Abstract
Cryptic female choice (CFC), a form of sexual selection during or post mating, describes processes of differential sperm utilization by females to bias fertilization outcomes towards certain males. In Chinook salmon (Oncorhynchus tshawytscha) the ovarian fluid surrounding the ova of a given female differently enhances the sperm velocity of males. Sperm velocity is a key ejaculate trait that determines fertilization success in externally fertilizing fishes, thus the differential effect on sperm velocity might bias male fertilization outcomes and represent a mechanism of CFC. Once sperm reach the oocyte, CFC could potentially be further facilitated by sperm–egg interactions, which are well understood in externally fertilizing marine invertebrates. Here, we explored the potential genetic basis of both possible mechanisms of CFC by examining whether the genotypic combinations of mates (amino-acid divergence, number of shared alleles) at the major histocompatibility complex (MHC) class I and II explain the variation in sperm velocity and/or male fertilization success that is not explained by sperm velocity, which might indicate MHC-based sperm–egg interactions. We recorded sperm velocity in ovarian fluid, employed paired-male fertilization trials and evaluated the fertilization success of each male using microsatellite-based paternity assignment. We showed that relative sperm velocity was positively correlated with fertilization success, confirming that the differential effect on sperm velocity may be a mechanism of CFC in Chinook salmon. The variation in sperm velocity was independent of MHC class I and II. However, the MHC class II divergence of mates explained fertilization success, indicating that this locus might influence sperm–egg interactions.
Introduction
In many species, ejaculates from several males are present during the time of fertilization, which creates the opportunity for sexual selection to continue post mating. There are two forms of sexual selection post mating: sperm competition and cryptic female choice (CFC). Although the former entails the competition among sperm to achieve fertilization of the ova (Parker, 1970), CFC comprises processes that bias fertilization success towards sperm from certain males in order to increase female reproductive success (Thornhill and Alcock, 1983; Eberhard, 1996; Birkhead, 1998).
There is evidence for two mechanisms of CFC in externally fertilizing species: one facilitated by sperm–ovarian fluid interactions, the other by interactions between the sperm and egg. Prior work in several fish species has shown that ovarian fluid, a viscous substance surrounding the spawned ova, enhances sperm longevity, per cent motility, path linearity and sperm velocity compared to water (for example, Litvak and Trippel, 1998; Turner and Montgomerie, 2002; Rosengrave et al., 2008; Galvano et al., 2013). The degree of stimulation of sperm velocity by the ovarian fluid is greatly explained by male–female interactions, which is indicative of the presence of CFC acting to promote gamete fusions of particular female–male combinations (Urbach et al., 2005; Rosengrave et al., 2008). Because sperm velocity is positively linked with fertilization success in fish (Gage et al., 2004; Boschetto et al., 2011; Evans et al., 2013; Rosengrave et al., 2016), it is possible that the differential stimulation of sperm velocity by ovarian fluid represents a mechanism of CFC (Rosengrave et al., 2008). Evidence for a second mechanism of CFC in external fertilizers via gamete interactions derives from studies of marine invertebrates, including tunicates (Scofield et al., 1982), sea urchins (Palumbi, 1999), abalone (Swanson et al., 2001), where proteins on the gamete surfaces facilitate non-random sperm–egg fusions. Similar gamete interactions likely exist in fish (Yeates et al., 2009), where sperm might interact with the inner canal walls of the micropyle (Hart, 1990).
To date, it is not fully known what individual specific component(s) may have the ability to govern the apparent sperm–ovarian fluid interactions and non-random gamete fusions in external fertilizers. However, genetic compatibility of mates appears to play a role in post-mating processes. In the guppy, sperm velocity in the ovarian fluid and fertilization success were higher for males unrelated to the female than for related (full-sib) males (Gasparini and Pilastro, 2011). Fertilization experiments in Arctic charr (Liljedal et al., 2008) revealed a higher fertilization success for males genetically less similar to the female, while in the Peron’s tree frog (Sherman et al., 2008) fertilization success was biased towards males genetically more similar to the female. In contrast, in Lake Trout, sperm velocity was lower for males unrelated to the female than for related (full-sib) males (Butts et al., 2012). Possibly, selection might have promoted alleles enabling an avoidance of genetically incompatible matings (Tregenza and Wedell, 2000). A major target in that respect are the genes of the major histocompatibility complex (MHC).
The MHC is a highly polymorphic, multi-gene family found in all jawed vertebrates and it is mediating an immune response (Kelley et al., 2005, for a review). The MHC consists of three subfamilies (classes) of which class I and II play important roles in the antigen presenting process (Kelley et al., 2005). The genes of these two classes code for glycoproteins that bind antigens and present these on the cell surface to be recognized by T-cells, which subsequently initiate an immune response cascade (Janeway et al., 2001; Bernatchez and Landry, 2003).
Studies of female choice prior to mating suggest that the MHC has also a function in sexual selection (for examples, see Potts et al., 1991; Wedekind et al., 1995). MHC-based sexual selection has two possible benefits. First, the MHC appears to have a role in kin recognition (Potts et al., 1991; Manning et al., 1992; Olsen et al., 1998) and thus, MHC-based sexual selection is thought to avoid inbreeding (Grob et al., 1997). Second, females may improve the immunological competence of their offspring by choosing mates with a MHC genotype compatible to their own (for example, Yamazaki et al., 1976; Eklund, 1997).
The MHC might also play a role in sexual selection post mating. There is some evidence for the expression of MHC class I and/or class II genes in male gametes of mice and humans (Fernandez et al., 1999, for a review) and Wedekind et al. (1996) proposed that oocytes might possess receptors to recognize a sperm’s MHC genotype. Potentially, MHC-dependent CFC could also arise after the sperm has entered the egg and before the pronucleus fusion (Yeates et al., 2009). MHC-based CFC was uncovered in the red jungle fowl (Løvlie et al., 2013) and in vitro fertilization experiments in MHC-congenic mice revealed an MHC-dependent gamete fusion (Wedekind et al., 1996; Rülicke et al., 1998).
Among external fertilizers, pacific salmon, such as the Chinook salmon (Oncorhynchus tshawytscha), are ideal organisms to study CFC for genetic benefits like MHC compatibility. Pacific salmon exhibit a non-resource based mating system, that is, females do not receive direct benefits (Foote, 1989). Species lacking forms of direct contribution to reproductive success especially are thought to sexually select mates for indirect (genetic) benefits (Searcy, 1982).
In in vitro fertilization experiments in externally fertilizing fish MHC-dependent (Yeates et al., 2009) and MHC-independent fertilization outcomes (Wedekind et al., 2004; Skarstein et al., 2005) were observed. These inconsistent results might have been due to different experimental designs used in these studies. Specifically, Yeates et al. (2009) used paired-male fertilization trials, which more accurately reflect natural spawning conditions in salmonids (Fleming, 1998), while other studies used single-male (no choice) fertilization trials (Wedekind et al., 2004; Skarstein et al., 2005). Furthermore, each of these studies examined the impact of only one MHC locus on fertilization outcome. To our knowledge, there is no study that explored the impact of the MHC on sperm velocity in ovarian fluid and on fertilization success and thus, it is yet impossible to draw conclusions on the mechanism by which the MHC might bias fertilization outcomes.
In this study, we first explored whether the differential sperm velocity of males in the ovarian fluid of a focal female predicts fertilization success in Chinook salmon and thus, may represent a mechanism of CFC. We then examined whether the sperm velocity in ovarian fluid is explained by the divergence of mates at MHC class I and II. Finally, to explore whether the MHC might play a role in gamete interactions, we tested whether the variation in fertilization success that could not be explained by the different sperm velocity of males, was explained by the MHC divergence of mates. For this, we have performed a series of paired-male fertilization trials. We tested the hypotheses that sperm velocity and fertilization success are biased towards males with whom the focal female would produce i) heterozygous offspring, which we assessed by the number of shared MHC alleles between mates and/or ii) offspring with the highest allelic divergence at MHC loci in order to improve the immunological competence of her offspring.
Based on cryptic preference for males more similar to the female at the MHC class I in Atlantic salmon (Yeates et al., 2009), we might find similar correlations in Chinook salmon. Neff et al. (2008) detected that females produced more offspring with MHC class II dissimilar males in Chinook salmon but since individuals were allowed to spawn without interference, it was impossible to disentangle female choice prior to and post mating. However, it allows to hypothesize that CFC might be biased towards MHC class II dissimilar males
Materials and methods
Fish sampling
Chinook salmon were caught over their annual spawning run (April-May 2010 and 2011) in a trap located on the Kaiapoi River, a tributary of the Waimakariri River system, Canterbury, New Zealand (Unwin et al., 2000). We collected sexually mature 3-year-old females and 2-year-old 'hooknose' males, that were then individually tagged and maintained in a river water hatchery raceway using standard husbandry procedures at Salmon Smolt NZ, Canterbury, New Zealand (Rosengrave et al., 2008; Evans et al., 2013). A small fin clip was taken from each fish and stored in 95% ethanol for DNA extraction. All animals were collected and maintained using protocols approved by the Animal Ethics Committee for the University of Otago, New Zealand.
