Abstract
Background:
High stability and disease-specific disarrangements suggest that microRNA molecules (miRNAs) present in body fluids are ideally suited for diagnostic applications, including gastric cancer (GC). However, the actual source of circulating miRNA biomarkers in GC has not been adequately evaluated, particularly in the Western populations that have some distinct characteristics compared with Asian patients.
Methods:
Twenty treatment-naive patients with GC along with 20 cancer-free controls were recruited. miRCURY LNA miRNA microarrays were used for miRNA expression profiling in primary tumours and adjacent healthy mucosa. Differentially expressed serum miRNAs were identified with a high throughput TaqMan OpenArray technology in tumour-draining veins of the portal system, as well as peripheral blood of the patients and controls.
Results:
Tissue profiling identified 108 sequences differentially expressed between primary tumours and adjacent mucosa (87 upregulated and 21 downregulated). Twenty miRNAs found in serum of GC patients showed expression levels higher than in controls. However, only seven of these molecules were overexpressed in primary tumours (miR-130a, miR-331, miR-19a, miR-223, miR-106a, miR-21, and miR-374). Moreover, expression of miR-331 and miR-21 was significantly higher in the peripheral circulation compared to tumour-draining veins of the portal system.
Conclusions:
The results indicate that the majority of potential serum miRNA biomarkers may originate from tissues other than the primary tumour.
Keywords: gastric cancer, microRNA, miRNA, serum, circulating miRNA, biomarker
Gastric cancer (GC) is the third leading cause of cancer death and a major public health care problem worldwide, including in Western populations (Ferlay et al, 2015). There are only limited diagnostic methods for early detection and no effective screening programs outside some Asian countries (Veitch et al, 2015). Consequently, most cases are diagnosed at an advanced stage when the prognosis is dismal (Van Cutsem et al, 2016). Thus, there is an urgent search for new, preferentially non-invasive, biomarkers to allow early detection of GC.
High stability and resistance to sample storage handling combined with tissue- and disease-specific disarrangements suggest that small non-coding RNAs (miRNAs), about 22 nucleotides long, present in body fluids are ideally suited for diagnostic applications (Cortez et al, 2011). Indeed, preliminary experiments using comprehensive expression profiling showed that serum miRNAs can serve as a potential source for biomarkers for the detection of human malignancies (Chen et al, 2008; Mitchell et al, 2008). Multiple subsequent clinical studies, including those carried out in patients with GC, suggested that the diagnostic performance of blood miRNAs signatures could outperform most previous serological markers. Nevertheless, despite intensive research with various profiling platforms, there are many unsolved questions addressing the origin and function of circulating miRNAs in cancer (Pritchard et al, 2012a).
Circulating miRNAs are believed to be actively secreted (Kosaka et al, 2010) or passively released from dead cancer cells and circulate packaged with RNA-binding proteins (Turchinovich et al, 2011), lipoproteins (Vickers et al, 2011), and extracellular vesicles (Arroyo et al, 2011). However, other sources of miRNAs circulating in cancer patients are also plausible, including immune and other blood cells (Pritchard et al, 2012b). Consequently, some studies demonstrated that only a small subset of circulating miRNAs has a tumour-specific origin (Wulfken et al, 2011). This phenomenon may be even more complex in malignancies of the alimentary system, as experimental studies suggested the importance of hepatic elimination of exogenously administered miRNAs (Bala et al, 2015; Imai et al, 2015). Consequently, the actual source of blood miRNA biomarkers in gastric cancer has not been adequately evaluated and little is known about their distribution, particularly in the Western populations.
To the best of our knowledge, this is the first study of comprehensive miRNome profiling aimed at evaluating the origins of serum microRNA biomarkers for GC. Herein, we first systematically examined miRNA expression profiles of primary gastric cancer and adjacent healthy tissues. Subsequently, serum miRNAs were identified to evaluate whether potential biomarkers found in peripheral blood corresponded to those identified in tumour-draining veins of the portal system.
Materials and methods
Study design and participants
The study was planned as a prospective, open label clinical trial. A total of 20 treatment-naive patients with histologically verified advanced non-cardia adenocarcinoma of the stomach scheduled for gastrectomy at the First Department of Surgery Jagiellonian University Medical College from January to December 2015 were recruited. A group of 20 patients subject to upper gastrointestinal endoscopy for complaints related to non-malignant conditions, mostly GERD and cholelithiasis, was selected as controls. Tumour typing and staging were adapted to the recent guidelines (Bosman et al, 2010; Edge et al, 2011). The study was approved by the Bioethics Committee of Jagiellonian University and all participants provided their written informed consent upon recruitment.
Sample processing
All blood and tissue samples were collected prior to starting any tumour-oriented treatment. Peripheral venous blood (5 ml) was collected into sterile BD Vacutainer tubes under fasting conditions immediately before surgery and from the control subjects. Another 5 ml was drawn intraoperatively from the gastroepiploic vein located in the vicinity of the tumour. After being allowed to clot at room temperature for 60 min, samples were centrifuged at 2000 g for 10 min at 4 °C. The serum was removed and stored at −80 °C until analysis.
Pairs of primary tumours and corresponding normal gastric mucosa were sampled from patients’ surgical specimens immediately after resection. The fresh specimens were collected into RNase free polypropylene tubes filled with RNAlater stabilisation solution (Qiagen, Valencia, CA, USA) and stored at −80 °C until RNA was extracted. Some portions of the collected specimens were used for routine histopathology to verify adequate cellularity of samples corresponding to tumour tissue and normal gastric mucosa.
