Abstract
Genetic variation in mitochondrial genes could underlie metabolic adaptations because mitochondrially encoded proteins are directly involved in a pathway supplying energy to metabolism. Macquarie perch from river basins exposed to different climates differ in size and growth rate, suggesting potential presence of adaptive metabolic differences. We used complete mitochondrial genome sequences to build a phylogeny, estimate lineage divergence times and identify signatures of purifying and positive selection acting on mitochondrial genes for 25 Macquarie perch from three basins: Murray–Darling Basin (MDB), Hawkesbury–Nepean Basin (HNB) and Shoalhaven Basin (SB). Phylogenetic analysis resolved basin-level clades, supporting incipient speciation previously inferred from differentiation in allozymes, microsatellites and mitochondrial control region. The estimated time of lineage divergence suggested an early- to mid-Pleistocene split between SB and the common ancestor of HNB+MDB, followed by mid-to-late Pleistocene splitting between HNB and MDB. These divergence estimates are more recent than previous ones. Our analyses suggested that evolutionary drivers differed between inland MDB and coastal HNB. In the cooler and more climatically variable MDB, mitogenomes evolved under strong purifying selection, whereas in the warmer and more climatically stable HNB, purifying selection was relaxed. Evidence for relaxed selection in the HNB includes elevated transfer RNA and 16S ribosomal RNA polymorphism, presence of potentially mildly deleterious mutations and a codon (ATP6113) displaying signatures of positive selection (ratio of nonsynonymous to synonymous substitution rates (dN/dS) >1, radical change of an amino-acid property and phylogenetic conservation across the Percichthyidae). In addition, the difference could be because of stronger genetic drift in the smaller and historically more subdivided HNB with low per-population effective population sizes.
Introduction
Understanding the mechanisms of local adaptation is important for sustainable management of populations facing rapidly changing environments (Sgrò et al., 2011). Environmental gradients (for example, temperature, oxygen content and food availability) can drive ecophysiological adaptation at the cellular level, and the mitochondrial genome (mitogenome) is a likely target for environmental selection (Gershoni et al., 2009). In vertebrates, the mitogenome comprises 37 genes, of which 13 encode protein subunits of the oxidative phosphorylation (OXPHOS) pathway, which produces cellular energy (adenosine triphosphate (ATP)) and heat, and 24 genes encode genes for the mitochondrial translational machinery, consisting of two ribosomal RNAs (rRNAs) and 22 transfer RNAs (tRNAs; Ballard and Whitlock, 2004). Four multi-subunit enzyme OXPHOS complexes (I, III, IV and V) are co-encoded in the nuclear and mitochondrial genomes and one (II) is encoded exclusively in the nuclear genome (Rand et al., 2004). Fixation of beneficial or slightly deleterious mutations in the mitochondrial or nuclear genomes can drive co-evolution of genes from the other genome (Rand et al., 2004).
Although mitochondrial genes evolve under strong purifying selection to maintain functionality of protein complexes (Ballard and Whitlock, 2004), evidence is accumulating for positive selection acting on mitochondrial gene products or particular amino acids, associated with thermal adaptations, size, diet, migration behaviour and swimming speed (Ballard et al., 2007; Jacobsen et al., 2015). Mitochondrial genetic variation is shown to affect respiration, ATP synthesis, mitochondrial content, longevity and disease resistance, along with other functions (Latorre-Pellicer et al., 2016), suggesting its strong potential role in adaptation. In fishes, adaptation to cold is associated with higher mitochondrial densities in muscle, incurring high energy demands for synthesis and maintenance of mitochondria (Johnston et al., 1998; Pörtner, 2002). Thus, in populations exposed to colder climates, selection may favour genotypes with high coupling efficiency between the electron transport chain and the ATP synthase activity (Gershoni et al., 2009). Mutations within the ATP6 gene have been associated with metabolic differences and selection on energetics (Kucharczyk et al., 2010; Jacobsen et al., 2015). Positively selected nonsynonymous substitutions in ND2, ND4 and ND5 in Atlantic herring Clupea harengus (Teacher et al., 2012) and in cytochrome b (Cytb) of European anchovy Engraulis encrasicolus (Silva et al., 2014) have been found to be associated with different thermal and aerobic conditions. Selection on the electrochemical gradient and proton pump efficiency have also been inferred from signatures of positive selection on ND2 and ND5 amino acids in Pacific salmon Oncorhynchus spp. (Garvin et al., 2011). Therefore, the mitochondrial genome is a tractable system for investigating local adaptation in organisms occurring across heterogeneous environments.
Eastern Australia represents an excellent region for such studies, because it is large, environmentally variable and had only minor glaciations despite significant climatic oscillations during the Pleistocene that enabled persistence of local lineages and provided opportunities for local adaptation (Hewitt, 2004; Byrne et al., 2011). Regional climates of eastern Australia are influenced by the Great Dividing Range, a mountain chain of modest elevation (up to 2228 m above sea level) running for over 3500 km along the eastern coastline (Figure 1). The rain-shadow effect of the Great Dividing Range results in a mesic coastal climate, and an arid or semiarid inland climate characterized by larger temperature fluctuations (Byrne et al., 2011). Phylogeographic studies using mitochondrial markers suggest that during the Pleistocene the Great Dividing Range has acted as a barrier for aquatic species, with rare opportunities for dispersal leading to the establishment of divergent lineages or speciation in fish and frogs (Symula et al., 2008; Faulks et al., 2015). Recent studies suggest that mitochondrial divergences between populations separated by the Great Dividing Range may be a result of not only isolation and genetic drift, but also positive selection in response to different environmental pressures (Morales et al., 2015; Harrisson et al., 2016). For example, in Maccullochella cods, mitolineages of coastal Mary River cod M. mariensis and the eastern freshwater cod M. ikei were inferred to have evolved under relaxed purifying selection, in contrast to their inland relatives, the Murray cod M. peelii and trout cod M. macquariensis, that appear to be under strong purifying selection (Harrisson et al., 2016). In addition, M. mariensis, from climatically more stable and warm habitats, showed signatures of positive selection on amino acids in six genes, ND1, ND5, ND6, CO1, CO2 and CO3 (Harrisson et al., 2016). Although a direct link between signatures of positive selection detected by these studies and fitness of the organisms has not been demonstrated, more cross-Great Dividing Range studies would help assess whether environmentally driven selection on mitochondrial genes is a general phenomenon, and whether there are commonalities among candidate genes. However, such studies of freshwater systems face an additional challenge: because of smaller sizes of the coastal basins and limited extent of past connectivity within them (Faulks et al., 2015), coastal populations are likely to have had lower historical effective population sizes (Ne) and experienced stronger genetic drift as compared with Murray–Darling Basin (MDB) populations. Fixation of slightly deleterious mutations by drift can leave similar signatures as relaxation of selective constraints, making it challenging to distinguish evolutionary forces driving mitogenome evolution (Wertheim et al., 2014).
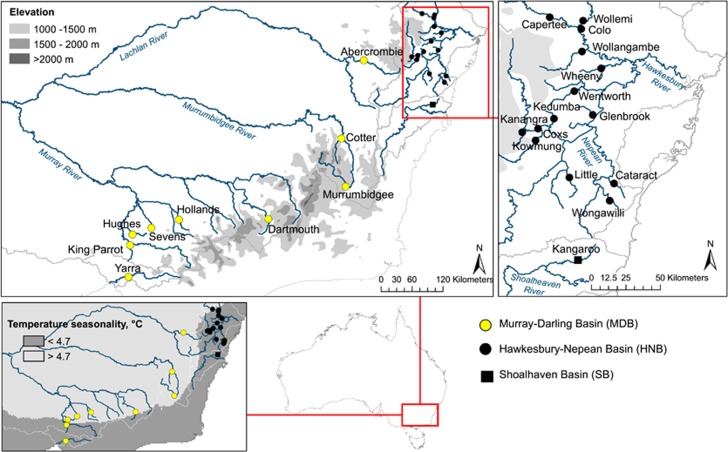
Figure 1.
Geographic distribution of Macquarie perch samples across Murray–Darling Basin (MDB; Yarra Basin population (Yarra) was established by translocations from the MDB), Hawkesbury–Nepean Basin (HNB) and Shoalhaven Basin (SB). Thin light grey lines show basin borders, shades of grey on top two maps show elevation of the Great Dividing Range. The bottom left map shows samples mapped over temperature seasonality (WorldClim variable BIO4/1000: s.d. of mean monthly temperatures). Cataract River contains native fish and fish translocated from the MDB; representative mitolineages of both are analysed here.