Experimental overview
To investigate whether the MHC genotypes of mates impact CFC, we performed paired-male fertilization trials, in which milt from two males was simultaneously added to a focal female’s batch of eggs. In order to test whether a difference in the males’ fertilization success can be attributed to a difference in sperm swimming speed, we recorded the sperm velocity of each male in the ovarian fluid of the female. All experiments were performed within four to six hours of gamete collection. The experiments were performed with a total of 37 fish; that is with 10 females (4 in season 2010, 6 in 2011) and 27 males (12 in 2010, 15 in 2011). Our experimental design was partially crossed, that is, some males were used in more than 1 female (10 males were used in only 1 female, 6 in 2, 9 in 3 and 2 in 4 females). However, each trial represents a unique combination of individuals. This partially crossed design, although strongly driven by the availability of fish, represents a compromise approach to two goals: to best distinguish the effect of the MHC from the effect of the genomic background on CFC, it is best to use males in only one female. However, to best distinguish the female effects, male effect and the female–male interaction on CFC, it is best to employ a fully-crossed design, that is, to re-use all males across all females. Hence, our partially crossed design, represents a middle course to examine the complex coherences of CFC.
Assessment of sperm velocity in ovarian fluid
Milt was collected by careful stripping of randomly chosen males. To prevent activation of the sperm by contact with water or urine prior to any experiments, males were dried around the cloaca. We measured the sperm velocity of six randomly collected males in the ovarian fluid from the focal female using a CEROS sperm tracker (v. 12, Hamilton Thorne Research, Beverly, MA, USA). Sperm velocity was determined as previously described (for example, Rosengrave et al., 2008; Evans et al., 2013) with minor modifications. In brief, milt was pipetted onto a 20 μl Leja slide (Leja Products BV, Nieuw-Vennep, The Netherlands) containing ovarian fluid of the focal female. We used the Average Path Velocity (VAP in μm/s), which estimates the average velocity of a sperm cell over 0.5 s over a smoothed cell path and was used to assess sperm velocity in related studies (for example, Rosengrave et al., 2008; Evans et al., 2013). Across all males, sperm recordings were taken at 10 s post activation of the spermatozoa in ovarian fluid, which is as soon as technically possible, as measurements later post activation might be confounded by varying sperm longevity across males. Sperm velocity of each male in a given female’s ovarian fluid was measured twice. Ovarian fluid is very viscous and fertilization takes place within a very limited time window, thus it is possible that the concentration of ovarian fluid is close to 100% near the egg surface, reducing with distance from the egg mass. There is likely no one concentration that best resembles natural spawning conditions, but we expect that sperm swim up a gradient of ovarian fluid when moving towards the egg. We examined sperm velocity in two different concentrations of ovarian fluid, that is in 50% and 100% in the years 2010 and 2011, respectively. We controlled for the difference in ovarian fluid concentrations and year in our analysis (see statistical analysis of sperm velocity).
Paired-male fertilization trials
In every female, we used 9 batches of eggs by sampling nine times 50 ml of the ova, which corresponds to ~100 eggs and 25 ml of ovarian fluid per batch. Each batch was fertilized with the sperm of two males, that is, we performed nine different fertilization trials per female using nine combinations of two males. For all ten females, this resulted in a total of 90 trials. To avoid the effects of CFC being potentially masked by sperm competition, we precautionary controlled for sperm number, a factor known to have a key role in sperm competition in the bluegill sunfish (Neff et al., 2003; Stoltz and Neff, 2006), but possibly not crucial in salmonids (Gage et al., 2004), by determining sperm densities using an improved Neubauer haemocytometer. We then added respective volumes of milt containing 1 × 108 of spermatozoa from each male (2.1–9.8 μl) to the given egg batch. Fertilizations were performed in a plastic beaker containing the focal female's egg batch. Sperm of each male was pipetted simultaneously but separately to the eggs in a stream of 250 ml river water to simulate natural spawning (corresponding to an ovarian fluid:water mixture of 1:10). The sperm–egg mixtures were left untouched for five minutes in a dark environment to ensure that the fertilization process took place undisturbed. Then the mixtures were transferred into a vertical incubation stack, in which fertilized egg batches were reared separately under conditions imitating the natural conditions: constant flowing river water at 12–12.5 °C and no exposure to sunlight (Billard and Jensen, 1996).
Single-male control trials. To ensure that no infertile male or sperm that may have been activated via contact with water during milt sampling confounds our results, we performed single-male control trials of each male and the female he was used in for our paired-male trials. We used a lower amount of sperm (1 × 105 sperm) to prevent fertilization rates being forced to 100% in spite of a potentially low proportion of viable spermatozoa. We found that all males had generally fertile sperm (fertilization rates 57%±4 (s.e.) of ~100 eggs) except for two males (fertilization rates of 1.5%) that have been excluded from further analysis. We therefore analyzed 84 of the initial 90 paired-male trials.
Microsatellite-based paternity assignment
DNA was extracted from whole embryos and from the dorsal fin clips of each parental fish using a standard Chelex-protocol (Walsh et al., 1991). A microsatellite-based paternity test was then performed on 48 randomly chosen offspring from each paired-male fertilization trial. Offspring were sampled when they reached the eyed stage of embryonic development; approximately five weeks post fertilization. For all 84 fertilization trials, a total of 4032 offspring were genotyped and assigned to a sire. The offspring and parental individuals were genotyped at nine microsatellite markers (Ocl-1 (Condrey and Bentzen, 1998), Omy-325, Ssa-85 ( Heath et al., 2002), Ots-101, Ots-104, Ots-107, Ots-2, Ots-3 (Beacham et al., 2009), Ssa197 (Banks et al., 1999)) and data were used to assign the offspring to either of the two competing males. The parentage assignment was performed using the maximum likelihood approach in the program CERVUS 3.0 ((Kalinowski et al., 2007), http://www.fieldgenetics.com) with a 95% confidence interval and an error rate 0.003.
MHC genotyping of parental individuals
MHC amplification
All 37 parental individuals were genotyped at the MHC class I A1 and class II B1 loci, which code for the α1- and β1-domain at each receptor, respectively. These domains contribute one of the two components constituting the peptide-binding region (PBR) and thus, functional changes would be expected in these regions. Locus-specific primers for MHC class I A1 and class II B1 and amplification conditions are described in Grimholt et al. (1993) and Hordvik et al. (1993), respectively. Briefly, MHC loci were amplified separately by PCR of genomic DNA in 30 μl reaction volume consisting of 20 ng DNA, 1 mm PCR buffer, 1.5 mm MgCl2, 0.2 nmol dNTPs, 0.3 μm of each primer, 4 mm Tetramethyl ammonium chloride and 0.2 units of Taq-Polymerase. PCR cycling conditions according to Miller and Withler (1997) were modified to 35 cycles: 30 s at 94 °C denaturation followed by 1 min at 50 °C for annealing and 1 min at 70 °C for elongation.
MHC sequencing
PCR products were verified on 1% agarose gels stained by SYBR Safe and then purified using AcroPrep Filter plates (Pall Corporation, New York, NY, USA). Purified PCR products were sequenced on an ABI 3730xl DNA Analyser (Applied Biosystems, Foster City, CA, USA) at the Genetic Analysis Services (University of Otago, New Zealand). For a subset of 10 individuals, three replicates from independent PCRs were sequenced to verify the repeatability of the sequencing results. In addition, purified PCR products for both MHC class I and II loci of seven individuals were cloned and Sanger sequenced at the Central Analytical Facility (Stellenbosch University, South Africa) and Macrogen, Inc. (Seoul, South Korea) to verify that a maximum of two alleles per individual are amplified, which indicates locus-specificity of the primer. Ten clones per amplicon were sequenced in both forward and reverse directions.
All sequences were analyzed with the software Sequencher 4.8 (Gene Code Corporation, Ann Arbor, MI, USA) and checked by an alignment to Chinook salmon MHC sequences available on GenBank using the BLAST alignment program (Altschul et al., 1997).
MHC haplotype reconstruction
The MHC haplotypes were reconstructed using the program PHASE 2.1 (Stephens et al., 2001; Stephens and Donnelly, 2003). SeqPhase (Flot, 2010) was used to prepare the input files and convert the PHASE output files into fasta formats. To increase the precision of phasing, we used haplotype information from unambiguous homozygous and cloned individuals.
MHC divergence of mates
MHC amino-acid divergence at the whole exon and at the PBR
The amino-acid divergence between mates at each MHC locus was estimated using a substitution model. Substitution models calculate the evolutionary distance between two sequences of interest. The divergence (D) between a mated female (for example, alleles a and b) and male (for example, alleles c and d) was estimated in a pairwise allelic manner, resulting in four values per mating pair (Dac, Dad, Dbc, Dbd). For each MHC locus, the amino-acid divergence of the whole exon was calculated using a suitable substitution model that was selected based on the Akaike Information Criterion (AIC) in ProtTest (Abascal et al., 2005). We used the Jones-Taylor-Thornton (JTT) +I+G substitution model (Jones et al., 1992) at MHC class I and the JTT at MHC class II, which were computed in protdist of the PHYLIP 3.67 package (Felsenstein, 1989, 1993) and in MEGA 5 (Tamura et al., 2011), respectively. The JTT model estimates the evolutionary distance of two sequences considering different probabilities of change from one amino-acid to another (Jones et al., 1992). The JTT+I+G models a proportion of invariant sites (+I) and gamma distributed substitution rates across sites (+G).