RNA extraction
Total tissue RNA was extracted using miRCURY RNA Isolation Kits (Exiqon, Vedbæk, Denmark) according to the manufacturer’s instructions. Prior to RNA extraction from serum samples, tubes were thawed completely on ice and 50 μl of each serum aliquot was spiked-in with 10 fmol synthetic C. elegans microRNA (cel-miR-39) (Applied Biosystems, Foster City, CA, USA). Serum RNA extraction for profiling experiments was carried out with TaqMan miRNA ABC Purification Kits Human Panel A and B (Applied Biosystems) containing superparamagnetic Dynabeads covalently bound to 754 anti-miRNA oligonucleotides selected for downstream applications. RNA concentrations were measured with NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA) and overall sample quality was determined by RNA integrity number (RIN) using Agilent Bioanalyser (Agilent Technologies, Palo Alto, CA, USA).
Tissue microRNA expression profiling
Total RNA (750 ng) isolated from tumour and healthy mucosa samples was labelled with Hy3 and Hy5 fluorescent labels, using the miRCURY LNA microRNA Hi-Power Labelling Kit, Hy3/Hy5 (Exiqon) following the manufacturer's instructions. Spike-in controls were added to evaluate the labelling reaction, hybridisation, and the performance of the array experiment. The labelled RNA samples were hybridised to the miRCURY LNA microRNA Array 7th Gen (Exiqon) containing capture probes targeting all microRNAs of human origin registered in the miRBASE 19.0. The hybridisation was performed using a Tecan HS4800 hybridisation station (Tecan, Salzburg, Austria) according to the manufacturer’s protocol. After hybridisation, the microarray slides were scanned using the Agilent G2565BA Microarray Scanner System (Agilent Technologies) and analysed with the ImaGene 9 software (BioDiscovery, El Segundo, CA, USA). The obtained signals were background corrected and data normalisation was performed with the Lowess (Locally Weighted Scatterplot Smoothing) algorithm. MicroRNAs with intensities above the detection threshold in less than 20% of the samples were removed from the final dataset used for the expression analysis. Microarray data were deposited in a Gene Expression Omnibus database (accession no. GSE93415).
Serum microRNA expression profiling
Serum miRNAs were profiled by qRT–PCR using the TaqMan OpenArray assays (Applied Biosystems). Briefly, 0.9–2.7 ng of isolated RNA was reverse transcribed in a 7.5 μl reaction volume using the TaqMan MicroRNA Reverse Transcription Kit and Megaplex RT Primers Human Pools A (v2.1) and B (v3.0) according to the manufacturer’s protocol (Applied Biosystems). The resulting cDNA (2.5 μl) was preamplified to increase the quantity of specific cDNA targets with Megaplex PreAmp Primers, Human Pools A (v2.1) and B (v3.0) and the TaqMan PreAmp Master Mix in a final volume of 25 μl (Applied Biosystems). The preamplified product was diluted 40 folds and mixed 1:1v/v with the TaqMan OpenArray Real-Time PCR Master Mix. 5 μl aliquots of the mixture were dispensed on an OpenArray 384-well sample plate. Subsequently, TaqMan OpenArray Human MicroRNA Panels were loaded with the OpenArray AccuFill System and the PCR reactions were carried out with an OpenArray System (Applied Biosystems) following the manufacturer's instructions. Values of Ct were processed and exported to DataAssist Software (Applied Biosystems).
Quantitative RT–PCR validation of tissue and serum miRNA profiling experiments
The expression of selected miRNAs was verified by qRT–PCR using TaqMan MicroRNA Assays (Applied Biosystems) according to the manufacturer’s instructions. Briefly, total RNA (10 ng) was reverse transcribed in a 15 μl reaction volume using miRNA-specific RT primers and reagents from the TaqMan MicroRNA Reverse Transcription Kit. The resulting cDNA was diluted 1:15 and assayed in 20 μl reaction volumes by qPCR using the corresponding miRNA assay primers and TaqMan Universal PCR Master Mix, No AmpErase UNG. The amplification was performed in 96-well plates with a ViiA 7 real-time PCR instrument (Applied Biosystems) in accordance with the manufacturer’s protocols. Three technical replicates were performed for each assay, and the obtained mean Ct values were exported for statistical analysis.
Normalisation for qRT–PCR microRNA expression assays
Since there is no consensus about an optimal reference for relative microRNA expression using the qRT–PCR assays, various controls were evaluated separately for serum and tissue samples. Three normalisation strategies for serum were examined with the NormFinder algorithm using most stably expressed miRNAs (endogenous controls), the mean expression of all the genes (global normalisation (Mestdagh et al, 2009)), and cel-miR-39 (exogenous control). Reference controls for tissues were evaluated using only the two former methods without exogenous spike-ins. Finally, the expression levels of miRNAs normalised to the most stable references (ΔCt) were calculated by subtracting the reference Ct values from the Ct values of the miRNA of interest (Livak and Schmittgen, 2001).
