Abstract
Background:
Pancreatic cancer is highly malignant and characterised by rapid and uncontrolled growth. While some of the important regulatory networks involved in pancreatic cancer have been determined, the cancer relevant genes have not been fully identified.
Methods:
We screened genes that may control proliferation in pancreatic cancer in seven pairs of matched pancreatic cancer and normal pancreatic tissue samples. We examined KIF15 expression in pancreatic cancer tissues and the effect of KIF15 on cell proliferation in vitro and in vivo. The mechanisms underlying KIF15 promotion of cell proliferation were investigated.
Results:
mRNA microarray and functional analysis identified 22 genes that potentially play an important role in the proliferation of pancreatic cancer. High-content siRNA screening evaluated whether silencing these 22 genes affected proliferation of pancreatic cancer. Notably, silencing KIF15 exhibited the most potent inhibition of proliferation compared with the rest of the 22 genes. KIF15 was upregulated in human pancreatic cancer tissues, and higher KIF15 expression levels correlated with shorter patient survival times. Upregulation KIF15 promoted pancreatic cancer growth. KIF15 upregulated cyclin D1, CDK2, and phospho-RB and also promoted G1/S transition in pancreatic cancer cells. KIF15 upregulation activated MEK–ERK signalling by increasing p-MEK and p-ERK levels. MEK–ERK inhibitors successfully inhibited cell cycle progression, and PD98059 blocked KIF15-mediated pancreatic cancer proliferation in vivo and in vitro.
Conclusions:
This study identified KIF15 as a critical regulator that promotes pancreatic cancer proliferation, broadening our understanding of KIF15 function in tumorigenesis.
Keywords: pancreatic cancer, KIF15, proliferation, G1/S phase transition
Pancreatic cancer (PC) is generally acknowledged as being difficult to diagnose and treat. Despite improvements in treatment, the low patient survival rate is partly attributed to early, extensive local tumour invasion and distant metastasis, as well as multiple chemoresistance (Rombouts et al, 2015). The development of PC is a multistep process based on the accumulation of genetic and epigenetic alterations. Therefore, obtaining a better understanding of the molecular mechanisms responsible for the rapid growth, distant metastasis and tumorigenic properties of PC is likely to lead to novel therapeutic strategies for this disease.
We screened for genes with abnormal overexpression in PC using seven matched pairs of PC and adjacent, normal pancreatic tissues with a gene expression profile microarray. We verified the function of the identified genes by high-content siRNA screening. Using this approach, we found that KIF15, a gene encoding a member of the kinesin family of proteins, may play a vital role in PC proliferation.
The kinesin superfamily comprises a group of proteins that share a highly conserved motor domain (Minakawa et al, 2013; Goulet et al, 2014). Most KIF proteins have ATP-dependent activity and can drive microtubule-dependent plus-end motion (Florian and Mayer, 2011). Kinesins participate in several essential cellular processes including mitosis, meiosis, and the transport of macromolecules (Liu et al, 2010). Increasing evidence indicates that kinesin proteins play critical roles in the genesis and development of human cancers (Yokota et al, 2012; Minakawa et al, 2013). Some kinesin proteins are associated with malignancy as well as with drug resistance in solid tumours (Buster et al, 2003). Thus, targeting KIF may be a promising anticancer strategy. KIF inhibitors such as kinesin spindle protein have been investigated as a monotherapy or in combination with other drugs in clinical trials. Other kinesins with potential anticancer characteristics are still being discovered (Song, 2015).
In this study, we demonstrate that KIF15 promotes PC proliferation via the MEK–ERK signalling pathway. This result provides insight into the molecular mechanisms of pancreatic carcinogenesis and suggests a novel therapeutic strategy for PC.
Materials and methods
Ethical statement
This study was approved by the Human Research Ethics Committees at the Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology (HUST), and was carried out in accordance with the principles embodied in the Declaration of Helsinki (more details can be found in the Supplementary Materials and Methods).
All in vivo animal experiments were approved by the Committee on the Ethics of Animal Experimentation of HUST (Permit no. 2015-S207). All treatments were carried out according to the US Public Health Service Policy on the Humane Care and Use of Laboratory Animals. All surgical procedures were conducted under sodium pentobarbital anaesthesia and every effort was made to minimise animal suffering.
Cell culture
MIA-PaCa-2 and PANC-1 cell lines were purchased from ATCC (Manassas, USA). The two cell lines were previously authenticated by ATCC via STR typing. MIA-PaCa-2 and PANC-1 cells were grown in DMEM medium (Gibco, Waltham, USA) supplemented with 10% fetal bovine serum (Gibco), 100 U ml−1 penicillin G, and 100 μg ml−1 streptomycin (Sigma, Shanghai, China) at 37 °C in a humidified atmosphere containing 5% CO2.
Human tissue samples
A PC tissue microarray (HPan-Ade180Sur-01) was obtained from Shanghai Outdo Biotech (Shanghai, China).
Surgical specimens of PCs and adjacent normal pancreatic tissue were obtained from 27 PC patients who underwent surgical resection from January 2013 to December 2015. The 27 PC patients comprised 12 men and 15 women with a mean age of 60.4 years (range, 45–79 years). All 27 cancer specimens were histologically classified as adenocarcinomas. This study was approved by the Human Research Ethics Committees at the Tongji Hospital, Tongji Medical College, HUST.
RNA microarrays
Total RNA extracted from seven PC tissue samples and matching normal pancreatic tissues were screened for differentially expressed genes using an Agilent RNA 6000 Nano Kit (Agilent, Santa Clara, CA, USA). In another experiment, total RNA was extracted from three KIF15 knocked-down (KIF15D) samples and three negative control (NC) samples using an Agilent RNA 6000 Nano Kit and a PrimeView Human GeneChip (Agilent) was used for microarray analysis. RNA labelling and hybridisation to Agilent miRNA microarray chips were performed with a GeneChip Hybridization Wash and Stain Kit (Agilent). Microarray data were deposited in the NCBI Gene Expression Omnibus public database (http://www.ncbi.nlm.nih.gov/geo/).
High-content screening and cell growth curve analysis
PANC-1 cells were transfected with a KIF15-KD or NC lentivirus (Supplementary Material) and seeded into 48-well plates. GFP expression was observed using a fluorescence microscope. When cells reached 80% confluency, they were collected for further experiments. A total of 2000 cells per well were analysed once a day using a Cellomics ArrayScan System. By adjusting the input parameters, cells could be quantified by measuring the green fluorescence signal in each well. Data were collected for statistical analysis using 5-day cell proliferation curves. Cells on the scanned image were counted using image analysis software. The number of cells at each time point was compared with the cell count on day 1 to obtain a cell proliferation ratio for each time point for each experimental group, and the fold change in proliferation was used to produce a cell growth curve. The cell proliferation ratio was calculated as follows: fold change (NC vs experimental group)=proliferation ratio on day 5 for the NC group/proliferation ratio on day 5 for the experimental group. A fold change in the proliferation ratio of two or more indicated that cell proliferation had slowed down sufficiently to allow the effect of RNAi lentivirus infection on cell proliferation to be measured.
