Abstract
Objective
Polycystic ovary syndrome (PCOS) is a condition of androgen excess and chronic anovulation frequently associated with insulin resistance. We combined a nontargeted and targeted metabolomics approach to identify pathways and metabolites that distinguished PCOS from metabolic syndrome (MetS).
Methods
Twenty obese women with PCOS were compared with 18 obese women without PCOS. Both groups met criteria for MetS but could not have diabetes mellitus or take medications that treat PCOS or affect lipids or insulin sensitivity. Insulin sensitivity was derived from the frequently sampled intravenous glucose tolerance test. A nontargeted metabolomics approach was performed on fasting plasma samples to identify differentially expressed metabolites, which were further evaluated by principal component and pathway enrichment analysis. Quantitative targeted metabolomics was then applied on candidate metabolites. Measured metabolites were tested for associations with PCOS and clinical variables by logistic and linear regression analyses.
Results
This multiethnic, obese sample was matched by age (PCOS, 37 ± 6; MetS, 40 ± 6 years) and body mass index (BMI) (PCOS, 34.6 ± 5.1; MetS, 33.7 ± 5.2 kg/m2). Principal component analysis of the nontargeted metabolomics data showed distinct group separation of PCOS from MetS controls. From the subset of 385 differentially expressed metabolites, 22% were identified by accurate mass, resulting in 19 canonical pathways significantly altered in PCOS, including amino acid, lipid, steroid, carbohydrate, and vitamin D metabolism. Targeted metabolomics identified many essential amino acids, including branched-chain amino acids (BCAA) that were elevated in PCOS compared with MetS. PCOS was most associated with BCAA (P = .02), essential amino acids (P = .03), the essential amino acid lysine (P = .02), and the lysine metabolite α-aminoadipic acid (P = .02) in models adjusted for surrogate variables representing technical variation in metabolites. No significant differences between groups were observed in concentrations of free fatty acids or vitamin D metabolites. Evaluation of the relationship of metabolites with clinical characteristics showed 1) negative associations of essential and BCAA with insulin sensitivity and sex hormone–binding globulin and 2) positive associations with homeostasis model of insulin resistance and free testosterone; metabolites were not associated with BMI or percent body fat.
Conclusions
PCOS was associated with significant metabolic alterations not attributed exclusively to androgen-related pathways, obesity, or MetS. Concentrations of essential amino acids and BCAA are increased in PCOS, which might result from or contribute to their insulin resistance.
Keywords: α-aminoadipic acid, branched-chain amino acids, insulin sensitivity, metabolic syndrome, vitamin D
1. Introduction
Polycystic ovary syndrome (PCOS), a condition of androgen excess and chronic anovulation, is the most common endocrine disorder among women of reproductive age (1, 2). Insulin resistance is highly prevalent in PCOS, even among lean women (3), and it is associated with a higher risk of type 2 diabetes mellitus and an increase in cardiovascular risk factors (4). Limited knowledge about the exact mechanisms underlying the pathophysiology of PCOS has resulted in few available or effective therapies that ameliorate symptoms of PCOS or improve fertility or the metabolic complications of insulin resistance. Genome-wide association studies have identified candidate genes associated with PCOS, but their role in the underlying pathophysiology of PCOS has not been delineated to date. In addition, the lower odds ratios of genetic associations suggest that candidate genes are likely relevant for only a subset of women with PCOS (5–8). Furthermore, heterogeneity in the diagnosis of PCOS, resulting from diagnostic criteria requiring 2 of 3 features, results in considerable phenotype overlap and variable response to antiandrogen therapies and metformin. This heterogeneity limits the progress of more traditional approaches to identify underlying causes and novel therapies for PCOS.
Genomic (9–11), proteomic (11, 12), and metabolomic (13–20) approaches to study the pathogenesis of PCOS have implicated various pathways, including oxidative stress, immune function, and lipid metabolism. However, the activation of these pathways might reflect the high prevalence of obesity in women with PCOS, rather than the underlying pathogenesis of insulin resistance associated with PCOS. Given the great potential for interaction or overlap of metabolic pathways in PCOS vs those of obesity, plasma metabolomics offers detailed profiling of small-molecule breakdown products downstream of genomic and proteomic expression to identify active metabolic pathways associated with specific PCOS phenotypes or response to therapy.
To date, metabolomic studies in PCOS have evaluated small samples of women or evaluated the effect of multidrug therapy on PCOS (14, 15, 17, 21). In addition, many studies could not examine or control for obesity or differences in insulin sensitivity (12–15, 18, 19). Therefore, any detected differences could have been due to obesity and obesity-related insulin resistance, rather than to PCOS. In addition, only 3 nontargeted metabolomics studies included a limited targeted quantitative approach (15, 17, 22) to further validate candidate pathways and correlations with PCOS characteristics.
In the current study, we sought to determine whether a metabolomics approach could identify a specific plasma metabolic fingerprint that could distinguish between obese women with PCOS and obese controls with metabolic syndrome (MetS). First, we conducted a nontargeted metabolomics analysis to determine candidate metabolic profiles and related pathways that were significantly altered between the two groups. Second, we completed a targeted metabolomics analysis of specific candidate pathways to explore metabolites that were quantitatively different between the two groups and to evaluate their association with clinical characteristics of PCOS.
2. Research Design and Methods
The study protocol was approved by the University of Texas Southwestern Medical Center Institutional Review Board, and all participants provided written informed consent to enroll in the study.
2.1. Study Participants
The plasma samples were previously collected for a study of PCOS comparing coronary function and cardiovascular performance in overweight or obese women with PCOS compared with overweight or obese women with MetS. Women with PCOS were recruited from an academic tertiary care center and county medical center. Controls with MetS were recruited from the same centers and from the Dallas Heart Study (23) to match PCOS participants by age and body mass index (BMI). The following inclusion and exclusion criteria were applied to both groups (women with PCOS and controls with MetS alone). Premenopausal women aged 30 to 50 years were required to have a BMI greater than 25 kg/m2 and evidence of prediabetes (elevated fasting glucose, 100–125 mg/dL) or MetS. Participants could not have a previous diagnosis of diabetes mellitus or hypertension or be receiving medication to treat PCOS or hypertension or to improve insulin sensitivity. Current tobacco users also were excluded. The control group reported regular menstrual cycles since adolescence and had no evidence of hyperandrogenism. Women in the control group without PCOS were studied between days 7 and 14 of the midfollicular phase.
2.2. Variable Definitions for the Diagnosis of PCOS and MetS
PCOS was diagnosed by using the Rotterdam criteria (24), requiring specifically the presence of both oligomenorrhea and hyperandrogenism. Oligomenorrhea was defined by fewer than 9 periods per year since menarche or since the age of 20 years when not pregnant, breastfeeding, or taking oral contraceptives. Hyperandrogenism was defined biochemically by an elevated total testosterone or dehydroepiandrosterone or symptoms of severe acne, androgenic alopecia, or Ferriman-Gallwey score ≥8.
MetS was defined according to the National Cholesterol Education Program Adult Treatment Panel criteria (at least 3 of 5 criteria): 1) high-density lipoprotein (HDL) cholesterol <50 mg/dL; 2) triglyceride ≥150 mg/dL; 3) impaired fasting glucose ≥100 mg/dL and <126 mg/dL; 4) hypertension or systolic blood pressure ≥130 mm Hg or diastolic blood pressure >85 mm Hg; and 5) waist circumference >88 cm (25).
2.3. Insulin Sensitivity
The Bergman minimal model (26) was used to calculate insulin sensitivity (SI), acute insulin response to glucose, disposition index, and glucose effectiveness from serum insulin and plasma glucose concentrations, obtained during the insulin-modified version of the frequently sampled intravenous glucose tolerance test. After obtaining fasting blood samples, dextrose (600 mg/kg; delivered as a 50% solution) was administered through an antecubital vein over a 2-minute period. During the next 3 hours, blood samples were obtained at 21 time points from an “arterialized” vein in a hand placed in a hot box warmed to approximately 60°C.
As a surrogate measure of insulin resistance, we also calculated the index of homeostasis model of insulin resistance (HOMA-IR) from the following formula (27):
2.4. Body Composition
Percent body fat was calculated from measurement of body density from underwater weight, as previously described (28).