Using the same models, we also calculated the amino-acid divergence of the putative peptide binding region (PBR) only. The putative PBR at MHC class I was previously determined in salmonid fish (Miller and Withler, 1998), while the PBR at MHC class II was only determined in humans (Brown et al., 1993). To locate the PBR in our Chinook salmon sequences (Supplementary Figure S1), the consensus sequence of the MHC class I and II derived from this study were aligned to the Chinook salmon MHC class I (GenBank accession: U80284.1) and to the human MHC class II homolog HLA-DRB1 (GenBank accession: AAB81176.1), respectively, using in Clustal Omega (Sievers et al., 2011).
Additionally, we calculated the MHC dissimilarity of mates by counting the number of different amino acids in a pairwise allelic manner. Finally, the four values of divergence at the whole exon and PBR were summed up as performed in Landry et al. (2001) to obtain one value between two mated individuals (D=Dac+Dad+Dbc+Dbd).
To distinguish MHC loci, the amino-acid divergence at the whole exon is denoted as AAI and AAII and PBRI and PBRII refers to the PBR.
Number of MHC alleles shared between mates
For each locus, the number of alleles shared between mates (Mxy) was calculated according to Blouin et al. (1996), whereby two diploid individuals can share 0, 1 or 2 alleles at one locus. To distinguish MHC loci, the number of shared alleles is denoted as MxyI and MxyII.
Statistical analysis
Statistical analyses were carried out in the statistical computing environment R v. 3.0.1. (R Core Team, 2013). The variation in sperm velocity and fertilization success were analyzed using the MCMCglmm package (Hadfield, 2010), which implements Markov Chain Monte Carlo (MCMC) routines to fit generalized linear mixed models. We ran all MCMCglmm models with the settings of 1,300,000 iterations, a thinning interval of 1000 and 300000 burn-in. The model fit was assessed using trace and posterior density plots. We ensured the non-independence between samples in the chain (autocorrelation) was below 0.1, indicating a good mixing. For all models, the R2 value was calculated according to Nakagawa and Schielzeth (2013); note that when random slope models were used, we report R2 values from corresponding random-intercept models. The individual fish, females and males, and the female–male interaction were modeled as random effects. There was no significant within-seasonal effect on sperm velocity (pMCMC=0.11–0.23 across models) and fertilization success (pMCMC=0.28–0.63 across models), nor was there a seasonal effect on fertilization success (pMCMC=0.19–0.6 across models) when the date of experiment within a season and the year were modeled as fixed effects and thus, we did not include these parameters in our final model to avoid overparameterization. We report the lower and upper bound of the 95% credible interval (CI) as an indication of the uncertainty around our estimates of regression coefficients and variance components. For all models, we used both inverse gamma priors and parameter-expanded priors for random effects (Hadfield, 2014). The results from the parameter-expanded prior were reported because the results were qualitatively identical and quantitatively similar between corresponding models using the two priors and also because the gamma inverse prior can incorrectly influence random effect estimates, especially when sample sizes are small to moderate.
Analysis of the impact of MHC divergence between mates on sperm velocity
In order to analyze the variation in sperm velocity, the two recorded values of sperm velocity per male measured in a focal female’s ovarian fluid were used as the response variable (with a Gaussian error with the identity link). To control for the season and the potential effect of sperm velocity measured in 50% and 100% ovarian fluid in field seasons 2010 and 2011, respectively, the seasons were modeled as a fixed effect. This allowed us to (i) distinguish how much of the variation in sperm velocity is attributed to the change in concentration and how much is explained by other variance components (female–male interaction and so on) and (ii) to pool the data from both seasons for analysis. At each MHC locus, each type of MHC divergence between focal female and male (AAI, PBRI, MxyI, AAII/PBRII and MxyII) was analyzed in a separate model and fitted as fixed effect.
Analysis of the impact of MHC divergence between mates on fertilization success
The variation in male fertilization success was analyzed by modeling the relative fertilization success of each male in a paired-male trial (n=84), that is, we used the number of eggs fertilized by a male as count data and the total number of eggs fertilized by both males in a focal female was modeled as fixed effect. We modeled a MCMCglmm with a Poisson error distribution with the logit link. The divergence between mates at each MHC locus (AAI, PBRI, MxyI, AAII/PBRII and MxyII) were modeled as a fixed effect in separate models. The relative sperm velocity of each male, that is, his velocity relative to his competitor, was modeled as a fixed effect (binary predictor SF for ‘slow male’ or ‘fast male’ in a paired-male trial). ‘SF’ was also modeled as random slope for each ‘male-male combination’ (Male–Male Pair) as some males were used in more than one female and thus, they can be the ‘slower male’ or ‘faster male’ in trials depending on their competitor's speed in the focal female. We additionally modeled the z-transformed velocity data for each season as fixed effect, which confirmed results found using SF. Furthermore, we analyzed the effect of the difference in sperm velocity between paired males, and their difference in MHC divergence to the female on their different fertilization success. Finally, we determined the MHC genotypic dissimilarity of mates by counting the number of amino acids in which the mates differ and analyzed its effect on fertilization success.
Results
MHC genotypes of parental fish
MHC genotyping of all 37 parental individuals revealed 20 nucleotide sequence variants of which each translated into a different amino-acid sequence at MHC class I. Nine of these 20 alleles were newly discovered in this study (GenBank accession numbers: KT156756–KT156764) and 11 alleles have been previously described in other Chinook salmon populations (Miller et al., 1997 (U80284.1, U80278.1); Miller et al., 1999 (AF162869.1); Garrigan and Hedrick, 2001 (AF362113.1, AF362117.1–AF362119.1, AF362123.1 and AF362125.1); Evans et al., 2009 (GU989264.1 and GU989272.1)). At MHC class II, we identified three nucleotide sequences that resulted in three amino-acid sequences; one has been previously identified (Miller and Withler, 1996 (U34719.1)), the other two were newly identified in this study (KT156765, KT156766). Average amino-acid divergence of mates considering the whole exon at MHC class I (76 amino acids) was AAI=642±25.6 (s.e.) and PBRI=1532±48 (s.e.) at the PBR (putative PBR: 14 amino acids). At MHC class I, 125 mates shared no allele, 43 shared 1 and no mates shared both MHC alleles. At the MHC class II (83 amino acids), the average amino-acid divergence is AAII/PBRII=0.08±0.002 (s.e.) (putative PBR: 20 amino acids) and 29 mates share no allele, 72 mates share 1 and 67 mates share both alleles.
Sources of variation in sperm velocity in ovarian fluid
The mean sperm velocity was 61um/s±1.86 (s.e.) and 79um/s±2.29 (s.e.) when measured in spawning season in the years 2010 (50% ovarian fuid) and 2011 (100% ovarian fluid), respectively. We accounted for the difference between seasons (that is, year and ovarian fluid concentrations) in our models and found it explained a significant amount of the variation in sperm velocity (Table 1). The variation not explained by season is explained by the following parameters: across all models, female identity explained 8–11%, while the male effect was stronger and explained 21–24% of the variation in sperm velocity. The female–male interaction explained 42–45% of the variance in sperm velocity and thus, was the major variance component (Table 1).
Table 1. The influence of MHC genotypic combination of mates on male sperm velocity in ovarian fluid in Chinook salmon.
| Estimate | Variance (%) | Lower 95% CI | Upper 95% CI | pMCMC | |
|---|---|---|---|---|---|
| Fixed effects | |||||
| Intercept | 59.58 | 41.08 | 74.73 | ||
| AAI | 0 | −0.016 | 0.014 | 0.952 | |
| Season | 21.37 | 6.190 | 35.612 | 0.008 | |
| Random effects | |||||
| Female (10) | 47.05 | 8.03 | <0.001 | 212.770 | |
| Male (27) | 133.95 | 22.86 | <0.001 | 269.263 | |
| Female–Male (57) | 266.25 | 45.42 | 113.135 | 427.263 | |
| Residual | 138.93 | 23.70 | 99.881 | 184.834 | |
| Summary statistics | |||||
| Marginal R2 | 0.13 | ||||
| Conditional R2 | 0.80 | ||||
| Fixed effects | |||||
| Intercept | 56.13 | 37.867 | 72.534 | ||
| AAII/PBRII | 29.39 | −101.089 | 150.775 | 0.662 | |
| Season | 21.49 | 6.327 | 38.960 | 0.012 | |
| Random effects | |||||
| Female (10) | 52.99 | 9.08 | <0.001 | 205.096 | |
| Male (27) | 131.03 | 22.45 | <0.001 | 278.514 | |
| Female–Male (57) | 259.28 | 44.43 | 107.408 | 413.900 | |
| Residual | 140.29 | 24.04 | 101.970 | 186.994 | |
| Summary statistics | |||||
| Marginal R2 | 0.13 | ||||
| Conditional R2 | 0.80 | ||||
| Fixed effects | |||||
| Intercept | 62.67 | 40.074 | 81.172 | ||
| PBRI | 0.00 | −0.012 | 0.006 | 0.52 | |
| Season | 22.00 | 5.298 | 36.864 | 0.008 | |
| Random effects | |||||
| Female (10) | 66.89 | 11.48 | <0.001 | 240.959 | |
| Male (27) | 123.15 | 21.14 | <0.001 | 263.199 | |
| Female–Male (57) | 253.21 | 43.47 | 113.123 | 419.656 | |
| Residual | 139.15 | 23.91 | 97.164 | 188.349 | |
| Summary statistics | |||||
| Marginal R2 | 0.13 | ||||
| Conditional R2 | 0.79 | ||||
| Fixed effects | |||||
| Intercept | 58.68 | 46.396 | 72.988 | ||
| MxyI | 2.26 | −9.320 | 14.469 | 0.68 | |
| Season | 21.08 | 5.536 | 35.962 | 0.014 | |
| Random effects | |||||
| Female (10) | 45.11 | 7.82 | <0.001 | 215.784 | |
| Male (27) | 129.94 | 22.53 | <0.001 | 280.324 | |
| Female–Male (57) | 264.46 | 45.86 | 95.492 | 432.000 | |
| Residual | 137.18 | 23.79 | 98.802 | 183.879 | |
| Summary statistics | |||||
| Marginal R2 | 0.13 | ||||
| Conditional R2 | 0.80 | ||||
| Fixed effects | |||||
| Intercept | 65.339 | 51.112 | 81.668 | ||
| MxyII | −5.18 | −11.660 | 1.641 | 0.136 | |
| Season | 22.77 | 5.433 | 37.791 | 0.01 | |
| Random effects | |||||
| Female (10) | 57.04 | 9.95 | <0.001 | 231.801 | |
| Male (27) | 137.89 | 24.07 | 24.964 | 276.052 | |
| Female–Male (57) | 241.61 | 42.28 | 95.605 | 393.166 | |
| Residual | 136.30 | 23.79 | 97.052 | 181.299 | |
| Summary statistics | |||||
| Marginal R2 | 0.13 | ||||
| Conditional R2 | 0.80 | ||||
Abbreviations: CI, confidence interval; MHC, major histocompatibility complex; PBR, peptide-binding region.