Animals
Female 6-week-old Balb/c nude mice were purchased from HFK Bioscience (Beijing, China). All mice were maintained under specific pathogen-free conditions in the Central Animal Laboratory, HUST.
Orthotopic transplantation of mouse PC cells in Balb/c nude mice
Female 8-week-old wild-type Balb/c nude mice were used in all experiments. For orthotopic implantation, hair was removed from the abdomen under pentobarbital sodium anesthesia. An abdominal longitudinal incision was then made to expose the pancreas, and a 20 μl cell suspension (approximate 2 × 106 cells) was injected directly into the pancreas. The incision was then closed with sutures. PD98059 was diluted in dimethyl sulfoxide (DMSO) solution to a concentration of 1.0 mg ml−1, and from day 1 onward, mice were subcutaneously injected with 40 μg kg−1·per day (Chen et al, 2015). All mice were routinely weighed and checked for signs of distress. Abdominal palpation was used to monitor tumour size. Mice were killed using CO2 narcosis followed by cervical dislocation at the end of the study period, or earlier if they appeared moribund or exhibited >15% weight loss. Tumours were evaluated both macroscopically and microscopically.
Subcutaneous transplantation of human PC cells in Balb/c nude mice
Female 6-week-old Balb/c nude mice were used in all experiments. A total of 2 × 106 transfected cells were subcutaneously injected into the right armpit of Balb/c nude mice. The weight of each mouse and the tumour diameter were measured every week. All mice were killed 9 weeks after initiation of treatment. Tumours were evaluated macroscopically and microscopically.
Statistical analyses
Results for continuous variables are presented as means±s.d. unless otherwise stated. Treatment groups were compared using independent sample t-tests. Pairwise multiple comparisons used one-way ANOVA (two-sided). A P value of<0.05 was considered statistically significant. All analyses were performed using IBM SPSS Statistics software version 17.0 (Chicago, IL, USA).
Results
Identification of KIF15 as a critical gene that promotes PC proliferation
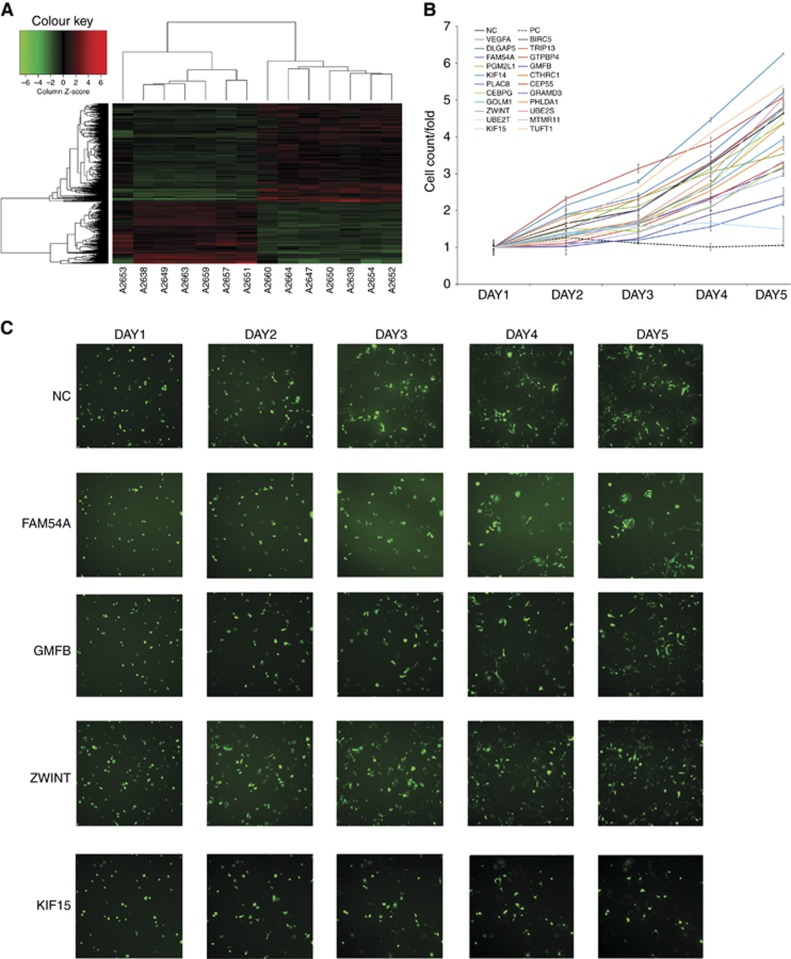
To identify genes with an essential role in PC tumorigenesis, we used mRNA microarray analysis to compare the mRNA expression profiles of seven pairs of matched PC and normal pancreatic tissue samples. The results identified 892 upregulated mRNAs and 568 downregulated mRNAs in the PC group compared with the control group (Figure 1A). On the basis of a functional analysis (Supplementary Table 1), 22 genes were found to potentially play an important role in the proliferation of PC. A literature review found that VEGFA and BIRC5, proteins encoded by two of the identified genes, were previously shown to promote PC cell proliferation (Lang et al, 2008; Glienke et al, 2010). All 22 candidate genes were then silenced in PANC-1 cells to examine the potential effect on the proliferation of PC cells in vitro (Figure 1B). Knockdown of four candidate genes, FAM54A, GMFB, KIF15, and ZWINT, in PANC-1 cells reduced the cell proliferation rate to a greater extent than VEGFA or BIRC5 knockdown (Figure 1B and C). KIF15 knockdown showed the greatest effect on reducing the proliferation rate and thus we focused on KIF15 in subsequent studies.
Figure 1.
Agilent mRNA microarray and high-content siRNA screening identified KIF15 as a critical gene in promoting PC proliferation. (A) Heat map showing gene expression profiles. Each row represents a gene and each column represents a sample. Red indicates high expression, whereas green indicates low expression. (B) A total of 22 genes were selected for validation by high-content screening. NC: negative control siRNA, PC: positive control siRNA targeting β-actin. (C) Representative fluorescence images of high-content siRNA screening for FAM54A, GMFB, KIF15, and ZWINT.
