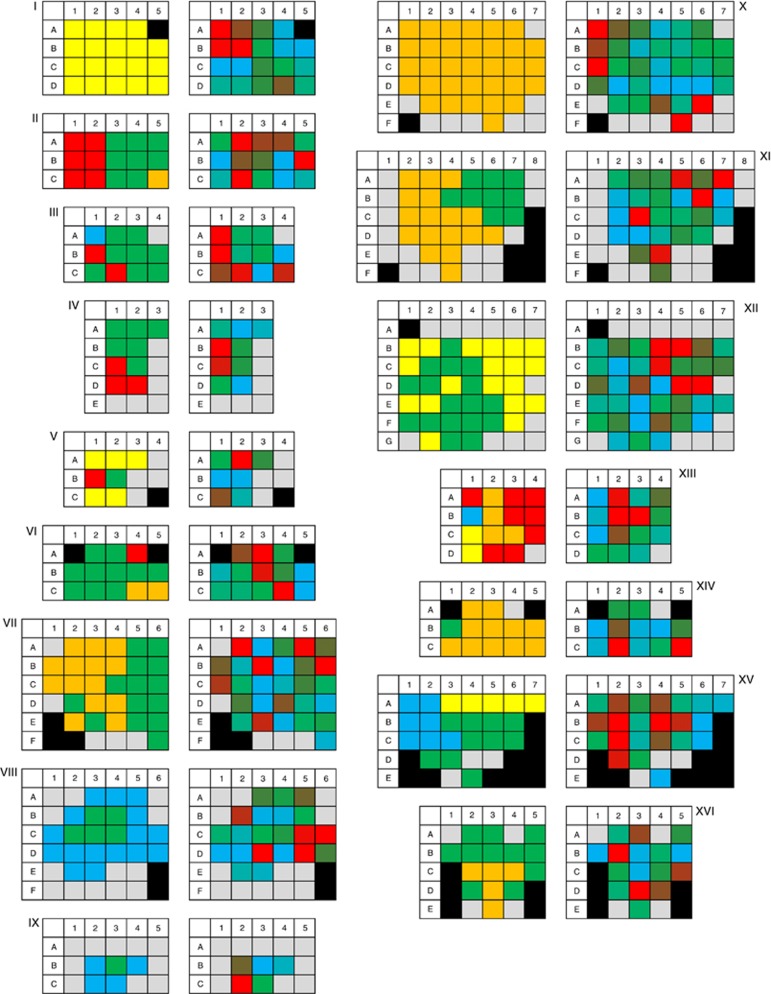
In order to analyse the spatial distribution of mtDNA copy numbers (mtDNA-CN) in ADCs, we determined mtDNA-CN stepwise (5 × 5 mm segments) in whole central sections of 16 ADCs and normalised it to the respective total DNA content. On average, sections were cut into 23 segments (minimum: 11; maximum: 41 segments). The image Supplementary 5 and the table Supplementary 6 provide an overview about the median, interquartile range, and the minimum and maximum values for each tumour. While most of the tumours showed median mtDNA copies per ng total DNA in a range of 60 000 to 126 000, three tumours (IV, II, III) had median values below 30 000 and three (XII, XI, I) over 200 000 mtDNA, respectively (P<0.01; compared to the overall average).