MCMCglmm analyses yielded significant pMCMC values (in bold) of fixed effects (season, MHC divergence). The amount of variation explained by fixed effects (marginal R2) and by the whole model (conditional R2) are reported. The variance (%) explained by each random effect refers to the total variance by all random effects (female identity (n=10), male identity (n=27), female–male interaction (n=58) and the residual variance. Following estimates of divergence between mates at MHC class I and II were tested for an impact on sperm velocity in ovarian fluid: i) divergence at the peptide sequence at the whole exon (denoted AAI and AAII), ii) divergence at the putative sites involved in peptide binding only (PBRI and PBRII) and iii) number of MHC alleles shared between mates (MxyI and MxyII). As all polymorphic sites at MHC class I are part of the PBR, divergence at the whole exon and the PBR are identical and thus, not listed separately.
We found that divergence among mates based on the amino acid sequence of the whole exon at MHC class I and II (denoted AAI and AAII) had no significant influence on the variation in sperm velocity by ovarian fluid (PAAI=0.95 and PAAII/PBRII=0.66). Also the divergence at the PBR for both MHC loci (denoted PBRI and PBRII) did not predict sperm velocity (PPBRI=0.52). Note that AAII and PBRII are identical as all variable sites in our population contribute to the putative PBR at MHC class II locus (Supplementary Figure S1). Finally, the number of MHC alleles shared between mates at either locus (denoted MxyI and MxyII) had also no impact on sperm velocity (PMxyI=0.68, PMxyII=0.14).
Analysis of the variation in fertilization success
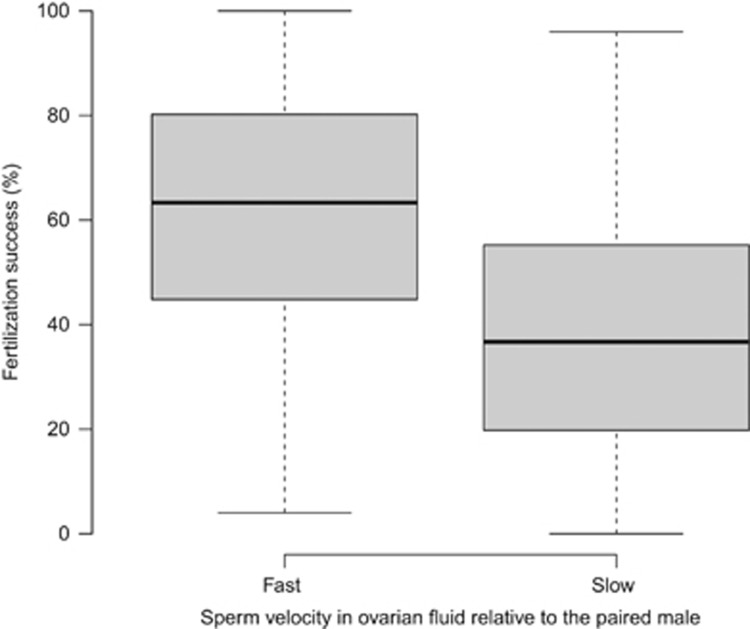
The total number of eggs in each batch used for our paired-male trials (n=84) was on average 115 (range 108–122 per batch). Of the 48 randomly sampled offspring per batch, on average 47 could be successfully assigned to a sire. The fertilization success of a male (n=27) in a paired-male trial is the proportion of the 48 offspring sired by the given male and ranged from 0 to 100%. Our MCMCglmm analysis revealed that a male's fertilization success depended significantly on his sperm velocity relative to his competitor (PSF=<0.001, Table 2), whereby the male with the higher sperm velocity achieved the greater fertilization success (Figure 1). The effect of relative sperm velocity on fertilization success was confirmed by additional analyses when the z-transformed velocity data (Supplementary Table S1) and the difference between males in sperm velocity (Supplementary Table S2 and Supplementary Figure S2) were modeled. Across models, we found that female and male identity explained only 0.87–4.6% of the variation in fertilization success (Table 2).
Table 2. The impact of MHC genotypic combination of mates on fertilization success in Chinook salmon was analyzed using MCMCglmms.
| Estimate | Variance (%) | Lower 95% CI | Upper 95% CI | pMCMC | |
|---|---|---|---|---|---|
| Fixed effects | |||||
| Intercept | 1.34 | 0.265 | 1.993 | ||
| AAI | <0.001 | −0.001 | <0.001 | 0.728 | |
| SF | 0.445 | 0.977 | 0.794 | <0.001 | |
| Random effects | |||||
| Female (10) | 0.004 | 0.87 | <0.001 | 0.045 | |
| Male (27) | 0.016 | 3.46 | <0.001 | 0.045 | |
| Male–Male Pair (84) | 0.439 | 95.02 | 0.269 | 0.689 | |
| SF (random slope) | 1.036 | 0.642 | 1.463 | ||
| Additive overdispersion | 0.003 | 0.65 | <0.001 | 0.007 | |
| Summary statistics | |||||
| Marginal R2 | 0.12 | ||||
| Conditional R2 | 0.39 | ||||
| Fixed effects | |||||
| Intercept | 1.416 | 0.514 | 2.435 | ||
| AAII/PBRII | −2.063 | −3.747 | −0.017 | 0.03 | |
| SF | 0.557 | 0.323 | 0.806 | <0.001 | |
| Random effects | |||||
| Female (10) | 0.008 | 1.74 | <0.001 | 0.029 | |
| Male (27) | 0.021 | 4.58 | <0.001 | 0.052 | |
| Male–Male Pair (84) | 0.427 | 93.03 | 0.269 | 0.598 | |
| SF (random slope) | 1.159 | 0.795 | 1.591 | ||
| Additive overdispersion | 0.003 | 0.65 | <0.001 | 0.008 | |
| Summary statistics | |||||
| Marginal R2 | 0.13 | ||||
| Conditional R2 | 0.43 | ||||
| Fixed effects | |||||
| Intercept | 1.218 | 0.152 | 2.275 | ||
| PBRI | <0.001 | −0.001 | <0.001 | 0.736 | |
| SF | 0.535 | 0.290 | <0.001 | <0.001 | |
| Random effects | |||||
| Female (10) | 0.008 | 1.66 | <0.001 | 0.032 | |
| Male (27) | 0.015 | 3.11 | <0.001 | 0.039 | |
| Male–Male Pair (84) | 0.457 | 94.62 | 0.290 | 0.626 | |
| SF (random slope) | 1.223 | 0.841 | 1.677 | ||
| Additive overdispersion | 0.003 | 0.62 | <0.001 | 0.009 | |
| Summary statistics | |||||
| Marginal R2 | 0.08 | ||||
| Conditional R2 | 0.42 | ||||
| Fixed effects | |||||
| Intercept | 1.227 | 0.243 | 2.239 | ||
| MxyI | 0.052 | −0.093 | 0.186 | 0.456 | |
| SF | 0.538 | 0.276 | 0.783 | <0.001 | |
| Random effects | |||||
| Female (10) | 0.008 | 1.68 | <0.001 | 0.029 | |
| Male (27) | 0.015 | 3.16 | <0.001 | 0.040 | |
| Male–Male Pair (84) | 0.449 | 94.53 | 0.286 | 0.636 | |
| SF (random slope) | 1.211 | 0.805 | 1.661 | ||
| Additive overdispersion | 0.003 | 0.63 | <0.001 | 0.008 | |
| Summary statistics | |||||
| Marginal R2 | 0.08 | ||||
| Conditional R2 | 0.39 | ||||
| Fixed effects | |||||
| Intercept | 1.141 | 0.199 | 2.151 | ||
| MxyII | 0.134 | 0.037 | 0.256 | 0.008 | |
| SF | 0.569 | 0.297 | 0.829 | <0.001 | |
| Random effects | |||||
| Female (10) | 0.017 | 3.67 | <0.001 | 0.064 | |
| Male (27) | 0.021 | 4.54 | <0.001 | 0.052 | |
| Male–Male Pair (84) | 0.422 | 91.14 | 0.264 | 0.599 | |
| SF (random slope) | 1.152 | 0.770 | 1.578 | ||
| Additive overdispersion | 0.003 | 0.65 | <0.001 | 0.008 | |
| Summary statistics | |||||
| Marginal R2 | 0.16 | ||||
| Conditional R2 | 0.50 | ||||
Abbreviations: CI, confidence interval; MHC, major histocompatibility complex; PBR, peptide-binding region.