KIF15 upregulation in PC tissues is associated with poor patient prognosis
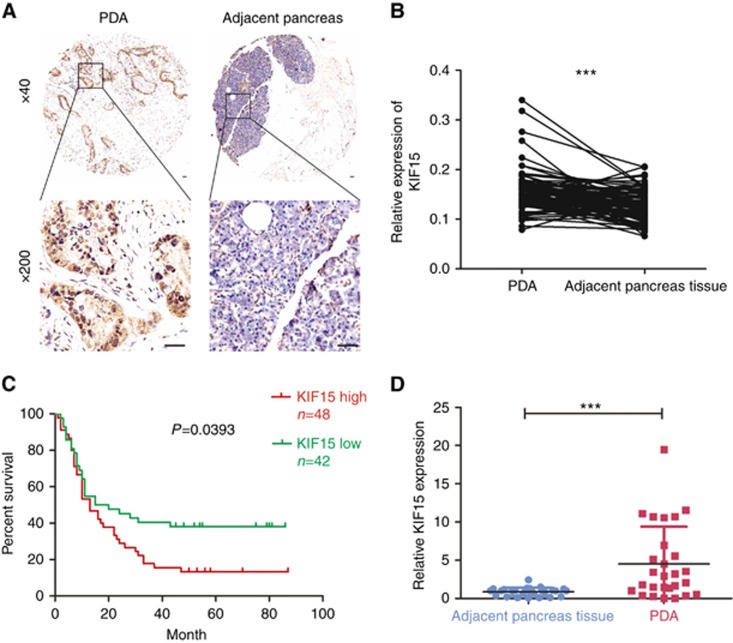
Immunohistochemical (IHC) staining of tissue microarrays shows that KIF15 was mainly localised to the cytoplasm of human PC cells (Figure 2A), whereas no KIF15 staining was present on adjacent, normal pancreatic tissue. Quantitative analysis indicated KIF15 was upregulated in human PC tissues compared with adjacent, normal pancreas tissues (Figure 2B). However, KIF15 expression levels did not vary significantly among different grades of human PC (Supplementary Table 2). Higher KIF15 expression (Supplementary Figure S1) was associated with poorer tumour differentiation (according to NCCN staging (Tempero et al, 2012), Supplementary Table 2) and shorter overall patient survival time (Figure 2C). Real-time PCR analysis of 27 pairs of matched PC and adjacent normal pancreatic tissue samples verified that the KIF15 mRNA expression was upregulated in PC tissues (Figure 2D). We have sequenced 20 cases of pancreatic cancer tissues and found no activating mutations in KIF15. According to the exon sequencing result, the observations in this study are not due to epiphenomenon. KIF15 in pancreatic cancer is not an epiphenomenon and is directly involved in pathogenesis of pancreatic cancer (Supplementary Figure S2).
Figure 2.
KIF15 is upregulated in PC tissues and KIF15 overexpression is associated with poor prognosis. (A) Immunohistochemical staining of KIF15 and representative images of one sample pair comprising PC tissue and adjacent, normal pancreas tissue. Magnification: × 40 (upper panel), × 200 (lower panel) (bar: 50 μm). (B) Quantitative analysis of KIF15 expression in PC tissues and adjacent, normal pancreas tissues. ***P<0.001. (C) By setting a relative KIF15 expression value of 0.5 as the demarcation point, PC patients were classified into high and low KIF15 expression groups. Kaplan–Meier analysis shows the correlation between KIF15 expression and overall survival of PC patients (P=0.0167). (D) PCR was used to quantify KIF15 mRNA expression in 27 pairs of PC tissue and adjacent, normal pancreas tissues. ***P<0.001.
KIF15 promotes PC cell proliferation in vitro and in vivo
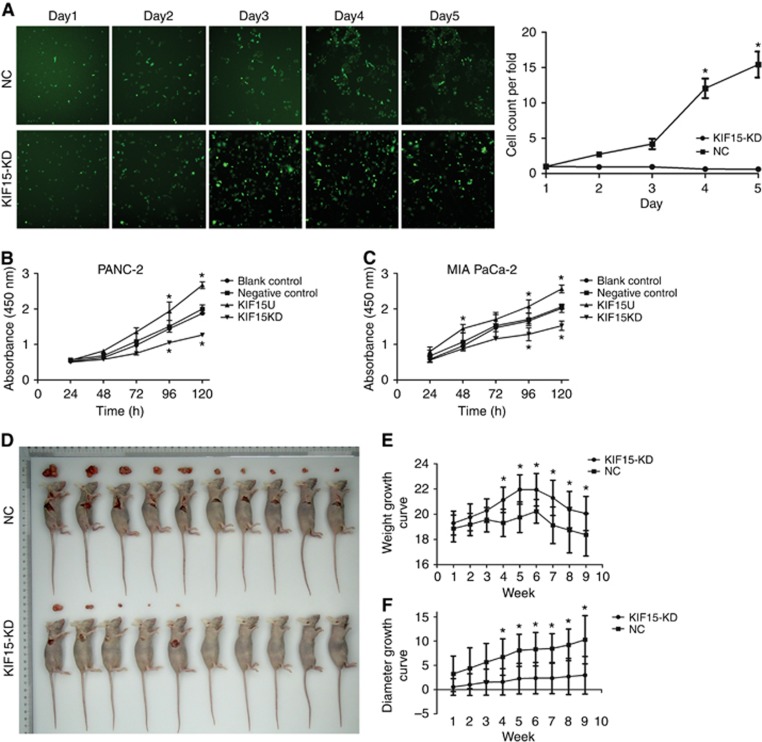
Stable KIF15 knockdown in the PANC-1 cell line was established using a lentiviral delivery system, and we confirmed downregulation of both KIF15 protein and mRNA in this cell line (Supplementary Figure S3). KIF15 knockdown suppressed PC cell proliferation, as measured by the CCK-8 assay and cell growth curve analysis using a fluorescence imaging system (Figure 3A–C). To further explore whether KIF plays a role in PC proliferation in vivo, a subcutaneous xenograft model of human PC cells in Balb/c nude mice was established. The KIF15 knockdown group showed a slower increase in tumour diameter and volume and less weight loss compared with controls (Figure 3D–F). In addition, Ki-67 staining was reduced in the KIF15 knockdown group compared with the control group (Supplementary Figure S4).
Figure 3.
KIF15 promotes PC cell proliferation in vivo and in vitro. (A) Cell growth curve analysis comparing KIF15 knockdown (KIF15KD) with negative control (NC) PC cells. n=3, *P<0.05. (B and C) CCK-8 assays comparing proliferation of KIF15KD, KIF15-overexpressing (KIF15U), NC and blank control (BC) PANC-1 (B) and MIA-PaCa-2 (C) cells. n=3, *P<0.05. (D) Images of the subcutaneous xenografts from the KIF15KD and NC groups. n=10. (E and F) Mice weight change curves (E) and tumour volume growth curves for subcutaneous xenografts. *P<0.05.