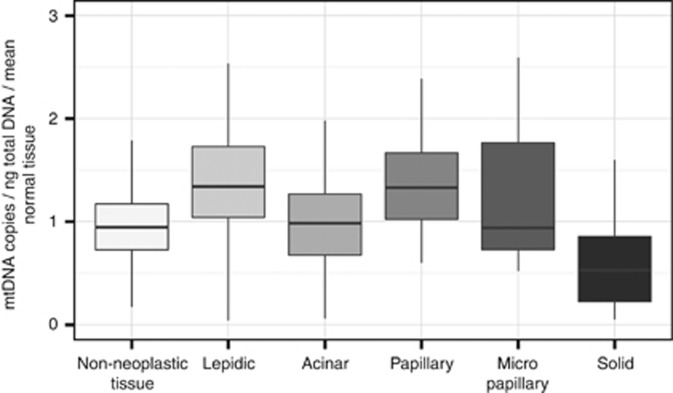
The analysis of all segments of each tumour revealed a heterogeneous distribution of mtDNA in all analysed specimens (Figure 3 and Supplementary 6); both in tumours with single and with multiple histological growth patterns. In order to investigate a potential correlation of mtDNA-CN to the predominant morphological growth patterns we normalised the mtDNA-CN values of the tumour segments to respective non-neoplastic tissue so that inter-patient differences were minimised (Figure 4). Owing to the availability of tumour-free segments this was possible for 10 (IV, V, VII–XII, XV, XVI) specimens (overall 283 single samples). On average tumour segments with a lepidic or a papillary predominant growth pattern had a significantly higher (P<0.01) mtDNA load compared to non-neoplastic tissue. In contrast, solid predominant segments showed significantly lower values (P<0.01). Samples with an acinar predominant growth pattern did not differ significantly from non-neoplastic tissue but from lepidic, papillary and solid segments (P<0.01). For micropapillary predominant segments we could not find significant differences to any other growth pattern or non-neoplastic tissue.
Figure 3.
Segmented analysis of a central tumour section of 16 adenocarcinoma (I–XVI). The left part of each diagram displays the predominant histological growth pattern in each tumour segment: blue – lepidic; green – acinar; yellow – papillary; orange – solid; red – micropapillary; grey – non-neoplastic tissue and black – no segment. The right side of the respective diagram represents the mtDNA copy numbers per nanogram total DNA in each tumour segment in gradual colour scales from blue (< 10% quantile) to green (50% quantile) to red (> 90% quantile); grey – non-neoplastic tissue and black – no segment. For absolute numbers see Supplementary 6.
Figure 4.
Correlation of mtDNA copy numbers to the predominant histological growth pattern in segmented central sections of 10 ADC (Figure 3: IV, V, VII–XII, XV, XVI). Boxplots showing the median, interquartile range, minimum and maximum mtDNA copy number per ng total DNA normalised to non-neoplastic tissue.
Quantification of mitochondria in ADC and SQCC
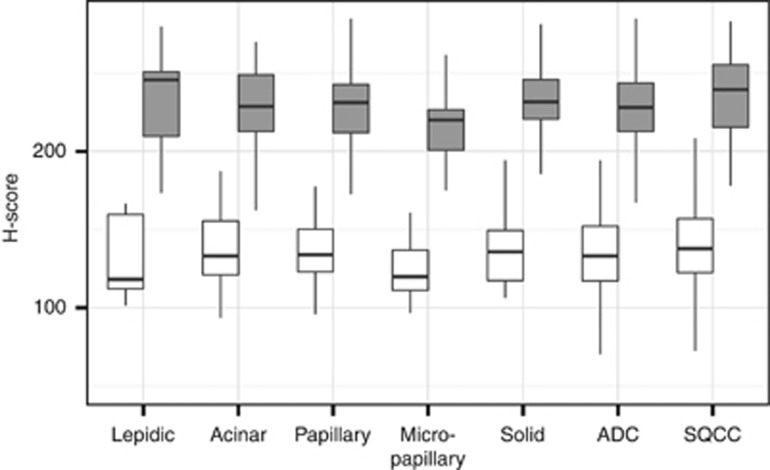
In a large-scale IHC approach, we determined the mitochondrial load in 171 ADC and 145 SQCC specimens (Ø 11,600 cells per tumour), discriminating between tumour and stroma cells (Figure 5 and Supplementary 7). In both entities the average H-scores of the tumour cells were significantly higher (P<0.01) than in stromal cells, which had comparable expression levels to non-neoplastic tissue. Comparing ADC with SQCC we found statistically significant (P<0.01) higher H-scores in SQCC tumour cells; however, the absolute difference of the H-scores was only minor (11.5). No significant differences were seen between stromal cells of both entities. A subdivision of the ADC group according to the predominant histological pattern did not reveal significant differences except for lower levels in the tumour cells of micropapillary predominant samples compared to acinar (P<0.05) or solid (P<0.01) predominant samples, respectively.