Fertilization success was the proportion of a total of 48 genotyped offspring sired by each male in a paired-male fertilization trial. For fixed effects (relative sperm velocity of paired males (SF for slower/faster male in a paired-male trial), MHC divergence), significant pMCMC values (in bold) and the amount of variation explained by fixed effects (marginal R2) and the whole model (conditional R2) are reported. The variance (%) explained by each random effect (female identity (n=10), male identity (n=27), combination of males of which sperm was used in a focal female, denoted Male–Male pair (n=84)) refers to the total variance by all random effects and the additive overdispersion. For different estimates of MHC similarities between mates analyzed see Table 1 legend.
Figure 1.
Boxplots of the relationship between relative sperm velocity and male fertilization success in 84 paired-male fertilization trials in Chinook salmon. Relative sperm velocity of the paired males in the focal female’s ovarian fluid significantly explained male fertilization success (pMCMC=<0.001, see Table 2 for MCMCglmm results). Males with a higher sperm velocity than their ‘competitor’ achieved a higher fertilization success. Fertilization success (%) is the proportion of 48 genotyped offspring sired by each male in a paired-male fertilization trial.
The amino acid divergence between mates at the whole exon (AA) or the PBR at MHC class I had no significant impact on male fertilization success (PAAI=0.73, PPBRI=0.74, Table 2). Neither did the number of shared alleles at MHC class I explain differences in fertilization success among mates (PMxyI=0.46).
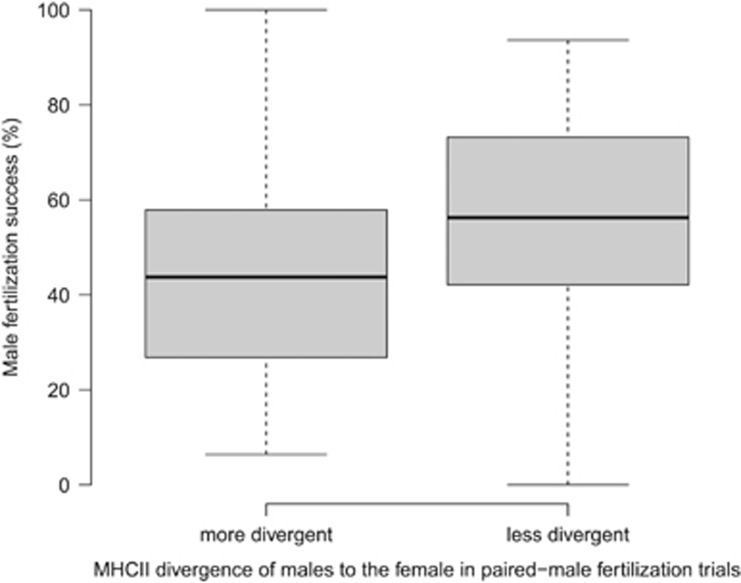
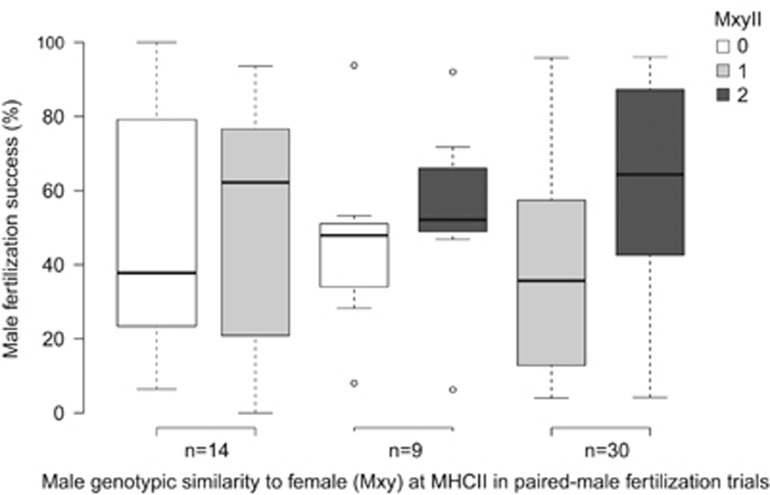
Amino-acid divergence of mates at MHC class II was negatively correlated with fertilization success (PAAII/PBRII=0.03, Table 2,Figure 2). AAII/PBRII and relative sperm velocity (SF) explained together 13% of the variation in fertilization success. The number of shared alleles between mates at MHC class II showed a significant positive correlation with fertilization outcomes (PMxyII=0.008) and explained together with the relative sperm velocity 16% of the variation (Table 2, Figure 3). Thereby, mates sharing a greater number of MHC class II alleles with the focal female had a higher fertilization success. Our results for both MHC loci were confirmed when the difference between males in their MHC divergence to the female was analyzed (Supplementary Table S2, Supplementary Figure S2) and when the MHC divergence between mates was assessed by counting the amino acids that differ between mates (Supplementary Table S3).
Figure 2.
Boxplots of the male fertilization success in paired-male fertilization trials (n=84, 36 trials shown) when paired males have a different amino-acid divergence to the female at MHC class II (MHCII) in Chinook salmon. Male fertilization success (%) is the proportion of 48 genotyped offspring sired by each male in a paired-male trial and is significantly higher for males less divergent to the female (right boxplot) at MHCII than the males they were paired with (left boxplot) (MCMCglmm analysis; pMCMC=0.03). Paired males that did not differ in their MHCII divergence to the female (48 trials) cannot be assigned to either boxplot and thus, are not shown but were included in the MCMCglmm analysis.
Figure 3.
Boxplots showing that male fertilization success in paired-male fertilization trials (n=84, 53 trials shown) is higher for the male sharing a greater number of MHC class II alleles (MxyII) with the female in Chinook salmon (MCMCglmm analysis; pMCMC=0.008). Male fertilization success (%) is the proportion of 48 genotyped offspring sired by each male in a paired-male trial. Paired males that did not differ in MxyII (31 trials) cannot be assigned to different boxplots and thus, are not shown but were included in the MCMCglmm analysis.
Discussion
Exploring the differential enhancement of sperm velocity by ovarian fluid, we found an impact of female and male identity on sperm velocity, which represents female and male quality and indicated that there were some males in our data that have a higher sperm velocity across females than others. These results are similar to previous studies in Arctic charr (Urbach et al., 2005) and Chinook salmon (Rosengrave et al., 2008).
Further, we found that the female–male interaction significantly determined sperm velocity and represented the major variance component, which is in line with previous findings in salmonid fish (Urbach et al., 2005; Rosengrave et al., 2008). In other words, a male’s sperm velocity can be relatively low in one female’s ovarian fluid and relatively high in another female. This suggested the presence of some form of mate compatibility, which allowed us to further explore whether it was characterized by the MHC genotypic combination of mates.
The MHC and sperm velocity in ovarian fluid
The MHC class I and II genotypic combination of mates does not appear to determine sperm velocity in ovarian fluid in Chinook salmon. The MHC allelic divergence at the whole exon and at the PBR as well as the number of shared alleles between mates did not explain a significant amount of the variation in sperm velocity. Thus, the observed mate compatibility, which explains roughly 44% of the variation in sperm velocity (42–45% across models), appears to be characterized by factors other than MHC, potentially by other peptides identified in the ovarian fluid of Chinook salmon (Johnson et al., 2014). MHC-dependent fertilization success was reported in mice (Wedekind et al., 1996; Rülicke et al., 1998) and the red jungle fowl (Løvlie et al., 2013), but the mechanisms by which such a non-random gamete fusion occurs are yet unknown. In this study, we observed an MHC class II-based fertilization success, but our data suggests that this bias is achieved via mechanisms other than MHC-based sperm velocity in ovarian fluid. In order to explore other mechanisms of CFC we examined the sources of variation in fertilization success among our crosses.
Fertilization success of males was significantly explained by the velocity of a male’s sperm relative to that of the male he was paired with. Males with a higher sperm velocity in a focal female’s ovarian fluid than the male they were paired with achieved a greater fertilization success. This result is in accordance with a recent study in Chinook salmon (Rosengrave et al., 2016). Our data confirm the suggestion that the impact of ovarian fluid on sperm velocity is indeed a mechanism of CFC to effectively bias male fertilization outcome in Chinook salmon (Rosengrave et al., 2008).
A previous study, however, found no sperm–ovarian fluid interaction effect on fertilization success when sperm velocity was measured in 10% ovarian fluid in Chinook salmon (Evans et al., 2013). Possibly, male competitive effects outweighed interaction effects since these might be less pronounced in 10% ovarian fluid than in 50% ovarian fluid (Evans et al., 2013) that was used in our study.
Similar to related studies (Urbach et al., 2005; Rosengrave et al., 2008) we have detected that sperm velocity can mainly be attributed to sperm–ovarian fluid interaction effects and to a smaller degree to male effects. Thus, in our experiment, female–male interaction effects appear to be the main driver of sperm velocity, which significantly impacts fertilization success, indicating the presence of CFC. Nevertheless, it might be interesting to test the sperm–ovarian fluid interaction effect at different concentrations of ovarian fluid, resembling the gradient of ovarian fluid encountered by sperm in natural spawnings.