Here we explore patterns of selection acting on a mitogenome of a native southeastern Australian freshwater fish, Macquarie perch Macquaria australasica (Perciformes: Percichthyidae). The historical range of Macquarie perch included three coastal basins, the Hawkesbury–Nepean (HNB), Shoalhaven (SB) and Georges, and one inland basin, the Murray–Darling Basin (MDB; Figure 1), presumed to be colonized from the HNB in the early Pleistocene (Lintermans, 2007; Faulks et al., 2010; Knight and Bruce, 2010). MDB populations are generally cooler and exposed to a wider range of air temperatures, as demonstrated by variation in WorldClim 1.4 bioclimatic variables (Hijmans et al., 2005): for the MDB vs HNB samples used here the range of temperature seasonality (BIO4, s.d. of mean monthly temperature) is 4.6 to 5.3 vs 3.9 to 4.9 °C, respectively (Figure 1), mean temperatures of coldest quarter (BIO11) 4.9 to 8.2 vs 8.5 to 10.7 °C and mean temperature of warmest quarter (BIO10) 17.4 to 20.8 vs 20.4 to 22.5 °C. Size differences between MDB and coastal individuals also suggest that fish in these basins may face different metabolic demands (Dalziel et al., 2015): Macquarie perch from coastal populations (HNB and SB) are smaller (25 vs 46 cm) and mature at smaller sizes (Dufty, 1986; Knight and Bruce, 2010). Given strong and numerous effects of mtDNA variation on metabolism (Latorre-Pellicer et al., 2016), cross-basin variation in mitogenomes might be adaptive.
Slightly deleterious mutations causing small defects of proteins should have larger fitness effect in populations exposed to greater climatic variation, and thus stronger functional constraints on protein-coding mitochondrial genes could be expected in the more climatically variable MDB as compared with the HNB where climates are milder and less variable (Nabholz et al., 2013). On the other hand, genetic drift might be stronger and purifying selection weaker in the HNB because of lower historical Ne values of HNB populations compared with MDB ones. Lower historical Nes for HNB vs MDB populations are suggested by estimates of control region haplotype diversity (mean 0.285 (minimum 0–maximum 0.855) vs 0.432 (0–0.876)), nucleotide diversity (0.0014 (0–0.0041) vs 0.0021 (0–0.0063)) and allelic richness for 19 microsatellites (2.694 (1.368–0.4) vs 3.045 (1.662–4.04)) (Pavlova et al., submitted).
Dramatic population declines over the past century resulted in the reduction of Macquarie perch to a few isolated headwater populations in the MDB and HNB and presumed extinction in the SB, from which the only known genetic sample was collected in 1998 (Faulks et al., 2010). These declines were reflected in extremely low estimates of contemporary per-population Ne (Pavlova et al., submitted). Given these population declines, Macquarie perch is listed as endangered under Australian federal legislation and three State/Territory Acts, and as Data Deficient on the IUCN (International Union for Conservation of Nature and Natural Resources) Red List. The species-level taxonomy requires re-examination because significant differentiation among Macquarie perch from the MDB, HNB and SB basins on the basis of external phenotypes and nuclear genetic loci has led to suggestions of the presence of cryptic species (Dufty, 1986; Faulks et al., 2011). However, the only phylogenetic hypothesis available (Faulks et al., 2010) is based on a fragment of mitochondrial control region and does not resolve HNB and MDB samples as monophyletic lineages. Testing for within-basin lineage monophyly and estimating divergence times while incorporating rate uncertainty using complete mitogenomes can improve our understanding of the evolutionary history and taxonomy of this important species because of greatly increased gene sampling compared with previous studies (DeFilippis and Moore, 2000; Morin et al., 2010).
In this paper we analyse complete mitogenome sequences for 25 Macquarie perch individuals sampled across three basins: MDB (N=10), HNB (14) and SB (1). We aim to: (1) test for reciprocal monophyly of mitolineages from the three basins, (2) improve estimates of divergence times between them and (3) identify hypotheses about possible role of purifying and positive selection on mitogenome evolution in different basins. To do this, we build a Bayesian phylogeny, estimate divergence times, explore mitochondrial protein-coding regions for signatures of selection using gene-based tests and codon-based models, quantify changes in amino-acid physicochemical properties and examine polymorphism within nonprotein-coding regions. To further investigate the nature of selection on candidate codons in Macquarie perch, we explore the evolution of candidate codons across the family Percichthyidae.
Materials and methods
Samples, mitogenome sequencing and annotations
Twenty-four new and one published mitogenome sequences (GenBank accession HG799088; Gan et al., 2014) were analysed (Figure 1, Supplementary Appendix 1). Individuals were chosen from 24 populations, found by Pavlova et al. (submitted) to be currently geographically isolated and genetically differentiated. A single known genetic sample was available from the SB (hence, this basin was not suitable for analyses of within-lineage variation). MDB and HNB individuals were selected to represent genetic and geographic diversity of control region haplotypes from population studies by Faulks et al. (2010) and Pavlova et al. (submitted; Supplementary Appendix 1). These studies inferred that within the MDB, the Lachlan River was historically isolated from tributaries of the Murrumbidgee and Murray rivers (Figure 1) that were connected by gene flow before arrival of European people in Australia, and that the northern HNB was historically isolated from the southern HNB. We refer to each individual by the population from which it was sampled, but note that most MDB haplotypes occur in multiple MDB populations, and some HNB haplotypes could be shared among neighbouring water bodies (Supplementary Appendix 1). Two individuals were sequenced from Cataract River: one of the endemic HNB lineage, and one of the introduced MDB lineage. Mitogenomes were sequenced, assembled and annotated as described in Supplementary Appendix 2.
Analyses of genetic diversity within and genetic divergence between lineages
Nucleotide diversity (π) and haplotype diversity (Hd) were calculated in DNASP 5.10.01 (Librado and Rozas, 2009) for each gene and complete mitogenome for the MDB and HNB. Between-lineage p-distances were calculated in MEGA 6.06 (Tamura et al., 2011).
Transfer RNA analyses
The secondary structure of most mitochondrial tRNAs has a cloverleaf-shaped base-pairing pattern with four stems (acceptor stem, D-stem, anticodon stem and T-stem) and three loops (D-loop, anticodon loop and T-loop) (Wittenhagen and Kelley, 2003). Because post-transcriptional modifications of the anticodon stem and loop create the ‘wobble-base’ necessary to enable the recognition of multiple codons (Agris et al., 2007), mutations in these regions might be functionally important. Mutations within stems can be deleterious when they disrupt Watson–Crick pairs and are not accompanied by a compensating substitution (Kern and Kondrashov, 2004). Locations of mutations within two-dimensional (2D) tRNA structures for polymorphic tRNA-coding genes were inferred by mapping Macquarie perch sequences on 2D tRNA models of Japanese Aucha perch Coreoperca kawamebari (Sinipercidae) available from the mitotRNAdb mitochondrial transfer RNA database (Juhling et al., 2009).
Phylogenetic analysis and estimation of time of lineage divergence
Bayesian phylogenetic analysis was performed on complete mitogenomes in MrBayes 3.2.2 (Ronquist et al., 2012) using golden perch Macquaria ambigua as an outgroup (GenBank accession AP014533; Lavoué et al., 2014). The best-fit partitioning scheme and nucleotide substitution models were determined using a heuristic search (greedy algorithm) under the Bayesian information criterion in PartitionFinder (Lanfear et al., 2012); parameters for all partitions were unlinked (Supplementary Appendix 3). Four replicate runs with four chains each were run for 2 million generations sampling every 1000 generations. Run convergence was confirmed using TRACER 1.6.0 (Rambaut and Drummond, 2007). Trees were summarized on a 50% majority rule tree after discarding 25% of trees as burn-in, and visualized using FigTree 1.4.2 (Rambaut, 2006).
Divergence times between major Macquarie perch lineages were estimated from complete mitogenomes using BEAST 1.8.2 (Drummond et al., 2012). The outgroup was not used, and hence a coalescent tree prior could be applied. Analyses were run using nine partitions found by PartitionFinder with unlinked substitution and clock models but linked tree topologies. Bayesian skyline plots with 10 time intervals and a piecewise linear skyline model and uniform (0–1000) prior for skyline PopSize were used as a population size prior. Because preliminary analysis using an uncorrelated lognormal relaxed clock model suggested lack of rate heterogeneity among lineages (95% highest posterior density interval of ucld.sdev included zero; Supplementary Appendix 4), a strict clock model was used. As fossil or geological calibrations were not available for our species, we used lognormal clock rate priors that encompass a range of mtDNA time-dependent rate calibrations for recent (<3 million years) divergences of galaxiid and other fishes (Burridge et al., 2008). For partitions involving the first and second codon positions, tRNAs and rRNAs, 95% of clock rates were sampled between 0.1 and 3% substitutions/site/million years, and for the third codon positions and the control region between 0.6 and 13.9% substitutions/site/million years (Supplementary Appendices 3 and 4). Six replicates were run for 200 million generations and sampled every 10 000 steps for the final analysis. Replicate runs were checked for convergence, combined and analysed in TRACER, discarding the first 10% as burn-in.