KIF15 promotes G1/S phase transition
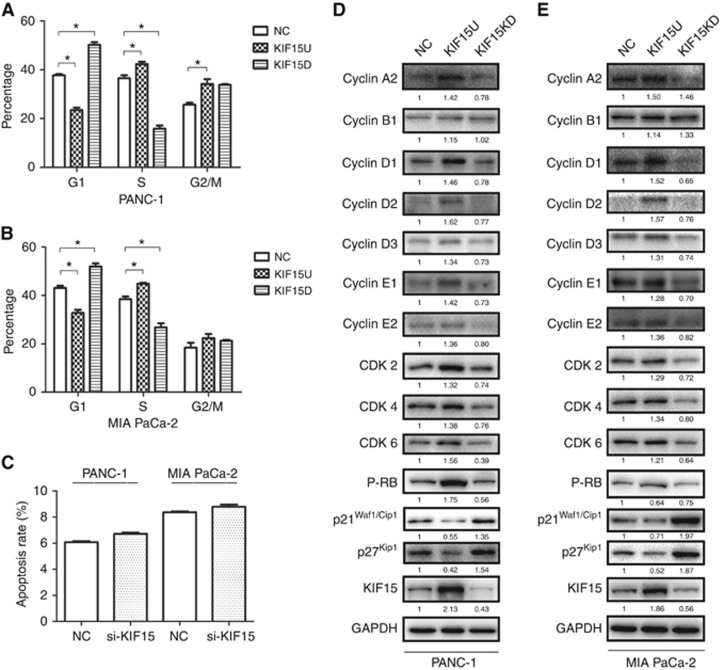
Flow cytometry was used to explore whether KIF15 promotes PC proliferation through regulating the cell cycle and/or apoptosis. KIF15 overexpression induced a dramatic alteration in cell cycle distribution in both PANC-1 and MIA-PaCa-2 cells: the fraction of cells in G1 phase decreased, while the fraction of cells in both S phase and G2/M phase increased compared with controls (Figure 4A and B). In both cell lines with KIF15 knockdown, the number of cells arrested in G1 phase increased compared with controls. However, changes in KIF15 levels had no effect on the apoptotic ratio (Figure 4C). Western blot analysis showed that KIF15 overexpression upregulated cyclin D1, CDK2 and p-RB, which promote cell cycle progression, while KIF15 knockdown had the opposite effect (Figure 4D and E).
Figure 4.
KIF15 regulates PC cell proliferation via promoting G1/S phase transition. (A and B) Flow cytometry analysis of the cell cycle in KIF15 knockdown (KIF15KD), KIF15-overexpressing (KIF15U), and negative control (NC) PANC-1 (A) and MIA-PaCa-2 (B) cells. (C) Flow cytometry analysis of apoptosis in KIF15KD, KIF15U and NC PANC-1 and MIA-PaCa-2 cells. (D and E) Western blotting shows cell cycle protein expression in KIF15KD, KIF15U, and NC PANC-1 (D) and MIA-PaCa-2 (E) cells. n=3. *P<0.05.
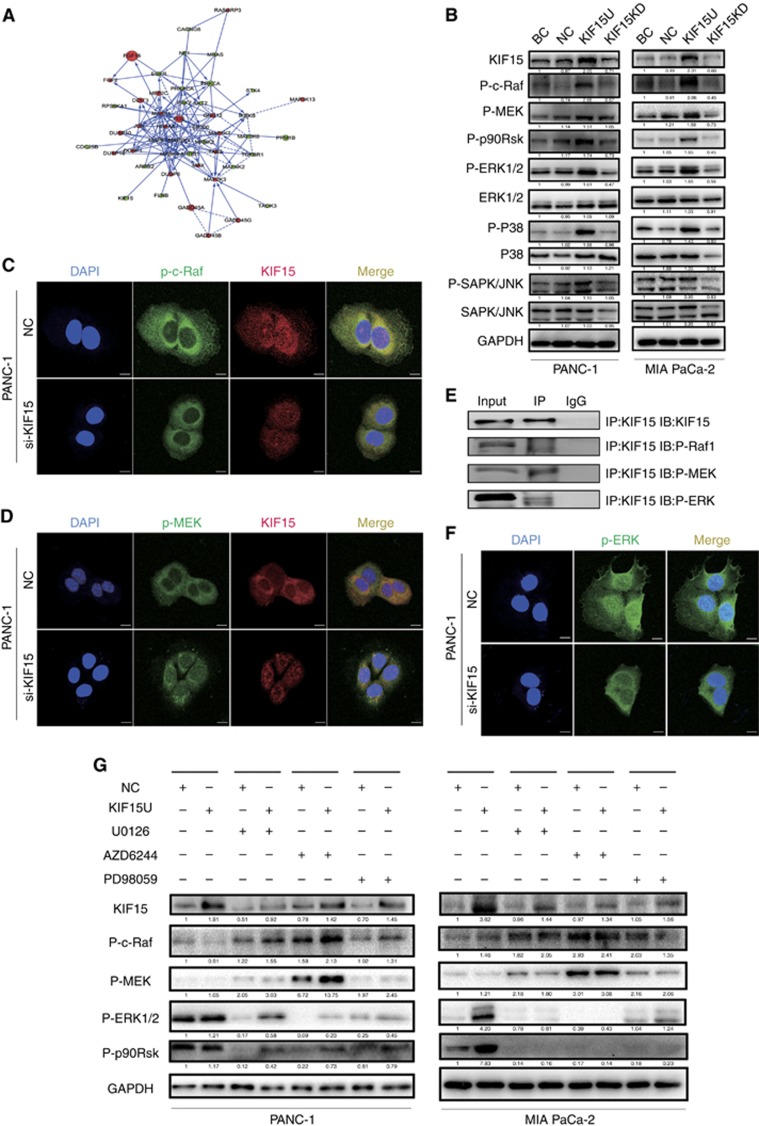
KIF15 activates the MEK–ERK pathway
To further elucidate the mechanisms underlying KIF15 promotion of PC proliferation, KIF15 knockdown cells and negative control cells were analysed by mRNA microarray. Surprisingly, many key genes in the MEK–ERK signalling pathway were downregulated in the KIF15 knockdown group compared with control cells. Pathway analysis confirmed that the MEK–ERK pathway was most closely connected with KIF15 (Figure 5A; Supplementary Figure S5A). We thus next investigated whether changes in KIF15 expression could affect activation of the MEK–ERK signalling pathway. Western blot and IHC analysis showed that KIF15 overexpression increased p-ERK expression, whereas KIF15 knockdown inhibited p-ERK expression in PC cells (Figure 5B; Supplementary Figure S6). Furthermore, KIF15 co-localised with p-c-Raf and p-MEK in PANC-1 (Figure 5C and D) and MIA-PaCa-2 cells (Supplementary Figure S5C and D). Moreover, KIF15 coimmunoprecipitated with both MEK and Raf (Figure 5E). KIF15 knockdown in PANC-1 and MIA-PaCa-2 cells suppressed p-ERK translocation from the plasma membrane to the nucleus (Figure 5F, Supplementary Figure S5B). MEK–ERK signalling pathway inhibitors U0126, AZD6244 and PD98059 inhibited KIF15 activation of the MEK–ERK signalling pathway (Figure 5G).
Figure 5.
KIF15 activated the MEK–ERK signalling pathway. (A) Network analysis of KIF15-associated signalling pathways in PC tissues using data obtained from mRNA microarrays. (B) Protein expression levels of key modulators of the MEK–ERK signalling pathway were analysed by western blotting in KIF15 knockdown (KIF15KD), KIF15-overexpressing (KIF15U), negative control (NC) and blank control (BC) PANC-1 and MIA-PaCa-2 cells. n=3. (C and D) Immunofluorescence staining shows p-c-Raf and p-MEK co-localisation with KIF15 in PANC-1 cells (bar: 10 μm). (E) KIF15 protein was immunoprecipitated with KIF15 antibody and levels of bound p-Raf, p-MEK, and p-ERK were detected by western blotting. (F) Immunofluorescence staining shows p-ERK expression in the nuclei of KIF15 knockdown (KIF15KD) and NC PANC-1 cells (bar: 10 μm). (G) Western blotting of protein lysates from NC, KIF15U, and KIF15U PANC-1 and MIA-PaCa-2 cells treated with three different MEK–ERK signalling pathway inhibitors: U0126(25 μM), AZD6244(25 μM), and PD98059(25 μM).