The MHC and fertilization success
The amount of variation in fertilization success that was not explained by sperm velocity, was not explained by any of our estimates of MHC class I divergence of mates. Based on the high diversity at MHC class I in our population, our data contained mates sharing no or one allele, but no mates shared both alleles. This might limit the power to detect a bias in CFC using the number of shared alleles approach. However, we found also no MHC class I-based fertilization success analyzing the degree of allelic divergence of mates, which is a more sensitive approach. Our results are not concordant with an earlier observation in Atlantic salmon, in which paired-male fertilization trials revealed a cryptic female preference for sperm from males more similar at MHC class I (Yeates et al., 2009). Possibly, the different observations across species could represent the diversity of strategies depending on the species-specific context. Wedekind et al. (2004) proposed that the MHC gene expression in sperm could depend on the infection status of the male and thus, might explain the controversial results of MHC expression in sperm (Fernandez et al., 1999, for a review). Such a condition dependent MHC expression in sperm could explain the presence or absence of MHC-based CFC found by Yeates et al. (2009) and this study, respectively. However, to date there is no evidence to support this explanation. If future studies uncover MHC-dependent and -independent CFC among populations within a species or within populations over time, such studies could, combined with the assessment of the parasitic load of individuals and MHC expression analyses, indicate the existence of a condition dependent MHC-based CFC. As the MHC is important for adaptive immunity, selection might have favored the flexibility in mate preferences within and between species rather than a certain preference, in order to ensure adaptive abilities.
In contrast to MHC class I, we found that MHC class II divergence of mates was significantly correlated with fertilization success. These different findings between MHC loci might result from their location on different linkage groups in teleost fish (Bingulac-Popovic et al., 1997; Hansen et al., 1999), which enables differential selection acting on these loci (Shum et al., 2001).
Since we can exclude that this MHC class II-based gamete fusion is driven by sperm velocity, our data suggests that other mechanisms, possibly sperm–egg interactions via proteins on the gamete surfaces as found in marine invertebrates (Palumbi, 1999; Swanson et al., 2001), might be present. The fusibility locus in Botryllus, a colonial tunicate, is thought to represent an ancestral MHC-like system (Scofield et al., 1982). The haploid expressed fusibility gene on the sperm was recognized by the egg and fusion with sperm sharing the same fusibility allele was avoided (Scofield et al., 1982). Considering some evidence for MHC expression in mammalian gametes (Fernandez et al., 1999) and the presence of chorion associated glycoproteins that appear to facilitate sperm entry through the micropyle in teleost fish (Yanagimachi et al., 2013), further investigation of the possibility of MHC expression in salmonid gametes might be worthwhile.
More specifically, we tested the ‘heterozygote advantage hypothesis’ by examining the number of shared MHC alleles between mates. This hypothesis was inferred from observations in mice (McClelland et al., 2003; Froeschke and Sommer, 2005) and fish (Hedrick et al., 2001; Arkush et al., 2002; Evans and Neff, 2009; Kekäläinen et al., 2009), in which individuals heterozygous at the MHC had higher survival rates and lower parasite loads than homozygotes. It is thought that the different MHC alleles in heterozygous individuals likely enable immunity against a wider range of pathogens (Agbali et al., 2010). Accordingly, females should prefer males with whom they do not share alleles and therefore likely produce heterozygous offspring. Our data, however, do not support this hypothesis in Chinook salmon. Instead, fertilization success was higher for mates with a higher number of MHC alleles shared with the female.
Similarly, the allelic divergence between mates in our study showed a significant negative correlation with fertilization success, that is, males less divergent to the female at the MHC class II locus had a higher fertilization success than those males that were more divergent to the female. Considering the allelic divergence of mates we tested the ‘divergent allele advantage hypothesis’ that predicts that a higher sequence divergence between MHC alleles gains a greater advantage in immunity than heterozygosity, as two different alleles could still be functionally similar (Wakeland et al., 1990). Consequently, females should prefer males with a high MHC sequence divergence to their own. Our data, however, indicate that CFC, via mechanisms other than differential sperm velocity enhancement, promotes offspring with low MHC class II allelic divergence in Chinook salmon. Our result is in line with the MHC class II-assortative fertilization success in Atlantic salmon (Weir et al., 2012). MHC class II-assortative CFC could be associated with some of the key life history traits of salmonids: Chinook salmon return to their natal freshwater stream and populations to spawn and thus, fish are thought to exhibit local adaptation to their breeding ground (Taylor, 1991). It has been suggested that the MHC class II confers local adaptation in Chinook salmon (Evans et al., 2010). Our findings suggest that the low MHC class II diversity found in this and other Chinook salmon populations (Evans and Neff, 2009; Evans et al., 2010) could be maintained via CFC, possibly to preserve adaptation to the parasite community at the spawning ground.
Studies in Atlantic salmon (Landry et al., 2001; Consuegra and de Leaniz, 2008) and Chinook salmon (Neff et al., 2008) revealed that reproductive success was positively correlated with MHC class II allelic distance of mates (disassortative). In these studies, females and males were allowed to spawn freely without interference and it is not possible to differentiate between the impact of female choice and male competitiveness pre- and post-spawning on the reproductive success. However, while these studies and our study appear not comparable and possibly contradictory, the observation of MHC-disassortative reproductive success does not imply that CFC has to be MHC-disassortative as well. This is because (i) female choice pre- and post-mating can be non-directional, that is, preferences can differ (Tregenza and Wedell, 2000; Birkhead and Pizzari, 2002) and (ii) CFC can only act within the frame set by pre-mating processes and thus might counteract, but not outweigh, paternity biases achieved by mating behaviors prior to spawning.
Our results are not in line with, or might not be comparable to, reports in Arctic charr (Skarstein et al., 2005) and whitefish (Wedekind et al., 2004), in which CFC was found to be independent of the MHC class II. In both studies, no-choice (single male) fertilization trials (in contrast to our paired-male trials) were performed, which might lead to a different male fertilization success (Geyer and Palumbi, 2005). Furthermore, different results of the role of MHC class II in CFC might reflect different adaptations to the species-specific life cycles and environments: for example, whitefish populations inhibit a lake and have been shown to have a high MHC class II diversity (Binz et al., 2001), while Chinook salmon populations migrate to sea and return to their natal stream to spawn.
Our results seem unlikely to be confounded by CFC mechanisms post fertilization. Rülicke et al. (1998) proposed a possible pregnancy termination based on MHC incompatibility of mates in mice. For an external fertilizer like Chinook salmon with no cost of pregnancy and rearing, this option does not seem plausible as the mortality of fertilized eggs represents a reduction of fitness for females as well as males.
In conclusion, we showed that fertilization success in Chinook salmon was significantly correlated with relative sperm velocity in ovarian fluid. Sperm velocity was mainly attributed to female–male interaction effects, indicating the presence of CFC via sperm–ovarian fluid interactions. Our data suggests that neither the MHC class I or II are involved in possible sperm–ovarian fluid interactions. Consequently, the exploration of other peptides in the ovarian fluid and the milt might be a promising field of research. Even though relative sperm velocity significantly explained fertilization success, some variation in fertilization success was not explained by sperm velocity. This variation could not be attributed to the MHC class I either. In contrast, MHC class II, which possibly plays a role in local adaptation (Evans et al., 2010), appears to be a significant driver of non-random gamete fusion in Chinook salmon. Since Chinook salmon are strongly adapted to their breeding ground, which is known to complicate the supplementation of endangered populations (Taylor, 1991), the MHC class II-based CFC could potentially be considered in targeted supportive breeding programs to augment and conserve endangered salmon populations.
Data Archiving
The MHC sequence data has been submitted to GenBank: accession numbers KT156756–KT156766.
Acknowledgments
We thank Dr Patrice Rosengrave, Janine Wing for their efforts at field work, Joanne Gillum and Sara Ferreira for their assistance with laboratory work and Dr Sheri Johnson for advice on the fertilization experiments. We express our gratitude to Genom NZ (Invermay, New Zealand) for the microsatellite genotyping. We are also very thankful to the staff at the Silverstream hatchery of Salmon Smolt NZ, in particular Ben Divett, Karl French, Errol White, Tom Gough and Luke Price. Finally, we thank anonymous reviewers for their comments and suggestions on an earlier version of this manuscript. This research was supported by Royal Society of New Zealand Marsden Fund grant UOO0913 awarded to NJG and a University of Otago Doctoral Scholarship awarded to CG.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
The authors declare no conflict of interest.