Tests of selection
For all analyses of protein-coding variation, sequences of 13 protein-coding genes were concatenated (ND6 sequence was reverse-complemented, as ND6 is encoded on the light strand) and stop codons removed. Overlapping regions (7 bp of ATP8/ATP6 and 4 bp of ND4L/ND4) were included in the alignment twice so to enable analyses of all 3800 codons.
McDonald–Kreitman tests
DNASP was used to calculate McDonald–Kreitman tests of directional or purifying selection (MK; McDonald and Kreitman, 1991) for each gene and complete mitogenome for each basin.
Codon-based ω ratios
The ratio of nonsynonymous to synonymous substitution rates (ω=dN/dS) is often used to detect selection, when ω=1 indicates neutral evolution, ω<1 purifying selection and ω>1 positive selection (Yang, 2007). We used four models that estimate ω for each codon in the data set based on a phylogeny, approximated here by the majority rule consensus tree built by MrBayes (the outgroup was pruned from the tree). The Fast Unconstrained Bayesian AppRoximation method (FUBAR; Murrell et al., 2013) was used to detect codons under pervasive purifying or diversifying selection under a model that evaluates the posterior probability (PP) that each codon belongs to each of three classes of ω: ω0<1, ω1=1 and ω2>1. Codons with PP>0.9 and ω<1 or ω>1 were inferred to have evolved under purifying and positive selection, respectively. A Mixed Effects Model of Evolution (meme; Murrell et al., 2012) was used to identify pervasive and episodic (that is, affecting only a subset of lineages) positive selection under a model that allows ω at each codon to vary across branches. Codons with P<0.05 were inferred to be under positive selection. FUBAR and MEME are implemented in the program HyPhy and were run via the Datamonkey web server (Delport et al., 2010) using the best-fit substitution models, also assessed using the web server.
Codeml, implemented in the software package PAML4.7 (Yang, 2007) and run via GUI PAMLX (Xu and Yang, 2013), was used to fit site-, branch- and branch-site models. Site models allow ω to vary among codons and assume one ω ratio for all branches (Nielsen and Yang, 1998; Yang et al., 2000). Five different site models were run using equal codon frequencies (results were the same for the analyses using observed codon frequencies): M0 (one ω ratio), M1a (nearly neutral; assumes two classes of codons, ω0<1 and ω1=1), M2a (positive selection; assumes three classes of codon, ω0<1, ω1=1, ω2>1), M7 (β sites are evolving under 0⩽ω⩽1 drawn from a β distribution) and M8 (β and ω as M7, but some codons evolve under ω>1; Table 1; Wong et al., 2004; Yang et al., 2005). A likelihood-ratio test was used to compare fit of the nested models: M0 vs M1a tested for a single ω ratio across codons, and M1a vs M2a and M7 vs M8 for presence of positive selection.
Table 1. Estimates of parameters of the six Codeml site models and P-values of likelihood-ratio tests (LRTs) comparing these models (P ΔLRT).
| Model | Model | No. of p | Log likelihood | Parameter estimates | Null model | P ΔLRT |
|---|---|---|---|---|---|---|
| Site model | M0 (one ratio) | 1 | −18 325 | ω=0.13 | ||
| M1a (nearly neutral) | 2 | −18 314 | ω0=0, p0=0.87; ω1=1, p1=0.13 | M0 | <0.001 | |
| M2a (positive selection) | 4 | −18 313 | ω0=0.00001, p0=0.90; ω1=1, p1=0.05; ω2=1.61, p2=0.05 | M1a | NS | |
| M7 (β) | 2 | −18 314 | p=0.01142; q=0.07027 | NS | ||
| M8 (β and ω>1) | 4 | −18 313 | p0=0.984, p=8.47, q=99.0; (p1=0.016), ω=3.582 | M7 | NS | |
| Branch model 2 | Three ω (HNB, SB, MDB) | 3 | −18 319 | ωHNB=0.23; ωSB=0.09; ωMDB=0.12 | M0 | P<0.01 |
| Branch-site model A modified, HNB as a foreground | Model A modified (estimated ωHNB) | 4 | −18 309 | p0=0.88, ωbackground=0, ωHNB=0; p1=0.1, ωbackground=1, ωHNB=1; | Model A null | NS |
| p2a=0.02, ωbackground=0, ωHNB=5.27; p2b=0.002, ωbackground=1, ωHNB=5.27 | M1a | P<0.01 | ||||
| Model A null (assumed ωHNB=1) | 3 | −18 310 | p0=0.80, ωbackground=0, ωHNB=0; p1=0.09, ωbackground=1, ωHNB=1; p2a=0.09, ωbackground=0, ωHNB=1; p2b=0.01, ωbackground=1, ωHNB=1 |
Abbreviations: HNB, Hawkesbury–Nepean Basin; MDB, Murray–Darling Basin; No. of p, number of free parameters in the model; NS, not significant (P>0.05); SB, Shoalhaven Basin.
Parameter estimates are estimates of ω=dN/dS ratios and proportion of codons that belong to each ω class (p and q for M7 and M8 are parameters of β distribution of ω under purifying selection). The Null model is a model against which the model in the second column was compared using likelihood-ratio test (LRT) test.
Branch model, which assumes different ω for different branches, was run to test whether fish in different basins (MDB, HNB and SB) evolved under different selective pressures. Three ω-values (MDB, HNB and SB) were estimated. To test for rate heterogeneity across lineages, branch model was compared with M0 (one ω ratio) using likelihood-ratio tests.
Branch-site model A modified (Zhang et al., 2005) was run to test for positive selection affecting particular sites in the HNB, a foreground lineage. This model assumes four categories of sites: sites under purifying selection (0<ω0<1); sites under neutrality (ω1=1); sites under positive selection ω2>1 in the foreground lineage but under purifying selection (0<ω0<1) in background lineages; and sites under positive selection ω2>1 in the foreground lineage but under neutrality (ω0=1) in background lineages. This model was compared with the null (fixed ω2=1) to test for presence of positive selection in the foreground lineage, and with model M1a (nearly neutral) to test whether positive or relaxed purifying selection has affected some sites in the foreground lineage (the third and/or fourth category have ω2>1 or ω2⩽1 in the foreground lineage, respectively). To identify codons under positive selection, posterior probabilities were calculated for models assuming positive selection (M2a, M8 and the branch-site model A modified) using Bayes empirical Bayes (Yang et al., 2005); codons with PP >0.7 were considered candidates for selection.
Finally, to test whether purifying selection in the HNB is relaxed compared with that in the MDB, we ran RELAX (Wertheim et al., 2014) via the Datamonkey web server. The RELAX model uses the selection intensity parameter k, which modulates the degree to which ω classes diverge from neutrality (ω=1), to determine whether the ω distribution inferred for the test set of branches represents selective relaxation (k<1) or intensification (k>1) relative to the reference set. Significance of the model is tested against the null (k=1) using likelihood-ratio test. If purifying selection in the HNB (test branches) is relaxed relative to that in the MDB (reference branches), we expect k<1 and ω in the HNB to be shifted towards larger ω<1 values compared with the MDB.
Radical changes in physicochemical properties of amino acids
TREESAAP (Woolley et al., 2003) was used to analyse the effect of amino-acid replacements on 20 physicochemical properties (those with >85% accuracy of detecting selection; McClellan and Ellison, 2010) over the evolutionary tree (as above) and to detect directional positive selection for these properties. TREESAAP should be more sensitive to detection of positive selection in recently diverged taxa than are codon models because it compares the observed physicochemical amino-acid changes inferred from a phylogenetic tree with the random distribution of changes expected under selective neutrality, and does not depend on sampling strategy or selection biases (Woolley et al., 2003). Only amino-acid changes with radical effects on properties (categories 6, 7 and 8) were considered. A global test was used to select properties under selection across all coding regions, and a sliding-window approach (window sizes 10 and 20 codons with 1-codon increment) was used to detect DNA regions (windows) affected by this selection. Significance, direction and amount of selection at every sliding-window were evaluated using z-values: z>0 indicate positive selection, z<0 purifying selection. Windows with z-scores of >3.09 and z <−3.09 (corresponding to P<0.001) were inferred to be significant.
Because only one individual from each location was sequenced, we could not establish whether the amino-acid replacements observed in a single sequence are unique to that sequence or fixed (potentially under selective pressure) in one or more locations. Thus, only codons with amino-acid replacements affecting two or more mitogenome sequences were considered candidates for positive selection. In addition, congruence between methods with different underlying assumptions (that is, codon models and TREESAAP) was assessed to reduce false inferences.