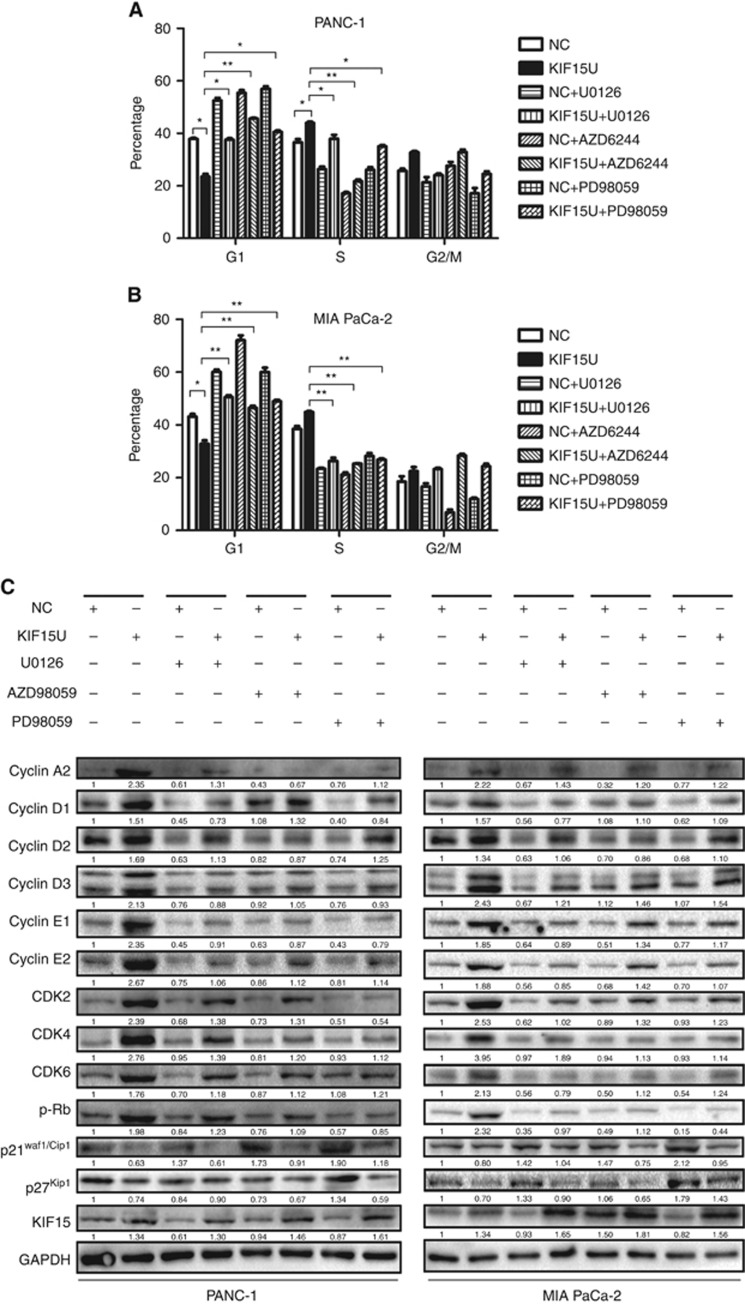
MEK–ERK pathway inhibitors block KIF15-dependent G1/S phase transition
We have shown that KIF15 promotes G1/S phase transition in PC cells. To investigate whether this effect is mediated by the MEK–ERK pathway, KIF15-overexpressing cell lines were treated with the MEK–ERK pathway inhibitors U0126, AZD6244, and PD98059 and effects on the cell cycle were examined. All the inhibitors blocked KIF15-dependent cell cycle transition (Figure 6A and B), indicating that KIF15-mediated inhibition of the G1/S phase transition occurs via the MEK–ERK pathway. Furthermore, KIF15-dependent increases in cyclin-D1, CDK2, and p-RB expression were blocked by treatment with U0126, AZD6244 and PD98059 (Figure 6C).
Figure 6.
KIF15 promotion of G1/S phase transition via the MEK–ERK signalling pathway is ablated by MEK–ERK pathway inhibitors. (A and B) Flow cytometry analysis of the cell cycle in negative control (NC), KIF15-overexpressing (KIF15U), and NC or KIF15U PANC-1 (A) and MIA-PaCa-2 (B) cells treated with three different MEK–ERK signalling pathway inhibitors: U0126(25 μM), AZD6244(25 μM), and PD98059(25 μM). (C) Western blotting shows cell cycle protein expression in NC, KIF15U and NC or KIF15U PANC-1 (left panel) and MIA-PaCa-2 (right panel) cells treated with three different MEK–ERK signalling pathway inhibitors: U0126; AZD6244; and PD98059. n=3. *P<0.05, **P<0.01.
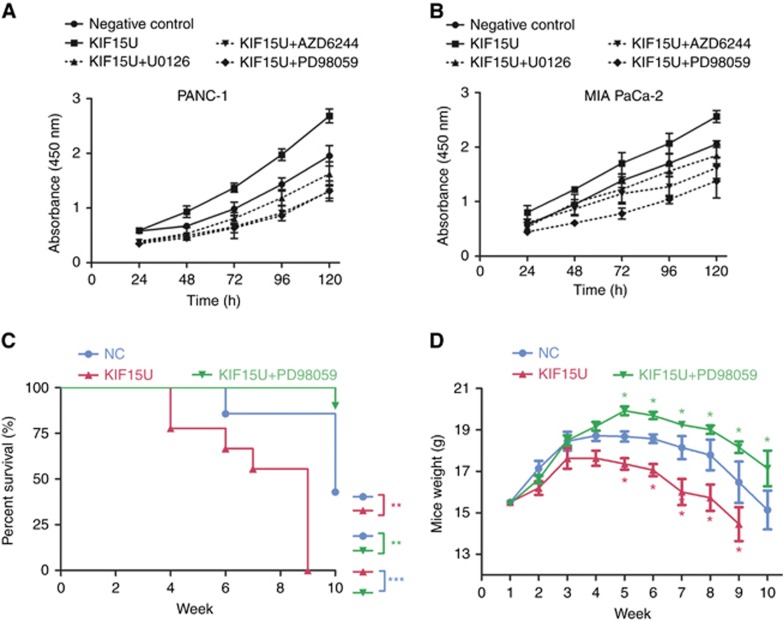
MEK–ERK inhibitors block KIF15-dependent PC proliferation in vivo and in vitro
We next examined whether the effect of KIF15 on promoting PC cell proliferation was mediated by the MEK–ERK pathway. We found that the enhancement of cell proliferation was counteracted to different extents by treatment with U0126, AZD6244 or PD98059 in CCK-8 assays (Figure 7A and B). Among the three inhibitors, PD98059 completely blocked KIF15-mediated PC cell proliferation.