Supplementary Material
References
- Abascal F, Zardoya R, Posada D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105. [DOI] [PubMed] [Google Scholar]
- Agbali M, Reichard M, Bryjová A, Bryja J, Smith C. (2010). Mate choice for nonadditive genetic benefits correlate with MHC dissimilarity in the rose bitterling (Rhodeus ocellatus). Evolution 64: 1683–1696. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkush KD, Giese AR, Mendonca HL, McBride AM, Marty GD, Hedrick PW. (2002). Resistance to three pathogens in the endangered winter-run chinook salmon (Oncorhynchus tshawytscha): effects of inbreeding and major histocompatibility complex genotypes. Can J Fish Aquat Sci 59: 966–975. [Google Scholar]
- Banks M, Blouin M, Baldwin B, Rashbrook VK, Fitzgerald HA, Blankenship SM et al. (1999). Isolation and inheritance of novel microsatellites in chinook salmon (Oncorhynchus tschawytscha). J Hered 90: 281–288. [Google Scholar]
- Beacham TD, Candy JR, Le KD, Wetklo M. (2009). Population structure of chum salmon (Oncorhynchus keta) across the Pacific Rim, determined from microsatellite analysis. Fish Bull 107: 244–260. [Google Scholar]
- Bernatchez L, Landry C. (2003). MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J Evol Biol 16: 363–377. [DOI] [PubMed] [Google Scholar]
- Billard R, Jensen JJO. (1996). Gamete removal, fertilization and incubation. Dev Aquacult Fish Sci 29: 291–364. [Google Scholar]
- Bingulac-Popovic J, Figueroa F, Sato A, Talbot WS, Johnson SL, Gates M et al. (1997). Mapping of mhc class I and class II regions to different linkage groups in the zebrafish, Danio rerio. Immunogenetics 46: 129–134. [DOI] [PubMed] [Google Scholar]
- Binz T, Largiader C, Müller R, Wedekind C. (2001). Sequence diversity of Mhc genes in lake whitefish. J Fish Biol 58: 359–373. [Google Scholar]
- Birkhead T. (1998). Cryptic female choice: criteria for establishing female sperm choice. Evolution 52: 1212–1218. [DOI] [PubMed] [Google Scholar]
- Blouin M, Parsons M, Lacaille V, Lotz S. (1996). Use of microsatellite loci to classify individuals by relatedness. Mol Ecol 5: 393–401. [DOI] [PubMed] [Google Scholar]
- Birkhead TR, Pizzari T. (2002). Postcopulatory sexual selection. Nat Rev Genet 3: 262–273. [DOI] [PubMed] [Google Scholar]
- Boschetto C, Gasparini C, Pilastro A. (2011). Sperm number and velocity affect sperm competition success in the guppy (Poecilia reticulata). Behav Ecol Sociobiol 65: 813–821. [Google Scholar]
- Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL et al. (1993). Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature 364: 33–39. [DOI] [PubMed] [Google Scholar]
- Butts IA, Johnson K, Wilson CC, Pitcher TE. (2012). Ovarian fluid enhances sperm velocity based on relatedness in lake trout, Salvelinus namaycush. Theriogenology 78: 2105–2109. e2101. [DOI] [PubMed] [Google Scholar]
- Condrey M, Bentzen P. (1998). Characterization of coastal cutthroat trout (Oncorhynchus clarki clarki) microsatellites and their conservation in other salmonids. Mol Ecol 7: 787–789. [PubMed] [Google Scholar]
- Consuegra S, de Leaniz CG. (2008). MHC-mediated mate choice increases parasite resistance in salmon. Proceedings of the Royal Society B: Biological Sciences 275: 1397–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard WG. (1996) Female Control: Sexual Selection by Cryptic Female Choice. Princeton University Press: Princeton, NJ, USA. [Google Scholar]
- Eklund A. (1997). The major histocompatibility complex and mating preferences in wild house mice (Mus domesticus). Behav Ecol 8: 630–634. [Google Scholar]
- Evans JP, Rosengrave P, Gasparini C, Gemmell NJ. (2013). Delineating the roles of males and females in sperm competition. Proc R Soc B Biol Sci 280: 20132047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M, Neff B, Heath D. (2009). MHC genetic structure and divergence across populations of Chinook salmon (Oncorhynchus tshawytscha). Heredity 104: 449–459. [DOI] [PubMed] [Google Scholar]
- Evans ML, Neff BD. (2009). Major histocompatibility complex heterozygote advantage and widespread bacterial infections in populations of Chinook salmon (Oncorhynchus tshawytscha). Mol Ecol 18: 4716–4729. [DOI] [PubMed] [Google Scholar]
- Evans ML, Neff BD, Heath DD. (2010). MHC-mediated local adaptation in reciprocally translocated Chinook salmon. Conserv Genet 11: 2333–2342. [Google Scholar]
- Felsenstein J. (1989). PHYLIP-Phylogeny Inference Package (Version 3.2) Cladistics 5: 164–166.
- Felsenstein J. (1993). PHYLIP (Phylogeny Inference Package) version 3.5 c. Distributed by the author, Department of Genetics, University of Washington, Seattle, USA, 1993.
- Fernandez N, Cooper J, Sprinks M, AbdElrahman M, Fiszer D, Kurpisz M et al. (1999). A critical review of the role of the major histocompatibility complex in fertilization, preimplantation development and feto-maternal interactions. Hum Reprod Update 5: 234–248. [DOI] [PubMed] [Google Scholar]
- Fleming IA. (1998). Pattern and variability in the breeding system of Atlantic salmon (Salmo salar), with comparisons to other salmonids. Can J Fish Aquat Sci 55: 59–76. [Google Scholar]
- Flot JF. (2010). SeqPHASE: a web tool for interconverting PHASE input/output files and FASTA sequence alignments. Mol Ecol Resour 10: 162–166. [DOI] [PubMed] [Google Scholar]
- Foote CJ. (1989). Female mate preference in Pacific salmon. Anim Behav 38: 721–723. [Google Scholar]
- Froeschke G, Sommer S. (2005). MHC class II DRB variability and parasite load in the striped mouse (Rhabdomys pumilio) in the southern Kalahari. Mol Biol Evol 22: 1254–1259. [DOI] [PubMed] [Google Scholar]
- Gage MJ, Macfarlane CP, Yeates S, Ward RG, Searle JB, Parker GA. (2004). Spermatozoal traits and sperm competition in Atlantic salmon: relative sperm velocity is the primary determinant of fertilization success. Curr Biol 14: 44–47. [PubMed] [Google Scholar]
- Galvano PM, Johnson K, Wilson CC, Pitcher TE, Butts IA. (2013). Ovarian fluid influences sperm performance in lake trout, Salvelinus namaycush. Reprod Biol 13: 172–175. [DOI] [PubMed] [Google Scholar]
- Garrigan D, Hedrick PW. (2001). Class I MHC polymorphism and evolution in endangered California Chinook and other Pacific salmon. Immunogenetics 53: 483–489. [DOI] [PubMed] [Google Scholar]
- Gasparini C, Pilastro A. (2011). Cryptic female preference for genetically unrelated males is mediated by ovarian fluid in the guppy. Proc R Soc Lond B Biol Sci 278: 2495–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer LB, Palumbi SR. (2005). Conspecific sperm precedence in two species of tropical sea urchins. Evolution 59: 97–105. [PubMed] [Google Scholar]
- Grimholt U, Hordvik I, Fosse VM, Olsaker I, Endresen C, Lie O. (1993). Molecular cloning of major histocompatibility complex class I cDNAs from Atlantic salmon (Salmo salar). Immunogenetics 37: 469–473. [DOI] [PubMed] [Google Scholar]
- Grob B, Knapp L, Martin R, Anzenberger G. (1997). The major histocompatibility complex and mate choice: inbreeding avoidance and selection of good genes. Exp Clin Immunogenet 15: 119–129. [DOI] [PubMed] [Google Scholar]
- Hadfield J. (2014). MCMCglmm course notes. Available at: http://cran.us.r-project.org/web/packages/MCMCglmm/vignettes/CourseNotes.pdf.