Comparative analysis of candidate codons across Percichthyidae
Because relaxed purifying selection and positive selection can have similar genomic signatures, we tested whether the candidate codons experienced multiple nonsynonymous substitutions over the phylogeny of the family Percichthyidae. Presence of multiple nonsynonymous changes across Percichthyidae phylogeny in the candidate codon was taken as evidence of relaxed functional constraint on that codon (Jacobsen et al., 2016), whose evolution in Macquarie perch was then interpreted as subject to relaxed purifying selection. For five genes harbouring six candidate codons (ND2, ND4, ND5, ATP6 and CO2, see Results), Percichthyidae sequences were downloaded from GenBank (one individual per species, 14–15 species per gene; details in Supplementary Appendix 5) and aligned in GENEIOUS. Number of nonsynonymous substitutions for each codon of each gene was calculated using HyPhy implemented in MEGA over the phylogenetic tree presented in Supplementary Appendix 5.
Results
Mitogenome annotations
A final alignment of Macquarie perch mitogenomes comprised 16 499 nucleotides. Length variation was present in 16S rRNA, tRNA-Lys and control region. Several annotation discrepancies compared with the published mitogenome were detected (Supplementary Appendix 6); new annotations are used here because MitoFish produces accurate predictions for fish mitogenomes (Iwasaki et al., 2013). Annotated mitogenomes were submitted to GenBank (accession KR152233–KR152256).
Phylogenetic analysis
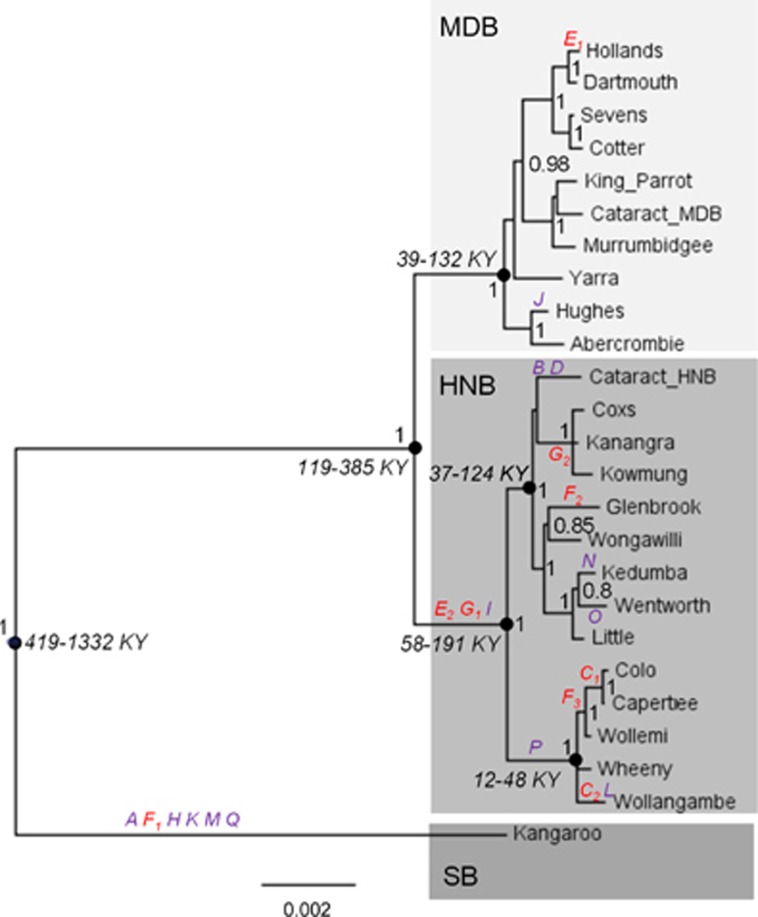
Bayesian phylogenetic analysis of mitogenomes in MrBayes showed three well-supported basin-based Macquarie perch lineages (Bayesian PP=1; Figure 2), monophyletic with respect to the outgroup: one comprised the SB individual, which was the sister to a clade comprising two sister clades, the MDB and the HNB. No phylogeographic structure was present in the MDB. In the HNB there were two well-supported (PP=1) clades, corresponding to the northern HNB (Colo, Capetree, Wollemi, Wheeny and Wollangambe) and southern HNB (remaining HNB samples).
Figure 2.
MrBayes majority rule consensus tree built from complete mitogenome sequences (rooted with golden perch Macquarie ambigua, not shown). Numbers are Bayesian posterior probability >0.8 for each node. Letters in italic are labels of amino-acid replacements inferred to be under selection by TREESAAP, MEME, FUBAR and/or Codeml models (Table 2); letters without numbers indicate replacements affecting a single lineage, and letters with numbers indicate codons with replacements in multiple lineages across the phylogeny. The branches over which the nonsynonymous replacements occurred were inferred by TreeSAAP analysis based on ancestral sequence reconstructed for each node. Numbers and letters in italic indicate BEAST 95% highest posterior density (HPD) estimates of time to most recent common ancestor for major lineages and sublineages (nodes indicated by black circles; KY, thousands of years).
Within-lineage genetic diversity and among-lineage divergence
Nucleotide diversity (π) and haplotype diversity (Hd) were lower in the MDB than in the HNB for complete mitogenomes, but varied across genes (Supplementary Appendix 7; note that the estimates for the HNB are influenced by pooling individuals from historically isolated northern and southern HNB). For complete mitogenomes and protein-coding regions respectively, average uncorrected between-lineage p-distances were 0.60 and 0.63% for MDB vs HNB, 1.86 and 2.10% for MDB vs SB, and 1.90 and 2.15% for HNB vs SB. The outgroup (golden perch) was >11.9 and >13.8% divergent from any Macquarie perch lineage. Between SB and MDB+HNB, 10 substitutions were fixed in 16S rRNA, 5 in 12S rRNA, 8 in tRNAs, 171 synonymous and 27 nonsynonymous in protein-coding genes and 25 in the control region. Between the MDB and HNB, 6 substitutions were fixed in 16S rRNA, 1 in tRNA-Glu, 32 synonymous and 4 nonsynonymous in protein-coding genes and 6 in the control region (Supplementary Appendix 7).
Time of lineage divergence
The time to the most recent common ancestor (tMRCA) of all Macquarie perch lineages was estimated by BEAST from complete mitogenomes at 812 (geometric mean; 95% highest posterior density 419–1332), of MDB+HNB lineages at 232 (119–385), of MDB at 79 (39–132) and of HNB at 113 (58–191) thousand years ago (KYA). The tMRCA for the southern HNB (that is, Cataract, Coxs, Glenbrook, Kedumba, Kanangra, Kowmung, Little, Wentworth and Wongawilli) was estimated at 74 (37–124) and 27 (12–48) KYA for the northern HNB (that is, Colo, Capertee, Wollemi, Wheeny and Wollangambe) (Figure 2 and Supplementary Appendix 4).
Ribosomal RNA diversity and divergence
For 12S rRNA, four haplotypes were restricted to the MDB, four to the HNB, one to the SB and one was shared between MDB and HNB individuals. MDB vs HNB haplotypes differed by 0–4 nucleotides, the SB haplotype was 6–8 nucleotides different from any other Macquarie perch haplotype. For 16S rRNA, there were three haplotypes in the MDB, eight in the HNB and one in the SB. Haplotypes were phylogenetically clustered according to basins (Supplementary Appendix 8). MDB vs HNB haplotypes differed by 7–12 nucleotides, SB haplotype was 12–16 nucleotides different from any other haplotype.
Transfer RNAs divergence and location of mutations within 2D structures of tRNAs
Macquarie perch tRNA sequences closely matched Aucha perch 2D tRNA models (Supplementary Appendix 9). The MDB and HNB shared haplotypes for all tRNAs, except tRNA-Glu, for which different haplotypes were fixed between basins. The SB lineage had unique haplotypes not shared with the other basins for 9 of 22 tRNAs. Within-lineage polymorphism was present in 9 of 22 tRNAs (4 in the MDB and 6 in the HNB). Variable positions were spread through all tRNA regions except the anticodon loop. Of the 11 substitutions within the stem regions, 7 disrupted Watson–Crick pairings and thus were putatively deleterious, whereas 4 restored Watson–Crick pairings, indicating potentially advantageous effects. The latter were substitutions in the anticodon stem of tRNA-Trp fixed in all MDB and HNB individuals, acceptor stem of tRNA-Arg in Wheeny (HNB), acceptor stem of tRNA-His fixed in the northern HNB (Colo, Capetree, Wollemi, Wheeny and Wollangambe) and the anticodon stem of tRNA-Thr in the SB lineage.
McDonald–Kreitman tests
An MK test of HNB vs MDB suggested that strong purifying selection influenced divergence between these basins (4 nonsynonymous/32 synonymous substitutions fixed between, and 45 nonsynonymous/117 synonymous polymorphisms within lineages; Fisher’s test P=0.035). Similarly, an MK test of HNB vs SB was significant (29/191 vs 32/68; P<0.001), but an MK test of MDB vs SB was not (33/189 vs 13/52; P>0.05). None of the MK tests performed on individual genes were significant.