Figure 7.
MEK–ERK signalling pathway inhibitors block KIF15-induced PC proliferation in vivo and in vitro. (A and B) Negative control (NC), KIF15-overexpressing (KIF15U) and KIF15U cells were treated with three different MEK–ERK signalling pathway inhibitors, U0126(25 μM), AZD6244(25 μM), and PD98059(25 μM), and proliferation rates were measured using the CCK-8 assay in PANC-1 (A) and MIA-PaCa-2 (B) cells. (C) Kaplan–Meier survival curves for different experimental mouse groups: NC, KIF15U, or KIF15U+PD98059 (10 mg kg−1), n=8, **P<0.01, ***P<0.001. (D) Mice weight changes in PANC-1 KIF15U and PANC-1 KIF15U+PD98059(10 mg kg−1) groups vs the NC group. *P<0.05.
To examine the relationship between KIF15 function and the MEK–ERK pathway in vivo, we established a Balb/c nude mice orthotopic transplantation model and examined three groups of mice: (1) a negative control (NC) group; (2) a group injected with KIF15-overexpressing cells (KIF15U); and (3) a group injected with KIF15-overexpressing cells plus a MEK–ERK pathway inhibitor (KIF15U+PD98059). Survival analyses revealed the mice in the KIF15U+PD98059 group had the longest survival time, whereas the KIF15U group had the shortest survival time (Figure 7C). The KIF15 overexpressing group showed a more rapid weight loss compared with the KIF15U+PD98059 group and NC group (Figure 7D). Together, these results suggest that MEK–ERK pathway inhibitors block KIF15-mediated PC proliferation both in vivo and in vitro.
Discussion
Although the structure and function of KIF15 have been studied for 10 years, the role of KIF15 in regulating the behavior of cancer cells has not yet been elucidated.
Kinesins are a superfamily of proteins with important roles in eukaryotic intracellular trafficking and cell division (Klejnot et al, 2014). The genomes of higher vertebrates contain as many as 45 genes encoding different kinesins (Florian and Mayer, 2011). Most of these molecular machines are implicated in intracellular transport, and one third of kinesin superfamily members play key roles at different stages of mitosis and cytokinesis (Messin and Millar, 2014). For example, the mitotic kinesin KIF11 drives glioblastoma invasion, proliferation and self-renewal (Venere et al, 2015). KIF1B promotes glioma migration and invasion via inducing the cell surface localisation of MT1-MMP (Chen et al, 2016). Silencing of KIF2A inhibits the proliferation and migration of breast cancer cells and correlates with an unfavourable prognosis for breast cancer patients (Wang et al, 2014). High levels of KIF18A expression are related to metastasis and significantly affect cancer progression (Shichijo et al, 2005; Nagahara et al, 2011; Rucksaken et al, 2012; Kasahara et al, 2016). KIF23 expression levels provide additional prognostic information for patients undergoing lung cancer surgery. KIF23 may also be a novel therapeutic target in this cancer type (Sun et al, 2015; Iltzsche et al, 2017; Kato et al, 2016; Sun et al, 2016). Therefore, several kinesin superfamily members have important effects on tumour behaviour.
Drechsler et al reported that KIF15 is a second tetrameric spindle motor (in addition to kinesin-5, Eg5) and reported the mechanisms by which hKIF15 and its inhibitor hTpx2 modulate spindle microtubule architecture (Tanenbaum et al, 2009; Drechsler et al, 2014; Eskova et al, 2014). The motor domain structure of KIF15 was captured in the ‘ATP-like’ configuration, with the neck linker docked onto the catalytic core. The interaction of KIF15 with microtubules was also investigated and structural differences between these two motors indicate profound differences in their modes of action, consistent with current models of microtubule crosslinking and sliding (Klejnot et al, 2014). KIF15 mediates plasma membrane localisation of the alternative clathrin adaptor Dab2, thus impinging on pathways that regulate α2-integrin internalisation. KIF15 can also drive centrosome separation during bipolar spindle assembly (Eskova et al, 2014). This activity requires both the KIF15 motor domain and interaction with TPX2 (Sturgill et al, 2014). KIF15 and TPX2 can crosslink and slide through two antiparallel microtubules, thereby driving centrosome separation. Studies on rodent brain development showed a pronounced enrichment of KIF15 in migratory neurons compared with other neurons. KIF15 opposes the capacity of other motors to generate independent microtubule movements within key regions of developing neurons (Buster et al, 2003; Drechsler et al, 2014). KIF15 also plays an important role in several tumours. KIF15 is required for maintenance of spindle bipolarity and is the breast cancer tumour antigen (Scanlan et al, 2001). KIF15 is essential for K5I resistance in HeLa cells, even in cases that necessitate additional factors such as the Eg5 rigor mutant (Sturgill et al, 2016). KIF15 is overexpressed in breast cancer cells and can have important values as both a prognostic factor and new therapeutic target for endocrine therapy-resistant breast cancer (Zou et al, 2014). KIF15 is also overexpressed in lung adenocarcinoma and may play a vital role in regulating the cell cycle (Bidkhori et al, 2013). In this study, we report for the first time that KIF15 promotes PC cell proliferation via the MEK–ERK pathway.
The MEK–ERK pathway plays pivotal role in cell proliferation (Park, 2014) and its deregulation is a signature of many epithelial cancers. The MEK–ERK signalling pathway is involved in controlling diverse cellular processes such as proliferation, survival, differentiation and motility (Giordano et al, 2015; Vajravelu et al, 2015). This pathway is often upregulated in human tumours and is thus an attractive target for the development of anticancer drugs (Hayashido et al, 2014). The mitogen-activated protein kinase 1/2 (MAP2K1/2) inhibitor PD98059 effectively inhibits ERK1/2 phosphorylation and enhances the radiosensitivity of rhabdomyosarcoma cells (Asati et al, 2016; Ding et al, 2016). Currently, dozens of MEK inhibitors targeting the MEK–ERK pathway have been included in clinical trials for cancer therapy (Johnson et al, 2014). The key step for quiescent cells to enter the cell cycle is the formation of an active cyclin D–CDK4/6 complex. ERK regulates cyclin D1 transcriptional induction via Fos family members and myc (Daksis et al, 1994). Furthermore, formation of the cyclin E/CDK2 complex seems to be indirectly regulated by ERK at two levels. First, ERK activity is required for nuclear translocation of CDK2. CDK2 is translocated to a nuclear compartment in which ERK is activated by threonine-160 phosphorylation via the CDK-activating kinase (CAK) and threonine-14 and tyrosine-16 dephosphorylation via the CDC25 phosphatase. However, blocking ERK activation does not modify the levels of cyclin E–CDK2 complexes: ERK activation is solely necessary for the nuclear localisation of CDK2. Second, ERK activity has been shown to regulate phosphorylation of CDK2 at threonine-160, an activating site. The pathways linking ERK activation to ERK-dependent CDK2 nuclear translocation and activating threonine-160 phosphorylation are not yet known. Further studies are necessary to resolve these questions. ERK activation plays a fundamental role in G1/S phase transition because it is required for the induction of cyclin D1 protein via several mechanisms; sustained activity of ERK is also required for the downregulation of many antiproliferative genes throughout the G1 phase of the cell cycle (Chambard et al, 2007).