2.5. Assays
Venous blood was collected in standard blood collection tubes containing ethylene diamine tetra-acetic acid for plasma and in serum separator tubes for serum. Plasma samples were immediately processed and aliquoted into 2-mL screw-top cryovials (Phoenix Research Products) and stored at −80°C. Samples from baseline fasting time points were thawed only once for the nontargeted and targeted metabolomics analyses.
2.6. Nontargeted Metabolomics
Plasma nontargeted metabolomics profiling was performed using liquid chromatography/time-of-flight mass spectrometry (6220 ToF MS; Agilent, Inc) operated both in positive and negative electrospray ionization modes, using a scan range of m/z 100–1200 at a resolution of 10,000, as described previously (20, 29). Small-molecule metabolites were extracted from 100 μL of plasma by deproteinization with 80% methanol. Before deproteinization, an internal standard solution (4 μL) of 13C6-phenylalanine (250 ng/μL) was added to each sample and plasma quality control (QC) samples to monitor recovery and reproducibility in metabolite extraction (29). Dried samples were stored at −20°C until analysis. Samples were reconstituted in running buffer and analyzed within 48 hours of reconstitution. A small fraction of supernatant from each sample was combined into a pool (pooled QC), and the remaining supernatant was split into 2 fractions to be used for polar hydrophilic interaction liquid chromatography and reverse-phase C18 ultra performance liquid chromatography separation. A separate plasma QC sample was analyzed with pooled QC to account for analytic and instrumental variability. Each QC sample was analyzed in duplicate, and chromatographic separation was achieved using hydrophilic interaction liquid chromatography and reverse phase (C18) liquid chromatography separately. The instrument settings were as follows: nebulizer gas temperature, 325°C; capillary voltage, 3.5 kV; capillary temperature, 300°C; fragmenter voltage, 150 V; skimmer voltage, 58 V; octapole voltage, 250 V; cycle time, 0.5 seconds; and run time, 15.0 minutes (20, 29).
2.6.1. Data Analysis
Metabolite peak intensities and differential regulation of metabolites between groups were determined as described previously (19, 28). Each sample was normalized to the median of the baseline and log2 transformed. Data alignment, filtering, and univariate and multivariate statistical and differential analysis were performed using Mass Profiler Professional software (Agilent Inc). Default settings were used, with the exception of the signal-to-noise ratio threshold, mass limit (0.0025 units), and time limit (9 seconds). Each metabolite was putatively identified on the basis of accurate mass (m/z) against the METLIN database using a detection window of ≤7 ppm. The identified metabolites are annotated as Chemical Abstracts Service, Kyoto Encyclopedia of Genes and Genomes, Human Metabolome Project database, and LIPID MAPS identifiers. Identification of selected metabolites was validated using the standards described in Supplemental Table 1.
2.6.2. Statistical Analysis
Metabolites detected in >50% of the samples in any of the study groups were selected for differential expression analyses (20). After normalization, univariate statistical analysis (unpaired t test analysis) was performed to compare the differentially expressed metabolites between PCOS and MetS (with false-discovery rates [FDRs] of ≤ 0.05) and was estimated by Q values (20, 29). Unsupervised principal component analysis (PCA) was performed to display variation between PCOS and MetS study groups for data visualization and whether group variation could be explained by these metabolite variables reducing the dimensionality of the qualitative data.
2.6.3. Pathway Analysis
Identified metabolites that were differentially expressed between groups were used for pathway enrichment using MetaCore (GeneGo) (14, 16). Metabolite identifiers (Chemical Abstracts Service and Kyoto Encyclopedia of Genes and Genomes) were used for each metabolite; identifiers included name, molecular weight, fold change, and differential P value. The P value from the hypergeometric test, generated by MetaCore, represents the enrichment of certain metabolites in a pathway. A P value <.05 indicates significant enrichment. The FDR of 0.15 was applied in the assessment of pathway enrichment to allow a greater number of pathways to be reviewed because of the smaller sample size of metabolites entered into the pathway analysis (20, 29). The issue of multiple testing is accounted for in the programming of the MetaCore tool (30).
2.7. Targeted Metabolomics
2.7.1. Quantitation of Free Fatty Acid and Amino Acids
Quantitative measurements of free fatty acid and 45 amino acid metabolites were performed by tandem mass spectrometry against 12-point calibration curves that underwent the same derivatization with internal standard as described previously (20, 29).
2.7.2. Quantitation of Vitamin D Metabolites
25-Hydroxyvitamin D2 [25(OH)D2], 25-hydroxyvitamin D3 [25(OH)D3], 1,25-dihydroxyvitamin D2 [1,25(OH)2D2]), and 1,25-dihydroxyvitamin D3 [1,25(OH)2D3] were extracted from serum samples by using deuterated stable isotopes (31). After derivatization, they were analyzed by liquid chromatography–mass spectrometry/mass spectrometry with multiple-reaction monitoring (31).
2.7.3. Statistical Analysis
To address technical variation in the metabolomics measurements, surrogate variables were created by using the R package sva (32). Briefly, variation in metabolites was compartmentalized into variation due to interperson variability and technical variation. The surrogate variables were created as principal components of the residual error of a linear model of the metabolites, adjusted for variables that would account for interperson variation (eg, PCOS, age, race, BMI, and 11 other measured characteristics) (33). These are then used in future analyses to account for any technical variation of the assay, assuming that this residual error is due to technical variation.
Because several metabolites had concentrations below the lower limit of quantification (LLOQ), the relationship of clinical variables and metabolites was modeled as 1) a linear regression of the van der Waerden rank–transformed metabolite on each clinical variable, individually adjusting for 2 surrogate variables for technical variation; and 2) a logistic regression of the metabolite concentration being above the LLOQ on each clinical variable, individually adjusting 2 surrogate variables for technical variation. To test the effect of a clinical variable on a metabolite, the minimum P value is observed between these 2 models, and the null distribution of the minimum P value is created empirically by randomly sorting the metabolite 1,000 times. For metabolites with quantifiable concentrations, Spearman partial correlation coefficients were calculated between the metabolites and clinical variables, including SI, HOMA-IR, fasting glucose, free and total testosterone, estrogen, progesterone, steroid hormone–binding globulin (SHBG), low-density lipoprotein (LDL) cholesterol, BMI, percent body fat, and age.
Again, because the concentrations of several metabolites were below the LLOQ, the effect of these metabolites on PCOS was modeled using logistic regression adjusted for 2 surrogate variables, with metabolites being modeled with 2 effects: 1) metabolite concentration above LLOQ and 2) an indicator of metabolite concentration being above LLOQ. To test the effect of the metabolite on PCOS, a likelihood ratio test with 2 degrees of freedom was used to test the 2 metabolite effects while simultaneously adjusting for the surrogate variables.
Groups of metabolites were tested for association with PCOS simultaneously by using random effects models implemented using the globaltest package (eg, essential amino acids, branched-chain amino acids [BCAAs], etc). Because of the small sample size, the effect of the metabolites could be tested in association with either the group term of PCOS vs MetS or individual clinical variables (described below). Additionally, because prior information indicated that phenylalanine and tyrosine were positively correlated, we conducted the more powerful test of summing phenylalanine and tyrosine and testing for the association in the same manner as for individual metabolites above. All metabolites tested in groups had no values below the LLOQ.
For the purposes of exploratory analysis, metabolites were further reviewed that had a P value <.05. Due to sample size, a limited number of covariates could be included in modeling PCOS (at most 2 to enable stable statistical modeling). It appeared that the largest source of variation in the measured metabolites was not explained by the measured variables in this study (these are assumed to contribute to the majority of interindividual variation in the actual metabolites, not the measured metabolites). Thus, these metabolite technical variation variables were included as the only covariates prioritized above the clinical variables. A stratified analysis of the metabolites and clinical variables by PCOS diagnosis would not be possible because the sample size would be cut in half, further exacerbating the sample size issue. Clinical variables were selected on the basis of known and observed differences between women with and without PCOS (age, BMI, insulin sensitivity, acute insulin response, HOMA-IR, fasting glucose, free testosterone, total testosterone, estradiol, progesterone, SHBG, LDL, percent body fat, and race/ethnicity). Therefore, metabolites were tested in association with clinical variables by pooling together women with PCOS and women with MetS only. In terms of association with clinical variables, using the Bonferroni correction, statistical significance was set at 0.004 for the evaluation of the 14 clinical factors.