Selection on protein-coding variation across all 13 mitochondrial protein-coding genes inferred by codon models
Evidence for pervasive purifying selection
FUBAR found 308 codons spread across all 13 genes with evidence of pervasive purifying selection (PP >0.9). The nearly neutral Codeml site model M1a best described the data (M0 vs M1a: likelihood-ratio test P<0.001); it estimated that 87% of all codons are under purifying selection, whereas the remaining codons are evolving neutrally. Site-models assuming positive selection (M2a and M8) were not significantly favoured over the models assuming that sites are evolving neutrally or under purifying selection (M1a and M7; both LR tests not significant; Table 1).
Evidence for relaxed purifying selection in the HNB
Branch- and branch-site Codeml models provided evidence for positive and/or relaxed purifying selection in the coastal HNB. Branch model assuming that ω for HNB was different than for MDB or SB (P<0.01) explained the data significantly better than M0 (which assumes a single ω for all branches) and estimated higher ω for HNB than for other basins (Table 1). Branch-site models (with HNB as a foreground clade) were the best models of all Codeml ones. The model assuming positive selection (model A modified) did not differ significantly from its null (that assumes relaxed purifying selection), but it differed significantly from M1a (that assumes that some codons evolve under purifying selection and some under neutrally; P<0.01) and estimated that 2% of sites were under positive selection in HNB and purifying selection in other basins, and 0.2% of sites were under positive selection in HNB and evolving neutrally in other basins (Table 1). Relaxed purifying selection in the HNB relative to MDB was supported by a significant RELAX test (k=0.71; P=0.039; LR=4.28). The alternative model, which showed a shift towards larger ω <1 values in the HNB compared with the MDB, was preferred over the null model that assumed that ω <1 values were the same in both basins. The null RELAX model (logL=−17533.9, 72 parameters) estimated that in both basins 93% of sites had ω1=0.119, 6.5% of sites had ω2=0.129 and 0.67% of sites had ω3=1. The alternative model (logL=−17531.8, 73 parameters) estimated that 87% of sites had ω1=0.154 in HNB and 0.072 in MDB, 13% of sites had ω2=0.176 in HNB and 0.087 in the MDB and 0.85% of sites had ω3=1 in both basins.
Codons under positive selection
Five codons were inferred to have evolved under positive selection by one of the codon models; all involved amino-acid replacements in the HNB (Table 2 and Figure 2). MEME analysis found two ND5 codons (labelled L and N in Table 2 and Figure 2) with significant evidence of episodic positive selection in two HNB tip lineages. FUBAR found a single codon in ATP6 (labelled F) with evidence of pervasive diversifying selection; the same amino-acid replacement evolved independently three times, in SB and two HNB lineages. Bayes empirical Bayes analysis of the Codeml branch-site model (model A modified) found two codons with evidence of positive selection in the HNB (C in ND2 and F in ATP6; Figure 2).
Table 2. List of 17 nonsynonymous codons encoding 22 amino acid replacements along Macquarie perch phylogeny inferred to be under positive selection by TREESAAP, MEME (P<0.05), FUBAR (PP >0.9) and/or Codeml BEB analysis under branch-site model with HNB in foreground (PP >0.8); see the list of all replacements in Supplementary Appendix 11.
| Gene | Codon position in a gene | Amino acid replacement label (Figure 1) | From codon | To codon | From amino acid | To amino acid |
TREESAAP |
Codon models | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cat α-helical tendencies | Amnt α-helical tendencies | Cat Equilibrium constant (ionization of COOH) | Amnt Equilibrium constant (ionization of COOH) | ||||||||
| ND1 | 159 | A | GCT | ACT | Ala | Thr | 6 | −0.59 | |||
| ND1 | 184 | B | GCC | ACC | Ala | Thr | 6 | −0.59 | |||
| ND2 | 299 | C1 | ATC | ACC | Ile | Thr | 6 | 0.74 | Codeml | ||
| ND2 | 299 | C2 | ATC | ACC | Ile | Thr | 6 | 0.74 | Codeml | ||
| CO1 | 71 | D | ATA | ACA | Met | Thr | 6 | −0.62 | |||
| CO2 | 150 | E1 | GTC | ATC | Val | Ile | 8 | −0.96 | |||
| CO2 | 150 | E2 | GTC | ATC | Val | Ile | 8 | −0.96 | |||
| ATP6 | 21 | F1 | GCC | ACC | Ala | Thr | 6 | −0.59 | FUBAR | ||
| ATP6 | 21 | F2 | GCC | ACC | Ala | Thr | 6 | −0.59 | FUBAR | ||
| ATP6 | 21 | F3 | GCC | ACC | Ala | Thr | 6 | −0.59 | FUBAR | ||
| ATP6 | 113 | G1 | ACC | GCC | Thr | Ala | 6 | 0.59 | Codeml | ||
| ATP6 | 113 | G2 | GCC | ACC | Ala | Thr | 6 | −0.59 | Codeml | ||
| ND4 | 163 | H | GTT | ATC | Val | Ile | 8 | −0.96 | |||
| ND4 | 186 | I | ATC | ACC | Ile | Thr | 6 | 0.74 | |||
| ND4 | 259 | J | AGC | ATC | Ser | Ile | 7 | −0.85 | |||
| ND4 | 439 | K | ACC | ATC | Thr | Ile | 6 | −0.74 | |||
| ND5 | 16 | L | TTC | ATC | Phe | Ile | MEME | ||||
| ND5 | 45 | M | GTT | ATT | Val | Ile | 8 | −0.96 | |||
| ND5 | 178 | N | CTT | ATT | Leu | Ile | 8 | −1 | MEME | ||
| ND5 | 308 | O | GTT | ATT | Val | Ile | 8 | −0.96 | |||
| ND5 | 511 | P | GCA | GGA | Ala | Gly | 8 | −0.85 | |||
| Cytb | 29 | Q | GTT | ATT | Val | Ile | 8 | −0.96 | |||
Abbreviations: Amnt, amount; BEB, Bayes empirical Bayes; Cat, category; HNB, Hawkesbury–Nepean Basin; PP, posterior probability.
Codons are labelled by letters (A–Q), multiple replacements of the same codon are numbered (see Figure 1). Amino-acid replacements of six codons (C, E, F, G, I and P, shaded grey) affected two or more sequences; these codons are candidates for positive selection. TreeSAAP: radical (categories 6–8) changes in amino-acid properties found to be significant by sliding window approach. Category and amount of change is given (negative values indicate decrease, whereas positive values indicate increase in property). Changes in Shoalhaven Basin (SB) lineage are in italics, and those basal to HNB lineage are in bold.
Evolution of amino-acid physicochemical properties inferred by TREESAAP
Global TREESAAP analysis detected two physicochemical properties (Equilibrium constant (ionization of COOH) and α-helical tendencies) with significant (z>3.09; P<0.001) radical changes, indicative of positive diversifying selection. None of the properties were inferred to be under significant negative selection (all z>−3.09).
Sliding window analyses of these two properties with a 10-codon window (Supplementary Appendix 10) showed that Equilibrium constant (ionization of COOH) was affected by amino-acid replacements in five genes (ND2, ND4, ND5, CO2 and Cytb) and α-helical tendencies were affected by replacements in four genes (ND1, ND5, CO1 and ATP6; Table 2). Close examination of significant windows showed that 11 and 6 codons, respectively, were involved in a radical shift in these properties (Table 2 and Supplementary Appendix 11).
Codon candidates for positive selection
Of 17 nonsynonymous codons inferred to be under positive selection by at least one of the methods (Table 2 and Figure 2), 11 codons had a replacement affecting a single tip lineage (replacements A, H, K, M, Q in SB lineage; B, D, L, N, O in the HNB; and J in the MDB) and 6 had replacements affecting more than one sequence (C, E, F, G, I and P; shaded in Table 2). The former group likely represents mildly deleterious mutations accumulated under relaxed purifying selection, whereas the latter set represents potential candidates for positive selection. Of the six candidates, three (C, G and F) were independently detected by methods with different underlying assumptions (that is, codon model and TREESAAP) and thus most likely were not false positives.
Evolution of candidate codons in Percichthyidae
Five of the six codon candidates for positive selection, ND2299, CO2150, ATP621, ND4186 and ND5511 (Macquarie perch codons C, E, F, I and P, respectively) were codons experiencing nonsynonymous substitutions on a Percichthyidae phylogeny (Supplementary Appendix 5), suggesting that their evolution is not strongly constrained by function and is likely subject to relaxed purifying selection in Macquarie perch. In contrast, ATP6113 (Macquarie perch codon G) was located in a conserved region of the ATP6 gene, and had zero nonsynonymous replacements across Percichthyidae, suggesting that nonsynonymous mutations of this codon could be functionally important. In Macquarie perch, an ATP6113 replacement of ancestral threonine (shared by all Percichthyidae) to alanine was fixed in the HNB clade, before being replaced back to threonine in the westernmost HNB subclade (G1 at the root of the HNB lineage, followed by G2 at the root of the clade Coxs+Kanangra+Kowmung).