Statistical analysis
The complete-linkage method and Euclidean distances were used for cluster analysis of microarray data. Comparative expression analysis was done in the software R/Bioconductor using mainly the limma package and moderated t-statistics (Huber et al, 2015). For OpenArray and qRT–PCR experiments, miRNAs with >10% undetectable expression data were excluded from the calculations; otherwise missing data were replaced by mean values. miRNAs with Ct value 5 amplification cycles lower than negative control and not exceeding 40 cycles were included for further analysis. The Mann–Whitney U-test was used to detect differences with regard to ΔCt values. Relative miRNA expression levels between groups were calculated by using the ΔΔCt method (Livak and Schmittgen, 2001). All raw P-values were adjusted for multiple testing by the Benjamini, Krieger, Yekutieli correction (Benjamini et al, 2006). Statistical analysis was performed using the IBM SPSS Statistics v.24 software package (IBM Corporation, Armonk, NY, USA).
Results
Demographic and clinical characteristics of the study subjects
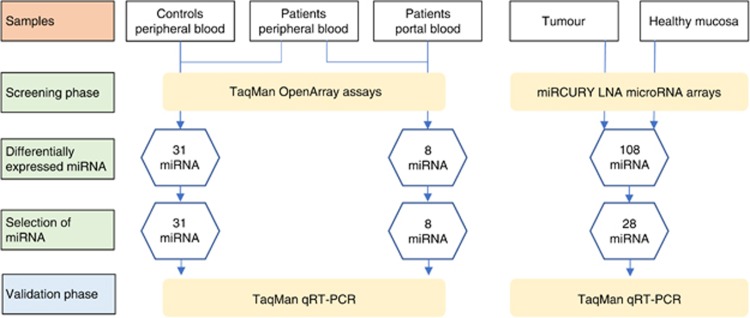
Clinical and demographic data of the selected population are summarised in Table 1. No significant difference in age, gender or comorbidities distribution was observed between patients and controls (P-values >0.05). Figure 1 shows a workflow for the subsequent analytical steps.
Table 1. Clinicopathological details of patients.
| Gastric cancer (n=20) | Controls (n=20) | |
|---|---|---|
| Age, median (range) | 64 (34–81) | 62 (32–78) |
| Female/male, n (%) | 9 (45)/11 (55) | 9 (45)/11 (55) |
| Comorbidities, n (%) | ||
| Circulatorya | 14 (70) | 12 (60) |
| Pulmonaryb | 2 (10) | 2 (10) |
| Diabetes mellitus | 4 (20) | 5 (25) |
| Location, n (%) | ||
| Upper third | 2 (10) | |
| Middle third | 8 (40) | |
| Distal third | 5 (25) | |
| Whole stomach | 5 (25) | |
| Differentiation | ||
| Well | 2 (10) | |
| Moderately | 3 (15) | |
| Poor | 15 (75) | |
| Lauren classification | ||
| Intestinal | 4 (20) | |
| Diffuse | 9 (45) | |
| Mixed | 7 (35) | |
| Stage | ||
| II | 2 (10) | |
| III | 13 (65) | |
| IV | 5 (25) |
Ischemic heart disease, arrhythmia, previous myocardial infarction, coronary artery disease.
Asthma, chronic obstructive pulmonary disease.
Figure 1.
Workflow showing the analytical strategy for the identification of microRNAs.
Identification of reference controls for serum and tissue qRT–PCR normalisation
As expected, tissue and blood samples showed distinct variability of miRNA expression patterns using the NormFinder algorithm. Tissue experiments identified miR-191-5p and miR-298 as the optimum reference controls (Supplementary Figure S1). However, the most stable controls for serum samples included the combination of cel-miR-39 spike-in and miR-191-5p.
Tissue microRNA of primary tumours and healthy gastric mucosa
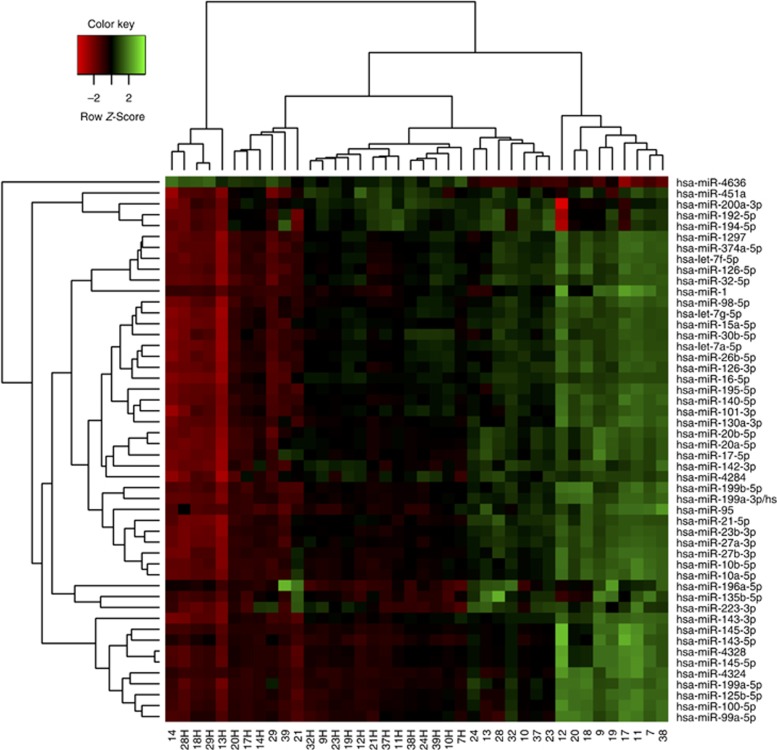
A total of 975 miRNAs were detected above background levels in at least two tumour samples, including a subset of 108 sequences where the absolute value of the log fold change compared to healthy mucosa was larger than 1 and the corrected P value was less than 0.05 (87 upregulated and 21 downregulated miRNAs; Supplementary Table S1). The heat map diagram shows the result of a two-way hierarchical clustering of microRNAs and samples using the complete-linkage method together with the Euclidean distance measure (Figure 2). Table 2 lists the 10 most significant miRNAs upregulated in tumour tissues and ranked by the relative fold change compared to healthy mucosa. Subsequently, arrays experiments for these 10 molecules were validated using qRT–PCR and expression data were normalised to the average of assays for miR-191-5p and miR-298. The results of qRT–PCR assays showed the same tendency as those of microarray experiments for all miRNAs tested, but fold-change values were generally lower (Table 2).