Figure 5.
Immunohistochemical analysis of the mitochondrial protein load in 171 adenocarcinoma (ADC) and 145 squamous cell carcinoma (SQCC) punch samples (white: tumour stroma, grey: tumour). ADC samples were discriminated according to the predominant growth pattern as follows: 7 lepidic, 62 acinar, 31 papillary, 55 solid and 16 micropapillary. For absolute numbers see Supplementary 7.
Discussion
Owing to their crucial role in the cell energy metabolism, the involvement in cellular signalling, survival pathways, aging, and apoptosis (Elmore, 2007), mitochondrial damage has been linked to various diseases including cancer. Over the last decades multiple studies have identified and investigated a wide variety of mitochondrial changes across human cancers such as mtDNA mutations (Chatterjee et al, 2006; Larman et al, 2012; van Gisbergen et al, 2015), mtDNA content (Lee et al, 2005; Dai et al, 2013; Reznik et al, 2016), mitochondrial protein expression and activity (Roberts and Thomas, 2013; van Gisbergen et al, 2015), but a detailed picture for lung cancer is missing so far. Somatic variations of the mitochondrial genome are commonly seen in tumours but it is unclear if they have an impact on tumorigenesis, similar to driver gene mutations. Our comprehensive analysis of mtDNA mutations in 352 NSCLCs did not reveal evident lung cancer-specific somatic mutations (Figure 2). The detected mutations were rather randomly distributed across the whole mitochondrial genome with very low occurrences. The most frequent mutation of the combined sample set was a G to A substitution at position 16 390 (1.7%), a known polymorphism and one of the defining mutations of the subequatorial African haplogroup L2 (6% gene bank frequency in MITOMAP). Despite a correlation with alterations in the gut microbiome (Ma et al, 2014) and as a risk factor for primary open-angle glaucoma (Collins et al, 2016) no association of this mutation to malignancy is described. Mutations at positions 3450, 4901, 8701, 10 398 have been postulated as potentially linked to lung cancer (Jakupciak et al, 2008; Choi et al, 2011); however, mutations at these positions were found only once or not at all in the comprehensive sample set, thus challenging their role in lung cancer carcinogenesis. Considering the numbers of somatic mutations per 500 bp, the most frequently mutated region in all analysed subgroups were identified between positions 16 000–16 569. This section contains the so-called hypervariable region I, probably the most polymorphic site of the complete mitochondrial genome (Meyer et al, 1999; Stoneking, 2000). Other putative hot spot regions were not evident. Therefore, our findings strengthen the results of Ju et al (2014), who compared somatic mtDNA alterations in 1675 tumours across 31 tumour types and found no evidence that the detected mitochondrial DNA mutations are significantly associated with the development or spread of cancer. By estimating dN/dS ratios (ratio of the number of nonsynonymous substitutions per non-synonymous site to the number of synonymous substitutions per synonymous site) for missense mutations they demonstrated that the frequency of the accumulation of somatic mtDNA mutations is very similar to what would be expected under neutrality. Therefore they conclude that most somatic mitochondrial mutations are passenger mutations without a tumour driving force. Our result does not challenge this consideration; however, it cannot be ultimately excluded that rare mutations might be associated with a malignant potential.