Group comparisons for baseline characteristics and vitamin D concentrations were made using Wilcoxon rank-sum test. Statistical analyses for these specific analyses were performed using JMP statistical software (SAS Institute Inc).
3. Results
3.1. Baseline Characteristics
Anthropometric characteristics and baseline biomarkers for the PCOS (n = 20) and MetS (n = 18) cohorts are shown in Table 1. Both groups were matched by age and BMI, and we observed no statistical difference in percent body fat or fasting glucose concentrations. Insulin sensitivity was more impaired in PCOS compared with MetS (P = .02), along with the disposition index, which is the product of insulin sensitivity and the acute insulin response (Table 1). As expected, total testosterone was significantly higher in PCOS (P = .04). Lipid concentrations (total cholesterol, triglycerides, and HDL cholesterol) were not statistically different between groups. The only exception was LDL cholesterol, which was higher in PCOS (P = .05).
Table 1.
Baseline Patient Characteristics, Stratified by PCOS vs MetS Status (N = 38)
| Characteristic | PCOS (n = 20) | MetS (n = 18) | P Value |
|---|---|---|---|
| Physical characteristics | |||
| Age, mean (SD), y | 37 (6) | 40 (6) | .06 |
| Body mass index, mean (SD), kg/m2 | 34.6 (5.1) | 33.7 (5.2) | .57 |
| Body fat, mean (SD), % | 39.0 (6.8) | 37.4 (6.2) | .44 |
| Race/ethnicity, No. (%) | .44 | ||
| White | 8 (40) | 5 (28) | |
| Black | 7 (35) | 10 (55) | |
| Hispanic | 5 (30) | 3 (17) | |
| Glucose metabolism, mean (SD) | |||
| Fasting glucose, mg/dL | 97 (6) | 95 (12) | .54 |
| Fasting insulin, μIU/mL | 11.1 (6.9) | 8.6 (8.5) | .08 |
| SI, min−1 × (μU/mL)−1 × 10−4 | 1.9 (1.0) | 3.0 (1.2) | .02 |
| AIRg, min × (μU/mL) | 701.6 (539.5) | 764.5 (388.6) | .68 |
| Disposition indexa | 1135.1 (643.4) | 2146.9 (1388) | .01 |
| Sg, min−1 × 100 | 1.7 (0.5) | 1.9 (0.98) | .36 |
| HOMA-IR | 2.8 (1.5) | 2.2 (2.7) | .45 |
| Sex steroid hormones, mean (SD) | |||
| Total testosterone, ng/dL | 45.6 (23.3) | 32.1 (15.3) | .04 |
| Free testosterone, ng/dL | 8.9 (6.4) | 5.6 (4.2) | .10 |
| Estradiol, pg/mL | 107.4 (70) | 118.4 (78) | .64 |
| Sex hormone-binding globulin, mcg/mL | 28.4 (15.6) | 35.4 (17.7) | .21 |
| Lipids, mean (SD), mg/dL | |||
| Total cholesterol | 191 (26) | 175 (39) | .15 |
| Low-density lipoprotein cholesterol | 120 (22) | 103 (30) | .05 |
| High-density lipoprotein cholesterol | 50 (8) | 51 (12) | .73 |
| Triglycerides | 105 (45) | 110 (53) | .78 |
Abbreviations: AIRg, acute insulin response to glucose; HOMA-IR, homeostasis model of insulin resistance; MetS, metabolic syndrome; PCOS, polycystic ovary syndrome; Sg, glucose effectiveness; SI, insulin sensitivity index.
Disposition index = SI × AIRg
3.2. Nontargeted Metabolomics Analysis
As shown in Figure 1, PCA showed PCOS and obese controls with MetS were distinctly separated on the basis of plasma metabolite differences. The first and second components in PCA explained 19.51% and 9 1% of variations, respectively. From the subset of 385 metabolites that significantly differed between PCOS and MetS, 85 (22%) were identified by accurate mass. Identification of 82 metabolites was validated by using standards (Supplemental Table 1). The identified metabolites characterized 19 significantly altered canonical pathways (P < 05; FDR, < 0.15) between PCOS and MetS controls (Figure 2), and metabolites were involved in networks for lipid, steroid, amino acid, carbohydrate, and vitamin metabolism (Supplemental Table 2). Among the significantly altered pathways were those for vitamin D, N-acylethanolamine, N-acyltransferase, prostaglandin biosynthesis and metabolism, regulation of cystic fibrosis transmembrane conductance regulator gating, BCAAs, and saturated fatty acids metabolism. Supplemental Table 3 demonstrates reproducibility of replicate sample values for components 1 and 2.
Figure 1.
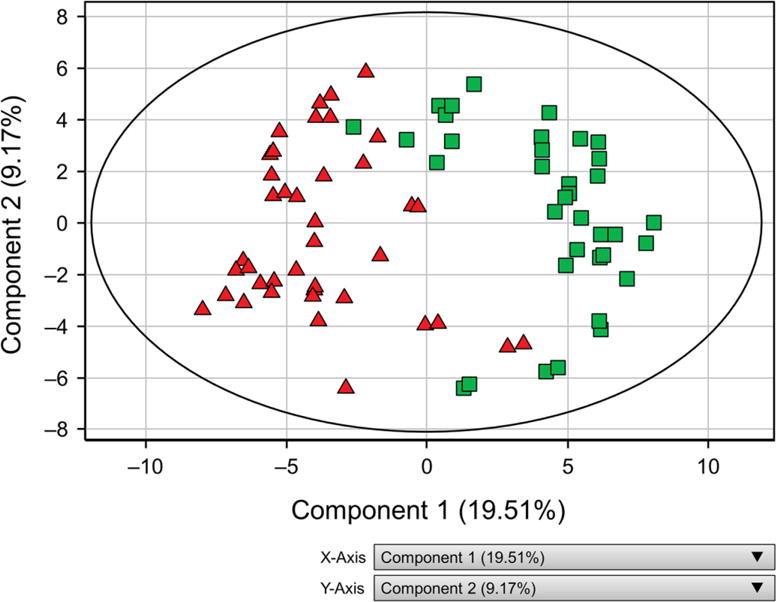
Two-dimensional Score Plot of Principal Component Analysis Showing Group Separation Between PCOS and MetS. Each person is represented by 2 replicate data points. PCOS and MetS groups can be seen in clusters separated on 2 components. The first and second components explained 18.51% and 9.67% of variations, respectively. MetS indicates controls with metabolic syndrome (green squares); PCOS, polycystic ovary syndrome (red triangles).
Figure 2.
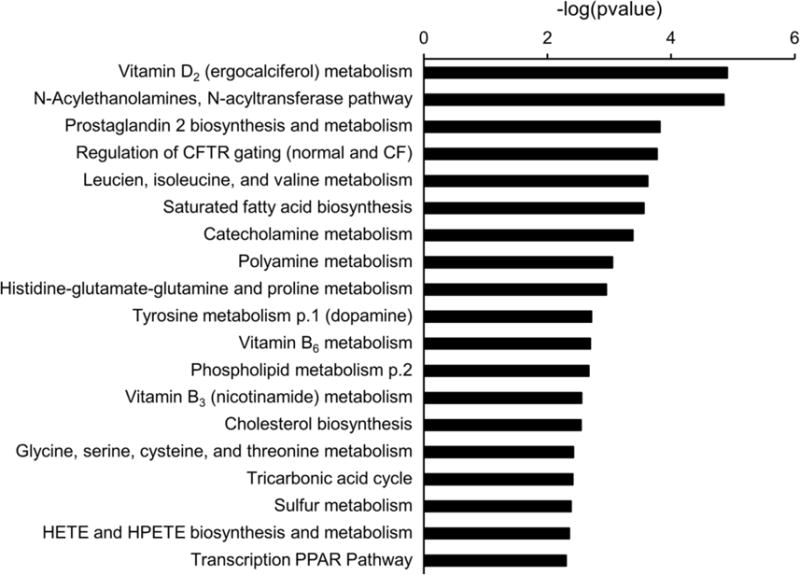
Metabolic Pathways Significantly Different in PCOS vs MetS. The significance of the pathways was evaluated using P values and a false-discovery rate <0.05. CF indicates cystic fibrosis; CFTR, cystic fibrosis transmembrane conductance regulator; HETE, hydroxyl eicosatetraenoic acid; HPETE, hydroperoxy eicosatetraenoic acid; PPAR, peroxisome proliferator-activated receptor.