Figure 2.
Heat map and unsupervised hierarchical clustering based on the 50 most differentially expressed miRNAs between primary tumours and adjacent healthy mucosa. The miRNA overexpression is in green, and lower miRNA expression is in red. Numbers correspond to paired samples from individual patients with ‘H’ denoting healthy mucosa.
Table 2. Ten most upregulated miRNAs in gastric cancer tissue when compared with normal gastric mucosa.
| Annotation |
microRNA array |
RT–PCR validation |
||
|---|---|---|---|---|
| fold change | P-valuea | fold change | P-valuea | |
| miR-199a-3p | 4.6 | 0.002 | 3.6 | 0.001 |
| miR-199a-5p | 3.7 | 0.001 | 2.7 | 0.002 |
| miR-223-3p | 3.7 | 0.002 | 3.1 | 0.005 |
| miR-125b-5p | 3.7 | 0.002 | 2.6 | 0.004 |
| miR-27a-3p | 3.5 | 0.002 | 2.8 | 0.001 |
| miR-196a-5p | 3.5 | 0.001 | 2.7 | 0.001 |
| miR-23b-3p | 3.5 | 0.002 | 3.1 | 0.002 |
| miR-21-5p | 3.2 | 0.001 | 2.6 | 0.004 |
| miR-23a-3p | 3.2 | 0.001 | 2.6 | 0.003 |
| miR-20a-5p | 3.0 | 0.001 | 2.0 | 0.001 |
Abbreviations: miRNA=microRNA; RT–PCR=reverse transcriptase PCR.
Benjamini, Krieger, and Yekutieli correction for multiple testing.
Serum miRNAs in gastric cancer and controls
Among the 434 miRNAs initially identified by TaqMan OpenArray assays, 31 sequences showed at least a twofold change in serum of gastric cancer patients compared to controls (all up-regulated; Supplementary Table S2). However, the subsequent qRT–PCR validation experiments for these 31 molecules showed that expression of only 20 miRNAs maintained their significance after correction for multiple comparisons with a fold change ranging from 4 (miR-30c) to 230 (miR-518f) (Table 3). There were no significant differences in these miRNAs in relation to the primary tumour (T2–T3 vs T4) or distant metastases (M0 vs M1).
Table 3. Upregulated serum miRNAs evaluated by qRT–PCR in the peripheral blood of patients with gastric cancer when compared with healthy controls.
| miRNAs | ΔCthealthy | ΔCtcancer | ΔΔCt | 2–ΔΔCt | Corrected Pa |
|---|---|---|---|---|---|
| miR-518f | −4.50±10.78 | −12.35±1.99 | −7.85 | 230.67 | 0.017 |
| miR-130a | 10.96±3.48 | 6.62±3.42 | −4.34 | 20.22 | 0.017 |
| miR-212 | 8.81±2.36 | 4.58±2.65 | −4.22 | 18.70 | 0.007 |
| miR-220 | 7.85±2.51 | 3.85±2.81 | −4.00 | 15.98 | 0.017 |
| miR-433 | 9.03±1.67 | 5.19±3.15 | −3.84 | 14.29 | 0.025 |
| miR-518d | 8.77±3.76 | 5.43±3.24 | −3.35 | 10.18 | 0.037 |
| miR-331 | 6.54±2.61 | 3.31±3.34 | −3.23 | 9.40 | 0.039 |
| miR-19a | 8.39±4.07 | 5.18±3.55 | −3.21 | 9.23 | 0.042 |
| miR-365 | 10.66±3.22 | 7.62±3.12 | −3.04 | 8.25 | 0.037 |
| miR-374 | 8.48±3.24 | 5.55±2.94 | −2.93 | 7.62 | 0.037 |
| miR-296 | 7.74±4.29 | 4.86±3.14 | −2.88 | 7.37 | 0.039 |
| miR-323-3p | 9.21±1.60 | 6.59±2.94 | −2.63 | 6.18 | 0.046 |
| miR-21 | 4.05±2.33 | 1.73±2.45 | −2.33 | 5.02 | 0.017 |
| miR-146b | 6.95±2.34 | 4.64±2.21 | −2.31 | 4.97 | 0.037 |
| miR-146a | 5.99±2.19 | 3.68±2.65 | −2.31 | 4.96 | 0.025 |
| miR-106a | 4.30±2.77 | 2.10±2.51 | −2.19 | 4.57 | 0.025 |
| miR-223 | −2.65±2.32 | −4.78±2.57 | −2.12 | 4.35 | 0.025 |
| miR-19b | 2.45±2.23 | 0.36±2.63 | −2.09 | 4.26 | 0.025 |
| miR-451 | 0.68±2.31 | −1.41±2.84 | −2.08 | 4.23 | 0.037 |
| miR-30c | 4.95±3.3 | 2.90±2.65 | −2.05 | 4.14 | 0.037 |
Abbreviations: miRNA=microRNA; qRT–PCR=quantitative reverse transcriptase PCR.