On this behalf, the question whether somatic mtDNA mutations that support tumour progression are indeed less likely to occur or if they are just not maintained in cancer cells arises. Respective issues were brought up by several groups (Alexeyev et al, 2013; Itsara et al, 2014; Ju et al, 2014; Stewart et al, 2015) and could be observed in our study as well. The majority of detected somatic mtDNA mutations seems not to be caused by ROS but from errors during mtDNA replication. Characteristic mutation types of oxidative damage, like G:C to T:A transversions (Cheng et al, 1992), were only seen in small numbers, while the great majority of mutations (86%, Supplementary 8) were G:C to A:T or T:A to C:G transitions. A possible explanation for this is described by Alexeyev et al (2013) and Shokolenko et al (2009), suggesting that oxidative stress may result in mutagenic mtDNA damage, but induces strand breaks with a much higher frequency. Those lesions could block transcription and replication of mtDNA and cause the degradation of effected mtDNA molecules, indicating a strong link between oxidative stress and mtDNA.
Following these considerations we focused our further investigations on alterations of mtDNA levels in ADC. While a reduced mtDNA content could be a consequence of mutagenic damage, an increase in the mtDNA content could result from a compensation for the increased energy need and/or a decreased capacity of oxidative phosphorylation in cancer cells (Lee et al, 2005). In other cancer types the mtDNA content has been postulated as potentially prognostic relevant as it correlates with the tumour progression, for example, in breast cancer (Weerts et al, 2016). However, the reports for lung cancer are quite contradictive, both increased as well as decreased mtDNA-CN levels have been reported (Lee et al, 2005; Dai et al, 2013; Lee et al, 2015; Reznik et al, 2016). Our systematic analysis of the mtDNA content in segmented central tumour sections could provide an explanation for these conflicting reports, as it revealed a great amount of inter- and intra-tumour heterogeneity (Figure 3 and Supplementary 8), pointing towards pitfalls of single region-based analysis. A comprehensive examination of the morphological characteristics of all segments revealed a correlation with the predominant histological pattern (Figure 4). The histological subtyping of ADC has a high prognostic (Warth et al, 2012) and predictive relevance (Tsao et al, 2015), with a gradual downgrading from lepidic (good prognosis) over acinar and papillary (moderate) to solid and micropapillary (poor). Thus, the histological patterns can be related to different stages of tumour progression (Weichert and Warth, 2014). Taking this into consideration, the observed correlation between histological growth patterns and mtDNA levels might be a stepwise reduction of mtDNA-CN during ADC evolution. Consenting results have been reported by Amuthan et al. for human lung carcinoma A549 cells, where mitochondrial stress induced by decreased mtDNA-CN led to phenotypic changes, tumour progression, and invasion (Amuthan et al, 2002).
The immunohistochemical quantification of mitochondria revealed a significant but fairly minor difference between ADC and SQCC tumour cells (Figure 5). The differences between tumour cells (increase) and associated stroma cells were significantly larger in all analysed samples, indicating an increased dependency on mitochondria of the tumour cells. In a comparable context for breast cancer cells, Martinez-Outschoorn et al (2011) postulated a potential metabolic coupling between oxidative active cancer cells and glycolytic stromal cells, which stands in contrast to the ‘Warburg effect’ (Warburg, 1956) and was therefore called ‘reversed Warburg effect’.
A limiting factor in our IHC approach exists in the applied antibody, although it has been used as a mitochondrial marker in over 30 publications (see manufacture’s site for references). Besides being a 60 kDa non-glycosylated protein component localised in human mitochondria, the recognised antigen has not been identified. It is also unknown if the antigen is nuclear or mtDNA encoded, in which the latter would be especially suitable to analyse correlations with mtDNA levels. Therefore, advanced studies using antibodies specific for mtDNA and nuclear encoded mitochondrial proteins, but also for mitochondrial metabolic markers (for example, oxidative activity) are needed to further investigate the background and the consequences of our data.
In conclusion, we systematically analysed the potential role of mitochondrial mutations and frequency in lung cancer. We demonstrate that somatic mtDNA mutations found in NSCLC are rather passenger mutations, thus challenging their suitability as relevant targets for anticancer therapies in NSCLC. Furthermore, we demonstrate that the spatial distribution of mtDNA levels in ADC has a high inter- and intra-tumour heterogeneity and provide first evidence for a depletion during ADC progression.