- Hadfield JD. (2010). MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J Stat Softw 33: 1–22.20808728 [Google Scholar]
- Hansen JD, Strassburger P, Thorgaard GH, Young WP, Du Pasquier L. (1999). Expression, linkage, and polymorphism of MHC-related genes in rainbow trout, Oncorhynchus mykiss. J Immunol 163: 774–786. [PubMed] [Google Scholar]
- Hart NH. (1990). Fertilization in teleost fishes: mechanisms of sperm-egg interactions. Int Rev Cytol 121: 1–66. [DOI] [PubMed] [Google Scholar]
- Heath DD, Bryden CA, Shrimpton JM, Iwama GK, Kelly J, Heath JW. (2002). Relationships between heterozygosity, allelic distance (d 2), and reproductive traits in chinook salmon, Oncorhynchus tshawytscha. Can J Fish Aquat Sci 59: 77–84. [Google Scholar]
- Hedrick PW, Kim TJ, Parker KM. (2001). Parasite resistance and genetic variation in the endangered Gila topminnow. Animal Conservation 4: 103–109. [Google Scholar]
- Hordvik I, Grimholt U, Fosse VM, Lie Ø, Endresen C. (1993). Cloning and sequence analysis of cDNAs encoding the MHC class II β chain in Atlantic salmon (Salmo salar). Immunogenetics 37: 437–441. [DOI] [PubMed] [Google Scholar]
- Janeway CA, Travers P, Walport M, Shlomchik MJ. (2001) Immunobiology: The Immune System in Health and Disease. Garland Science: New York, NY, USA. [Google Scholar]
- Johnson SL, Villarroel M, Rosengrave P, Carne A, Kleffmann T, Lokman PM et al. (2014). Proteomic analysis of Chinook salmon (Oncorhynchus tshawytscha) ovarian fluid. PLoS One 9: e104155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. (1992). The rapid generation of mutation data matrices from protein sequences. CABIOS 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Kalinowski ST, Taper ML, Marshall TC. (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16: 1099–1106. [DOI] [PubMed] [Google Scholar]
- Kekäläinen J, Vallunen JA, Primmer CR, Rättyä J, Taskinen J. (2009). Signals of major histocompatibility complex overdominance in a wild salmonid population. Proc R Soc B Biol Sci 276: 3133–3140. rspb20090727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley J, Walter L, Trowsdale J. (2005). Comparative genomics of major histocompatibility complexes. Immunogenetics 56: 683–695. [DOI] [PubMed] [Google Scholar]
- Landry C, Garant D, Duchesne P, Bernatchez L. (2001). ‘Good genes as heterozygosity’: the major histocompatibility complex and mate choice in Atlantic salmon (Salmo salar). Proc R Soc Lond B: Biol Sci 268: 1279–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljedal S, Rudolfsen G, Folstad I. (2008). Factors predicting male fertilization success in an external fertilizer. Behav Ecol Sociobiol 62: 1805–1811. [Google Scholar]
- Litvak MK, Trippel EA. (1998). Sperm motility patterns of Atlantic cod (Gadus morhua) in relation to salinity: effects of ovarian fluid and egg presence. Can J Fish Aquat Sci 55: 1871–1877. [Google Scholar]
- Løvlie H, Gillingham MA, Worley K, Pizzari T, Richardson DS. (2013). Cryptic female choice favours sperm from major histocompatibility complex-dissimilar males. Proc R Soc B Biol Sci 280: 20131296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning C, Potts W, Wakeland E. (1992). Communal nesting patterns in mice implicate MHC gene in kin recognition. Nature 360: 581–584. [DOI] [PubMed] [Google Scholar]
- McClelland EE, Penn DJ, Potts WK. (2003). Major histocompatibility complex heterozygote superiority during coinfection. Infect Immun 71: 2079–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Withler RE. (1997). Mhc diversity in pacific salmon: Population structure and trans‐species sllelism. Hereditas 127: 83–95. [DOI] [PubMed] [Google Scholar]
- Miller K, Withler R, Beacham T. (1997). Molecular evolution at Mhc genes in two populations of chinook salmon Oncorhynchus tshawytscha. Mol Ecol 6: 937–954. [DOI] [PubMed] [Google Scholar]
- Miller KM, Ming T, Schulze A, Withler RE. (1999). Denaturing gradient gel electrophoresis (DGGE): a rapid and sensitive technique to screen nucleotide sequence variation in populations. Biotechniques 27: 1016–1031. [DOI] [PubMed] [Google Scholar]
- Miller KM, Withler RE. (1996). Sequence analysis of a polymorphic Mhc class II gene in Pacific salmon. Immunogenetics 43: 337–351. [DOI] [PubMed] [Google Scholar]
- Miller KM, Withler RE. (1998). The salmonid class I MHC: limited diversity in a primitive teleost. Immunol Rev 166: 279–293. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Schielzeth H. (2013). A general and simple method for obtaining R2 from generalized linear mixed‐effects models. Methods Ecol Evol 4: 133–142. [Google Scholar]
- Neff B, Garner S, Heath J, Heath D. (2008). The MHC and non-random mating in a captive population of Chinook salmon. Heredity 101: 175–185. [DOI] [PubMed] [Google Scholar]
- Neff BD, Fu P, Gross MR. (2003). Sperm investment and alternative mating tactics in bluegill sunfish (Lepomis macrochirus). Behav Ecol 14: 634–641. [Google Scholar]
- Olsen KH, Grahn M, Lohm J, Langefors Å. (1998). MHC and kin discrimination in juvenile Arctic charr, Salvelinus alpinus (L.). Anim Behav 56: 319–327. [DOI] [PubMed] [Google Scholar]
- Palumbi SR. (1999). All males are not created equal: fertility differences depend on gamete recognition polymorphisms in sea urchins. Proc Natl Acad Sci USA 96: 12632–12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker GA. (1970). Sperm competition and its evolutionary consequences in the insects. Biol Rev 45: 525–567. [Google Scholar]
- Potts WK, Manning CJ, Wakeland EK. (1991). Mating patterns in seminatural populations of mice influenced by MHC genotype. Nature 352: 619–621. [DOI] [PubMed] [Google Scholar]
- R Core Team. (2013) A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Rosengrave P, Gemmell NJ, Metcalf V, McBride K, Montgomerie R. (2008). A mechanism for cryptic female choice in chinook salmon. Behav Ecol 19: 1179–1185. [Google Scholar]
- Rosengrave P, Montgomerie R, Gemmell N. (2016). Cryptic female choice enhances fertilization success and embryo survival in chinook salmon. Proc Biol Sci 283: 20160001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rülicke T, Chapuisat M, Homberger FR, Macas E, Wedekind C. (1998). MHC-genotype of progeny influenced by parental infection. Proc R Soc Lond B Biol Sci 265: 711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield VL, Schlumpberger JM, West LA, Weissman I. (1982). Protochordate allorecognition is controlled by a MHC-like gene system. Nature 295: 499–502. [DOI] [PubMed] [Google Scholar]
- Searcy WA. (1982). The evolutionary effects of mate selection. Annu Rev Ecol System 13: 57–85. [Google Scholar]
- Sherman C, Wapstra E, Uller T, Olsson M. (2008). Males with high genetic similarity to females sire more offspring in sperm competition in Peron's tree frog Litoria peronii. Proc R Soc Lond B Biol Sci 275: 971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum BP, Guethlein L, Flodin LR, Adkison MA, Hedrick RP, Nehring RB et al. (2001). Modes of salmonid MHC class I and II evolution differ from the primate paradigm. J Immunol 166: 3297–3308. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W et al. (2011). Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstein F, Folstad I, Liljedal S, Grahn M. (2005). MHC and fertilization success in the Arctic charr (Salvelinus alpinus). Behav Ecol Sociobiol 57: 374–380. [Google Scholar]
- Stephens M, Donnelly P. (2003). A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73: 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. (2001). A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68: 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltz J, Neff B. (2006). Sperm competition in a fish with external fertilization: the contribution of sperm number, speed and length. J Evol Biol 19: 1873–1881. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Aquadro CF, Vacquier VD. (2001). Polymorphism in abalone fertilization proteins is consistent with the neutral evolution of the egg's receptor for lysin (VERL) and positive Darwinian selection of sperm lysin. Mol Biol Evol 18: 376–383. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor EB. (1991). A review of local adaptation in Salmonidac, with particular reference to Pacific and Atlantic salmon. Aquaculture 98: 185–207. [Google Scholar]
- Thornhill R, Alcock J. (1983) The Evolution of Insect Mating Systems. Harvard University Press: Cambridge, MA, USA. [Google Scholar]
- Tregenza T, Wedell N. (2000). Genetic compatibility, mate choice and patterns of parentage: invited review. Mol Ecol 9: 1013–1027. [DOI] [PubMed] [Google Scholar]
- Turner E, Montgomerie R. (2002). Ovarian fluid enhances sperm movement in Arctic charr. J Fish Biol 60: 1570–1579. [Google Scholar]
- Unwin M, Quinn T, Kinnison M, Boustead N. (2000). Divergence in juvenile growth and life history in two recently colonized and partially isolated chinook salmon populations. J Fish Biol 57: 943–960. [Google Scholar]
- Urbach D, Folstad I, Rudolfsen G. (2005). Effects of ovarian fluid on sperm velocity in Arctic charr (Salvelinus alpinus). Behav Ecol Sociobiol 57: 438–444. [Google Scholar]
- Wakeland EK, Boehme S, She JX, Lu CC, McIndoe RA, Cheng I et al. (1990). Ancestral polymorphisms of MHC class II genes: divergent allele advantage. Immunol Res 9: 115–122. [DOI] [PubMed] [Google Scholar]
- Walsh PS, Metzger DA, Higuchi R. (1991). Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10: 506–513. [PubMed] [Google Scholar]
- Wedekind C, Chapuisat M, Macas E, Rulicke T. (1996). Non-random fertilization in mice correlates with the MHC and something else. Heredity 77: 400–409. [DOI] [PubMed] [Google Scholar]
- Wedekind C, Seebeck T, Bettens F, Paepke AJ. (1995). MHC-dependent mate preferences in humans. Proc R Soc Lond B Biol Sci 260: 245–249. [DOI] [PubMed] [Google Scholar]
- Wedekind C, Walker M, Portmann J, Cenni B, Müller R, Binz T. (2004). MHC‐linked susceptibility to a bacterial infection, but no MHC‐linked cryptic female choice in whitefish. J Evol Biol 17: 11–18. [DOI] [PubMed] [Google Scholar]
- Weir LK, Hutchings JA, Heath DD, Grant JW. (2012). Influence of density and major histocompatibility genotype on sexual selection in a salmonid alternative mating strategy. Can J Fish Aquat Sci 69: 670–680. [Google Scholar]
- Yamazaki K, Boyse E, Mike V, Thaler HT, Mathieson BJ, Abbott J et al. (1976). Control of mating preferences in mice by genes in the major histocompatibility complex. J Exp Med 144: 1324–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagimachi R, Cherr G, Matsubara T, Andoh T, Harumi T, Vines C et al. (2013). Sperm attractant in the micropyle region of fish and insect eggs. Biol Reprod 88: 47. [DOI] [PubMed] [Google Scholar]
- Yeates SE, Einum S, Fleming IA, Megens HJ, Stet RJ, Hindar K et al. (2009). Atlantic salmon eggs favour sperm in competition that have similar major histocompatibility alleles. Proc R Soc B Biol Sci 276: 559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.