Values are expressed as means±s.d.
ΔCt=Ct sample−Ct average expression of cel-miR-39 and miR-191-5p.
ΔΔCt=ΔCt cancer−ΔCt healthy.
Benjamini, Krieger, and Yekutieli correction for multiple testing.
Portal and peripheral blood miRNAs
Paired serum samples obtained from peripheral and gastroepiploic veins were evaluated to determine whether miRNA biomarkers found in peripheral blood corresponded to those identified in tumour-draining veins of the portal system. Eight of the evaluated miRNAs showed significantly increased levels in the peripheral circulation by TaqMan OpenArray cards and these sequences were subsequently validated with qRT–PCR assays (Table 4). Moreover, the up-regulated group included miR-331 and miR-21, which were earlier identified as potential biomarkers of gastric cancer comparing miRNA expression in peripheral blood of patients and controls (Table 3).
Table 4. Dysregulated miRNAs in peripheral circulation when compared with portal blood by qRT–PCR.
| miRNAs | ΔCtperipheraL | ΔCtportal | ΔΔCt | 2–ΔΔCt | P-value | Corrected P-valuea |
|---|---|---|---|---|---|---|
| miR-21 | 1.73±2.45 | 5.28±3.09 | −3.55 | 11.71 | 0.001 | 0.004 |
| miR-16 | 0.36±3.42 | 3.81±3.91 | −3.45 | 10.94 | 0.045 | 0.045 |
| miR-99a | 1.13±2.37 | 4.29±1.18 | −3.15 | 8.88 | 0.016 | 0.032 |
| miR-95 | 6.96±2.71 | 10.04±2.21 | −3.08 | 8.46 | 0.012 | 0.032 |
| miR-532 | 5.42±1.22 | 8.48±1.21 | −3.06 | 8.34 | 0.029 | 0.033 |
| miR-891a | 2.77±1.79 | 5.80±1.66 | −3.03 | 8.19 | 0.001 | 0.004 |
| miR-331 | 3.31±3.34 | 5.40±2.37 | −2.09 | 4.26 | 0.028 | 0.033 |
| miR-92a | 0.19±1.89 | 2.26±2.02 | −2.07 | 4.20 | 0.027 | 0.033 |
Abbreviations: miRNA=microRNA; qRT–PCR=quantitative reverse transcriptase PCR.
Values are expressed as means±s.d.
ΔCt=Ct sample−Ct average expression of cel-miR-39 and miR-191-5p
ΔΔCt=ΔCt peripheral−ΔCt portal.
Benjamini, Krieger, and Yekutieli correction for multiple testing.
Correlation of tissue and blood miRNAs
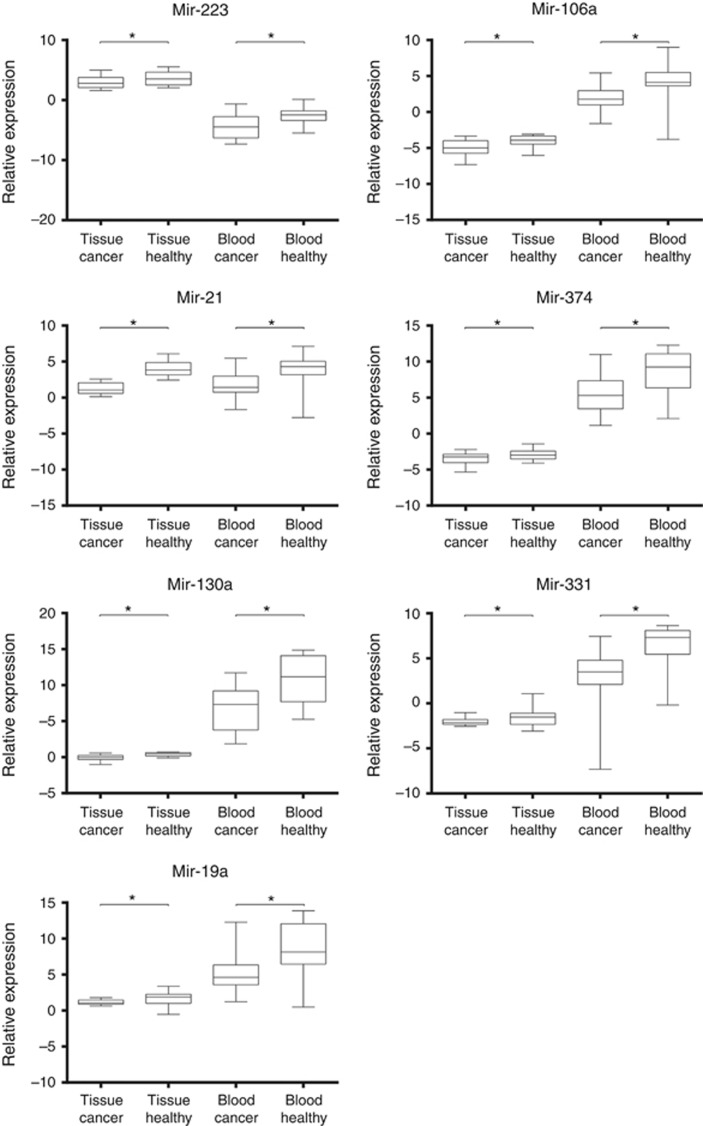
To correlate profiles of tissue and circulating miRNAs, we compared expression levels of primary tumours and healthy gastric mucosa with peripheral blood of patients and healthy controls. Among 20 miRNAs upregulated in the peripheral blood of cancer patients, only seven (Figure 3) demonstrated higher levels in tumours compared to adjacent normal mucosa (miR-130a, miR-331, miR-19a, miR-223, miR-106a, miR-21, and miR-374).