3.3. Targeted Metabolomics Analysis
Targeted metabolomics analysis was performed to confirm associations of PCOS with leading candidate pathways: 1) amino acids and their metabolites and 2) fatty acids and their metabolites. Vitamin D concentrations were also quantitatively measured with mass spectrometry. Essential amino acids (histidine, methionine, threonine, lysine, valine, isoleucine, leucine, phenylalanine, tryptophan) (Figure 3A) and the subgroup of BCAA (valine, isoleucine, leucine) (Figure 3B) were most strongly associated with PCOS. Within these groups, further analysis of individual amino acids, adjusted for surrogate variables, showed that the metabolites significantly elevated in PCOS were BCAAs, lysine (median PCOS vs control, 167.7 vs 154.1 μM; P = .02) (Supplemental Table 4), and the lysine metabolite α-aminoadipic acid (α-AA) (median PCOS vs control, 0.9 vs 0.7 μM; P = .02). When we analyzed the sum of the concentrations of phenylalanine and tyrosine (aromatic amino acids), they were higher in PCOS compared with MetS (median PCOS vs control, 109.5 vs 104.2 μM; P = .0497). Individually, phenylalanine and tyrosine were not significantly associated with PCOS (P = 16 and .06, respectively).
Figure 3.
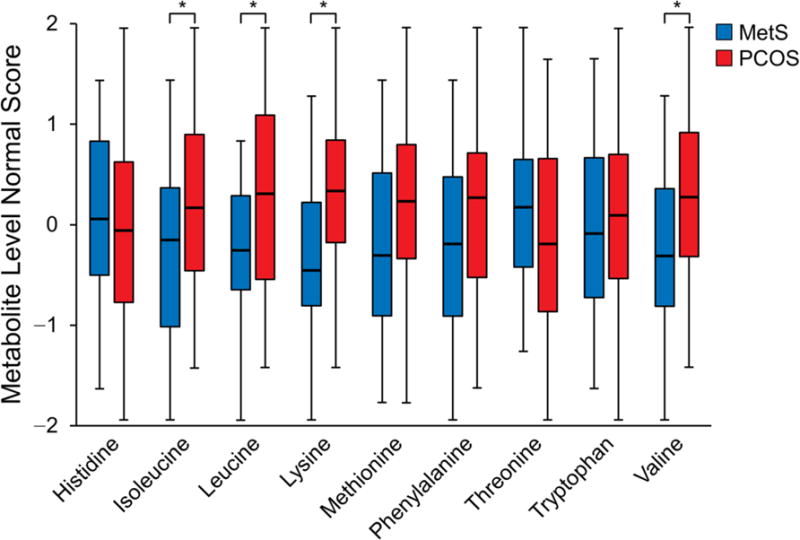
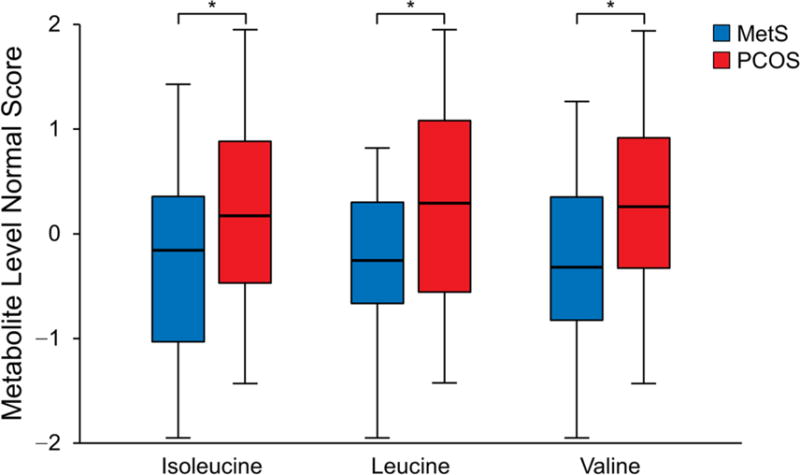
Concentrations of Amino Acids Normalized to Account for Technical Variation in Metabolites. A, Essential amino acids as a group were higher in PCOS (P = .03). Among the individual essential amino acids, aside from the observed differences in branched-chain amino acids, only the lysine concentration was higher in PCOS (P = .02). B, Branched-chain amino acid concentrations as a group were higher in PCOS (P = .02). Concentrations of individual branched-chain amino acids were also higher in the PCOS group compared with MetS controls: isoleucine (P = .03), leucine (P = .02), and valine (P = .03). The asterisk indicates P < 05; MetS, metabolic syndrome; PCOS, polycystic ovary syndrome.
Free fatty acids and lipid metabolites, when evaluated individually or after grouping by specific characteristics (eg, saturated fatty acids, unsaturated fatty acids, ceramides), were not significantly different between PCOS and MetS. Linoleic acid was the only lipid metabolite associated with PCOS (P = .04). Vitamin D concentrations were not significantly different between PCOS and MetS (Table 2).
Table 2.
Vitamin D Concentrations in PCOS vs MetS
| Vitamin | PCOS, median (IQR) |
MetS, median (IQR) |
P Value |
|---|---|---|---|
| 25(OH)D3, ng/mL | 19 (14–24) | 14 (9–21) | .11 |
| 25(OH)D2, ng/mL | 0 (0–4.5)a | 0 (0–53) | .97 |
| 1,25(OH)2D3, pg/mL | 46 (41–61) | 47 (37–54) | .42 |
| 1,25(OH)2D2, pg/mL | 0 (0–15)a | 0 (0–36) | .83 |
Abbreviations: 1,25(OH)2D2, 1,25-dihydroxyvitamin D2; 1,25(OH)2D3, 1,25-dihydroxyvitamin D3; 25(OH)D2, 25-hydroxyvitamin D2; 25(OH)D3, 25-hydroxyvitamin D3; MetS, metabolic syndrome; PCOS, polycystic ovary syndrome.
The entire range reported here.
3.4. Association of Amino Acids and Lipid Metabolites With Clinical Characteristics
Because of the differences in SI and androgens between PCOS and MetS, we questioned whether the observed associations with amino acid metabolites could be associated with measures of SI, body composition, or sex hormones among the entire participant cohort. SI was negatively associated with essential amino acids, including lysine, α-AA, and the BCAAs valine and leucine (Table 3). α-AA was also positively associated with fasting glucose and HOMA-IR. Among the remaining amino acid metabolites measured, asparagine and glycine were negatively associated with SI and positively associated with HOMA-IR, and tyrosine was positively associated with HOMA-IR (Supplemental Table 5).
Table 3.
| BMI
|
SIc
|
HOMA-IR
|
Free Testoster one
|
SHBG
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Amino Acid | ρ | P Value | ρ | P Value | ρ | P Value | ρ | P Value | ρ | P Value |
| Essential | ||||||||||
| Histidine | 0.07 | .950 | −0.02 | .737 | 0.11 | .982 | 0.24 | .682 | −0.21 | .209 |
| Methionine | −0.03 | .736 | −0.31 | .108 | 0.37 | .022 | 0.20 | .999 | −0.16 | .222 |
| Threonine | 0.05 | .951 | −0.15 | .874 | 0.02 | .661 | 0.11 | .684 | −0.02 | .372 |
| Lysine | 0.20 | .212 | −0.46 | .008 | 0.43 | .008 | 0.33 | .361 | −0.44 | .019 |
| α-Aminoadipic acidd | 0.17 | .290 | −0.37 | .015 | 0.42 | .007 | 0.16 | .347 | −0.34 | .036 |
| Branched chain | ||||||||||
| Valine | 0.13 | .412 | −0.25 | .029 | 0.21 | .217 | 0.26 | .021 | −0.47 | .023 |
| Isoleucine | 0.18 | .301 | −0.44 | .010 | 0.43 | .007 | 0.29 | .033 | −0.46 | .069 |
| Leucine | 0.18 | .389 | −0.30 | .043 | 0.29 | .086 | 0.26 | .034 | −0.60 | .019 |
| Aromatic | ||||||||||
| Phenylalanine | 0.20 | .472 | −0.07 | .422 | 0.11 | .212 | −0.09 | .558 | −0.19 | .728 |
| Tryptophan | 0.15 | .270 | 0.05 | .970 | 0.06 | .639 | 0.06 | .935 | −0.28 | .013 |
Abbreviations: BMI, body mass index; HOMA-IR, homeostasis model of insulin resistance; SHBG, sex hormone-binding globulin; SI, insulin sensitivity.