Discussion
We used mitogenome data to investigate whether basin-based differences could have affected the mitogenome evolution of Macquarie perch, to test for reciprocal monophyly of mitolineages and estimate divergence times between them. Overall, our results show that purifying selection and genetic drift were the most prominent forces shaping the Pleistocene evolution of Macquarie perch mitogenome. Purifying selection appears to have been relaxed in the coastal HNB, putatively associated with milder climatic conditions and/or smaller Ne. Fixation of a nonsynonymous substitution at ATP6113 codon in the HNB and/or its reversal in northern HNB could have occurred under positive selection, although the effect of genetic drift cannot be rejected. The pattern of relaxed purifying selection in coastal but not inland basins strongly mirrors that found in Maccullochella cods, another lineage of Percichthyidae with Pleistocene cross-Great-Dividing-Range divergence (Harrisson et al., 2016), and suggests that evolution of other similarly distributed aquatic species could be driven by similar constraints on metabolism or effective population sizes.
Divergence of Macquarie perch lineages
Our phylogenetic analyses based on complete mitochondrial DNA sequences of Macquarie perch found that basin-based lineages form well-supported clades. This is a new result compared with the earlier mitochondrial phylogeny (Faulks et al., 2010) that showed the MDB clade embedded within the HNB clade. Although sequence divergence among clades is low (<1% between the MDB and HNB and ~2% between SB and either MDB or HNB), together with cross-basin differentiation based on morphological phenotypes, allozymes and microsatellites (Dufty, 1986; Faulks et al., 2011; Pavlova et al., submitted), reciprocal monophyly of mitolineages indicates that the three basins harbour incipient or undescribed species. Analysis of genome-wide markers of the Cataract River population, where the endemic HNB and translocated MDB populations are known to hybridize (Faulks et al., 2011; Pavlova et al., submitted), provides an opportunity to test for the presence of reproductive isolation between HNB and MDB.
According to our analyses of divergence time, the initial early- to mid-Pleistocene (419–1331 KYA) split between the SB and the ancestral MDB+HNB lineages was followed by the mid- to late-Pleistocene divergence between the MDB and HNB (119–385 KYA), and later by divergence of northern and southern HNB sublineages (58–191 KYA). These estimates are more recent compared with those based on partial control region (~2 MYA for SB vs rest, 657 KYA for MDB vs HNB and 130 KYA for northern vs southern HNB) (Faulks et al., 2010). The discrepancy is likely because of the contribution of other loci to the estimate of splits (48 × more data were analysed here) rather than difference in clock rates used: our clock rate estimate for control region was 3–11.7%/MY (Supplementary Appendix 4) that includes the 3.6%/MY rate used earlier. As Faulks et al. (2010), we acknowledge the limitations of using clock rates calibrated for different species. Faulks et al. (2010) hypothesized that the HNB-to-MDB colonization by Macquarie perch occurred through topographically low parts of the Great Dividing Range, such as Lake George and/or Lake Bathurst and Breadalbane Plain area, that have numerous relatively low relief borders with the Murrumbidgee, Lachlan, Hawkesbury-Nepean and Shoalhaven catchments. Our estimate of within-MDB lineage divergence (39–132 KYA) is consistent with Macquarie perch lineage diversification before colonization of the MDB during westward overflow of Lake George 21–27 KYA into a tributary of the Murrumbidgee and/or Lachlan rivers (Coventry, 1976). Our estimate of divergence time between the inland MDB and coastal HNB lineages of Macquarie perch (119–385 KYA) is also consistent with cross-Great Dividing Range divergence of Maccullochella species of freshwater cods (220–710 KY; Harrisson et al., 2016), suggesting that geologic and climatic changes in the region may have facilitated cross-basin colonization in different aquatic species (Faulks et al., 2015).
Purifying selection acting across coding genes of Macquarie perch violates a major assumption of the molecular clock (neutral evolution). Nevertheless, taxa for which mitochondrial clocks are calibrated will generally be under similar functional constraints. Thus, as long as a clock was calibrated on a similar scale of divergence, any biases introduced by purifying selection on clock rate estimates should be similar in related taxa, assuming their effective population sizes are comparable (Ho and Larson, 2006; Burridge et al., 2008). Given that signal of positive selection is low, it is unlikely that positive selection had a strong impact on Macquarie perch divergence estimates, because only a single codon may have been affected, rate heterogeneity across lineages was rejected and strict as well as relaxed clock analyses yielded very similar results (Supplementary Appendix 4). If ATP6113 was indeed fixed in the HNB under positive selection, our tMRCA of HNB could represent an underestimate, but other tMRCA estimates should not be affected.
Differences between basin-based mitolineages
During the divergence of HNB and MDB lineages, three nonsynonymous mutations became fixed in each, and those fixed in the HNB (E2, G1 and I) were inferred to change amino-acid properties radically (Table 2 and Supplementary Appendix 11), suggesting potential differences in protein function between basins. In addition, differences in gene expression among the basins are possible given the presence of fixed differences in tRNA-Glu and 16S rRNA between the MDB and HNB and substitutions restoring Watson–Crick pairings in tRNA-Trp anticodon stem of the MDB+HNB lineage that could influence tRNA stability and efficiency of post-transcriptional modifications (Wittenhagen and Kelley, 2003; Agris et al., 2007). Fixed differences between MDB and HNB lineages were also observed in the control region, containing the main regulatory elements for mtDNA transcription and replication (Fernández-Silva et al., 2003). Neither the origin of heavy-strand replication located in the conserved sequence block II, which in Macquarie perch was identical to that of salmonids (Shedlock et al., 1992) and some other fish (Lee et al., 1995), nor transcriptional promoters (light-strand promoter and heavy-strand promoter 1) located upstream of conserved sequence block II in the domain III of the control region had any sequence variation within or between Macquarie perch lineages, implying that RNA synthesis and ability to modify mtDNA replication in response to physiological demands are unaffected by lineage divergence (Fish et al., 2004; Bonawitz et al., 2006). Similarly, tRNA-Phe, where the transcriptional promoter heavy-strand promoter 2 is located, had no sequence variation (except a single substitution in the Wollangambe sequence). Determining whether the between-basin differences are linked to metabolic differences could be achieved by comparing metabolic performance of HNB and MDB Macquarie perch in common and different environments. The Cataract River population, containing endemic HNB as well as translocated MDB lineages and their hybrids, is an excellent study system for doing this. Evaluating fitness of pure forms (that is, HNB or MDB mitolineages in pure HNB and pure MDB nuclear backgrounds, respectively) and mitolineages in opposite or hybrid nuclear backgrounds in the Cataract River will also reveal whether hybridization of HNB and MDB fish results in outbreeding depression and whether mitochondrial-associated selection plays a role.
Because the SB was represented by a single individual, whether the diverged haplotypes for both rRNAs, nine tRNAs and control region found in the SB lineage were fixed in the now-extinct population will remain unknown unless other samples are discovered, hence inferences are unwarranted.
Contribution of selection and drift to inland–coast divergence of Macquarie perch mitogenomes
Purifying selection appears to be the main evolutionary force affecting Macquarie perch mitogenome evolution: 87% of codons were inferred to be under strong purifying selection, and 13% either under moderate purifying selection (RELAX results) or neutrality (nearly neutral site model; Table 1). Strong purifying selection to preserve functionality of mitochondrial proteins is expected, and was previously inferred for many organisms (Nabholz et al., 2013) including Maccullochella cods in the region of the present study (Harrisson et al., 2016). Nevertheless, several candidates for positive selection were detected by various analyses of selection (Table 2). Signatures of positive selection can resemble relaxed purifying selection, and the test designed to distinguish them (Wertheim et al., 2014) supported relaxed purifying selection in HNB Macquarie perch. This suggests that the majority of amino-acid replacements shown in Table 2 were likely slightly deleterious mutations fixed by drift (Hughes, 2007). These results support our expectations that purifying selection was relaxed in the HNB, because functional constraints were ameliorated in the HNB owing to milder and less variable climates, and/or because HNB populations had lower historical effective population sizes (Pavlova et al., submitted) and hence were more affected by genetic drift. Less stringent selective pressures and/or stronger drift in the coastal HNB was supported by higher within-lineage polymorphism for tRNA and 16S rRNA genes in the HNB. The latter suggests that higher variation in efficiency of mitochondrial gene expression might be tolerated in the HNB (Montooth et al., 2009). Therefore, genetic drift was also a prominent driver of Macquarie perch mitogenome evolution. Studies of domesticated animals suggest that relaxation of selection or reduction in population size, operating over a short timescale, can cause significant changes to the patterns of mitochondrial molecular evolution, including higher ratios of nonsynonymous to synonymous changes (Cruz et al., 2008; Moray et al., 2014). Distinguishing whether milder selective pressures or smaller Ne facilitated drift is challenging, particularly given that they are not mutually exclusive. Estimates of past population dynamics from nuclear sequence data may help resolve this: times of fixation of mutations in particular clades corresponding to temporary population bottlenecks would support lower Ne as major reason for fixation of mildly deleterious mutations (Drummond et al., 2005).