Figure 3.
Box plots of relative miRNA expression levels in tissues and peripheral blood. The statistical significance of differences was calculated using the Mann–Whitney U-test (*P<0.05). The line inside the boxes indicates median values with the upper and lower limits corresponding to the 75th and 25th percentiles, respectively. The upper and lower horizontal whiskers denote the 95th and 5th percentiles respectively.
Discussion
Circulating miRNAs found in all body fluids may potentially serve as clinically relevant biomarkers of human malignancies. Herein, it was found that serum levels of miRNAs detected in tumour-draining veins of the portal system poorly corresponded to molecules abundantly expressed by primary tumours of gastric cancer. Moreover, some potential biomarkers were significantly up-regulated in peripheral blood suggesting that these miRNAs are not released from the tumour.
The remarkable stability of miRNAs under various environmental conditions initiated extensive research to validate serum and plasma profiling in search of putative cancer biomarkers (Cortez et al, 2011). Such attempts were undertaken also for gastric cancer, suggesting that assays using blood miRNAs could markedly outperform currently available solutions (Zhu et al, 2014a; Zhou et al, 2015). However, the results of previous studies are highly inconsistent for most malignancies and question any physiological role of miRNAs. One of the potential explanations for this discrepancy is the suspicion that potential biomarker candidates in fact originate from other sources than the primary tumour, such as normal tissues of other organs or blood cells (Pigati et al, 2010; Ribeiro-dos-Santos et al, 2010).
Comprehensive expression profiling of miRNAs in the current study based on a well-defined population of Western patients with gastric cancer provides a unique opportunity to study the origins of potential serum biomarkers. Initially, 20 of 434 miRNA sequences identified in peripheral blood serum were significantly up-regulated in cancer patients. However, only seven of them showed higher levels in primary tumours compared to adjacent normal mucosa. Moreover, two potential biomarkers (miR-331 and miR-21) demonstrated higher expression in peripheral blood than in the tumour-draining vein. Altogether, our results strongly suggest non-tumour origins for the majority of circulating miRNA biomarkers. To our knowledge no previous study has evaluated the correlation between potential miRNA biomarkers of gastric cancer in tumour tissue with reference to portal and peripheral blood.
Early experiments have demonstrated that miRNAs originating from human cancer xenografts enter the circulation and can be readily measured in plasma (Mitchell et al, 2008). However, in the mice model of gastric cancer, the comparison of tissue and serum samples showed that only some miRNAs differentiating cancer and normal tissues were overexpressed in serum (Rotkrua et al, 2013). Human observations supporting such animal data are scarce as even though some human studies correlated tissue and serum/plasma levels of miRNAs, such assays were often carried out in separate sets of patients and only a few reported results on a within-patient basis. One such study examined levels of miRNAs identified as significantly elevated (miR-223 and miR-21) and decreased (miR-218) in plasma of gastric cancer compared to healthy controls in 8 pairs of primary tumours and adjacent normal tissue (Li et al, 2012). The expression of miR-223 and miR-21 was higher in seven and eight, respectively, of eight cancer tissues than in controls, whereas miR-218 was lower in seven of eight patients. A similar experimental procedure was carried out in four pairs of primary tumours and healthy adjacent tissues with miR-378 identified as significantly elevated (threefold) in serum of cancer patients compared to controls (Liu et al, 2012). However, contrary to the expectations, tissue levels of miR-378 were significantly lower in primary tumours than in healthy tissues. Another study examined miR-451 and miR-486 identified as diagnostic biomarkers for gastric cancer based on pre- and post-operative paired plasma samples (Konishi et al, 2012). Despite high diagnostic sensitivity and specificity of 96% and 100%, respectively, for detecting GC compared to healthy controls, the miR-451 expression was found to be significantly lower in primary tumours than in surrounding normal tissues, while miR-486 expression was comparable. Altogether, these results imply that the circulating pool of miRNA biomarkers of gastric cancer originates not only from the primary tumour but also may reflect a yet unknown biological phenomenon elicited by the presence of cancer.
It is still poorly understood why miRNAs with a relatively low expression in primary tumours are abundantly expressed in plasma or serum of GC patients compared to healthy controls. There are at least two likely explanations. First, previous observations in some other malignancies, such as breast cancer, showed that only a few differentially expressed miRNAs were common to tumours and serum and the potential biomarkers were selectively released to circulation (Chan et al, 2013; Zhu et al, 2014b). Second, circulating miRNAs might also originate from non-tumour tissues. Such a mechanism could explain our observations related to miR-21 and miR-331 that achieved markedly higher expression levels in peripheral blood than in the vein draining the primary tumour. The plausibility of this phenomenon is further substantiated by experiments carried out with animal models and clinical observations of breast cancer where some of extracellular miRNA was derived from normal epithelial cells (Pigati et al, 2010).