Acknowledgments
AW was supported by an excellence grant from the Else Kröner-Fresenius Foundation. We thank Gloria Laukemper, Veronika Geißler and Terence Osere for excellent technical assistance, and the Tissue Bank of the National Centre for Tumor Diseases, Heidelberg.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Alexeyev M, Shokolenko I, Wilson G, LeDoux S (2013) The maintenance of mitochondrial DNA integrity – critical analysis and update. Cold Spring Harb Perspect Biol 5(5): a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amuthan G, Biswas G, Ananadatheerthavarada HK, Vijayasarathy C, Shephard HM, Avadhani NG (2002) Mitochondrial stress-induced calcium signaling, phenotypic changes and invasive behavior in human lung carcinoma A549 cells. Oncogene 21(51): 7839–7849. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Mambo E, Sidransky D (2006) Mitochondrial DNA mutations in human cancer. Oncogene 25(34): 4663–4674. [DOI] [PubMed] [Google Scholar]
- Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA (1992) 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G——T and A——C substitutions. J Biol Chem 267(1): 166–172. [PubMed] [Google Scholar]
- Choi SJ, Kim SH, Kang HY, Lee J, Bhak JH, Sohn I, Jung SH, Choi YS, Kim HK, Han J, Huh N, Lee G, Kim BC, Kim J (2011) Mutational hotspots in the mitochondrial genome of lung cancer. Biochem Biophys Res Commun 407(1): 23–27. [DOI] [PubMed] [Google Scholar]
- Collins DW, Gudiseva HV, Trachtman B, Bowman AS, Sagaser A, Sankar P, Miller-Ellis E, Lehman A, Addis V, O'Brien JM (2016) Association of primary open-angle glaucoma with mitochondrial variants and haplogroups common in African Americans. Mol Vis 22: 454–471. [PMC free article] [PubMed] [Google Scholar]
- Dai JG, Zhang ZY, Liu QX, Min JX (2013) Mitochondrial genome microsatellite instability and copy number alteration in lung carcinomas. Asian Pac J Cancer Prev 14(4): 2393–2399. [DOI] [PubMed] [Google Scholar]
- Dasgupta S, Soudry E, Mukhopadhyay N, Shao C, Yee J, Lam S, Lam W, Zhang W, Gazdar AF, Fisher PB, Sidransky D (2012) Mitochondrial DNA mutations in respiratory complex-I in never-smoker lung cancer patients contribute to lung cancer progression and associated with EGFR gene mutation. J Cell Physiol 227(6): 2451–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Yung RC, Westra WH, Rini DA, Brandes J, Sidransky D (2009) Following mitochondrial footprints through a long mucosal path to lung cancer. PloS One 4(8): e6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore S (2007) Apoptosis: a review of programmed cell death. Toxicol Pathol 35(4): 495–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnie G, Sotgia F, Lisanti MP (2015) High mitochondrial mass identifies a sub-population of stem-like cancer cells that are chemo-resistant. Oncotarget 6(31): 30472–30486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, Jen J, Sidransky D (2000) Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science 287(5460): 2017–2019. [DOI] [PubMed] [Google Scholar]
- Itsara LS, Kennedy SR, Fox EJ, Yu S, Hewitt JJ, Sanchez-Contreras M, Cardozo-Pelaez F, Pallanck LJ (2014) Oxidative stress is not a major contributor to somatic mitochondrial DNA mutations. PLoS Genet 10(2): e1003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakupciak JP, Maragh S, Markowitz ME, Greenberg AK, Hoque MO, Maitra A, Barker PE, Wagner PD, Rom WN, Srivastava S, Sidransky D, O'Connell CD (2008) Performance of mitochondrial DNA mutations detecting early stage cancer. BMC Cancer 8: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakupciak JP, Wang W, Markowitz ME, Ally D, Coble M, Srivastava S, Maitra A, Barker PE, Sidransky D, O'Connell CD (2005) Mitochondrial DNA as a cancer biomarker. J Mol Diagn 7(2): 258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Zhang J, Gao Y, Ding K, Wang N, Zhou D, Jen J, Cheng S (2007) Relationship between mitochondrial DNA mutations and clinical characteristics in human lung cancer. Mitochondrion 7(5): 347–353. [DOI] [PubMed] [Google Scholar]