The Spearman partial correlation (ρ) adjusted for surrogate variables representing technical variation in metabolites.
Boldface text shows associations with P < 05.
The insulin sensitivity index (10−4 × min−1 × μU−1 × mL−1) was derived from frequently sampled intravenous glucose tolerance tests.
A metabolite of lysine.
BCAAs and essential amino acids were not associated with BMI or percent body fat. Isoleucine was the only amino acid positively associated with LDL cholesterol (P = .02). No associations were identified between these groups of amino acids and hormones such as estradiol, progesterone, and total testosterone. However, free testosterone was positively associated with BCAA (Table 3). SHBG was inversely correlated with lysine, α-AA, valine, leucine, and tryptophan (Table 3).
Among the lipid metabolites, linoleic acid was negatively associated with SI and LDL, and stearic acid was positively associated with BMI and percent body fat. Among the ceramides, C16-ceramide was negatively associated with SI and SHBG and positively associated with fasting glucose; C18-ceramide was positively associated with LDL, total and free testosterone, and estradiol; and C22-ceramide was positively associated with fasting glucose and total testosterone (Supplemental Table 5).
4. Discussion
In the current study, we identified 385 metabolites and 19 metabolic pathways that distinguished PCOS from age- and BMI-matched controls with MetS. We extended this nontargeted approach by performing a confirmatory, quantitative, targeted metabolomics analysis and showed that PCOS is strongly associated with groups of amino acid metabolites compared with obese women with MetS. Specifically, greater concentrations of BCAA and essential amino acids seemed to distinguish PCOS from MetS.
Comparisons to previous studies of plasma metabolomics profiles in PCOS are limited by small sample sizes, further diluted by subgroups of BMI and insulin resistance status (14, 15, 21), as well as by different analytic techniques, including less-sensitive nuclear magnetic resonance or nontargeted metabolomics and lack of BMI matching in control groups (13, 14, 16, 18). Most prior studies used the Rotterdam criteria for PCOS and did not restrict study participants to those with the more severe phenotype of oligomenorrhea and anovulation. Consequently, those studies might include women with a milder metabolic phenotype and less insulin resistance than this cohort (34). Together, these factors may account for the conflicting findings of prior studies that reported varying increases and decreases in fatty acid and amino acid metabolites in PCOS vs controls (35). Our results share the greatest agreement with the largest previously published study that confirmed significantly different metabolites from its nontargeted analysis with standards and reported increased BCAA and lysine in PCOS compared with controls (18). However, this group also described changes in other amino acids that were not corroborated by our data; these discordant findings could be attributable to their PCOS group not being overweight and to using a control group with a markedly lower BMI than our controls. None of the prior studies performed subsequent quantitative targeted analysis of amino acid profiles, as was done in this study.
Extensive evidence from non-PCOS studies suggest that increased BCAAs and essential amino acids are associated with obesity, insulin resistance, and type 2 diabetes mellitus (36, 37). Multiple cohort studies using metabolomics have demonstrated a stronger association of obesity and insulin resistance with BCAA and related metabolites than with lipid metabolites (38–40). Furthermore, elevated essential and BCAAs predict the development of diabetes mellitus in the Framingham and Malmo cohorts, even after adjusting for insulin resistance (7).
With our detailed characterization of insulin sensitivity and percent body fat, we have demonstrated that obese women with PCOS have greater impairment of insulin sensitivity than obese women with MetS, and this difference could not be explained by percent body fat or androgens. These results suggest that greater insulin resistance, rather than obesity or elevated androgens, is a key factor for the altered metabolomic consequences of PCOS.
To date, the proposed models for increased BCAA concentrations in obesity, insulin resistance, and type 2 diabetes mellitus suggest decreased BCAA catabolism due to decreased expression or action of BCAA catabolic enzymes in adipose and skeletal muscle tissue (36). Increases in BCAA might also contribute to glucose intolerance by increasing the supply of BCAA metabolites glutamate and alanine for gluconeogenesis. Although we did not observe significant differences in glutamate and alanine concentrations between PCOS and MetS controls, our controls were matched by BMI and had similar percent body fat; possibly, the previously observed differences in obese cohorts might have been attributable to differences in adiposity (38). Increases in circulating BCAA might interfere with insulin signaling in skeletal muscle and further promote insulin resistance, especially in the context of obesity and a high-fat diet (36, 38, 41). Interestingly, improved glucose homeostasis has been reported for animals fed a diet specifically enriched in leucine (42). In a cohort of healthy adults, leucine ingested with glucose acted synergistically to stimulate insulin secretion (43). In our cohort of obese individuals, leucine concentrations were not associated with the acute insulin response during the intravenous glucose tolerance test (Supplemental Table 5).
In the present study, we observed greater circulating concentrations of lysine and α-AA in PCOS compared with MetS. The essential amino acid lysine and its metabolite α-AA were negatively associated with SI and positively associated with HOMA-IR. α-AA was also positively associated with fasting glucose. Consistent with these findings, previous studies have identified α-AA as a novel biomarker for type 2 diabetes mellitus and a potential modulator of β-cell function. In the Framingham cohort, individuals in the highest quartile for α-AA concentrations had greater than a 4-fold increase in risk for type 2 diabetes mellitus during a 12-year period, even in individuals with normal glucose tolerance (44). A previous interventional study in PCOS identified lysine as a metabolite with marked changes after polytherapy that included insulin sensitizers and an antiandrogen (15). In adults with glucose intolerance, plasma concentrations of lysine and α-AA were significantly reduced in response to metformin and pioglitazone treatment (45). Although the mechanisms are not well understood, experimental data have shown that α-AA increased insulin secretion from pancreatic β cells in cell culture and lowered fasting plasma glucose in mice (44). Hyperglycemia also increased α-AA production in endothelial cell culture (46). Future human studies are needed to better understand the mechanisms for increased lysine and α-AA and their potential role in glucose homeostasis.
The aromatic amino acids phenylalanine and tyrosine have also been implicated in insulin resistance and metabolic disease (7, 36, 45), as well as in PCOS (18). Although we did not observe a strong association with phenylalanine, plasma concentrations of tyrosine were associated with PCOS, and the sum of phenylalanine and tyrosine concentrations was greater in PCOS compared with MetS.
Our nontargeted metabolomics analysis identified the saturated fatty acid biosynthesis pathway as being significantly altered in the PCOS group. However, when we examined free fatty acid metabolites in the plasma, only linoleic acid was associated with PCOS. Escobar-Morreale et al (15) demonstrated that obese women with PCOS had a different profile of fatty acid metabolites suggestive of increased lipolysis compared with nonobese women with PCOS who demonstrated suppression of lipolysis. When comparing obese women with PCOS to nonobese women with PCOS or to BMI-matched controls without PCOS, long-chain fatty acids such as linoleic acid were elevated only in the obese women with PCOS (15); this finding was corroborated by other groups that did not match control groups by BMI (19, 21). The lack of significant differences in the lipid metabolite profile in the current study may be attributable to matching our control group for both obesity and MetS. In the present study, linoleic acid was also positively associated with SI and negatively associated with LDL cholesterol; these clinical variables were both different in the women with PCOS compared with controls.