Aberrant expression of miR-21 was identified in tissue and blood of various malignancies, suggesting its potential role as a diagnostic and prognostic biomarker (Yin et al, 2015). This was likely related to the oncogenic activity of miR-21 that can stimulate proliferation, migration, and invasion of cancer cells, including gastric cancer (Sekar et al, 2016). In the latter case, the molecule has been demonstrated to modulate the expression of multiple genes, such as epidermal growth factor receptor (EGFR), phosphatase and tensin homologue (PTEN), programmed cell death 4 (PDCD4), Fas ligand (FASLG), and reversion-inducing cysteine-rich protein with kazal motifs (RECK). Contrary to miR-21, miR-331 has been much less extensively studied. Therefore, the actual function of the molecule is somehow unclear as previous experiments suggested its ability to promote proliferation and metastasis (Chang et al, 2014), as well as inhibit cell growth and promote apoptosis (Zhao et al, 2016). Nevertheless, it was reported as a circulating biomarker for several tumours, including hepatocellular carcinoma (Chen et al, 2015), laryngeal cancer (Ayaz et al, 2013), and colorectal adenomas (Kanaan et al, 2013). In the current study, the exact significance of increased expression levels for miR-21 and miR-331 found in peripheral blood compared to tumour-draining veins is unclear, but suggests that some diagnostic miRNA biomarkers may actually originate from tissues distant to the primary tumour.
Despite a moderate sample size, our results provide an important framework for further studies focused on blood miRNome of gastrointestinal malignancies to elucidate the origins of cancer-associated biomarker miRNAs. However, some important limitations should be considered. First, an appropriate normalisation strategy is a key step for the accurate relative quantification of miRNA expression levels with qRT–PCR. However, there are no generally accepted internal controls for plasma/serum miRNA measurement to date, and the discussion over selection of the most appropriate reference genes in miRNA profiling studies is ongoing (Mestdagh et al, 2009; Kok et al, 2015). To minimise the potential bias of using inadequate controls we have tested all currently recommended normalisation strategies (endogenous and exogenous controls, global normalisation) and accordingly selected the most suitable approach. Second, we have evaluated only serum miRNome without molecules present in plasma or bound to membrane vesicles. Although some minor differences in plasma and serum pools of miRNAs have been demonstrated, they seem unlikely to affect identification of potential biomarkers. Moreover, the selection of serum could be more relevant for clinical applications. Third, no laser microdissection was used to selectively isolate cancer cells for subsequent miRNA profiling, but microscopic validation was used to confirm representative components of primary tumours. Although the former method is more specific for isolation of cancer cells, the latter more likely reflects the heterogeneity of cellular components of the primary tumours and their miRNA contents. Finally, differences in miRNA expression patents within the primary tumour that were not evaluated in the present study could potentially affect our results.
In summary, we conducted a comprehensive miRNA profiling of primary gastric cancer and adjacent healthy tissues combined with serum samples of portal and peripheral blood to investigate the potential diagnostic biomarkers. Poor correlations between differentially expressed miRNAs in tissues and blood suggest that a marked proportion of potential biomarker molecules originate from tissues other than the primary tumour, reflecting a yet unknown biological phenomenon. This has important implications for further studies to elucidate the role and origins of circulating cancer-associated miRNAs, and should be validated by other research groups.
Acknowledgments
This study was supported by the Polish National Science Centre, grants Nos. DEC-2012/05/B/NZ4/02661 and DEC-2012/07/B/NZ5/02743, and the Leading National Research Centre (KNOW) of the Medical Faculty, Jagiellonian University Medical College.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
The authors declare no conflict of interest.
Supplementary Material
References
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M (2011) Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA 108: 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz L, Görür A, Yaroğlu HY, Ozcan C, Tamer L (2013) Differential expression of microRNAs in plasma of patients with laryngeal squamous cell carcinoma: potential early-detection markers for laryngeal squamous cell carcinoma. J Cancer Res Clin Oncol 139: 1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bala S, Csak T, Momen-Heravi F, Lippai D, Kodys K, Catalano D, Satishchandran A, Ambros V, Szabo G (2015) Biodistribution and function of extracellular miRNA-155 in mice. Sci Rep 5: 10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Krieger AM, Yekutieli D (2006) Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93: 491–507. [Google Scholar]
- Bosman FT, Carneiro F, Hruban RH, Theise ND (2010) WHO Classification of Tumours of the Digestive System 4th edn. World Health Organization: Lyon, France. [Google Scholar]
- Chan M, Liaw CS, Ji SM, Tan HH, Wong CY, Thike AA, Tan PH, Ho GH, Lee AS (2013) Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res 19: 4477–4487. [DOI] [PubMed] [Google Scholar]
- Chang RM, Yang H, Fang F, Xu JF, Yang LY (2014) MicroRNA-331-3p promotes proliferation and metastasis of hepatocellular carcinoma by targeting PH domain and leucine-rich repeat protein phosphatase. Hepatology 60: 1251–1263. [DOI] [PubMed] [Google Scholar]
- Chen L, Chu F, Cao Y, Shao J, Wang F (2015) Serum miR-182 and miR-331-3p as diagnostic and prognostic markers in patients with hepatocellular carcinoma. Tumour Biol 36: 7439–7447. [DOI] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY (2008) Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18: 997–1006. [DOI] [PubMed] [Google Scholar]
- Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA (2011) MicroRNAs in body fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol 8: 467–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edge S, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A (2011) AJCC cancer staging manual 7th edn. Springer: Chicago, USA. [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136: E359–E386. [DOI] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oles AK, Pages H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M (2015) Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Takahashi Y, Nishikawa M, Kato K, Morishita M, Yamashita T, Matsumoto A, Charoenviriyakul C, Takakura Y (2015) Macrophage-dependent clearance of systemically administered B16BL6-derived exosomes from the blood circulation in mice. J Extracell Vesicles 4: 26238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaan Z, Roberts H, Eichenberger MR, Billeter A, Ocheretner G, Pan J, Rai SN, Jorden J, Williford A, Galandiuk S (2013) A plasma microRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer. Ann Surg 258: 400–408. [DOI] [PubMed] [Google Scholar]
- Kok MG, Halliani A, Moerland PD, Meijers JC, Creemers EE, Pinto-Sietsma SJ (2015) normalisation panels for the reliable quantification of circulating microRNAs by RT-qPCR. FASEB J 29: 3853–3862. [DOI] [PubMed] [Google Scholar]
- Konishi H, Ichikawa D, Komatsu S, Shiozaki A, Tsujiura M, Takeshita H, Morimura R, Nagata H, Arita T, Kawaguchi T, Hirashima S, Fujiwara H, Okamoto K, Otsuji E (2012) Detection of gastric cancer-associated microRNAs on microRNA microarray comparing pre- and post-operative plasma. Br J Cancer 106: 740–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T (2010) Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem 285: 17442–17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BS, Zhao YL, Guo G, Li W, Zhu ED, Luo X, Mao XH, Zou QM, Yu PW, Zuo QF, Li N, Tang B, Liu KY, Xiao B (2012) Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS ONE 7: e41629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhu L, Liu B, Yang L, Meng X, Zhang W, Ma Y, Xiao H (2012) Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett 316: 196–203. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J (2009) A novel and universal method for microRNA RT-qPCR data normalisation. Genome Biol 10: R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105: 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, Hastings ML, Duelli DM (2010) Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One 5: e13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CC, Cheng HH, Tewari M (2012. a) MicroRNA profiling: approaches and considerations. Nat Rev Genet 13: 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, Tait JF, Tewari M (2012. b) Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 5: 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-dos-Santos A, Khayat AS, Silva A, Alencar DO, Lobato J, Luz L, Pinheiro DG, Varuzza L, Assumpcao M, Assumpcao P, Santos S, Zanette DL, Silva WA Jr, Burbano R, Darnet S (2010) Ultra-deep sequencing reveals the microRNA expression pattern of the human stomach. PLoS ONE 5: e13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotkrua P, Shimada S, Mogushi K, Akiyama Y, Tanaka H, Yuasa Y (2013) Circulating microRNAs as biomarkers for early detection of diffuse-type gastric cancer using a mouse model. Br J Cancer 108: 932–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar D, Krishnan R, Thirugnanasambantham K, Rajasekaran B, Islam VI, Sekar P (2016) Significance of microRNA 21 in gastric cancer. Clin Res Hepatol Gastroenterol 40: 538–545. [DOI] [PubMed] [Google Scholar]
- Turchinovich A, Weiz L, Langheinz A, Burwinkel B (2011) Characterization of extracellular circulating microRNA. Nucleic Acids Res 39: 7223–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H (2016) Gastric cancer. Lancet 388: 2654–2664. [DOI] [PubMed] [Google Scholar]
- Veitch AM, Uedo N, Yao K, East JE (2015) Optimizing early upper gastrointestinal cancer detection at endoscopy. Nat Rev Gastroenterol Hepatol 12: 660–667. [DOI] [PubMed] [Google Scholar]
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT (2011) MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 13: 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulfken LM, Moritz R, Ohlmann C, Holdenrieder S, Jung V, Becker F, Herrmann E, Walgenbach-Brunagel G, von Ruecker A, Muller SC, Ellinger J (2011) MicroRNAs in renal cell carcinoma: diagnostic implications of serum miR-1233 levels. PLoS ONE 6: e25787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C, Zhou X, Dang Y, Yan J, Zhang G (2015) Potential role of circulating MiR-21 in the diagnosis and prognosis of digestive system cancer: a systematic review and meta-analysis. Medicine (Baltimore) 94: e2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Sui Y, Zheng X (2016) MiR-331-3p inhibits proliferation and promotes apoptosis by targeting HER2 through the PI3K/Akt and ERK1/2 pathways in colorectal cancer. Oncol Rep 35: 1075–1082. [DOI] [PubMed] [Google Scholar]
- Zhou X, Zhu W, Li H, Wen W, Cheng W, Wang F, Wu Y, Qi L, Fan Y, Chen Y, Ding Y, Xu J, Qian J, Huang Z, Wang T, Zhu D, Shu Y, Liu P (2015) Diagnostic value of a plasma microRNA signature in gastric cancer: a microRNA expression analysis. Sci Rep 5: 11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Ren C, Han J, Ding Y, Du J, Dai N, Dai J, Ma H, Hu Z, Shen H, Xu Y, Jin G (2014. a) A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br J Cancer 110: 2291–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zheng Z, Wang J, Sun J, Wang P, Cheng X, Fu L, Zhang L, Wang Z, Li Z (2014. b) Different miRNA expression profiles between human breast cancer tumors and serum. Front Genet 5: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.