- Ju YS, Alexandrov LB, Gerstung M, Martincorena I, Nik-Zainal S, Ramakrishna M, Davies HR, Papaemmanuil E, Gundem G, Shlien A, Bolli N, Behjati S, Tarpey PS, Nangalia J, Massie CE, Butler AP, Teague JW, Vassiliou GS, Green AR, Du MQ, Unnikrishnan A, Pimanda JE, Teh BT, Munshi N, Greaves M, Vyas P, El-Naggar AK, Santarius T, Collins VP, Grundy R, Taylor JA, Hayes DN, Malkin D Group IBC Group ICMD Group IPC Foster CS, Warren AY, Whitaker HC, Brewer D, Eeles R, Cooper C, Neal D, Visakorpi T, Isaacs WB, Bova GS, Flanagan AM, Futreal PA, Lynch AG, Chinnery PF, McDermott U, Stratton MR, Campbell PJ (2014) Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. eLife 3: eLife.02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan A, Wells RB, Sivakumar S, Komatsu S, Singh KP, Samten B, Philley JV, Sauter ER, Ikebe M, Idell S, Gupta S, Dasgupta S (2016) Mitochondrial reprogramming regulates breast cancer progression. Clin Cancer Res 22(13): 3348–3360. [DOI] [PubMed] [Google Scholar]
- Kavlick MF, Lawrence HS, Merritt RT, Fisher C, Isenberg A, Robertson JM, Budowle B (2011) Quantification of human mitochondrial DNA using synthesized DNA standards*. J Forensic Sci 56(6): 1457–1463. [DOI] [PubMed] [Google Scholar]
- Larman TC, DePalma SR, Hadjipanayis AG, Cancer Genome Atlas Research Network, Protopopov A, Zhang J, Gabriel SB, Chin L, Seidman CE, Kucherlapati R, Seidman JG (2012) Spectrum of somatic mitochondrial mutations in five cancers. Proc Natl Acad Sci USA 109(35): 14087–14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Lee JH, Kim DK, Keum DY (2015) Nuclear and mitochondrial DNAs microsatellite instability and mitochondrial DNA copy number in adenocarcinoma and squamous cell carcinoma of lung: a pilot study. APMIS 123(12): 1048–1054. [DOI] [PubMed] [Google Scholar]
- Lee HC, Yin PH, Lin JC, Wu CC, Chen CY, Wu CW, Chi CW, Tam TN, Wei YH (2005) Mitochondrial genome instability and mtDNA depletion in human cancers. Ann NY Acad Sci 1042: 109–122. [DOI] [PubMed] [Google Scholar]
- Liu L, Tan D, Wong LJ (2004) [Somatic mutation detection in complete mitochondrial DNA of lung cancer patients]. Zhongguo fei ai za zhi 7(2): 125–129. [DOI] [PubMed] [Google Scholar]
- Ma J, Coarfa C, Qin X, Bonnen PE, Milosavljevic A, Versalovic J, Aagaard K (2014) mtDNA haplogroup and single nucleotide polymorphisms structure human microbiome communities. BMC Genom 15: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Outschoorn UE, Pavlides S, Howell A, Pestell RG, Tanowitz HB, Sotgia F, Lisanti MP (2011) Stromal-epithelial metabolic coupling in cancer: integrating autophagy and metabolism in the tumor microenvironment. Int J Biochem Cell Biol 43(7): 1045–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Weiss G, von Haeseler A (1999) Pattern of nucleotide substitution and rate heterogeneity in the hypervariable regions I and II of human mtDNA. Genetics 152(3): 1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2016) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. Available from http://www.R-project.org/.