4.1. Vitamin D
In the nontargeted metabolomics analysis, the vitamin D2 (ergocalciferol) metabolism network was the most significantly altered pathway in PCOS compared with MetS (Figure 2). The 25(OH)D2 concentrations measured in PCOS and MetS were indicative of vitamin D deficiency (<20 ng/mL), as defined by the Institute of Medicine (47) and seen in other studies of PCOS (21, 48, 49). Vitamin D deficiency has also been associated with metabolic disturbances such as obesity, insulin resistance, and dyslipidemia (50, 51). Although 25(OH)D2 has been associated with insulin resistance in PCOS, as measured by HOMA-IR, these associations were not significant after adjusting for BMI (49, 52). In agreement with other studies (53, 54), we did not find any differences in plasma concentrations of vitamin D [25(OH)D2 or 1,25(OH)2D2] between PCOS and BMI-matched controls with MetS. This finding suggests that low vitamin D might be associated with obesity, rather than with PCOS. Although we did not quantify all the intermediate vitamin D metabolites, it is unlikely that we had the power to detect these smaller changes if 25(OH)D2 and 1,25(OH)2D2 concentrations already were similar. Another explanation for the discrepancy between the pathways analysis and the quantitative measurements relates to the analytic technique. Pathway analysis represents the enrichment of certain metabolites in a metabolic pathway. The P value from the hypergeometric test in pathway analysis is generated from the number of metabolites present in a given pathway, instead of from significantly changing metabolites in the pathway. In this manner, the vitamin D pathway might be selected due to enrichment of vitamin D metabolites and its derivatives that are putatively identified by accurate mass but cannot be measured quantitatively because of the lack of availability of standards. The quantitative analysis targeted clinically relevant and routinely tested metabolites of vitamin D, which did not confirm the nontargeted analysis. This illustrates the importance of follow up targeted analysis.
4.2. Strengths and Limitations
Because of the limited number of participants, our results should be considered exploratory until they can be confirmed in a larger cohort. Recognizing the limitations of the sample size, strengths of this study include model development for the targeted analysis, which accounted for technical and interindividual variation. The approach of validating the nontargeted analysis results with quantitative targeted metabolomics is another strength of this study. However, because of the limited sample size, we could not further evaluate the associations of clinical variables with specific metabolites. Here, we reduced the confounding effects of obesity, dyslipidemia, and insulin resistance by choosing obese women with MetS as the control group. However, the obesity and MetS in our cohort might explain the lack of differences between PCOS and controls in free fatty acids, ceramides, and sphingolipids that have been previously reported with insulin resistance (38) and obesity (15, 18). A few ceramide metabolites were associated with SI but among the entire sample of women, irrespective of PCOS status. Sample size may have limited our ability to discern associations of BCAA metabolites glutamate, glutamine, and alanine that were described previously (36, 38). Participants were studied after a 12-hour overnight fast as outpatients. They did not have a standardized diet before samples were obtained for metabolomics analysis, which could increase variability in the measured metabolites and decrease our ability to observe differences. However, these limitations should not have affected the observed associations with PCOS or correlations with clinical variables. Although women with PCOS did not have an induced withdrawal period for timing of blood samples, they were selected because they had a higher likelihood of anovulatory cycles. Because the women without PCOS were studied in the midfollicular phase, differences in estrogen and progesterone concentrations should be minimized. Estradiol and progesterone concentrations were also evaluated and were not associated with the metabolites (Supplemental Table 5).
Although it would be desirable to examine the PGE2 and N-acyltransferase pathways, because of the lack of availability of standards, we could not measure these metabolites. Experiments in the future might explore the functional aspects of these important pathways in PCOS and offer new insights regarding the pathophysiology of PCOS.
In this cross-sectional study, we were unable to determine causality (ie, were metabolic changes caused by PCOS or did they cause the development of PCOS features). As discussed above, the selection of an obese comparison group with MetS and detailed phenotyping suggests that impaired insulin sensitivity is a key factor in the metabolic consequences or contributors to PCOS.
4.3. Conclusions
In summary, PCOS was associated with significant metabolic alterations when compared with age- and BMI-matched controls with MetS. Although we used a relatively small group of subjects, we observed that greater concentrations of BCAA, essential amino acids, and the lysine metabolite α-AA seemed to distinguish PCOS from MetS. How these specific amino acid elevations in PCOS might contribute or result from PCOS remains to be determined. The translational implication is that further study of amino acid metabolism in PCOS might determine the underlying pathophysiology of insulin resistance in PCOS and target the development of new therapies. Because BCAA and lysine metabolism is increased in other insulin-resistant individuals without PCOS, future study might also be relevant for the prevention and treatment of type 2 diabetes mellitus. Finally, the current study indicates that a nontargeted metabolomics approach is feasible for identifying candidate pathways characterizing PCOS, and it highlights the importance of a targeted approach for follow-up in the discovery of biomarkers.
Supplementary Material
Acknowledgments
None.
Funding
Research reported in this publication was supported by the Mayo Clinic Metabolomics Core through grant number U24DK100469 from the National Institute of Diabetes and Digestive and Kidney Diseases, and grant number UL1 TR000135 from the National Center for Advancing Translational Sciences (NCATS), the American Heart Association (Fellow-to-Faculty Transition Award to A.Y.C.), and the Office of Women’s Health Research (Building Interdisciplinary Careers in Women’s Health Award K12HD065987 to A.Y.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
The sponsors of the study had no role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Abbreviations
- 1,25(OH)2D2
1,25-dihydroxyvitamin D2
- 1,25(OH)2D3
1,25-dihydroxyvitamin D3
- 25(OH)D2
25-hydroxyvitamin D2
- 25(OH)D3
25-hydroxyvitamin D3
- α-AA
α-aminoadipic acid
- BCAA
branched-chain amino acid
- BMI
body mass index
- FDR
false-discovery rate
- HDL
high-density lipoprotein
- HOMA-IR
homeostasis model of insulin resistance
- LDL
low-density lipoprotein
- LLOQ
lower limit of quantification
- MetS
metabolic syndrome
- PCA
principal component analysis
- PCOS
polycystic ovary syndrome
- QC
quality control
- SHBG
steroid hormone–binding globulin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: None.
Publisher: To expedite proof approval, send proof via email to scipubs@mayo.edu.
Disclosure Statement
The authors declare no potential conflicts of interests.
Author Contributions
A.Y.C. designed the study, collected data, analyzed data, interpreted data, and wrote the manuscript. She is the guarantor of this work.
A.L. wrote the manuscript and contributed to data analysis and interpretation.
G.D.J. analyzed data and contributed to interpretation and manuscript writing.
T.D. analyzed data and contributed to interpretation and manuscript writing.
R.E.C. analyzed data and contributed to interpretation and manuscript writing.
R.J.S. contributed to data analysis and interpretation.
K.S.N. contributed to the design of the study, interpretation of data, and manuscript writing.
Contributor Information
Dr Alice Y. Chang, Division of Endocrinology, Diabetes, Metabolism, and Nutrition, Mayo Clinic, Rochester, Minnesota.
Dr Antigoni Z. Lalia, Division of Endocrinology, Diabetes, Metabolism, and Nutrition, Mayo Clinic, Rochester, Minnesota.
Mr Gregory D. Jenkins, Division of Biomedical Statistics and Informatics, Mayo Clinic, Rochester, Minnesota.
Dr Tumpa Dutta, Division of Endocrinology, Diabetes, Metabolism, and Nutrition, Mayo Clinic, Rochester, Minnesota.
Dr Rickey E. Carter, Division of Biomedical Statistics and Informatics, Mayo Clinic, Rochester, Minnesota.
Dr Ravinder J. Singh, Division of Clinical Biochemistry and Immunology, Mayo Clinic, Rochester, Minnesota.
Dr K. Sreekumaran Nair, Division of Endocrinology, Diabetes, Metabolism, and Nutrition, Mayo Clinic, Rochester, Minnesota.