This study thus reports a novel role for KIF15 in promoting cancer cell proliferation. Previous research efforts have focused on KIF15 functions related to microtubules and mitosis. This study reveals that KIF15 also promotes cancer proliferation and identifies a potential link between its cancer-promoting effects and the MEK–ERK signalling pathway (Supplementary Figure S7).
In conclusion, our study provides evidence that supports KIF15 as a key regulator that promotes the proliferation of PC. This is accomplished by promoting G1/S phase transition via regulating the MEK–ERK signalling pathway. Thus, MEK–ERK signalling pathway inhibitors may form the basis of novel therapies to block cancer promotion by KIF15.
Acknowledgments
This study was funded by The National Natural Science Foundation of China (grant no. 81160311 and 81572429 to JJ, and grant no. 81602475 to XG).
Author contributions
Conception and design: XG, JJ. Development of methodology: XG, JW. Acquisition of the data: JW, XG, JJ. Analysis and interpretation of data: JW, XG, JJ. Writing, review, and/or revision of the manuscript: XG, CX. Study supervision: JJ.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Asati V, Mahapatra DK, Bharti SK (2016) PI3K/Akt/mTOR and Ras/Raf/MEK/ERK signalling pathways inhibitors as anticancer agents: Structural and pharmacological perspectives. Eur J Med Chem 109: 314–341. [DOI] [PubMed] [Google Scholar]
- Bidkhori G, Narimani Z, Hosseini AS, Moeini A, Nowzari-Dalini A, Masoudi-Nejad A (2013) Reconstruction of an integrated genome-scale co-expression network reveals key modules involved in lung adenocarcinoma. PLoS ONE 8: e67552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buster DW, Baird DH, Yu W, Solowska JM, Chauviere M, Mazurek A, Kress M, Baas PW (2003) Expression of the mitotic kinesin Kif15 in postmitotic neurons: implications for neuronal migration and development. J Neurocytol 32: 79–96. [DOI] [PubMed] [Google Scholar]
- Chambard JC, Lefloch R, Pouyssegur J, Lenormand P (2007) ERK implication in cell cycle regulation. Biochim Biophys Acta 1773: 1299–1310. [DOI] [PubMed] [Google Scholar]
- Chen S, Han M, Chen W, He Y, Huang B, Zhao P, Huang Q, Gao L, Qu X, Li X (2016) KIF1B promotes glioma migration and invasion via cell surface localization of MT1-MMP. Oncol Rep 35: 971–977. [DOI] [PubMed] [Google Scholar]
- Chen XY, Cai HZ, Wang XY, Chen QY, Yang H, Chen YJ, Tang YP (2015) Application of the ERK signalling pathway inhibitor PD98059 in long-term in vivo experiments. Genet Mol Res 14: 18325–18333. [DOI] [PubMed] [Google Scholar]
- Daksis JI, Lu RY, Facchini LM, Marhin WW, Penn LJ (1994) Myc induces cyclin D1 expression in the absence of de novo protein synthesis and links mitogen-stimulated signal transduction to the cell cycle. Oncogene 9: 3635–3645. [PubMed] [Google Scholar]
- Ding C, Luo J, Li L, Li S, Yang L, Pan H, Liu Q, Qin H, Chen C, Feng J (2016) Gab2 facilitates epithelial-to-mesenchymal transition via the MEK/ERK/MMP signalling in colorectal cancer. J Exp Clin Cancer Res 35: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler H, McHugh T, Singleton MR, Carter NJ, McAinsh AD (2014) The Kinesin-12 Kif15 is a processive track-switching tetramer. eLife 3: e01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskova A, Knapp B, Matelska D, Reusing S, Arjonen A, Lisauskas T, Pepperkok R, Russell R, Eils R, Ivaska J, Kaderali L, Erfle H, Starkuviene V (2014) An RNAi screen identifies KIF15 as a novel regulator of the endocytic trafficking of integrin. J Cell Sci 127: 2433–2447. [DOI] [PubMed] [Google Scholar]
- Florian S, Mayer TU (2011) Modulated microtubule dynamics enable Hklp2/Kif15 to assemble bipolar spindles. Cell Cycle 10: 3533–3544. [DOI] [PubMed] [Google Scholar]
- Giordano G, Febbraro A, Tomaselli E, Sarnicola ML, Parcesepe P, Parente D, Forte N, Fabozzi A, Remo A, Bonetti A, Manfrin E, Ghasemi S, Ceccarelli M, Cerulo L, Bazzoni F, Pancione M (2015) Cancer-related CD15/FUT4 overexpression decreases benefit to agents targeting EGFR or VEGF acting as a novel RAF-MEK-ERK kinase downstream regulator in metastatic colorectal cancer. J Exp Clin Cancer Res 34: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glienke W, Maute L, Wicht J, Bergmann L (2010) Curcumin inhibits constitutive STAT3 phosphorylation in human pancreatic cancer cell lines and downregulation of survivin/BIRC5 gene expression. Cancer Invest 28: 166–171. [DOI] [PubMed] [Google Scholar]
- Goulet A, Major J, Jun Y, Gross SP, Rosenfeld SS, Moores CA (2014) Comprehensive structural model of the mechanochemical cycle of a mitotic motor highlights molecular adaptations in the kinesin family. Proc Natl Acad Sci USA 111: 1837–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashido Y, Kitano H, Sakaue T, Fujii T, Suematsu M, Sakurai S, Okamoto T (2014) Overexpression of integrin alphav facilitates proliferation and invasion of oral squamous cell carcinoma cells via MEK/ERK signalling pathway that is activated by interaction of integrin alphavbeta8 with type collagen. Int J Oncol 45: 1875–1882. [DOI] [PubMed] [Google Scholar]
- Iltzsche F, Simon K, Stopp S, Pattschull G, Francke S, Wolter P, Hauser S, Murphy DJ, Garcia P, Rosenwald A, Gaubatz S (2017) An important role for Myb-MuvB and its target gene KIF23 in a mouse model of lung adenocarcinoma. Oncogene 36(1): 110–121. [DOI] [PubMed] [Google Scholar]
- Johnson GL, Stuhlmiller TJ, Angus SP, Zawistowski JS, Graves LM (2014) Molecular pathways: adaptive kinome reprogramming in response to targeted inhibition of the BRAF-MEK-ERK pathway in cancer. Clin Cancer Res 20: 2516–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, Nagahara M, Nakagawa T, Ishikawa T, Sato T, Uetake H, Sugihara K (2016) Clinicopathological relevance of kinesin family member 18A expression in invasive breast cancer. Oncol Lett 12: 1909–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Wada H, Patel P, Hu HP, Lee D, Ujiie H, Hirohashi K, Nakajima T, Sato M, Kaji M, Kaga K, Matsui Y, Tsao MS, Yasufuku K (2016) Overexpression of KIF23 predicts clinical outcome in primary lung cancer patients. Lung Cancer 92: 53–61. [DOI] [PubMed] [Google Scholar]