Codon models could produce false positive and negative results when used on closely related taxa because of increased stochastic effects (Mugal et al., 2013), and performing several tests of selection on the same data could multiply this effect. Our inference of prevalence of relaxed purifying selection in the HNB compared with the MDB, supported by a statistical test (RELAX), was robust to the exclusion of codons supported by a single methodological approach or occurring in a single sequence (Table 2 and Figure 2).
Codon candidates for positive selection in the HNB
Overall, support for positive selection in Macquarie perch was limited. Results of our analyses of positive selection on Macquarie perch mitogenomes showed little congruence across different approaches (Table 2), suggesting the possibility of false positive results. Across six candidate codons with replacements in two or more lineages, only three (C, F and G) were detected by TreeSAAP plus one of the codon models, and only one (G: ATP6113) passed a phylogenetic conservation test.
Given the conservation of ATP6113 amino acid across the Percichthyidae, its forward and reversed replacements in Macquarie perch have likely affected protein function. It is possible that the forward ATP6113 replacement was originally fixed in the HNB because it imparted higher fitness at the time of fixation. Previous inferences of population expansion after colonization of the HNB (Faulks et al., 2010) suggest that at the time colonization Ne might have been low, thus positive selection for the original ATP6113 replacements would have to be strong to overcome the effect of drift. However, the reverse replacement of ATP6113 in the western part of the southern HNB supports the alternative explanation, where the original slightly deleterious replacement has been fixed by drift in the ancestral HNB population, and the reverse ATP6113 replacement to the ancestral state was favoured by positive selection. Demonstrations of fitness effects incurred by nonsynonymous changes in mitochondrial genes are rare and often limited to lab experiments (Dowling et al., 2008). Nevertheless, links between metabolism-related phenotypes and positive selection at ATP6 were previously suggested for various taxa (Garvin et al., 2015). In freshwater eels, number of amino-acid changes at five positively selected ATP6 codons and their differences in physicochemical properties were positively correlated with larval phase duration and migratory distance (Jacobsen et al., 2015), although ATP6113 did not feature among those selected codons (ATP6 positions 43, 52, 83, 105 and 195). Because non-mammalian vertebrates possess nucleated red blood cells that retain functional mitochondria (Stier et al., 2015), metabolic functioning of Macquarie perch with different ATP6113 variants could be measured with low-impact blood sampling. Fish with a positively selected variant would be expected to have better metabolic performance in local environments.
Management considerations
Current management of the endangered Macquarie perch precludes mixing of the MDB and HNB lineages; however, our data suggest only limited evidence of non-neutrality of mitolineage divergence across basins. Nevertheless, relaxed purifying selection in the HNB indicates presence of deleterious mutations that could contribute to genetic load and lower fitness in small populations (Gemmell et al., 2004). Our data suggest that fish translocations for genetic supplementation within the MDB need not account for the presence of mitochondrial adaptation. A potential candidate for positive selection in the HNB warrants investigation and caution while designing translocations among southern HNB populations.
Data archiving
Sequence data have been submitted to GenBank: accession numbers KR152233–KR152256.
Acknowledgments
This work was supported by the Australian Research Council Grant LP110200017 to Monash University, Flinders University of South Australia and the University of Canberra, with Partner Organization University of Montana. Funding and other support was contributed by industry Partner Organizations ACTEW Corporation, Department of Sustainability and Environment (Victoria; now within DELWP (Department of Environment, Land, Water and Planning)) and Fisheries Victoria (now within DEDJTR (Department of Economic Development, Jobs, Transport and Resources)), Melbourne Water. Additional funding was provided to AP and PS by Recreational Fishing Grants program (DELWP) and to DMG by NSW Department of Primary industries. We thank L Beheregaray, P Bishop, T Curmi, A Gonçalves da Silva, K Harrisson, P Hesse and S Kaminskas for useful discussions, and A Bruce, L Faulks, J Kearns, A McDonald, J O'Mahony, A Pickworth, S Raymond and Z Tonkin for sample collection.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
The authors declare no conflict of interest.
Supplementary Material
References
- Agris PF, Vendeix FA, Graham WD. (2007). tRNA’s wobble decoding of the genome: 40 years of modification. J Mol Biol 366: 1–13. [DOI] [PubMed] [Google Scholar]
- Ballard JWO, Whitlock MC. (2004). The incomplete natural history of mitochondria. Mol Ecol 13: 729–744. [DOI] [PubMed] [Google Scholar]
- Ballard JWO, Melvin RG, Katewa SD, Maas K. (2007). Mitochondrial DNA variation is associated with measurable differences in life-history traits and mitochondrial metabolism in Drosophila simulans. Evolution 61: 1735–1747. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Clayton DA, Shadel GS. (2006). Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell 24: 813–825. [DOI] [PubMed] [Google Scholar]
- Burridge CP, Craw D, Fletcher D, Waters JM. (2008). Geological dates and molecular rates: fish DNA sheds light on time dependency. Mol Biol Evol 25: 624–633. [DOI] [PubMed] [Google Scholar]
- Byrne M, Steane DA, Joseph L, Yeates DK, Jordan GJ, Crayn D et al. (2011). Decline of a biome: evolution, contraction, fragmentation, extinction and invasion of the Australian mesic zone biota. J Biogeogr 38: 1635–1656. [Google Scholar]
- Coventry R. (1976). Abandoned shorelines and the late quaternary history of lake George, New South Wales. J Geol Soc Aust 23: 249–273. [Google Scholar]
- Cruz F, Vilà C, Webster MT. (2008). The legacy of domestication: accumulation of deleterious mutations in the dog genome. Mol Biol Evol 25: 2331–2336. [DOI] [PubMed] [Google Scholar]
- Dalziel AC, Martin N, Laporte M, Guderley H, Bernatchez L. (2015). Adaptation and acclimation of aerobic exercise physiology in Lake Whitefish ecotypes (Coregonus clupeaformis. Evolution 69: 2167–2186. [DOI] [PubMed] [Google Scholar]
- DeFilippis VR, Moore WS. (2000). Resolution of phylogenetic relationships among recently evolved species as a function of amount of DNA sequence: an empirical study based on woodpeckers (Aves: Picidae). Mol Phylogenet Evol 16: 143–160. [DOI] [PubMed] [Google Scholar]
- Delport W, Poon AF, Frost SD, Kosakovsky Pond SL. (2010). Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26: 2455–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling DK, Friberg U, Lindell J. (2008). Evolutionary implications of non-neutral mitochondrial genetic variation. Trends Ecol Evol 23: 546–554. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus O. (2005). Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22: 1185–1192. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufty S. (1986). Genetic and morphological divergence between populations of Macquarie perch (Macquaria australasica east and west of the Great Dividing Range. Unpublished BSc(Hons) thesis, University of New South Wales, Kensington.