References
- 1.Carmina E, Azziz R. Diagnosis, phenotype, and prevalence of polycystic ovary syndrome. Fertil Steril. 2006 Jul;86(Suppl 1):S7–8. doi: 10.1016/j.fertnstert.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Ehrmann DA, Liljenquist DR, Kasza K, Azziz R, Legro RS, Ghazzi MN, PCOS/Troglitazone Study Group Prevalence and predictors of the metabolic syndrome in women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2006 Jan;91(1):48–53. doi: 10.1210/jc.2005-1329. Epub 2005 Oct 25. [DOI] [PubMed] [Google Scholar]
- 3.Dunaif A, Segal KR, Futterweit W, Dobrjansky A. Profound peripheral insulin resistance, independent of obesity, in polycystic ovary syndrome. Diabetes. 1989 Sep;38(9):1165–74. doi: 10.2337/diab.38.9.1165. [DOI] [PubMed] [Google Scholar]
- 4.Legro RS, Arslanian SA, Ehrmann DA, Hoeger KM, Murad MH, Pasquali R, et al. Endocrine Society. Diagnosis and treatment of polycystic ovary syndrome: an Endocrine Society clinical practice guideline. J Clin Endocrinol Metab. 2013 Dec;98(12):4565–92. doi: 10.1210/jc.2013-2350. Epub 2013 Oct 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen ZJ, Zhao H, He L, Shi Y, Qin Y, Shi Y, et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011 Jan;43(1):55–9. doi: 10.1038/ng.732. Epub 2010 Dec 12. [DOI] [PubMed] [Google Scholar]
- 6.Goodarzi MO, Jones MR, Li X, Chua AK, Garcia OA, Chen YD, et al. Replication of association of DENND1A and THADA variants with polycystic ovary syndrome in European cohorts. J Med Genet. 2012 Feb;49(2):90–5. doi: 10.1136/jmedgenet-2011-100427. Epub 2011 Dec 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011 Apr;17(4):448–53. doi: 10.1038/nm.2307. Epub 2011 Mar 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welt CK, Styrkarsdottir U, Ehrmann DA, Thorleifsson G, Arason G, Gudmundsson JA, et al. Variants in DENND1A are associated with polycystic ovary syndrome in women of European ancestry. J Clin Endocrinol Metab. 2012 Jul;97(7):E1342–7. doi: 10.1210/jc.2011-3478. Epub 2012 Apr 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007 Sep;3(9):1724–35. doi: 10.1371/journal.pgen.0030161. Epub 2007 Aug 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corton M, Botella-Carretero JI, Benguria A, Villuendas G, Zaballos A, San Millan JL, et al. Differential gene expression profile in omental adipose tissue in women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2007 Jan;92(1):328–37. doi: 10.1210/jc.2006-1665. Epub 2006 Oct 24. [DOI] [PubMed] [Google Scholar]
- 11.Corton M, Botella-Carretero JI, Lopez JA, Camafeita E, San Millan JL, Escobar-Morreale HF, et al. Proteomic analysis of human omental adipose tissue in the polycystic ovary syndrome using two-dimensional difference gel electrophoresis and mass spectrometry. Hum Reprod. 2008 Mar;23(3):651–61. doi: 10.1093/humrep/dem380. Epub 2007 Dec 22. [DOI] [PubMed] [Google Scholar]
- 12.Insenser M, Martinez-Garcia MA, Montes R, San-Millan JL, Escobar-Morreale HF. Proteomic analysis of plasma in the polycystic ovary syndrome identifies novel markers involved in iron metabolism, acute-phase response, and inflammation. J Clin Endocrinol Metab. 2010 Aug;95(8):3863–70. doi: 10.1210/jc.2010-0220. Epub 2010 May 19. [DOI] [PubMed] [Google Scholar]
- 13.RoyChoudhury S, Mishra BP, Khan T, Chattopadhayay R, Lodh I, Datta Ray C, et al. Serum metabolomics of Indian women with polycystic ovary syndrome using (1)H NMR coupled with a pattern recognition approach. Mol Biosyst. 2016 Oct 18;12(11):3407–3416. doi: 10.1039/c6mb00420b. [DOI] [PubMed] [Google Scholar]
- 14.Atiomo W, Daykin CA. Metabolomic biomarkers in women with polycystic ovary syndrome: a pilot study. Mol Hum Reprod. 2012 Nov;18(11):546–53. doi: 10.1093/molehr/gas029. Epub 2012 Jul 18. [DOI] [PubMed] [Google Scholar]
- 15.Escobar-Morreale HF, Samino S, Insenser M, Vinaixa M, Luque-Ramírez M, Lasuncion MA, et al. Metabolic heterogeneity in polycystic ovary syndrome is determined by obesity: plasma metabolomic approach using GC-MS. Clin Chem. 2012 Jun;58(6):999–1009. doi: 10.1373/clinchem.2011.176396. Epub 2012 Mar 16. [DOI] [PubMed] [Google Scholar]
- 16.Haoula Z, Ravipati S, Stekel DJ, Ortori CA, Hodgman C, Daykin C, et al. Lipidomic analysis of plasma samples from women with polycystic ovary syndrome. Metabolomics. 2015;11(3):657–666. doi: 10.1007/s11306-014-0726-y. Epub 2014 Aug 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whigham LD, Butz DE, Dashti H, Tonelli M, Johnson LK, Cook ME, et al. Metabolic evidence of diminished lipid oxidation in women with polycystic ovary syndrome. Curr Metabolomics. 2014;2(4):269–278. doi: 10.2174/2213235X01666131203230512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Fu L, Li R, Wang LN, Yang Y, Liu NN, et al. Metabolic profiles characterizing different phenotypes of polycystic ovary syndrome: plasma metabolomics analysis. BMC Med. 2012 Nov 30;:10–153. doi: 10.1186/1741-7015-10-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X, Xu F, Qi B, Hao S, Li Y, Li Y, et al. Serum metabolomics study of polycystic ovary syndrome based on liquid chromatography-mass spectrometry. J Proteome Res. 2014 Feb 7;13(2):1101–11. doi: 10.1021/pr401130w. Epub 2014 Jan 24. [DOI] [PubMed] [Google Scholar]
- 20.Dutta T, Chai HS, Ward LE, Ghosh A, Persson XM, Ford GC, et al. Concordance of changes in metabolic pathways based on plasma metabolomics and skeletal muscle transcriptomics in type 1 diabetes. Diabetes. 2012 May;61(5):1004–16. doi: 10.2337/db11-0874. Epub 2012 Mar 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong F, Deng D, Chen H, Cheng W, Li Q, Luo R, et al. Serum metabolomics study of polycystic ovary syndrome based on UPLC-QTOF-MS coupled with a pattern recognition approach. Anal Bioanal Chem. 2015 Jun;407(16):4683–95. doi: 10.1007/s00216-015-8670-x. Epub 2015 Apr 10. [DOI] [PubMed] [Google Scholar]
- 22.Wang W, Wang S, Tan S, Wen M, Qian Y, Zeng X, et al. Detection of urine metabolites in polycystic ovary syndrome by UPLC triple-TOF-MS. Clin Chim Acta. 2015 Aug 25;448:39–47. doi: 10.1016/j.cca.2015.06.008. Epub 2015 Jun 18. [DOI] [PubMed] [Google Scholar]
- 23.Victor RG, Haley RW, Willett DL, Peshock RM, Vaeth PC, Leonard D, et al. Dallas Heart Study Investigators The Dallas Heart Study: a population-based probability sample for the multidisciplinary study of ethnic differences in cardiovascular health. Am J Cardiol. 2004 Jun;93(12):15. 1473–80. doi: 10.1016/j.amjcard.2004.02.058. [DOI] [PubMed] [Google Scholar]
- 24.Rotterdam ESHRE/ASRM-Sponsored PCOS consensus workshop group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS) Hum Reprod. 2004 Jan;19(1):41–7. doi: 10.1093/humrep/deh098. [DOI] [PubMed] [Google Scholar]
- 25.Chen K, Lindsey JB, Khera A, De Lemos JA, Ayers CR, Goyal A, et al. Independent associations between metabolic syndrome, diabetes mellitus and atherosclerosis: observations from the Dallas Heart Study. Diab Vasc Dis Res. 2008 Jun;5(2):96–101. doi: 10.3132/dvdr.2008.016. [DOI] [PubMed] [Google Scholar]
- 26.Pacini G, Bergman RN. MINMOD: a computer program to calculate insulin sensitivity and pancreatic responsivity from the frequently sampled intravenous glucose tolerance test. Comput Methods Programs Biomed. 1986 Oct;23(2):113–22. doi: 10.1016/0169-2607(86)90106-9. [DOI] [PubMed] [Google Scholar]
- 27.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985 Jul;28(7):412–9. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 28.Abate N, Garg A, Peshock RM, Stray-Gundersen J, Grundy SM. Relationships of generalized and regional adiposity to insulin sensitivity in men. J Clin Invest. 1995 Jul;96(1):88–98. doi: 10.1172/JCI118083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dutta T, Kudva YC, Persson XM, Schenck LA, Ford GC, Singh RJ, et al. Impact of long-term poor and good glycemic control on metabolomics alterations in type 1 diabetic people. J Clin Endocrinol Metab. 2016 Mar;101(3):1023–33. doi: 10.1210/jc.2015-2640. Epub 2016 Jan 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Applying the power of MetaCore to Next Generation Sequencing Data: Uncovering potential mechanisms for Interferon alpha induced Thyroiditis: case study [Internet] 2012:8. [cited 2016 Jul 13]; Available from: http://thomsonreuters.com/content/dam/openweb/documents/pdf/pharma-life-sciences/case-study/metacore-ngs-casestudy-cin-en.pdf.