- Klejnot M, Falnikar A, Ulaganathan V, Cross RA, Baas PW, Kozielski F (2014) The crystal structure and biochemical characterization of Kif15: a bifunctional molecular motor involved in bipolar spindle formation and neuronal development. Acta Crystallogr D Biol Crystallogr 70: 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang SA, Schachtschneider P, Moser C, Mori A, Hackl C, Gaumann A, Batt D, Schlitt HJ, Geissler EK, Stoeltzing O (2008) Dual targeting of Raf and VEGF receptor 2 reduces growth and metastasis of pancreatic cancer through direct effects on tumor cells, endothelial cells, and pericytes. Mol Cancer Ther 7: 3509–3518. [DOI] [PubMed] [Google Scholar]
- Liu M, Nadar VC, Kozielski F, Kozlowska M, Yu W, Baas PW (2010) Kinesin-12, a mitotic microtubule-associated motor protein, impacts axonal growth, navigation, and branching. J Neurosci 30: 14896–14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messin LJ, Millar JB (2014) Role and regulation of kinesin-8 motors through the cell cycle. Syst Synth Biol 8: 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakawa Y, Kasamatsu A, Koike H, Higo M, Nakashima D, Kouzu Y, Sakamoto Y, Ogawara K, Shiiba M, Tanzawa H, Uzawa K (2013) Kinesin family member 4A: a potential predictor for progression of human oral cancer. PLoS ONE 8: e85951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahara M, Nishida N, Iwatsuki M, Ishimaru S, Mimori K, Tanaka F, Nakagawa T, Sato T, Sugihara K, Hoon DS, Mori M (2011) Kinesin 18A expression: clinical relevance to colorectal cancer progression. Int J Cancer 129: 2543–2552. [DOI] [PubMed] [Google Scholar]
- Park JI (2014) Growth arrest signalling of the Raf/MEK/ERK pathway in cancer. Front Biol (Beijing) 9: 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombouts SJ, Vogel JA, van Santvoort HC, van Lienden KP, van Hillegersberg R, Busch OR, Besselink MG, Molenaar IQ (2015) Systematic review of innovative ablative therapies for the treatment of locally advanced pancreatic cancer. Br J Surg 102: 182–193. [DOI] [PubMed] [Google Scholar]
- Rucksaken R, Khoontawad J, Roytrakul S, Pinlaor P, Hiraku Y, Wongkham C, Pairojkul C, Boonmars T, Pinlaor S (2012) Proteomic analysis to identify plasma orosomucoid 2 and kinesin 18A as potential biomarkers of cholangiocarcinoma. Cancer Biomark 12: 81–95. [DOI] [PubMed] [Google Scholar]
- Scanlan MJ, Gout I, Gordon CM, Williamson B, Stockert E, Gure AO, Jager D, Chen YT, Mackay A, O’Hare MJ, Old LJ (2001) Humoral immunity to human breast cancer: antigen definition and quantitative analysis of mRNA expression. Cancer Immun 1: 4. [PubMed] [Google Scholar]
- Shichijo S, Ito M, Azuma K, Komatsu N, Maeda Y, Ishihara Y, Nakamura T, Harada M, Itoh K (2005) A unique gene having homology with the kinesin family member 18A encodes a tumour-associated antigen recognised by cytotoxic T lymphocytes from HLA-A2+ colon cancer patients. Eur J Cancer 41: 1323–1330. [DOI] [PubMed] [Google Scholar]
- Song M (2015) Progress in discovery of KIF5B-RET kinase inhibitors for the treatment of non-small-cell lung cancer. J Med Chem 58: 3672–3681. [DOI] [PubMed] [Google Scholar]
- Sturgill EG, Das DK, Takizawa Y, Shin Y, Collier SE, Ohi MD, Hwang W, Lang MJ, Ohi R (2014) Kinesin-12 Kif15 targets kinetochore fibers through an intrinsic two-step mechanism. Curr Biol 24: 2307–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill EG, Norris SR, Guo Y, Ohi R (2016) Kinesin-5 inhibitor resistance is driven by kinesin-12. J Cell Biol 213: 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhang C, Yang Z, Wu Y, Wang H, Bao Z, Jiang T (2016) KIF23 is an independent prognostic biomarker in glioma, transcriptionally regulated by TCF-4. Oncotarget 7: 24646–24655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Jin Z, Song X, Wang J, Li Y, Qian X, Zhang Y, Yin Y (2015) Evaluation of KIF23 variant 1 expression and relevance as a novel prognostic factor in patients with hepatocellular carcinoma. BMC Cancer 15: 961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Janssen A, Geers EF, Alvarez-Fernandez M, Medema RH (2009) Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr Biol 19: 1703–1711. [DOI] [PubMed] [Google Scholar]
- Tempero MA, Arnoletti JP, Behrman SW, Ben-Josef E, Benson AR, Casper ES, Cohen SJ, Czito B, Ellenhorn JD, Hawkins WG, Herman J, Hoffman JP, Ko A, Komanduri S, Koong A, Ma WW, Malafa MP, Merchant NB, Mulvihill SJ, Muscarella PN, Nakakura EK, Obando J, Pitman MB, Sasson AR, Tally A, Thayer SP, Whiting S, Wolff RA, Wolpin BM, Freedman-Cass DA, Shead DA (2012) Pancreatic adenocarcinoma, version 2.2012: featured updates to the NCCN Guidelines. J Natl Compr Canc Netw 10: 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajravelu BN, Hong KU, Al-Maqtari T, Cao P, Keith MC, Wysoczynski M, Zhao J, Moore JT, Bolli R (2015) C-Kit promotes growth and migration of human cardiac progenitor cells via the PI3K-AKT and MEK-ERK pathways. PLoS One 10: e0140798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venere M, Horbinski C, Crish JF, Jin X, Vasanji A, Major J, Burrows AC, Chang C, Prokop J, Wu Q, Sims PA, Canoll P, Summers MK, Rosenfeld SS, Rich JN (2015) The mitotic kinesin KIF11 is a driver of invasion, proliferation, and self-renewal in glioblastoma. Sci Transl Med 7: 304ra143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ma S, Ma R, Qu X, Liu W, Lv C, Zhao S, Gong Y (2014) KIF2A silencing inhibits the proliferation and migration of breast cancer cells and correlates with unfavorable prognosis in breast cancer. BMC Cancer 14: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota K, Sasaki H, Okuda K, Shimizu S, Shitara M, Hikosaka Y, Moriyama S, Yano M, Fujii Y (2012) KIF5B/RET fusion gene in surgically-treated adenocarcinoma of the lung. Oncol Rep 28: 1187–1192. [DOI] [PubMed] [Google Scholar]
- Zou JX, Duan Z, Wang J, Sokolov A, Xu J, Chen CZ, Li JJ, Chen HW (2014) Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Mol Cancer Res 12: 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.