- Faulks L, Gilligan D, Beheregaray LB. (2015) ‘Ragged mountain ranges, droughts and flooding rains’: the evolutionary history and conservation of Australian freshwater fishes. In: Stow A, Maclean N, Holwell GI (eds). Austral Ark: The State of Wildlife in Australia and New Zealand. Cambridge University Press: Cambridge, UK. [Google Scholar]
- Faulks LK, Gilligan DM, Beheregaray LB. (2010). Evolution and maintenance of divergent lineages in an endangered freshwater fish, Macquaria australasica. Conserv Genet 11: 921–934. [Google Scholar]
- Faulks LK, Gilligan DM, Beheregaray LB. (2011). The role of anthropogenic vs. natural in-stream structures in determining connectivity and genetic diversity in an endangered freshwater fish, Macquarie perch (Macquaria australasica. Evol Appl 4: 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Silva P, Enriquez JA, Montoya J. (2003). Replication and transcription of mammalian mitochondrial DNA. Exp Physiol 88: 41–56. [DOI] [PubMed] [Google Scholar]
- Fish J, Raule N, Attardi G. (2004). Discovery of a major D-loop replication origin reveals two modes of human mtDNA synthesis. Science 306: 2098–2101. [DOI] [PubMed] [Google Scholar]
- Gan HM, Tan MH, Austin CM. (2014). The complete mitogenome of the Macquarie perch, Macquaria australasica Cuvier, 1830 (Teleostei: Percichthyidae). Mitochondrial DNA 27: 383–384. [DOI] [PubMed] [Google Scholar]
- Garvin MR, Bielawski JP, Gharrett AJ. (2011). Positive Darwinian selection in the piston that powers proton pumps in complex I of the mitochondria of Pacific salmon. PLoS One 6: e24127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin MR, Bielawski JP, Sazanov LA, Gharrett AJ. (2015). Review and meta-analysis of natural selection in mitochondrial complex I in metazoans. J Zool Syst Evol Res 53: 1–17. [Google Scholar]
- Gemmell NJ, Metcalf VJ, Allendorf FW. (2004). Mother's curse: the effect of mtDNA on individual fitness and population viability. Trends Ecol Evol 19: 238–244. [DOI] [PubMed] [Google Scholar]
- Gershoni M, Templeton AR, Mishmar D. (2009). Mitochondrial bioenergetics as a major motive force of speciation. Bioessays 31: 642–650. [DOI] [PubMed] [Google Scholar]
- Harrisson KA, Pavlova A, Gan HM, Lee YP, Austin CM, Sunnucks P. (2016). Pleistocene divergence across a mountain range and the influence of selection on mitogenome evolution in threatened Australian freshwater cod species. Heredity 116: 506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt GM. (2004). Genetic consequences of climatic oscillations in the Quaternary. Philos Trans R Soc Lond B Biol Sci 359: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. (2005). Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25: 1965–1978. [Google Scholar]
- Ho S, Larson G. (2006). Molecular clocks: when times are a-changin'. Trends Genet 22: 79–83. [DOI] [PubMed] [Google Scholar]
- Hughes AL. (2007). Looking for Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity 99: 364–373. [DOI] [PubMed] [Google Scholar]
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP et al. (2013). MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol 30: 2531–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen MW, Pujolar JM, Hansen MM. (2015). Relationship between amino acid changes in mitochondrial ATP6 and life-history variation in anguillid eels. Biol Lett 11: 20150014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen MW, da Fonseca RR, Bernatchez L, Hansen MM. (2016). Comparative analysis of complete mitochondrial genomes suggests that relaxed purifying selection is driving high nonsynonymous evolutionary rate of the NADH2 gene in whitefish (Coregonus ssp.). Mol Phylogenet Evol 95: 161–170. [DOI] [PubMed] [Google Scholar]
- Johnston I, Calvo J, Guderley H, Fernandez D, Palmer L. (1998). Latitudinal variation in the abundance and oxidative capacities of muscle mitochondria in perciform fishes. J Exp Biol 201: 1–12. [DOI] [PubMed] [Google Scholar]
- Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. (2009). tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37: D159–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern AD, Kondrashov FA. (2004). Mechanisms and convergence of compensatory evolution in mammalian mitochondrial tRNAs. Nat Genet 36: 1207–1212. [DOI] [PubMed] [Google Scholar]
- Knight J, Bruce A. (2010). Threatened fish profile: ‘Eastern’ macquarie perch Macquaria australasica cuvier 1830. Aust Soc Fish Biol Newsl 40: 76–78. [Google Scholar]
- Kucharczyk R, Ezkurdia N, Couplan E, Procaccio V, Ackerman SH, Blondel M et al. (2010). Consequences of the pathogenic T9176C mutation of human mitochondrial DNA on yeast mitochondrial ATP synthase. Biochim Biophys Acta 1797: 1105–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SY, Guindon S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29: 1695–1701. [DOI] [PubMed] [Google Scholar]
- Latorre-Pellicer A, Moreno-Loshuertos R, Lechuga-Vieco AV, Sánchez-Cabo F, Torroja C, Acín-Pérez R et al. (2016). Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature 535: 561–565. [DOI] [PubMed] [Google Scholar]
- Lavoué S, Nakayama K, Jerry DR, Yamanoue Y, Yagishita N, Suzuki N et al. (2014). Mitogenomic phylogeny of the Percichthyidae and Centrarchiformes (Percomorphaceae): comparison with recent nuclear gene-based studies and simultaneous analysis. Gene 549: 46–57. [DOI] [PubMed] [Google Scholar]
- Lee W-J, Conroy J, Howell WH, Kocher T. (1995). Structure and evolution of teleost mitochondrial control regions. J Mol Evol 41: 54–66. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Lintermans M. (2007) Fishes of the Murray-Darling Basin: An Introductory Guide. Murray-Darling Basin Commission: Canberra, ACT. [Google Scholar]
- McClellan DA, Ellison DD. (2010). Assessing and improving the accuracy of detecting protein adaptation with the TreeSAAP analytical software. Int J Bioinform Res Appl 6: 120–133. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. (1991). Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- Montooth KL, Abt DN, Hofmann JW, Rand DM. (2009). Comparative genomics of Drosophila mtDNA: novel features of conservation and change across functional domains and lineages. J Mol Evol 69: 94–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales H, Pavlova A, Joseph L, Sunnucks P. (2015). Positive and purifying selection in mitochondrial genomes of a bird with mitonuclear discordance. Mol Ecol 24: 2820–2837. [DOI] [PubMed] [Google Scholar]
- Moray C, Lanfear R, Bromham L. (2014). Domestication and the mitochondrial genome: comparing patterns and rates of molecular evolution in domesticated mammals and birds and their wild relatives. Genome Biol Evol 6: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin PA, Archer FI, Foote AD, Vilstrup J, Allen EE, Wade P et al. (2010). Complete mitochondrial genome phylogeographic analysis of killer whales (Orcinus orca) indicates multiple species. Genome Res 20: 908–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugal CF, Wolf JB, Kaj I. (2013). Why time matters: codon evolution and the temporal dynamics of dN/dS. Mol Biol Evol 31: 212–231, mst192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Pond SLK. (2012). Detecting individual sites subject to episodic diversifying selection. PLoS Genet 8: e1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Pond SLK et al. (2013). FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol 30: 1196–1205, mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabholz B, Ellegren H, Wolf JB. (2013). High levels of gene expression explain the strong evolutionary constraint of mitochondrial protein-coding genes. Mol Biol Evol 30: 272–284. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. (1998). Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148: 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova A, Beheregaray LB, Coleman R, Gilligan D, Harrisson KA, Ingram BA et al. (submitted). Severe consequences of habitat fragmentation on genetic diversity of an endangered Australian freshwater fish: a call for assisted gene flow. Evol Appl. EVA-2016-176-OA, submitted 14/12/2016. [DOI] [PMC free article] [PubMed]
- Pörtner H-O. (2002). Climate variations and the physiological basis of temperature dependent biogeography: systemic to molecular hierarchy of thermal tolerance in animals. Comp Biochem Physiol A Mol Integr Physiol 132: 739–761. [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2006–2009). Available at http://tree.bio.ed.ac.uk/software/figtree/.
- Rambaut A, Drummond AJ. (2007). Available at http://beast.bio.ed.ac.uk/Tracer.
- Rand DM, Haney RA, Fry AJ. (2004). Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol 19: 645–653. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgrò CM, Lowe AJ, Hoffmann AA. (2011). Building evolutionary resilience for conserving biodiversity under climate change. Evol Appl 4: 326–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedlock AM, Parker JD, Crispin DA, Pietsch TW, Burmer GC. (1992). Evolution of the salmonid mitochondrial control region. Mol Phylogenet Evol 1: 179–192. [DOI] [PubMed] [Google Scholar]
- Silva G, Lima FP, Martel P, Castilho R. (2014). Thermal adaptation and clinal mitochondrial DNA variation of European anchovy. Proc R Soc B Biol Sci 281: 20141093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stier A, Reichert S, Criscuolo F, Bize P. (2015). Red blood cells open promising avenues for longitudinal studies of ageing in laboratory, non-model and wild animals. Exp Gerontol 71: 118–134. [DOI] [PubMed] [Google Scholar]
- Symula R, Keogh JS, Cannatella DC. (2008). Ancient phylogeographic divergence in southeastern Australia among populations of the widespread common froglet, Crinia signifera. Mol Phylogenet Evol 47: 569–580. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teacher AG, André C, Merilä J, Wheat CW. (2012). Whole mitochondrial genome scan for population structure and selection in the Atlantic herring. BMC Evol Biol 12: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Pond SLK, Scheffler K. (2014). RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol 32: 820–832, msu400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenhagen LM, Kelley SO. (2003). Impact of disease-related mitochondrial mutations on tRNA structure and function. Trends Biochem Sci 28: 605–611. [DOI] [PubMed] [Google Scholar]
- Wong WS, Yang Z, Goldman N, Nielsen R. (2004). Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168: 1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley S, Johnson J, Smith MJ, Crandall KA, McClellan DA. (2003). TreeSAAP: selection on amino acid properties using phylogenetic trees. Bioinformatics 19: 671–672. [DOI] [PubMed] [Google Scholar]
- Xu B, Yang Z. (2013). PAMLX: A graphical user interface for PAML. Mol Biol Evol 30: 2723–2724. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R. (2005). Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22: 1107–1118. [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Nielsen R, Goldman N, Pedersen AMK. (2000). Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155: 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. (2005). Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol 22: 2472–2479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.