- 31.Netzel BC, Cradic KW, Bro ET, Girtman AB, Cyr RC, Singh RJ, et al. Increasing liquid chromatography-tandem mass spectrometry throughput by mass tagging: a sample-multiplexed high-throughput assay for 25-hydroxyvitamin D2 and D3. Clin Chem. 2011 Mar;57(3):431–40. doi: 10.1373/clinchem.2010.157115. Epub 2011 Jan 18. [DOI] [PubMed] [Google Scholar]
- 32.Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004 Jan 1;20(1):93–9. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 33.Karpievitch YV, Nikolic SB, Wilson R, Sharman JE, Edwards LM. Metabolomics data normalization with EigenMS. PLoS One. 2014 Dec 30;9(12):e116221. doi: 10.1371/journal.pone.0116221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moran L, Teede H. Metabolic features of the reproductive phenotypes of polycystic ovary syndrome. Hum Reprod Update. 2009 Jul-Aug;15(4):477–88. doi: 10.1093/humupd/dmp008. Epub 2009 Mar 11. [DOI] [PubMed] [Google Scholar]
- 35.Murri M, Insenser M, Escobar-Morreale HF. Metabolomics in polycystic ovary syndrome. Clin Chim Acta. 2014 Feb 15;429:181–8. doi: 10.1016/j.cca.2013.12.018. Epub 2013 Dec 22. [DOI] [PubMed] [Google Scholar]
- 36.Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012 May;15(5):2. 606–14. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batch BC, Shah SH, Newgard CB, Turer CB, Haynes C, Bain JR, et al. Branched chain amino acids are novel biomarkers for discrimination of metabolic wellness. Metabolism. 2013 Jul;62(7):961–9. doi: 10.1016/j.metabol.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huffman KM, Shah SH, Stevens RD, Bain JR, Muehlbauer M, Slentz CA, et al. Relationships between circulating metabolic intermediates and insulin action in overweight to obese, inactive men and women. Diabetes Care. 2009 Sep;32(9):1678–83. doi: 10.2337/dc08-2075. Epub 2009 Jun 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009 Apr;9(4):311–26. doi: 10.1016/j.cmet.2009.02.002. Erratum in: Cell Metab. 2009 Jun;9(6):565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tai ES, Tan ML, Stevens RD, Low YL, Muehlbauer MJ, Goh DL, et al. Insulin resistance is associated with a metabolic profile of altered protein metabolism in Chinese and Asian-Indian men. Diabetologia. 2010 Apr;53(4):757–67. doi: 10.1007/s00125-009-1637-8. Epub 2010 Jan 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krebs M, Krssak M, Bernroider E, Anderwald C, Brehm A, Meyerspeer M, et al. Mechanism of amino acid-induced skeletal muscle insulin resistance in humans. Diabetes. 2002 Mar;51(3):599–605. doi: 10.2337/diabetes.51.3.599. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Guo K, LeBlanc RE, Loh D, Schwartz GJ, Yu YH. Increasing dietary leucine intake reduces diet-induced obesity and improves glucose and cholesterol metabolism in mice via multimechanisms. Diabetes. 2007 Jun;56(6):1647–54. doi: 10.2337/db07-0123. Epub 2007 Mar 14. [DOI] [PubMed] [Google Scholar]
- 43.Kalogeropoulou D, Lafave L, Schweim K, Gannon MC, Nuttall FQ. Leucine, when ingested with glucose, synergistically stimulates insulin secretion and lowers blood glucose. Metabolism. 2008 Dec;57(12):1747–52. doi: 10.1016/j.metabol.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 44.Wang TJ, Ngo D, Psychogios N, Dejam A, Larson MG, Vasan RS, et al. 2-Aminoadipic acid is a biomarker for diabetes risk. J Clin Invest. 2013 Oct;123(10):4309–17. doi: 10.1172/JCI64801. Epub 2013 Sep 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irving BA, Carter RE, Soop M, Weymiller A, Syed H, Karakelides H, et al. Effect of insulin sensitizer therapy on amino acids and their metabolites. Metabolism. 2015 Jun;64(6):720–8. doi: 10.1016/j.metabol.2015.01.008. Epub 2015 Jan 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan W, Zhang J, Li S, Edwards JL. Amine metabolomics of hyperglycemic endothelial cells using capillary LC-MS with isobaric tagging. J Proteome Res. 2011 Nov;10(11):4. 5242–50. doi: 10.1021/pr200815c. Epub 2011 Oct 17. [DOI] [PubMed] [Google Scholar]
- 47.Dietary reference intakes for calcium and vitamin D [Internet] Washington (DC): Institute of Medicine of the National Academies; Nov, 2010. [cited 2016 Apr 4]. Available from: https://www.nationalacademies.org/hmd/∼/media/Files/Report%20Files/2010/Dietary-Reference-Intakes-for-Calcium-and-Vitamin-D/Vitamin%20D%20and%20Calcium%202010%20Report%20Brief.pdf. [Google Scholar]
- 48.Garg G, Kachhawa G, Ramot R, Khadgawat R, Tandon N, Sreenivas V, et al. Effect of vitamin D supplementation on insulin kinetics and cardiovascular risk factors in polycystic ovarian syndrome: a pilot study. Endocr Connect. 2015 Jun;4(2):108–16. doi: 10.1530/EC-15-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krul-Poel YH, Snackey C, Louwers Y, Lips P, Lambalk CB, Laven JS, et al. The role of vitamin D in metabolic disturbances in polycystic ovary syndrome: a systematic review. Eur J Endocrinol. 2013 Oct;169(6):23. 853–65. doi: 10.1530/EJE-13-0617. [DOI] [PubMed] [Google Scholar]
- 50.Ford ES, Ajani UA, McGuire LC, Liu S. Concentrations of serum vitamin D and the metabolic syndrome among U.S. adults. Diabetes Care. 2005 May;28(5):1228–30. doi: 10.2337/diacare.28.5.1228. [DOI] [PubMed] [Google Scholar]
- 51.Anderson JL, May HT, Horne BD, Bair TL, Hall L, Carlquist JF, et al. Intermountain Heart Collaborative (IHC) Study Group Relation of vitamin D deficiency to cardiovascular risk factors, disease status, and incident events in a general healthcare population. Am J Cardiol. 2010 Oct;106(7):1. 963–8. doi: 10.1016/j.amjcard.2010.05.027. Epub 2010 Aug 11. [DOI] [PubMed] [Google Scholar]
- 52.Wehr E, Pilz S, Schweighofer N, Giuliani A, Kopera D, Pieber TR, et al. Association of hypovitaminosis D with metabolic disturbances in polycystic ovary syndrome. Eur J Endocrinol. 2009 Oct;161(4):575–82. doi: 10.1530/EJE-09-0432. Epub 2009 Jul 23. [DOI] [PubMed] [Google Scholar]
- 53.Kim JJ, Choi YM, Chae SJ, Hwang KR, Yoon SH, Kim MJ, et al. Vitamin D deficiency in women with polycystic ovary syndrome. Clin Exp Reprod Med. 2014 Jun;41(2):80–5. doi: 10.5653/cerm.2014.41.2.80. Epub 2014 Jun 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Figurova J, Dravecka I, Javorsky M, Petrikova J, Lazurova I. Prevalence of vitamin D deficiency in Slovak women with polycystic ovary syndrome and its relation to metabolic and reproductive abnormalities. Wien Klin Wochenschr. 2016 Sep;128(17–18):641–8. doi: 10.1007/s00508-015-0768-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