- Reznik E, Miller ML, Senbabaoglu Y, Riaz N, Sarungbam J, Tickoo SK, Al-Ahmadie HA, Lee W, Seshan VE, Hakimi AA, Sander C (2016) Mitochondrial DNA copy number variation across human cancers. eLife 5: e10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts ER, Thomas KJ (2013) The role of mitochondria in the development and progression of lung cancer. Comput Struct Biotechnol J 6: e201303019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF (2009) Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res 37(8): 2539–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JB, Alaei-Mahabadi B, Sabarinathan R, Samuelsson T, Gorodkin J, Gustafsson CM, Larsson E (2015) Simultaneous DNA and RNA mapping of somatic mitochondrial mutations across diverse human cancers. PLoS Genet 11(6): e1005333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneking M (2000) Hypervariable sites in the mtDNA control region are mutational hotspots. Am J Hum Genet 67(4): 1029–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JH, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, Geisinger K, Hirsch FR, Ishikawa Y, Kerr KM, Noguchi M, Pelosi G, Powell CA, Tsao MS, Wistuba I Panel WHO (2015) The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 10(9): 1243–1260. [DOI] [PubMed] [Google Scholar]
- Tsao MS, Marguet S, Le Teuff G, Lantuejoul S, Shepherd FA, Seymour L, Kratzke R, Graziano SL, Popper HH, Rosell R, Douillard JY, Le-Chevalier T, Pignon JP, Soria JC, Brambilla EM (2015) Subtype classification of lung adenocarcinoma predicts benefit from adjuvant chemotherapy in patients undergoing complete resection. J Clin Oncol 33(30): 3439–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gisbergen MW, Voets AM, Starmans MH, de Coo IF, Yadak R, Hoffmann RF, Boutros PC, Smeets HJ, Dubois L, Lambin P (2015) How do changes in the mtDNA and mitochondrial dysfunction influence cancer and cancer therapy? Challenges, opportunities and models. Mutat Res Rev Mutat Res 764: 16–30. [DOI] [PubMed] [Google Scholar]
- Warburg O (1956) On the origin of cancer cells. Science 123(3191): 309–314. [DOI] [PubMed] [Google Scholar]
- Warth A, Endris V, Stenzinger A, Penzel R, Harms A, Duell T, Abdollahi A, Lindner M, Schirmacher P, Muley T, Dienemann H, Fink L, Morresi-Hauf A, Pfarr N, Weichert W (2016) Genetic changes of non-small cell lung cancer under neoadjuvant therapy. Oncotarget 7(20): 29761–29769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth A, Muley T, Herpel E, Meister M, Herth FJ, Schirmacher P, Weichert W, Hoffmann H, Schnabel PA (2012) Large-scale comparative analyses of immunomarkers for diagnostic subtyping of non-small-cell lung cancer biopsies. Histopathology 61(6): 1017–1025. [DOI] [PubMed] [Google Scholar]
- Weerts MJ, Sieuwerts AM, Smid M, Look MP, Foekens JA, Sleijfer S, Martens JW (2016) Mitochondrial DNA content in breast cancer: impact on in vitro and in vivo phenotype and patient prognosis. Oncotarget 7(20): 29166–29176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert W, Warth A (2014) Early lung cancer with lepidic pattern: adenocarcinoma in situ, minimally invasive adenocarcinoma, and lepidic predominant adenocarcinoma. Curr Opin Pulm Med 20(4): 309–316. [DOI] [PubMed] [Google Scholar]
- Yan B, Dong L, Neuzil J (2016) Mitochondria: an intriguing target for killing tumour-initiating cells. Mitochondrion 26: 86–93. [DOI] [PubMed] [Google Scholar]
- Yang Y, Karakhanova S, Hartwig W, D'Haese JG, Philippov PP, Werner J, Bazhin AV (2016) Mitochondria and mitochondrial ROS in cancer: novel targets for anticancer therapy. J Cell Physiol 231(12): 2570–2581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.